Abstract
Signal transducers and activators of transcription (STATs) reside in a latent state in the cytoplasm of the cell, but accumulate in the nucleus in response to cytokines or growth factors. Localization in the nucleus occurs following STAT tyrosine phosphorylation and dimerization. In this report we demonstrate a direct interaction of importin-α5 with tyrosine-phosphorylated STAT1 dimers, and provide evidence that a nuclear localization signal (NLS) exists in an inactive state within a STAT1 monomer. A mutation in STAT1 leucine 407 (L407A) is characterized, which generates a protein that is accurately tyrosine phosphorylated in response to interferon, dimerizes and binds DNA, but does not localize to the nucleus. The import defect of STAT1(L407A) appears to be a consequence of the inability of this protein to be recognized by its import shuttling receptor. In addition, we demonstrate that STAT1 binding to specific target DNA effectively blocks importin-α5 binding. This result may play a role in localizing STAT1 to its destination in the nucleus, and in releasing importin-α5 from STAT1 for recycling back to the cytoplasm.
Keywords: gene expression/interferon/signal transduction
Introduction
Regulated cellular localization of a transcription factor can serve as a biological switch in response to extracellular cues such as cytokines or growth factors. Members of the family of signal transducers and activators of transcription (STAT) exemplify such molecular switches. The STAT transcription factors normally reside in a latent state in the cytoplasm of the cell, but following tyrosine phosphorylation by activated kinases such as the Janus kinase (JAK) family they dimerize via SH2 phosphotyrosine interactions and subsequently translocate to the nucleus (Gilmour and Reich, 1995; Ihle et al., 1995; Darnell, 1997; Leonard and O’Shea, 1998; Stark et al., 1998). In the nucleus, the STAT dimers bind to specific DNA targets and induce gene transcription that effects appropriate physiological responses. Accurate dynamic localization is essential for STATs to function as sensors in the cytoplasm and as mediators of transcription in the nucleus.
Facilitated transport of proteins into the nucleus requires the presence of signal motifs that are recognized by specific soluble shuttling receptors of the importin/karyopherin family (Mattaj and Englmeier, 1998; Pemberton et al., 1998; Adam, 1999; Gorlich and Kutay, 1999; Macara, 2001). Motifs that direct proteins to the nucleus have been designated nuclear localization signals (NLSs) (Jans et al., 2000). The best characterized NLSs are rich in basic amino acids and these classical NLSs are recognized in the cytoplasm by importin-α receptors, which heterodimerize with the importin-β receptor (Jans et al., 2000; Hodel et al., 2001). The importin-β receptor facilitates nuclear translocation by binding to nuclear pore proteins and Ran GTPase in conjunction with other proteins leading to nuclear import. Non-classical NLSs that diverge from the basic consensus have also been identified and these are transported directly by other members of the importin-β superfamily. To date, an autonomous NLS has not been identified in STAT factors. Studies have noted a potential role of the DNA binding domain of STATs in nuclear accumulation, and one study suggested the import function of a cluster of basic residues (Herrington et al., 1999; Melen et al., 2001). To evaluate accurately STAT dynamic redistribution, it is critical to evaluate STATs that retain the ability to dimerize and bind DNA. Our previous results indicate that a STAT1 protein defective for DNA binding does not accumulate in the nucleus only because it is effectively exported to the cytoplasm via a nuclear export signal within the DNA binding domain (McBride et al., 2000).
Although the STAT1 NLS has not been defined, the molecular mechanism that regulates STAT1 nuclear import has been shown to involve the function of a member of the importin-α receptor family: importin-α5/karyopherin-α1/NPI-1/SRP1 (Sekimoto et al., 1997). Importin-α5 was shown to bind tyrosine-phosphorylated STAT1 isolated from interferon-γ (IFN-γ)-treated cell lysates. Mapping the region of importin-α5 that binds STAT1 produced the intriguing result that STAT1 binds to a domain of importin-α5 distinct from that of classical NLS motifs. Classical NLSs bind to central armadillo repeats in the importin-α receptors; however, tyrosine-phosphorylated STAT1 binds to a C-terminal domain that does not overlap with the central armadillo repeats (Sekimoto et al., 1997; Conti et al., 1998; Fontes et al., 2000). STAT1 is the only known imported substrate that interacts with the C-terminus of importin-α. This finding supports the hypothesis that the STAT1 NLS does not conform to a classical NLS.
The mechanisms that regulate STAT nuclear– cytoplasmic localization are critical to effecting proper cytokine signal transduction. In this report we demonstrate that an element within the DNA binding domain of STAT1 is essential both for nuclear import and for direct association with the importin-α5 shuttling receptor. Introduction of a specific mutation in the DNA binding domain of STAT1 disrupts nuclear import coincident with disruption of importin-α5 binding, but it does not impair tyrosine phosphorylation, dimerization or DNA binding. We have thereby functionally separated the ability of STAT1 to bind importin-α5 from its ability to bind DNA. Domains that regulate nuclear import and DNA binding have been found to overlap in other transcription factors and may represent the outcome of functional co-evolution (LaCasse and Lefebvre, 1995; Cokol et al., 2000). In this report we demonstrate that specific DNA binding effectively displaces importin-α5 from STAT1 dimers. This interplay of DNA binding and shuttling receptor binding with STAT1 may be a mechanism regulating cellular localization that is shared with other DNA binding factors. STATs are distinct, however, in that they are directly tyrosine phosphorylated in the cytoplasm and appear to gain an unconventional NLS only following their dimerization.
Results
Mutational analyses effect nuclear import
Cytokine activation results in a dynamic redistribution of STAT1 from the cytoplasm to the nucleus. Since an NLS is required for transport of proteins larger than ∼40 kDa into the nucleus, the function of this signal motif in STAT1 must be regulated to effect correct localization (Davis, 1995; Nigg, 1997). To visualize cellular localization of STAT1 by fluorescent microscopy we linked the gene encoding green fluorescent protein (GFP) downstream of the STAT1 gene (STAT1–GFP), and transfected the fusion into cells that lack endogenous STAT1 (U3A cells) (Muller et al., 1993; McBride et al., 2000). In response to IFN-γ, this STAT1–GFP fusion was tyrosine phosphorylated, translocated to the nucleus, bound target DNA sites, and activated specific gene transcription (McBride et al., 2000). Our previous analyses revealed the presence of a constitutive nuclear export signal within the DNA binding domain of STAT1, but an NLS within the molecule remains to be identified (McBride et al., 2000). Since many DNA binding proteins have been found to possess an NLS located within their DNA binding domain, we generated a number of site-directed mutations within the DNA binding domain of STAT1–GFP and examined dynamic localization following IFN-γ treatment (Figure 1) (LaCasse and Lefebvre, 1995; Cokol et al., 2000).
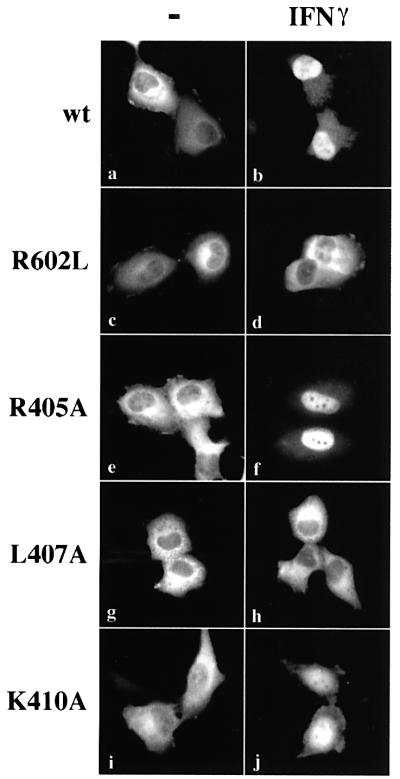
Fig. 1. Leucine 407 is critical for STAT1 nuclear accumulation. U3A cells expressing STAT1–GFP (a–b), STAT1–GFP(R602L) (c–d), STAT1–GFP(R405A) (e–f), STAT1–GFP(L407A) (g–h) or STAT1– GFP(K410A) (i–j) were untreated (a, c, e, g and i) or treated with IFN-γ for 30 min (b, d, f, h and j). Cells were fixed and STAT1–GFP cellular localization was examined by fluorescent microscopy.
