Abstract
Acetylation is a prominent post-translational modification of nucleosomal histone N-terminal tails, which regulates chromatin accessibility. Accordingly, histone acetyltransferases (HATs) play major roles in processes such as transcription. Here, we show that the HAT Tip60, which is involved in DNA repair and apoptosis following γ irradiation, is subjected to proteasome-dependent proteolysis. Furthermore, we provide evidence that Mdm2, the ubiquitin ligase of the p53 tumour suppressor, interacts physically with Tip60 and induces its ubiquitylation and proteasome-dependent degradation. Moreover, a ubiquitin ligase-defective mutant of Mdm2 had no effect on Tip60 stability. Our results indicate that Mdm2 targets both p53 and Tip60, suggesting that these two proteins could be co-regulated with respect to protein stability. Consistent with this hypothesis, Tip60 levels increased significantly upon UV irradiation of Jurkat cells. Collectively, our results suggest that degradation of Tip60 could be part of the mechanism leading to cell transformation by Mdm2.
Keywords: histone acetyltransferase/Mdm2/proteasome/Tip60/ubiquitylation
Introduction
Cellular processes that require access to the DNA double helix have to deal with chromatin structure. Therefore, the proteins that regulate chromatin structure play major roles in processes such as transcription, and DNA replication and repair. At the nucleosome level, two main mechanisms of chromatin remodelling have been described, one relying on ATP hydrolysis and catalysed by the ATPases of the SWI/SNF family, the other involving covalent modifications of nucleosomal histones. Histones are modified mainly within their N-terminal tails, which protrude from the nucleosomes. These tails can be modified by acetylation, phosphorylation, methylation and ADP ribosylation. Histone acetylation largely correlates with transcriptional activation and has generally been thought to create an ‘open’ chromatin structure, more accessible to transcription factors. Indeed, acetylation of histones increases the affinity of TATA box binding protein (TBP) for nucleosomal DNA (Sewack et al., 2001). An alternative, not exclusive, hypothesis is that histone acetylation functions as a signal regulating protein binding to histone N-terminal tails (Strahl and Allis, 2000). Consistent with this possibility, acetylated lysines are recognized by the so-called ‘bromodomain’, a domain shared by many proteins involved in chromatin function (Jacobson et al., 2000).
Whatever the mechanism, enzymes that control histone acetylation are critical for the proper control of gene expression. Histone acetyltransferases (HATs) are generally transcriptional co-activators, whereas histone deacetylases (HDACs) are usually involved in transcriptional repression (Turner, 2000). They are believed to be targeted to specific promoters through physical interactions with promoter-specific transcription factors. Through their effect on the transcription of specific genes, HATs and HDACs are involved in the control of many cellular processes, such as cell cycle progression, terminal differentiation and apoptosis (Marmorstein and Roth, 2001).
Recently, the HAT Tip60, identified as a cellular protein interacting with the HIV Tat protein (Kamine et al., 1996), has been shown to be required for DNA repair and apoptosis following irradiation of human cells (Ikura et al., 2000). Tip60 is a member of the MYST family of HATs, a family that is conserved from yeast to mammals (Sterner and Berger, 2000). Members of this family are characterized by a conserved catalytic domain, the MYST domain. In addition to the MYST domain, Tip60 contains a chromodomain at its N-terminus. Chromodomains are found in many chromatin-related proteins, and have been proposed to mediate protein–RNA interaction (Akhtar and Becker, 2001) or binding to methylated lysines (Bannister et al., 2001; Lachner et al., 2001).
Many studies have shown that Tip60 can regulate transcription, either positively or negatively depending on the cell type or the promoter, but the role of its HAT activity in these processes remains to be elucidated (Brady et al., 1999; Creaven et al., 1999; Hlubek et al., 2001). Interestingly, the HAT activity of Tip60 is modulated by Tat, suggesting that it could be an important cellular target of Tat (Creaven et al., 1999). The functional importance of Tip60 is also suggested by the fact that many alternatively spliced variants of Tip60 have been described (Ran and Pereira-Smith, 2000; Hlubek et al., 2001), perhaps allowing a strict regulation of its activity.