Latent STAT1–GFP localizes to the cytoplasm of unstimulated cells and accumulates in the nucleus of cells following IFN-γ treatment (Figure 1a and b). Nuclear accumulation of STAT1 coincides with its specific tyrosine phosphorylation on residue 701 and subsequent dimerization via SH2–phosphotyrosine interactions. The mutation of a critical arginine residue within the SH2 domain of STAT1 (R602L) disrupts both IFN-γ-stimulated phosphorylation and subsequent dimerization, and results in a protein that does not accumulate in the nucleus (Figure 1c and d) (Improta et al., 1994; Shuai et al., 1994). Introduction of point mutations within a region of the DNA binding domain produced STAT1 proteins that either accumulated in the nucleus following IFN-γ or remained in the cytoplasm. For example, mutation of arginine 405 to alanine (R405A) had no effect on the STAT1 response to IFN-γ (Figure 1e and f), whereas mutation of leucine 407 to alanine (L407A) inhibited accumulation in the nucleus (Figure 1g and h), and mutation of lysine 410 to alanine (K410A) had only a slight effect on STAT1 nuclear accumulation (Figure 1i and j).
Several STAT1 mutations that disrupted nuclear accumulation were characterized and leucine 407 was found to play a critical role in IFN-γ-induced nuclear import. Analyses of this mutation distinguished between the ability of a STAT1 protein to dimerize following tyrosine phosphorylation and to translocate to the nucleus. The phosphorylation status of STAT1–GFP(L407A) was investigated following transfection of STAT1- deficient U3A cells. Either wild-type (wt)STAT1– GFP-expressing cells or cells expressing STAT1–GFP (L407A) were treated with IFN-γ, and STAT1 tyrosine phosphorylation was evaluated by immunoprecipitation of cell lysates with anti-STAT1 antibody and western blot analysis with STAT1 phosphotyrosine-701-specific antibody (Figure 2A). Both wtSTAT1–GFP and STAT1– GFP(L407A) were reactive with phosphotyrosine-701-specific antibody, indicating that STAT1–GFP(L407A) is phosphorylated on the proper tyrosine residue. It is well established that tyrosine-phosphorylated STAT1 binds to its DNA target as a dimer (Chen et al., 1998). To ensure that STAT1–GFP(L407A) does not disrupt STAT1 dimer formation, we determined whether tyrosine-phosphorylated STAT1–GFP(L407A) could bind a DNA target. Whole-cell lysates were prepared from control U3A cells or cells expressing STAT1–GFP(L407A), and DNA binding was evaluated by an electrophoretic mobility-shift assay (EMSA) with DNA corresponding to an IFN-γ-activated site (GAS) (Figure 2B) (McBride et al., 2000). A specific STAT1–GFP(L407A) DNA binding complex was evident in IFN-γ-treated cells, as demonstrated by the inhibitory effect of antibodies to STAT1 (Figure 2B, lanes 4 and 5).
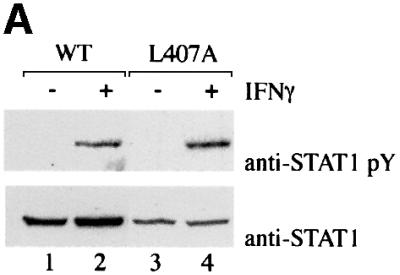
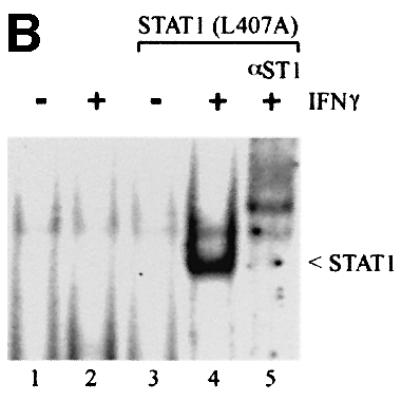
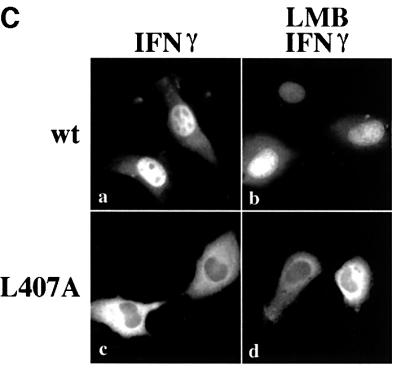
Fig. 2. STAT1–GFP(L407A) is tyrosine phosphorylated and binds DNA but does not translocate to the nucleus. (A) U3A cells expressing STAT1–GFP (lanes 1 and 2) or STAT1–GFP(L407A) (lanes 3 and 4) were untreated (–) or treated with IFN-γ for 30 min (+). Proteins from cell lysates were immunoprecipitated with anti-STAT1 antibody and western blots were performed with anti-STAT1 phosphotyrosine antibody (anti-STAT1 pY) (upper panel) or anti-STAT1 antibody (lower panel). (B) EMSA was performed with the IRF-1 GAS probe and whole-cell lysates from U3A cells (lanes 1 and 2) or U3A cells expressing STAT1–GFP (L407A) (lanes 3–5) were either untreated (–) or treated with IFN-γ for 30 min (+). Anti-STAT1 antibody (lane 5) was added to the DNA binding reaction. (C) U3A cells expressing wtSTAT1–GFP or STAT1–GFP(L407A) were treated with IFN-γ for 30 min, with or without pre-treatment with LMB. Cellular localization was examined by fluorescent microscopy.
These studies confirmed proper tyrosine phosphorylation and dimerization of STAT1–GFP(L407A), and yet this mutant did not accumulate in the nucleus following IFN-γ stimulation. It remained possible that the L407A mutant was transported into the nucleus, but did not accumulate due to effective constitutive nuclear export. We and others demonstrated previously that STAT1 is exported from the nucleus by the CRM1/exportin1 shuttling receptor (Begitt et al., 2000; McBride et al., 2000; Mowen and David, 2000). To block the CRM1-mediated export pathway we evaluated the effect of a specific inhibitor of CRM1, leptomycin B (LMB) (Wolff et al., 1997; Kudo et al., 1998). U3A cells expressing either wtSTAT1–GFP or STAT1–GFP(L407A) were treated with IFN-γ in the presence or absence of LMB. wtSTAT1–GFP accumulated in the nucleus normally following IFN-γ treatment; however, STAT1–GFP (L407A) did not accumulate in the nucleus even when nuclear export was blocked by LMB (Figure 2C). These results indicated that STAT1–GFP(L407A) remains localized to the cytoplasm because it does not translocate to the nucleus, even following tyrosine phosphorylation and dimerization.