Recently, Tip60 was proposed to function as the HAT subunit of a large complex (Ikura et al., 2000). This complex is likely to be the mammalian homologue of the yeast NuA4 complex, which contains the MYST acetyltransferase ESA1 (Allard et al., 1999). Whereas purified Tip60 acetylates nucleosomal substrates poorly, the Tip60 complex can readily acetylate histones H4 and H2A assembled in nucleosomes. The Tip60 complex also contains human Tip49a and Tip49b, two helicases related to the Escherichia coli RuvB protein (Ikura et al., 2000). This might suggest that the Tip60 complex could be involved in repair of DNA double strand breaks by homologous recombination. Although the involvement of the Tip60 complex in the process of homologous recombination is still unproven, it has been shown that Tip60 HAT activity is required for DNA repair and apoptosis following DNA double strand breaks induced by γ irradiation (Ikura et al., 2000).
DNA damage-induced apoptosis is a process that is, at least in part, under the control of the p53 tumour suppressor (Soussi, 2000). In the absence of appropriate signalling, p53 activity is inhibited by Mdm2, which binds directly to the transactivation domain of p53 and blocks its activity (Momand et al., 1998). In addition, Mdm2 also functions as an E3 ubiquitin ligase for p53 (Honda et al., 1997). Since Mdm2-induced ubiquitylation of p53 targets p53 for proteasome-dependent degradation (Haupt et al., 1997; Kubbutat et al., 1997), the p53 protein, and accordingly its activity, are maintained at very low levels under normal growth conditions. However, upon DNA damage (for a recent review see Zhou and Elledge, 2000), p53 is no longer subjected to proteolytic degradation, resulting in its accumulation and stimulation of its negative growth-regulatory properties.
In this study, we report that, similar to p53, Tip60 is regulated by the ubiquitin–proteasome pathway. Further more, Mdm2 binds to Tip60 and induces its ubiquitylation in vivo and in vitro, indicating that Mdm2 functions as a ubiquitin ligase for Tip60. Moreover, overexpression of Mdm2 induces the proteasome-dependent degradation of Tip60 in living cells. Taken together, these results suggest that Mdm2 links the regulation of Tip60 to that of p53. Consistent with that, we found that UV irradiation strongly induces Tip60 expression.
Results
Mdm2 expression decreases Tip60 levels
The HAT p300 has been shown to be required for p53 accumulation following DNA damage (Yuan et al., 1999). We tested the hypothesis that the effect of Tip60 on apoptosis could be mediated by a similar mechanism, involving the control of p53 expression. We assayed the effect of Tip60 overexpression on p53 expression in transient transfection experiments, either in the presence or absence of exogenous Mdm2. Although these experiments were not conclusive (data not shown), we consistently noticed that, in the presence of Mdm2, the amount of overexpressed Tip60 was reduced. We thus intended to study whether this effect was specific for Tip60 and was not due to any effect on transcription or translation from the Tip60 expression vector. We transfected U2OS cells with CMV-driven expression vectors for HA-tagged Tip60, HA-tagged Ku80 as an internal control and luciferase reporter protein, either in the presence or absence of Mdm2. After standardization using luciferase activity for transfection efficiency and any non-specific effects on the CMV promoter, we analysed total cell extracts for the expression levels of Tip60 and Ku80 by an anti-HA western blot (Figure 1A). We found that, in the presence of exogenous Mdm2, the amount of HA-Tip60 was significantly reduced, whereas the amount of HA-Ku80 was unaffected. Since both cDNAs were cloned in the same vector, which expresses proteins with an N-terminal tag of ∼70 amino acids, this result suggests that Mdm2 overexpression affects the half-life of ectopically expressed Tip60 rather than its transcription/translation rate.
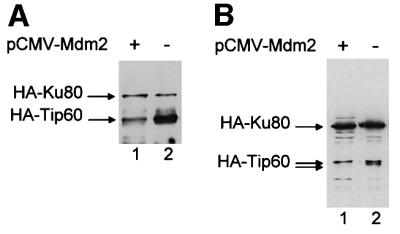
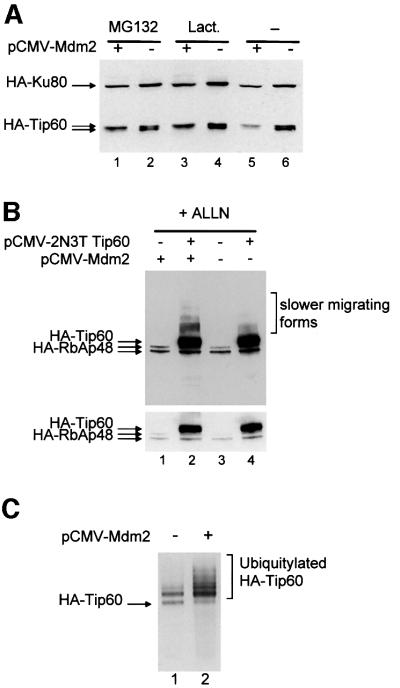
Fig. 1. Co-expression of Mdm2 decreases Tip60 expression. (A) U2OS cells (1 × 105) were transfected with 100 ng of pCMV luciferase reporter vector, 1 µg of pCMV 2N3T Tip60, 1 µg of pCMV 2N3T Ku80, in the presence (lane 1) or absence (lane 2) of 2 µg of pCMV NeoBam Mdm2. Ku80 is a component of the DNA end-binding Ku heterodimer. The amount of CMV promoter in the transfection was kept constant by the addition of empty vectors. Luciferase activity was measured 24 h after transfection,. After standardization for luciferase activity, total cell extracts (obtained by boiling cells in Laemmli sample buffer) were loaded directly and subjected to an anti-HA western blot to detect exogenous Tip60 and Ku80, as indicated. (B) Same experiment as in (A), except that we used pCDNA3 HA-Tip60 instead of pCMV 2N3T Tip60. Note that this exogenous Tip60 migrates as two bands in the absence of Mdm2, because of phosphorylation (unpublished results).
The HA-tagged Tip60 that we used in Figure 1A contained two heterologous nuclear localization signals. To check whether the decrease in the presence of Mdm2 was not due to an aberrant subcellular localization of the exogenous protein, we used in a similar experiment an expression vector for Tip60 tagged with only one HA tag (Figure 1B). This shorter protein resolves better on a SDS–PAGE gel and migrates as a doublet, with the higher form corresponding to phosphorylated Tip60 (our unpublished results). In the presence of exogenous Mdm2, we observed that the lower form disappeared almost completely, whereas the higher form was largely unaffected. Note that in other experiments (e.g. Figure 6B), both bands were affected by the presence of exogenous Mdm2. This variation probably reflected a difference in the relative levels of expression of exogenous Mdm2 and Tip60. Subsequent experiments (see Figures 3A and 6B), as well as the use of a non-phosphorylated Tip60 mutant (Supplementary figure 1, available at The EMBO Journal Online), indicated that phosphorylation of Tip60 is unlikely to regulate its degradation by Mdm2. Neverthe less, this result confirmed that the presence of Mdm2 affected Tip60 expression.
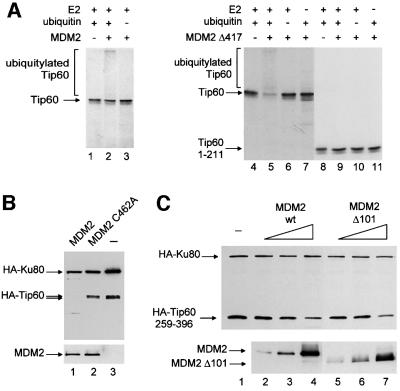
Fig. 6. Ubiquitylation of Tip60 by Mdm2 is involved in Tip60 degradation. (A) 35S-labelled, in vitro-translated, full-length Tip60 (lanes 1–7) or Tip60 1–211 (lanes 8–11) were subjected to an in vitro ubiquitylation reaction by GST–Mdm2 or GST–Mdm2 Δ417, where indicated. Ubiquitin was omitted in reactions loaded in lanes 3, 6 and 10 whereas UbcH5 (E2) was omitted in lanes 7 and 11. Note the appearance in the presence of Mdm2 (lane 2) and Mdm2Δ417 (lane 5) of bands of lower mobility, which indicate mono- and polyubiquitylation, and the concomitant decrease in the amount of unmodified Tip60. (B) U2OS cells were transfected with 100 ng of pCMV luciferase reporter vector, 1 µg of pCDNA3 HA-Tip60, 1 µg of pCMV 2N3T Ku80, in the absence (lane 