STAT1(L407A)-phosphorylated dimers do not bind importin-α5
The importin-α family of shuttling receptors bind classical NLS motifs and facilitate nuclear import in concert with the nuclear import machinery. It was previously reported that importin-α5 interacts with tyrosine-phosphorylated STAT1 isolated from cell lysates (Sekimoto et al., 1997). Importin-α5 shares conserved domains of other importin- α family members, including an N-terminal importin-β binding (IβB) domain, central armadillo repeat motifs and a C-terminal acidic-rich domain (Figure 3A) (Cortes et al., 1994; Gorlich et al., 1996; Moroianu et al., 1996; Weis et al., 1996). Structural and biochemical studies have revealed that it is the central armadillo repeats that bind classical NLSs (Conti et al., 1998; Dingwall and Laskey, 1998; Fontes et al., 2000). However, STAT1 was found to bind the C-terminal region encompassing amino acid residues 425–538 of importin-α5 (Sekimoto et al., 1997).
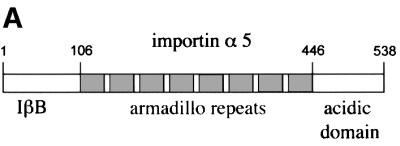
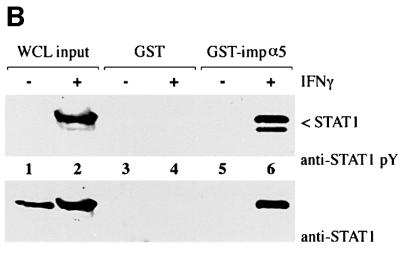
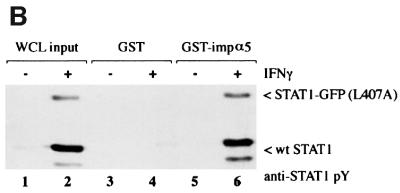
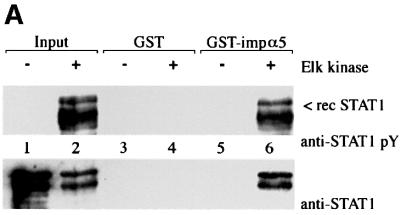
Fig. 3. STAT1, but not STAT1–GFP(L407A), binds importin-α5 following IFN-γ stimulation. (A) A linear diagram of the importin-α5 family member. Amino acids are noted (Kohler et al., 1999). (B) An importin-α5 interaction assay was performed with lysates from HeLa cells either untreated (–) or treated with IFN-γ for 30 min (+). Input is displayed in lanes 1 and 2. Lysates were incubated with either GST (lanes 3 and 4) or GST fused to the C-terminus of importin-α5 (GST-impα5) (lanes 5 and 6) bound to glutathione beads. Western blots were performed with anti-STAT1 phosphotyrosine antibody (anti-STAT1 pY) (upper panel) or anti-STAT1 antibody (lower panel). (C) The importin-α5 interaction assay was performed as in (B), with lysates from U3A cells expressing STAT1–GFP(L407A).
To investigate the ability of STAT1–GFP(L407A) to interact with the importin-α5 receptor, we first established an importin-α5 interaction assay with tyrosine-phosphorylated wtSTAT1. The importin-α5 protein encoding residues 404–538 was expressed as a fusion protein linked to glutathione S-transferase (GST) and purified from bacteria. Whole-cell lysates from untreated HeLa cells or cells treated with IFN-γ were incubated with GST–importin-α5 bound to glutathione–agarose beads. Proteins bound to GST–importin-α5 or control GST were eluted and analyzed by western blotting for the presence of STAT1 (Figure 3B). Two splice products of STAT1 (p91 and p84) specifically bound to GST–importin-α5 only following tyrosine phosphorylation in response to IFN-γ (Figure 3B, lane 6, upper panel). Antibodies that recognize the C-terminus of STAT1 (p91) confirm that only STAT1 from cells treated with IFN-γ can associate with importin-α5 (Figure 3B, lane 6, lower panel). To determine whether tyrosine-phosphorylated STAT1–GFP(L407A) interacts with importin-α5, a similar analysis was performed (Figure 3C). Cell lysates were prepared from U3A cells expressing STAT1–GFP(L407A), either untreated or treated with IFN-γ and incubated with GST or GST–importin-α5. Although STAT1–GFP(L407A) was tyrosine phosphorylated following IFN-γ, no interaction with importin-α5 was detected. These results indicate that STAT1–GFP(L407A) cannot translocate to the nucleus because it is not recognized by importin-α5.
Evidence that the STAT1 NLS resides within a monomer
Since the NLS of STAT1 becomes functional only following dimerization, the NLS may be created by the juxtaposition of residues from each STAT1 monomer. Alternatively, a single STAT1 monomer may contain the NLS motif necessary for importin-α recognition, but requires a conformational change that follows tyrosine phosphorylation and dimerization. To distinguish between these possibilities we evaluated the behavior of heterologous dimers composed of the STAT1(L407A) mutant and wtSTAT molecules.
HT1080 cells expressing endogenous STAT1 were transfected with either wtSTAT1–GFP or STAT1– GFP(L407A), and cellular localization of the GFP-tagged proteins was visualized (Figure 4A). wtSTAT1–GFP behaved as expected and accumulated in the nucleus following IFN-γ stimulation (Figure 4A, a and b). STAT1–GFP(L407A) also responded to IFN-γ with detectable nuclear accumulation (Figure 4A, c and d). The nuclear accumulation of STAT1–GFP(L407A) in HT1080 cells is distinct from its defective nuclear translocation in U3A cells that lack endogenous wtSTAT1 (Figure 1). The individual cell transiently transfected with STAT1–GFP(L407A) can form three types of STAT1 dimers: wtSTAT1:wtSTAT1, wtSTAT1: STAT1–GFP(L407A) and STAT1–GFP(L407A):STAT1– GFP(L407A). The nuclear presence of STAT1–GFP (L407A) following IFN-γ stimulation is consistent with the ability of the STAT1:STAT1–GFP(L407A) heterodimers to translocate to the nucleus. Since the mutant STAT1–GFP(L407A) homodimers are defective for nuclear translocation, they are probably responsible for the cytoplasmic fluorescent signal. The ability of wtSTAT1 to restore STAT1–GFP(L407A) nuclear accumulation was also substantiated by co-transfection of wtSTAT1 and STAT1–GFP(L407A) in U3A cells (data not shown). The results suggest that a monomer of STAT1 with a functional NLS can dimerize and rescue a defective STAT1–GFP(L407A), supporting the existence of an NLS on an individual STAT1 monomer.
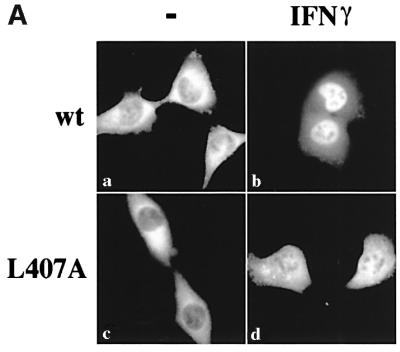
Fig. 4. wtSTAT1 rescues STAT1(L407A)-deficient nuclear accumulation. (A) HT1080 cells expressing wtSTAT1–GFP (a and b) or STAT1–GFP(L407A) (c and d) were untreated (a and c) or treated with IFN-γ for 30 min (b and d). Cells were fixed and STAT1–GFP cellular localization was examined by fluorescent microscopy. (B) The importin- α5 interaction assay was performed on whole-cell lysates from STAT1–GFP(L407A) expressing HT1080 cells either untreated (–) or treated with IFN-γ (+). Input is displayed in lanes 1 and 2. Cell lysates were incubated with GST (lanes 3 and 4) or GST–importin-α5 (lane 5 and 6) bound to glutathione beads. The interacting proteins were analyzed by western blot with anti-STAT1 phosphotyrosine antibody and the positions of endogenous wtSTAT1 and STAT1–GFP(L407A) are noted.
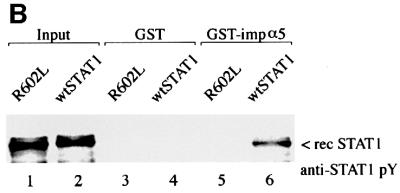
Accumulation of STAT1–GFP(L407A) in the nucleus of cells expressing wtSTAT1 in response to IFN-γ predicts that the STAT1:STAT1–GFP(L407A) heterodimer can interact with the importin-α receptor. To examine this possibility, an importin-α interaction assay was evaluated. A culture of HT1080 cells was transiently transfected with the gene encoding STAT1–GFP(L407A) and was untreated or treated with IFN-γ. Cell lysates were prepared from the population of cells and the ability of endogenous STAT1 and transfected STAT1–GFP(L407A) to bind importin-α5 was examined (Figure 4B). Both endogenous STAT1 and STAT1–GFP(L407A) were found to associate with importin-α5 following tyrosine phosphorylation. The relative protein signals in the western blot do not represent levels of mutant STAT1–GFP(L407A) or endogenous STAT1 present in the individual cells evaluated in Figure 4A. Since the transfection efficiency of a culture is not complete, the lysate contains proteins from both untransfected and successfully transfected cells in the population. Since STAT1–GFP(L407A) homodimers cannot bind importin-α5 (Figure 3C), these results indicate that heterodimers of STAT1:STAT1–GFP(L407A) are able to interact with importin-α5, and suggest that a monomer of STAT1 with a functional NLS can compensate for the inability of STAT1–GFP(L407A) to bind importin-α5.
To determine whether other STAT molecules can rescue the import defect of STAT1–GFP (L407A), the effect of a known dimerization partner of STAT1, STAT2, was evaluated (Figure 5A). The cytokine IFN-α induces tyrosine phosphorylation of both STAT1 and STAT2, which subsequently heterodimerize in association with the interferon regulatory factor-9/p48 to form a multimeric complex that translocates to the nucleus and binds a distinct DNA target site (Fu et al., 1992; Schindler et al., 1992; Veals et al., 1992; Darnell, 1997; Martinez-Moczygemba et al., 1997). STAT1:STAT1 homodimers also form in response to IFN-α, but functional STAT2 homodimers do not form. U3A cells that lack STAT1 but express endogenous STAT2 were used to evaluate localization of transfected wtSTAT1–GFP (top) or STAT1–GFP(L407A) (bottom). Localization of endogenous STAT2 was detected by immunofluorescence with rhodamine-labeled antibodies (STAT2, right panels), and compared with the localization of wtSTAT1–GFP or STAT1–GFP(L407A) (STAT1, left panels) in the same cell. In untreated cells expressing wtSTAT1–GFP, both STAT1 and STAT2 were primarily cytoplasmic (Figure 5A, a and b). Following IFN-α, wtSTAT1–GFP accumulated in the nucleus (Figure 5A, c); however, endogenous STAT2 only accumulated in the nucleus if the cells expressed wtSTAT1–GFP (Figure 5A, d, arrow). This result clearly shows that STAT2 accumulation in the nucleus requires STAT1:STAT2 dimerization, as has been suggested (Improta et al., 1994).
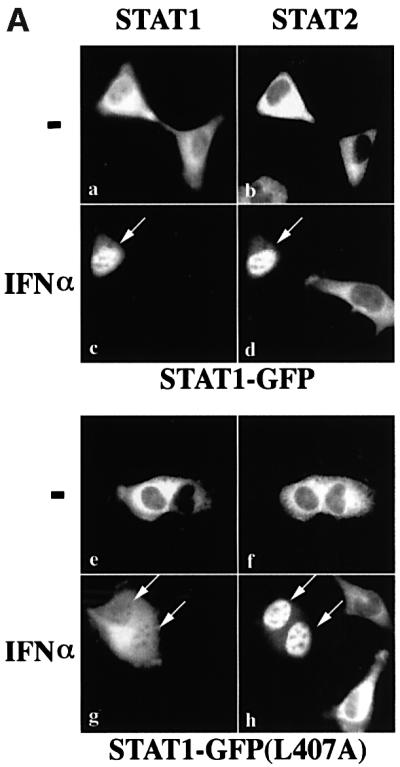
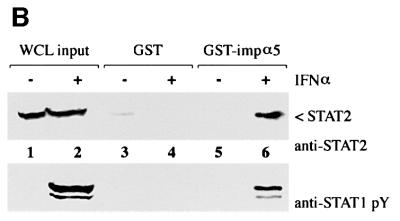
Fig. 5. STAT2 rescues STAT1(L407A) nuclear accumulation in response to IFN-α. (A) U3A cells transfected with STAT1–GFP (top panels) or STAT1–GFP(L407A) (bottom panels) were untreated (–) or treated with IFN-α for 45 min. Cells were fixed and STAT1–GFP fluorescence (left panels) or STAT2 immunofluorescence (right panels) was evaluated by fluorescent microscopy. Arrows point to cells expressing both endogenous STAT2 and transfected STAT1. (B) The importin-α5 interaction assay was performed on whole-cell lysates from HeLa cells that were either untreated (–) or treated with IFN-α (+). Input is displayed in lanes 1 and 2. Lysates were incubated with either GST (lanes 3 and 4) or GST–importin-α5 (lane 5 and 6) bound to glutathione beads. Western blots were performed with anti-STAT2 antibody (upper panel) or anti-STAT1 phosphotyrosine antibody (anti-STAT1 pY) (lower panel).
We next evaluated STAT1–GFP(L407A) localization in the U3A cells expressing endogenous STAT2. In the absence of IFN-α, STAT1–GFP(L407A) and STAT2 are cytoplasmic (Figure 5A, e and f). Following IFN-α, STAT1–GFP(L407A) fluorescence is apparent in the nucleus (Figure 5A, g), and STAT2 is nuclear in cells expressing STAT1–GFP(L407A) (Figure 5A, h, arrows). In response to IFN-α, both homodimers of STAT1–GFP(L407A):STAT1–GFP(L407A) and heterodimers of STAT1–GFP(L407A):STAT2 can form. The STAT1–GFP(L407A) nuclear fluorescence is probably due to STAT1–GFP(L407A):STAT2 heterodimers, since STAT1–GFP(L407A) homodimers are defective for nuclear translocation. The defective STAT1–GFP (L407A) homodimers can produce the residual cytoplasmic signal. Since STAT2 requires dimerization with STAT1 for nuclear translocation, it appears that dimerization of STAT1–GFP(L407A) with STAT2 enables translocation to the nucleus (Figure 5A, h, arrows). The results shown in Figures 4A and 5A are consistent with the premise that a functional NLS on a STAT1 or STAT2 monomer is sufficient for nuclear translocation of STAT1–GFP(L407A). To determine whether STAT1: STAT2 heterodimers can bind to the same importin-α5 receptor as STAT1:STAT1 homodimers, we performed an in vitro binding assay with lysates from cells expressing endogenous STAT1 and STAT2 (Figure 5B). Following cell treatment with IFN-α, both STAT2 (upper panel) and STAT1 (lower panel) interacted specifically with the C-terminus of importin-α5. This finding indicates that a similar unconventional NLS in STAT1 homodimers or STAT1:STAT2 heterodimers can be recognized by importin-α5.