3) or presence of either 2 µg of pXJ Mdm2 (lane 1) or 2 µg of pXJ Mdm2 C462A (lane 2). The amount of promoters in the transfection was kept constant by the addition of empty vectors. Luciferase activity was measured 24 h after transfection. Total cell extracts were loaded directly and subjected to an anti-HA western blot to detect exogenous Tip60 and Ku80, as indicated (upper panel). In the lower panel, 10 µl of total cell extracts were loaded directly, and probed with the anti-Mdm2 antibody. (C) U2OS cells were transiently transfected with 100 ng of pCMV luciferase reporter vector, 1 µg of pCDNA3 Tip60 259–396, 1 µg of pCMV 2N3T Ku80 and increasing amounts of either pCMV Neo Bam Mdm2 or pCMV Mdm2 Δ101 (0.5, 1 or 2 µg as indicated by the height of the open triangle). Total cell extracts were loaded directly and subjected to an anti-HA western blot to detect exogenous Tip60 and Ku80, as indicated (upper panel). In the lower panel, 10 µl of total cell extracts were loaded directly, and probed with the anti-Mdm2 antibody.
Fig. 3. Mdm2 induces the ubiquitylation and proteasome-mediated degradation of exogenous Tip60. (A) U2OS cells were transfected as in Figure 1B with pCMV luciferase, pCDNA3 HA-Tip60, pCMV 2N3T Ku80 and with or without pCMVNeoBam Mdm2. Twenty-four hours after transfection, the proteasome inhibitor indicated (Lact., lactacystin) was added (lanes 1–4) or not (lanes 5–6). After 6 h, total cell extracts were analysed as described in Figure 1B. (B) U2OS cells were transfected with 100 ng of pCMV luciferase reporter vector, 1 µg of pCMV 2N3T RbAp48, 1 µg of pCMV 2N3T Tip60 where indicated and 2 µg of pCMV NeoBam Mdm2 as indicated. The amount of promoters in the transfection was kept constant using empty vectors. ALLN was added for 6 h, 24 h after transfection,. Total cell extracts were analysed by western blotting using the anti-HA antibody. The top panel shows a long exposure of the blot. Note the increase in the amount of HA-reactive more slowly migrating forms in the presence of Mdm2 (compare lane 2 with lane 4). The bottom panel shows a short exposure of the same blot. (C) U2OS cells transfected with 10 µg pCMV 2N3T Tip60, 10 µg pSG5 His-ubiquitin and 10 µg pCMV NeoBam Mdm2 where indicated, were analysed as in Figure 2C.
Tip60 is degraded by the ubiquitin–proteasome pathway
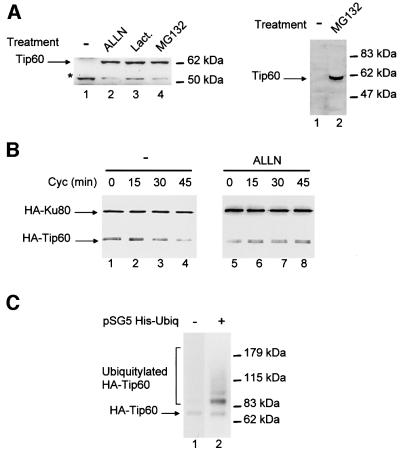
Since Mdm2 is a ubiquitin ligase (Honda et al., 1997) that is known to induce the degradation of p53 through the proteasome (Haupt et al., 1997; Kubbutat et al., 1997), we next tested whether Tip60 expression could also be subjected to proteasome-mediated proteolysis. We found that the treatment of various human cell lines by the proteasome inhibitor ALLN, lactacystin or MG132 resulted in the appearance of a strong band at 60 kDa (Figure 2A; data not shown), which most likely represents Tip60, since it was detected by two independent anti-Tip60 antibodies (Figure 2A). This result indicates that endogenous Tip60 is degraded through the proteasome pathway.
Fig. 2. Tip60 is subjected to proteasome-mediated degradation. (A) Jurkat cells (left panel) or HeLa cells (right panel) were treated or not with the proteasome inhibitor indicated (Lact., lactacystine). Total cell extracts were then tested for the presence of Tip60 by western blotting using either the anti-Tip60 DT antibody (left panel) or the anti-Tip60 SK antibody (right panel). The band indicated Tip60 co-migrates with the exogenous HA-Tip60 produced from pCDNA3 (data not shown). The asterisk (*) indicates a non-specific band recognized by the anti-Tip60 DT antibody. (B) U2OS cells were transfected as in Figure 1B. Twenty-four hours after transfection, the proteasome inhibitor ALLN was added where indicated (lanes 5–8). After 2 h, cycloheximide was added and cells were harvested after treatment for the length of time indicated. Total cell extracts were analysed as described in Figure 1A. (C) U2OS cells were transfected with 10 µg of pCMV 2N3T Tip60 and 10 µg of either pSG5 His-ubiquitin (lane 2) or pSG5 HA-ubiquitin (lane 1). Twenty-four hours after transfection, cells were lysed as described in Materials and methods and His-tagged proteins were purified by nickel chromatography. Nickel binding proteins were subjected to a western blot using the anti-HA antibody. The arrow points to the unmodified HA-Tip60 protein that harboured background binding to nickel beads.
To strengthen the notion that Tip60 is degraded in a proteasome-dependent manner, we next investigated whether proteasome inhibition would increase the half-life of Tip60. Unfortunately, we could not reproducibly detect endogenous Tip60 in the absence of proteasome inhibitors (see e.g. Figure 2A, lane 1). Using pulse–chase labelling, we found that transfected HA-Tip60 was a highly unstable protein, with a half-life of between 15 and 30 min depending on the experiment (Supplementary figure 2). However, we consistently noticed that, in these experiments, only a very low proportion of Tip60 was extracted (<5%), the remainder being completely insoluble in any immunoprecipitation-compatible buffer tested (data not shown and see also legend to Figure 5). Consequently, this calculated half-life is correct for only the extractible fraction of the exogenous Tip60.
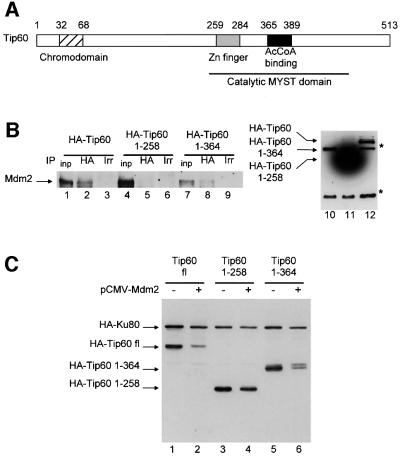
Fig. 5. Mdm2 induces Tip60 degradation through their physical interaction. (A) Schematic representation of the Tip60 protein. The hatched box represents the chromodomain, the grey box the putative Zn finger and the black box the AcCoA binding site. Also indicated is the HAT domain, which is conserved among proteins of the MYST family. (B) U2OS cells were transfected with 10 µg of pCMV NeoBam Mdm2 and either 10 µg of pCDNA3 HA-Tip60 fl, pcDNA3 HA-Tip60 1–258 or pCDNA3 HA-Tip60 1–364. Whole-cell extracts were prepared 24 h after transfection, and subjected to immunoprecipitation with either the anti-HA antibody (lanes 2, 5 and 8) or an irrelevant antibody (anti-myc 9E10, lanes 3, 6 and 9), as indicated. Immunoprecipitates were tested for the presence of Mdm2 by western blotting. In lanes labelled inp (input), 10 µl of whole-cell extracts were loaded directly. In lanes 10–12, 10 µl of whole-cell extracts were subjected to an anti-HA western blot, to monitor the quantity of the various mutants in the extracts. Note that Tip60 1–258 is present in far larger amounts than Tip60 fl or 1–364, because these latter proteins are very poorly extractible from cells (compare with C). The asterisks indicate two non-specific bands. HA-Tip60 1–364 migrates just below a non-specific band. (C) U2OS cells were transfected as in Figure 1B with expression vectors of the Tip60 deletion mutant indicated (in pCDNA3-HA, 1 µg), pCMV 2N3T Ku80 (1 µg), pCMV luciferase reporter vector (100 ng), in the presence or absence of pCMV NeoBam Mdm2 (2 µg), as indicated. The amount of promoters in the transfection was kept constant using empty vectors. Luciferase activity was measured 24 h after transfection. The steady-state levels of HA-tagged proteins were assessed by western blotting.