Direct interaction of STAT1 with importin-α5
It remained possible that the interaction of STAT1 with importin-α5 was indirect and mediated by another cellular protein that served as an NLS bridging adapter. To test this possibility we isolated STAT1 from bacteria that express an inducible Elk-1 tyrosine kinase. Unphosphorylated STAT1 and tyrosine-phosphorylated STAT1 were prepared from bacteria and incubated with GST or GST–importin-α5 (Figure 6A). Only tyrosine-phosphorylated STAT1 was found to interact specifically with importin-α5 (lane 6). The interaction of importin-α5 with tyrosine-phosphorylated STAT1 in the absence of other mammalian cellular proteins indicates that binding is direct. Although the binding to importin-α5 is dependent on STAT1 tyrosine phosphorylation, the specific role that dimerization of STAT1 plays has not been demonstrated directly. To evaluate the requirement of dimerization, a mutant of STAT1 was tested that contains an alteration in the SH2 domain (R602L) and cannot dimerize (Figure 6B). This SH2 mutation expressed in mammalian cells is not tyrosine phosphorylated and does not accumulate in the nucleus following IFN-γ (Figure 1) (Improta et al., 1994; Shuai et al., 1994; Mowen and David, 1998). We used this mutant to distinguish between the contribution that phosphorylation or dimerization provides to importin-α recognition of STAT1. The STAT1 SH2–(R602L) mutation that cannot dimerize was expressed and isolated from bacteria with the activated Elk-1 tyrosine kinase. The mutant protein was accurately phosphorylated on tyrosine as evidenced by recognition with specific antibodies; however, in comparison with wtSTAT1 it could not bind importin-α5 (Figure 6B, lane 5 versus lane 6). These results demonstrate a requirement not only for tyrosine phosphorylation, but for dimerization of STAT1 to reveal the NLS inherent in a monomer.
Fig. 6. Tyrosine phosphorylated dimers of STAT1 interact directly with importin-α5. (A) Recombinant (rec) wtSTAT1 was expressed in TKX1 bacteria either unphosphorylated (–) or tyrosine phosphorylated (+) by the Elk kinase. Purified rec STAT1 was incubated with GST (lanes 3 and 4) or GST–importin-α5 (lane 5 and 6) bound to glutathione beads. The binding of rec STAT1 was detected by western blots performed with anti-STAT1 phosphotyrosine antibody (anti-STAT1 pY) (upper panel) or anti-STAT1 antibody (lower panel). (B) Recombinant (rec) tyrosine-phosphorylated wtSTAT1 or STAT1 SH2–(R602L) was purified from TKX1 bacteria expressing Elk kinase and evaluated in an importin-α5 binding assay. The binding of the rec STAT1 proteins was detected by western blot performed with anti-STAT1 phosphotyrosine antibody (anti-STAT1 pY).
DNA target sites effectively compete with importin-α for binding STAT1
The DNA binding domain of STAT1 spans amino acids 316–488 (Chen et al., 1998). The requirement of leucine 407 for STAT1 nuclear import suggests that this region may have a dual function in DNA contact and nuclear import. If the DNA binding domain functions in both recognition of DNA and of importin-α5, the STAT1 dimer may not be able to accommodate occupancy by both DNA and importin-α5 simultaneously. To investigate whether there is competition between DNA and importin-α5 for binding to STAT1, we evaluated the influence of each of them in binding assays.
Tyrosine-phosphorylated STAT1 dimers isolated from IFN-γ-treated HeLa cells were incubated with DNA corresponding to the specific IFN-γ activated site (GAS), or to non-specific DNA. An interaction assay was then performed with GST–importin-α5 and the presence of STAT1 bound to importin-α5 was detected by western blot (Figure 7A). STAT1 dimers clearly interacted with importin-α5 in the absence of DNA (Figure 7A, lane 1), but preincubation of STAT1 with DNAs corresponding to the GAS target sites of the IRF-1 gene or the FcγRI gene effectively ablated STAT1 interaction with importin-α5 (lanes 2 and 3) (Gutch et al., 1992; Kotanides et al., 1995). Non-specific DNA corresponding to a mutated GAS site or the interferon-stimulated response element (ISRE) did not impair the ability of STAT1 to bind importin-α5 (Figure 7A, lanes 4 and 5).

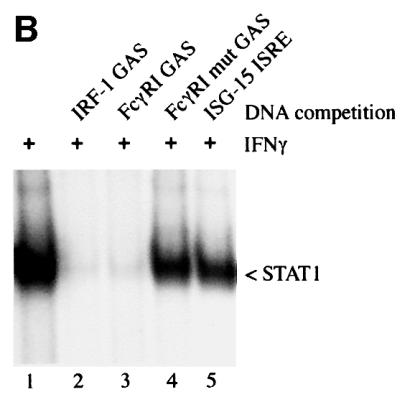
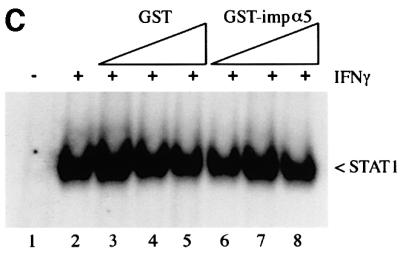
Fig. 7. Target DNA competes with importin-α5 for binding to STAT1 dimers. (A) Whole-cell lysates from IFN-γ-treated HeLa cells were preincubated with dsDNA oligonucleotides corresponding to the GAS element from the promoter of the IRF-1 gene (lane 2), the GAS element from the FcγRI gene (lane 3), a mutated oligonucleotide version of the FcγRI GAS (lane 4) or the ISRE from the promoter of the ISG15 gene (lane 5). The importin-α5 interaction assay was then performed by incubating the lysates with GST–importin-α5 bound to glutathione beads. Interacting STAT1 was detected by immunoblotting with anti-STAT1 antibody. (B) EMSA was performed with the radiolabeled IRF-1 GAS probe and extracts from HeLa cells treated with IFN-γ for 30 min. The extracts were preincubated for 20 min with 100-fold molar excess of unlabeled DNA corresponding to the IRF-1 GAS (lane 2), FcγRI GAS (lane 3), mutated FcγRI GAS (lane 4) or the ISG15 GAS (lane 5) prior to addition of probe. (C) EMSA was performed with the IRF-1 GAS probe and extracts from untreated HeLa cells (lane 1) or cells treated for 30 min with IFN-γ (lanes 2–8). The extracts were preincubated for 20 min with 0.05 µg (lanes 3 and 6), 0.5 µg (lanes 4 and 7) or 5 µg (lanes 5 and 8) of GST (lanes 3–5) or GST–importin-α5 (lanes 6–8).
The specific interaction of tyrosine-phosphorylated STAT1 dimers with the DNAs was confirmed by an EMSA (Figure 7B). Nuclear extract from IFN-γ treated cells was incubated with a radiolabeled GAS probe of the IRF-1 gene, and the appearance of the STAT1:DNA complex was readily detected (Figure 7B, lane 2). DNAs used in the importin-α5 binding assay were evaluated for their ability to bind STAT1 by competing with the radiolabeled DNA probe. Excess unlabeled DNA corresponding to the GAS sites of the IRF-1 or FcγRI gene competed with the radiolabeled DNA probe for STAT1 binding (lanes 2 and 3), but the mutant GAS and the ISG-15 ISRE did not (lanes 4 and 5). The results shown in Figure 7A and B indicate that importin-α5 cannot bind to tyrosine-phosphorylated STAT1 if it is bound to DNA, and suggest that importin-α5 and DNA have overlapping recognition sites within the STAT1 DNA binding domain.
To determine whether importin-α5 could influence the ability of STAT1 to bind DNA, the effect of importin-α5 was evaluated by an EMSA (Figure 7C). Nuclear extracts from IFN-γ-treated cells containing STAT1 dimers were incubated with increasing amounts of bacterially expressed GST–importin-α5 prior to incubation with DNA (Figure 7C). The prior binding of importin-α5 to STAT1 did not influence the ability of DNA to associate with STAT1. Even a molar concentration of GST–importin-α5 at approximately three orders of magnitude greater than DNA had no effect (Figure 7C, lane 8). This result indicates that DNA has a higher relative affinity for STAT1 dimers than importin-α5.