Therefore, we determined the half-life of ectopically expressed Tip60 using cycloheximide, which is a potent inhibitor of protein translation. We transfected U2OS cells with a HA-Tip60 expression vector, and the amount of exogenous Tip60 remaining after various durations of cycloheximide treatment was analysed (Figure 2B). This experiment confirms that transfected Tip60 has a half-life of ∼30 min (left panel). In addition, this half-life was increased significantly in the presence of a proteasome inhibitor (right panel, ALLN). Taken together, these data strongly indicate that HA-Tip60 is degraded by the proteasome.
Ubiquitylation has often be linked to degradation through the proteasome. To test whether Tip60 can be ubiquitylated in living cells, we transfected U2OS cells with expression vectors for His-tagged ubiquitin and for HA-tagged Tip60. Ubiquitylated proteins were then purified by nickel affinity chromatography, and the presence of Tip60 in the retained fraction was tested by anti-HA western blotting (Figure 2C). We found that in the presence of both His-tagged ubiquitin and HA-tagged expression vector (lane 2), many HA-reactive bands were detected. These bands were specific, since they were not detected in the absence of the His-tagged ubiquitin expression vector (lane 1) nor in the absence of the HA-Tip60 expression vector (data not shown), and most likely correspond to mono-ubiquitylated and polyubiquitylated HA-Tip60. Thus, Tip60 can be ubiquitylated in living cells.
Mdm2 induces the ubiquitylation and proteasome-mediated degradation of Tip60
Since the results presented above clearly indicate that Tip60 is a substrate of the ubiquitin–proteasome system, we next tested whether the effect of Mdm2 on Tip60 expression (Figure 1) also involves the proteasome. Indeed, we found that in the presence of proteasome inhibitors (Figure 3A, lanes 1–4), the effect of Mdm2 on HA-Tip60 expression (lanes 5–6) was abolished. This result indicates that the presence of Mdm2 induced the degradation of Tip60 in a proteasome-dependent manner. When a similar experiment with a smaller internal control (an HA-tagged version of the retinoblastoma binding protein RbAp48) was exposed for a longer period of time, HA-reactive bands that migrated more slowly than Tip60 were detected in the presence of proteasome inhibitors (Figure 3B, upper panel). These bands most likely correspond to ubiquitylated forms of Tip60 (Figure 2C), since they are not detected in the absence of HA-Tip60 (lanes 1 and 3). Interestingly, they were stronger in the presence of exogenous Mdm2 (Figure 3B, upper panel, lane 2), although the expression levels of Tip60 were similar (Figure 3B, lower panel), suggesting that Mdm2 could induce Tip60 ubiquitylation.
This result led us to test directly whether Mdm2 induced the ubiquitylation of Tip60. We transfected U2OS cells with expression vectors for His-tagged ubiquitin and HA-tagged Tip60 in the presence or absence of exogenous Mdm2. The presence of exogenous Mdm2 resulted in a significant increase in the amount of ubiquitylated Tip60 (Figure 3C, compare lane 2 with lane 1). Furthermore, polyubiquitylation was more prominent in the presence of exogenous Mdm2. Taken together, these results clearly indicate that Mdm2 can induce both the ubiquitylation and the proteasome-mediated degradation of Tip60 in living cells.