Discussion
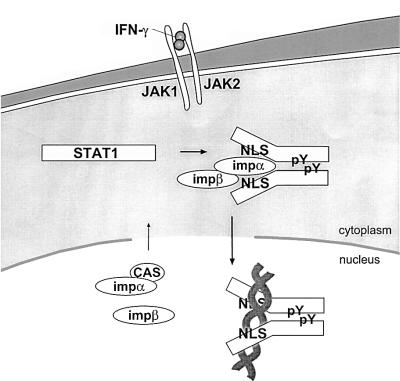
The ability of STAT transcription factors to redistribute from the cytoplasm to the nucleus is critical for their proper function as signaling molecules. In this study we investigated the mechanisms that regulate STAT1 dynamic redistribution in response to the IFN-γ cytokine. A conceptual illustration of STAT1 transport to the nucleus is presented in Figure 8, based on results from this and other studies. The latent STAT1 monomer that resides in the cytoplasm contains an NLS in an inactive state. Our previous studies demonstrated that STAT1 translocates to the nucleus by a gain-of-function mechanism, it is not anchored in the cytoplasm and does not constitutively shuttle in and out of the nucleus (McBride et al., 2000). Following specific tyrosine phosphorylation, STAT1 dimerizes via intermolecular SH2–phosphotyrosine interactions and this dimerization appears to trigger a conformational change in the STAT1 molecule that reveals a functional NLS.
Fig. 8. Conceptual model of STAT1 nuclear translocation. Latent STAT1 exists in the cytoplasm as a monomer. Following IFN-γ stimulation, STAT1 is tyrosine phosphorylated and dimerizes via intermolecular phosphotyrosine (pY)-SH2 domains. Dimerization results in a conformational change that allows the STAT1 NLS on each monomer to become functional. The importin-α5 (impα) shuttling receptor recognizes and binds to the NLS on the STAT1 dimer and effects translocation into the nucleus in association with importin-β (impβ). In the nucleus, STAT1 recognition of a specific DNA target leads to dissociation of importin-α5. Importin-α5 can be recycled to the cytoplasm by the cellular apoptosis susceptibility protein (CAS) export receptor.
We determined whether the NLS is intrinsic to the STAT1 protein or whether nuclear translocation of STAT1 dimers depends on association with an adapter protein. Since bacteria lack nuclear transport proteins, they were used as a source to prepare tyrosine-phosphorylated STAT1 dimers. These recombinant STAT1 dimers were shown to bind purified, bacterially expressed importin-α5 in vitro, supporting the concept that the NLS is intrinsic to the STAT1 protein (Figure 6A). However, tyrosine phosphorylation alone is not enough to trigger STAT1 nuclear translocation. Our in vitro analyses with the STAT1 SH2 mutation verify that nuclear localization is dependent on protein dimerization. Although the STAT1 SH2 mutation is accurately tyrosine phosphorylated by the Elk kinase in bacteria, the protein cannot dimerize via SH2 domain interactions, and it cannot bind to the importin-α5 shuttling receptor in vitro (Figure 6B). In addition, using complementary approaches others have demonstrated nuclear accumulation of STAT molecules following artificial dimerization in the absence of tyrosine phosphorylation (Bromberg et al., 1999; Milocco et al., 1999). These studies together indicate that dimerization is required to facilitate the functional appearance of an NLS on STATs.
Dimerization promotes not only nuclear translocation, but the ability of STATs to bind DNA. There is a growing number of characterized proteins that possess NLS motifs embedded within their DNA binding domains, and for this reason we introduced point mutations in the DNA binding domain of STAT1 to evaluate effects on nuclear translocation. The STAT1(L407A) mutation met the criterion of a mutation with a defect in NLS function. STAT1(L407A) was accurately tyrosine phosphorylated following IFN-γ stimulation and could subsequently dimerize and bind DNA; however, it did not translocate to the nucleus (Figures 1 and 2). Evaluation of the ability of STAT1(L407A) to bind importin-α5 revealed that the dimers were defective in their ability to bind the import shuttling receptor, indicating this as the basis for the lack of nuclear import (Figure 3).
A previous report described a double site mutation in STAT1, KK410/413AA, that lacked the ability to accumulate in the nucleus in response to IFN-γ, indicating that a region of the DNA binding domain was involved in nuclear import (Melen et al., 2001). However, this mutant protein also lacked the ability to bind DNA. The location of the L407A mutation in our study also suggests that the NLS is within the STAT1 DNA binding domain, but it is possible that the mutation causes a conformational change to inhibit NLS function at a distant location in the molecule. We speculated that if this region of L407 functioned as an NLS, it could bind either importin-α5 or target DNA, but would not be able to accommodate binding of both simultaneously. Our analyses clearly demonstrated that STAT1 binding to specific target DNA effectively blocks importin-α5 binding (Figure 7). These results support the proposition that this region of the DNA binding domain has a direct role in STAT1 interaction with both DNA and importin-α5. Our results also provide evidence that each STAT1 monomer possesses a functional NLS. Although the STAT1(L407A) mutant protein is tyrosine phosphorylated and dimerizes in response to IFN-γ, homodimers are not translocated to the nucleus. However, heterodimers of STAT1(L407A) with either wtSTAT1 or STAT2 can be translocated to the nucleus (Figures 4 and 5). These results suggest that one STAT monomer containing a functional NLS is sufficient to direct nuclear translocation of the dimer. In vitro physical association of wtSTAT1:STAT1(L407A) heterodimers with importin-α5 support this premise. The STAT1 NLS may represent a non-conventional NLS structure since the requirement of leucine 407 does not conform to the classical basic amino acid composition of an NLS (Dingwall and Laskey, 1991). In addition, the C-terminal region of the importin-α5 molecule that interacts with STAT1 is distinct from the armadillo repeat region that binds conventional NLSs (Sekimoto et al., 1997; Conti et al., 1998; Fontes et al., 2000).
The ability of specific DNA sequences to compete effectively with importin-α5 for binding to STAT1 dimers may play a role in appropriate targeting of STAT1 dimers to their destination in the nucleus, and in releasing the importin-α5 receptor from STAT1. Following importin- α5 release from its STAT1 cargo, it can be recognized by the export shuttling receptor, cellular apoptosis susceptibility protein (CAS), in association with Ran (Kutay et al., 1997). The CAS export receptor binds to the C-terminal region of importin-α5, as does the STAT1 dimer, and mediates importin-α5 transport back to the cytoplasm (Herold et al., 1998). The association of the STAT1 dimer with DNA is transient and following tyrosine dephosphorylation in the nucleus, it is exported back to the cytoplasm by the CRM1 export pathway (Haspel and Darnell, 1999; Begitt et al., 2000; McBride et al., 2000; Mowen and David, 2000). We demonstrated previously that the nuclear export signal (NES) of STAT1 is located within its DNA binding domain (amino acids 400–409) and the NES is masked when STAT1 is bound to DNA (McBride et al., 2000). The DNA binding domain of STAT1 appears to have evolved with NLS and NES motifs that may overlap and serve to direct the STAT1 protein to its proper cellular localization. The position of the NLS and the NES within the DNA binding domain of STAT1 suggests an elegant mechanism to regulate cellular localization of a signal transducer and activator of transcription.