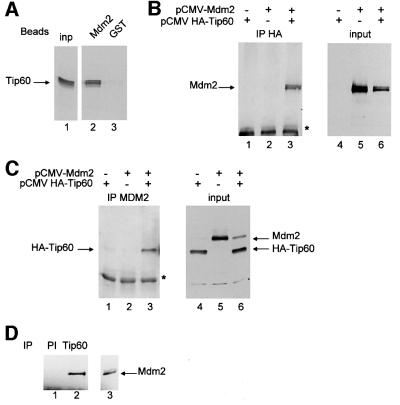
Mdm2 interacts physically with Tip60 in vitro and in living cells
Mdm2 induces the degradation of p53 through direct binding (Haupt et al., 1997; Kubbutat et al., 1997). We therefore tested the possibility of a physical interaction between Mdm2 and Tip60. 35S-labelled in vitro-translated Tip60 was subjected to a glutathione S-transferase (GST) pull-down experiment using beads harbouring either bacterially expressed GST–Mdm2 or GST as a control. We observed that Tip60 was efficiently retained on the Mdm2 beads (Figure 4A, lane 2), but not on control GST beads (lane 3). This indicates that Mdm2 and Tip60 interact physically in vitro. To test whether they can also interact in cells, we performed co-immunoprecipitation experiments from transfected cells extracts. We transfected U2OS cells with expression vectors for Mdm2 and/or HA-tagged Tip60. Whole-cell extracts were subjected to immunoprecipitation with either an anti-Mdm2 or an anti-HA antibody. Immunoprecipitation of HA-Tip60 led to the co-immunoprecipitation of transfected Mdm2 (Figure 4B, left panel, lane 3). This co-immunoprecipitation was specific since it was not seen in the absence of the HA-Tip60 expression vector (lane 2), although Mdm2 was produced at equivalent levels (right panel, lanes 5 and 6). Similarly, immunoprecipitation of Mdm2 led to the specific co-immunoprecipitation of HA-Tip60 (Figure 4C). Thus, Mdm2 and Tip60 are present within the same complex in cells.
Fig. 4. Mdm2 interacts physically with Tip60. (A) GST pull-down with 35S-labelled in vitro-translated Tip60 using beads harbouring bacterially produced GST–Mdm2 (lane 2) or control GST (lane 3) proteins. Bound proteins were analysed by SDS–PAGE followed by autoradiography. In lane 1, 2 µl of the in vitro translation reaction products were loaded directly. (B) U2OS cells were transfected with 10 µg of pCMV 2N3T Tip60 and/or 10 µg of pCMV NeoBam Mdm2, as indicated. ALLN was added 12 h after transfection. Whole-cell extracts were prepared 24 h after transfection, and subjected to immunoprecipitation with the anti-HA antibody. Immunoprecipitates were tested for the presence of Mdm2 by western blotting using the SMP14 antibody (left panel). In the right panel, 10 µl of whole-cell extracts were loaded directly and subjected to an anti-Mdm2 western blot. The asterisk indicates a band corresponding to the immunoglobulin light chain of the immunoprecipitating antibody. (C) As in (B), except that whole-cell extracts were immunoprecipitated with an anti-Mdm2 antibody (SMP14). Immuno precipitates were tested for the presence of exogenous Tip60 by western blotting using the anti-HA antibody (left panel). In the right panel, 10 µl of whole-cell extracts were loaded directly, and subjected to a western blotting with both the anti-HA and the anti-Mdm2 antibody. The asterisk indicates a band corresponding to the immunoglobulin light chain of the immunoprecipitating antibody. (D) HeLa nuclear extracts (Computer Cell Culture Center) were immunoprecipitated with the anti-Tip60 DT antibody (lane 2), or control pre-immune antibody (PI, lane 1) as a control. Immunoprecipitates were tested for the presence of Mdm2 by western blotting using the anti-Mdm2 antibody SMP14. In lane 3, 5 µl of extracts of U2OS transfected with the Mdm2 expression vector were loaded directly.
Since the above experiments relied on overexpressed proteins, we intended to demonstrate that endogenous Tip60 and Mdm2 can also interact. We immunoprecipitated Tip60 from HeLa nuclear extracts, and tested for the presence of co-immunoprecipitated Mdm2 by western blotting. We detected a band co-migrating with Mdm2 using the anti-Tip60 antiserum as the immunoprecipitating antibody (Figure 4D, lane 2). This interaction was specific, since this band was not detected when pre-immune serum was used (PI, lane 1), suggesting that endogenous proteins are associated physically in cells. Taken together, the results shown in Figure 4 indicate that Tip60 interacts physically with Mdm2.