Materials and methods
Cell culture and reagents
HeLa cells, HT1080 cells (ATCC) and U3A (STAT1–/–) (gift from G.R.Stark, Cleveland Clinic Foundation Research Institute) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 8% fetal bovine serum. Recombinant human IFN-α and IFN-γ was provided by Hoffman-LaRoche (Nutley, NJ) and used at 1000 U/ml. LMB was a gift from Barbara Wolff-Winiski (Novartis Research Institute, Austria) and used at 10 nM. DNA transfections were performed with Fugene-6 (Roche) or the calcium phosphate method (Wigler et al., 1979). The antibodies anti-STAT2 (Santa Cruz), anti-STAT1 phosphotyrosine (New England Biolabs) and anti-STAT1 antibody were as described previously (McBride et al., 2000).
Plasmid constructs
STAT1–GFP expressing human STAT1α was cloned into the green fluorescent protein EGFP-N1 plasmid (Clontech) as described previously (McBride et al., 2000). The STAT1–GFP (L407A), (K410A), (R405A) and (R602L) mutations were constructed by site-directed mutagenesis (Quikchange kit, Stratagene) and confirmed by sequencing the entire coding region of STAT1. wtSTAT1α was subcloned into the ProEX vector (Invitrogen) to construct a bacterial expression His-tagged STAT1. The construct GST–importin-α5 expressing amino acid residues 404–538 of the murine importin-α5 gene was generated by polymerase chain reaction (PCR) amplification of the importin-α5 gene followed by cloning in-frame with PGEX-KG (Guan and Dixon, 1991; Kumar et al., 2000).
Importin-α5 interaction assay
Whole-cell lysates were prepared from mammalian cells either untreated or treated with IFN-γ or IFN-α for 30–45 min. Briefly, cells were incubated in buffer A (20 mM HEPES pH 7.9, 1 mM EDTA, 10 µM sodium vanadate, 2 mM 2-mercaptoethanol and protease inhibitors) for 20 min at 4°C. Cells were dounce homogenized, and the cytosol was separated from the nuclear fraction by centrifugation. Nuclear proteins were extracted in buffer B (20 mM HEPES pH 7.9, 420 mM KCl, 20% glycerol, 1 mM EDTA, 2 mM 2-mercaptoethanol, 10 µM sodium vanadate and protease inhibitors) for 20 min at 4°C. Nuclei were pelleted and discarded and the nuclear extracts were combined with the cytosolic extracts to generate whole-cell lysates. The interaction assay was performed with 500 µg of protein from whole-cell lysates incubated with 20 µg of bacterially expressed and purified GST or GST– importin-α5 bound to glutathione–agarose beads pre-blocked with 1% bovine serum albumin (BSA) for 1 h at 4°C. For the DNA competition experiments, the indicated double-stranded (ds) DNA oligonucleotides were added at a concentration of 1 µg/ml. Input STAT protein represents anti-STAT immunopreciptation performed with 500 µg protein of whole-cell lysates (Martinez-Moczygemba et al., 1997).
Bacterial STAT1 with a His6 tag at the N-terminus was expressed in TKX1 bacteria (Stratagene) using the suggested protocols to produce either unphosphorylated or tyrosine-phosphorylated protein. Bacteria were sonicated and STAT1 was purified using Talon resin (Clontech), eluted with EDTA and dialyzed in Tris-buffered saline pH 7.5 with 10% glycerol. The GST–importin-α5 interaction assay was performed with 1 µg of bacterially expressed and purified STAT1 in 0.5 ml of buffer A with 110 mM potassium acetate, 1 mg/ml of BSA, 0.1% Tween-20.
Fluorescent microscopy
Cells were plated on glass coverslips and transfected with Fugene-6 (Roche) 48 h prior to evaluation. Where indicated cells were pretreated for 30 min with LMB. The cells were then treated with IFN-γ for 30 min or IFN-α for 45 min, and fixed with 4% formaldehyde solution. GFP localization was evaluated using a Zeiss Axioskop (Carl Zeiss) equipped for epifluorescence with a GFP filter set (Chroma Technology). STAT2 localization was detected by permeablizing cells with 0.2% Triton X-100, immunostaining with anti-STAT2 antibody and rhodamine-conjugated secondary antibody. The fluorescence was detected with a rhodamine filter set (Chroma Technology). Images were captured with a Diagnostic Instruments Spot 2 CCD camera (Sterling Heights, MI) and presented in Adobe Photoshop 4.0.
Protein analysis and DNA binding assay
STAT1 immunoprecipitation was performed on whole-cell lysates with anti-STAT1 monoclonal antibody (Martinez-Moczygemba et al., 1997). Immunocomplexes were collected on protein G–agarose beads, separated by SDS–PAGE and transferred to Immobilon-P membrane (Millipore). The membrane was reacted with appropriate primary antibody, and protein was detected using secondary antibody (Amersham) and the enhanced chemiluminescence system (Dupont NEN).
The EMSAs were performed as described previously using the IRF-1 GAS element as a radiolabeled probe (Kotanides and Reich, 1993; McBride et al., 2000). The STAT1–GFP and STAT1–GFP(L407A) proteins were prepared from whole-cell lysates by incubating cells in buffer B with 40 µg/ml digitonin for 30 min. Cell debris was pelleted and discarded, and 50 µg protein lysate was used per reaction. The DNA binding reactions were preincubated with anti-STAT1 antibody prior to the addition of a radiolabeled IRF-1 GAS dsDNA oligonucleotide. For competition experiments, 100 ng of dsDNA oligonucleotides were preincubated with extracts prior to addition of DNA probe: IRF-1 GAS (5′-GCCTGATTTCCCCGAAATGACGG), 5′-FcγRI GAS (5′-GTATTTCCCAGAAAAGGAAC), FcγRI mutant GAS (5′-GTATTTCCCCAGAAAAGGAACAT), ISG15 ISRE (5′-GGGAAAGGGAAACCGAAACTGAA).
Acknowledgments
Acknowledgements
The authors thank members of the laboratory for their advice and support, particularly Sarah Van Scoy for her technical assistance. The collective input of our colleagues Dafna BarSagi, Patrick Hearing, Michael Hayman and W.Todd Miller is appreciated. We thank James E.Darnell, Jr (Rockefeller University) for the gift of the STAT1α cDNA, Colin Dingwall (Glaxo Smith Kline) for the gift of full-length importin-α5, George R.Stark (Cleveland Clinic Foundation Research Institute) for the gift of U3A cells and Barbara Wolff-Winiski (Novartis Research Institute) for the gift of LMB. This work was supported by grants from the National Institutes of Health (RO1CA50773 and PO1CA28146) to N.C.R. and in part by a Carol M.Baldwin Cancer Award.
References
- Adam S.A. (1999) Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr. Opin. Cell Biol., 11, 402–406. [DOI] [PubMed] [Google Scholar]
- Begitt A., Meyer,T., van Rossum,M. and Vinkemeier,U. (2000) Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl Acad. Sci. USA, 97, 10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J.F., Wrzeszczynska,M.H., Devgan,G., Zhao,Y., Pestell,R.G., Albanese,C. and Darnell,J.E.,Jr (1999) Stat3 as an oncogene [published erratum appears in Cell (1999) 99, 239]. Cell, 98, 295–303. [DOI] [PubMed] [Google Scholar]
- Chen X., Vinkemeier,U., Zhao,Y., Jeruzalmi,D., Darnell,J.E.,Jr and Kuriyan,J. (1998) Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell, 93, 827–839. [DOI] [PubMed] [Google Scholar]
- Cokol M., Nair,R. and Rost,B. (2000) Finding nuclear localization signals. EMBO rep., 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E., Uy,M., Leighton,L., Blobel,G. and Kuriyan,J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell, 94, 193–204. [DOI] [PubMed] [Google Scholar]
- Cortes P., Ye,Z.S. and Baltimore,D. (1994) RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc. Natl Acad. Sci. USA, 91, 7633–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E. Jr, (1997) STATs and gene regulation. Science, 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- Davis L.I. (1995) The nuclear pore complex. Annu. Rev. Biochem., 64, 865–896. [DOI] [PubMed] [Google Scholar]
- Dingwall C. and Laskey,R.A. (1991) Nuclear targeting sequences—a consensus? Trends Biochem. Sci., 16, 478–481. [DOI] [PubMed] [Google Scholar]
- Dingwall C. and Laskey,R.A. (1998) Nuclear import: a tale of two sites. Curr. Biol., 8, R922–R924. [DOI] [PubMed] [Google Scholar]
- Fontes M.R., Teh,T. and Kobe,B. (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol., 297, 1183–1194. [DOI] [PubMed] [Google Scholar]
- Fu X.Y., Schindler,C., Improta,T., Aebersold,R. and Darnell,J.E.,Jr (1992) The proteins of ISGF-3, the interferon α-induced transcriptional activator, define a gene family involved in signal transduction. Proc. Natl Acad. Sci. USA, 89, 7840–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour K.C. and Reich,N.C. (1995) Signal transduction and activation of gene transcription by interferons. Gene Expr., 5, 1–18. [PMC free article] [PubMed] [Google Scholar]
- Gorlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Henklein,P., Laskey,R.A. and Hartmann,E. (1996) A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J., 15, 1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Guan K.L. and Dixon,J.E. (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem., 192, 262–267. [DOI] [PubMed] [Google Scholar]
- Gutch M.J., Daly,C. and Reich,N.C. (1992) Tyrosine phosphorylation is required for activation of an α interferon-stimulated transcription factor. Proc. Natl Acad. Sci. USA, 89, 11411–11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haspel R.L. and Darnell,J.E.,Jr (1999) A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl Acad. Sci. USA, 96, 10188–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A., Truant,R., Wiegand,H. and Cullen,B.R. (1998) Determination of the functional domain organization of the importin α nuclear import factor. J. Cell Biol., 143, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington J., Rui,L., Luo,G., Yu-Lee,L.Y. and Carter-Su,C. (1999) A functional DNA binding domain is required for growth hormone-induced nuclear accumulation of Stat5B. J. Biol. Chem., 274, 5138–5145. [DOI] [PubMed] [Google Scholar]
- Hodel M.R., Corbett,A.H. and Hodel,A.E. (2001) Dissection of a nuclear localization signal. J. Biol. Chem., 276, 1317–1325. [DOI] [PubMed] [Google Scholar]
- Ihle J.N., Witthuhn,B.A., Quelle,F.W., Yamamoto,K. and Silvennoinen,O. (1995) Signaling through the hematopoietic cytokine receptors. Annu. Rev. Immunol., 13, 369–398. [DOI] [PubMed] [Google Scholar]
- Improta T., Schindler,C., Horvath,C.M., Kerr,I.M., Stark,G.R. and Darnell,J.E.,Jr (1994) Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc. Natl Acad. Sci. USA, 91, 4776–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans D.A., Xiao,C.Y. and Lam,M.H. (2000) Nuclear targeting signal recognition: a key control point in nuclear transport? BioEssays, 22, 532–544. [DOI] [PubMed] [Google Scholar]
- Kohler M., Speck,C., Christiansen,M., Bischoff,F.R., Prehn,S., Haller,H., Gorlich,D. and Hartmann,E. (1999) Evidence for distinct substrate specificities of importin α family members in nuclear protein import. Mol. Cell. Biol., 19, 7782–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotanides H. and Reich,N.C. (1993) Requirement of tyrosine phosphorylation for rapid activation of a DNA binding factor by IL-4. Science, 262, 1265–1267. [DOI] [PubMed] [Google Scholar]
- Kotanides H., Moczygemba,M., White,M.F. and Reich,N.C. (1995) Characterization of the interleukin-4 nuclear activated factor/STAT and its activation independent of the insulin receptor substrate proteins. J. Biol. Chem., 270, 19481–19486. [DOI] [PubMed] [Google Scholar]
- Kudo N., Wolff,B., Sekimoto,T., Schreiner,E.P., Yoneda,Y., Yanagida,M., Horinouchi,S. and Yoshida,M. (1998) Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res., 242, 540–547. [DOI] [PubMed] [Google Scholar]
- Kumar K.P., McBride,K.M., Weaver,B.K., Dingwall,C. and Reich,N.C. (2000) Regulated nuclear–cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol., 20, 4159–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Gorlich,D. (1997) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- La Casse E.C. and Lefebvre,Y.A. (1995) Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res., 23, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard W.J. and O’Shea,J.J. (1998) Jaks and STATs: biological implications. Annu. Rev. Immunol., 16, 293–322. [DOI] [PubMed] [Google Scholar]
- Macara I.G. (2001) Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev., 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Moczygemba M., Gutch,M.J., French,D.L. and Reich,N.C. (1997) Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-α-stimulated transcription factor ISGF3. J. Biol. Chem., 272, 20070–20076. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- McBride K.M., McDonald,C. and Reich,N.C. (2000) Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J., 19, 6196–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K., Kinnunen,L. and Julkunen,I. (2001) Arginine/lysine-rich structural element is involved in IFN-induced nuclear import of STATs. J. Biol. Chem., 276, 16447–16455. [DOI] [PubMed] [Google Scholar]
- Milocco L.H., Haslam,J.A., Rosen,J. and Seidel,H.M. (1999) Design of conditionally active STATs: insights into STAT activation and gene regulatory function. Mol. Cell. Biol., 19, 2913–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J., Blobel,G. and Radu,A. (1996) The binding site of karyopherin α for karyopherin β overlaps with a nuclear localization sequence. Proc. Natl Acad. Sci. USA, 93, 6572–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen K. and David,M. (1998) Role of the STAT1-SH2 domain and STAT2 in the activation and nuclear translocation of STAT1. J. Biol. Chem., 273, 30073–30076. [DOI] [PubMed] [Google Scholar]
- Mowen K. and David,M. (2000) Regulation of STAT1 nuclear export by Jak1. Mol. Cell. Biol., 20, 7273–7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Laxton,C., Briscoe,J., Schindler,C., Improta,T., Darnell,J.E.,Jr, Stark,G.R. and Kerr,I.M. (1993) Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J., 12, 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. (1997) Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature, 386, 779–787. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Blobel,G. and Rosenblum,J.S. (1998) Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol., 10, 392–399. [DOI] [PubMed] [Google Scholar]
- Schindler C., Fu,X.Y., Improta,T., Aebersold,R. and Darnell,J.E.,Jr (1992) Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon α. Proc. Natl Acad. Sci. USA, 89, 7836–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T., Imamoto,N., Nakajima,K., Hirano,T. and Yoneda,Y. (1997) Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J., 16, 7067–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K., Horvath,C.M., Huang,L.H., Qureshi,S.A., Cowburn,D. and Darnell,J.E.,Jr (1994) Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell, 76, 821–828. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Veals S.A., Schindler,C., Leonard,D., Fu,X.Y., Aebersold,R., Darnell,J.E.,Jr and Levy,D.E. (1992) Subunit of an α-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol., 12, 3315–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K., Ryder,U. and Lamond,A.I. (1996) The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J., 15, 1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Sweet,R., Sim,G.K., Wold,B., Pellicer,A., Lacy,E., Maniatis,T., Silverstein,S. and Axel,R. (1979) Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell, 16, 777–785. [DOI] [PubMed] [Google Scholar]
- Wolff B., Sanglier,J.J. and Wang,Y. (1997) Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol., 4, 139–147. [DOI] [PubMed] [Google Scholar]