Mdm2-induced degradation of Tip60 involves their physical interaction
To elucidate the relationship between Mdm2 binding and degradation of Tip60, we decided to map the site of Mdm2 binding within Tip60. We constructed Tip60 deletion mutants and we transfected U2OS cells with expression vectors for some of these mutants, together with the Mdm2 expression vector. Immunoprecipitation of full-length Tip60 (Figure 5B, lane 2) or Tip60 1–364 (lane 8), but not Tip60 1–258 (lane 5), led to the co-immunoprecipitation of Mdm2. This result indicates that the region between amino acid residues 258 and 364 of Tip60 is involved in binding to Mdm2. In a similar experiment, we found that a small protein harbouring Tip60 amino acids 259–396 could also co-immunoprecipitate Mdm2 (see Supple mentary figure 3), indicating that this region is also sufficient for Mdm2 binding. To test whether physical binding was involved in Mdm2-induced degradation of Tip60, we determined the expression levels of several Tip60 deletion mutants in the presence or absence of Mdm2 expression vector. We found that the region between 285 and 396 was necessary and sufficient for Mdm2-induced degradation of Tip60 (Figure 5C; Supplementary figure 4). Since this region is identical to the Mdm2 binding site, this strongly suggests that Mdm2-induced degradation of Tip60 requires their physical interaction.
Tip60 is targeted by the ubiquitin ligase activity of Mdm2 in vitro and in living cells
The results presented above suggest that Mdm2 serves as a ubiquitin ligase for Tip60. To test this hypothesis directly, in vitro ubiquitylation assays were performed. We found that bacterially produced full-length Mdm2 can induce ubiquitylation of in vitro-translated Tip60 (Figure 6A). However, the efficiency was rather low. This suggests that additional factors may be required for Mdm2-induced ubiquitylation of Tip60 that are missing in the in vitro system, or that Tip60 or Mdm2 need to be properly modified for an efficient ubiquitylation to occur. None the less, the Mdm2-mediated ubiquitylation of Tip60 was specific since a deletion mutant of Tip60 (Tip60 1–211) that does not interact with Mdm2 and is not degraded by Mdm2 in transfected cells (Figure 5B), was not ubiquitylated in this in vitro system (data not shown). In addition, a hyperactive deletion mutant of Mdm2 that still binds Tip60 (data not shown), efficiently ubiquitylated full-length Tip60, as indicated by the appearance of bands of lower mobility that were dependent on the presence of exogenously added E2 and ubiquitin (Figure 6A). Again, this ubiquitylation reaction was specific in that Tip60 1–211 was not ubiquitylated. Taken together, these results indicate that Tip60 is a specific substrate of the Mdm2 E3 ubiquitin ligase activity in vitro.
To test whether Mdm2 is a ubiquitin ligase for Tip60 also in living cells, we used a point mutant of Mdm2 (Mdm2 C462A) that is mutated in the RING finger domain, and thus does not harbour any enzymatic activity (Argentini et al., 2000). This mutation does not change the ability of Mdm2 to bind Tip60 (data not shown). In the presence of the mutated Mdm2 (Figure 6B, upper panel, lane 2), Tip60 expression was comparable to its expression in the absence of Mdm2 (lane 3), whereas wild-type (wt) Mdm2 induced a decrease in Tip60 expression (lane 1). Since the expression levels of mutated or wt Mdm2 were similar (lower panel), this result strongly indicates that the ubiquitin ligase activity of Mdm2 is required for degradation of Tip60.
The main known target of the ubiquitin ligase activity of Mdm2 is p53. Mdm2 contacts p53 through its N-terminus. To demonstrate that the effect of Mdm2 on Tip60 is not due to any indirect consequence of p53 ubiquitylation, we constructed an expression vector for a Mdm2 deletion mutant unable to interact with p53 (Mdm2 Δ101). We found that Tip60 259–396 expression was decreased as efficiently by the expression of Mdm2 Δ101 (Figure 6C, upper panel, lane 7) as by wt Mdm2 (lane 4). Mdm2 Δ101 was produced at levels similar to those of wt (lower panel), and interacted with Tip60 similarly to wt (data not shown). This result thus indicates that the effect of Mdm2 on Tip60 was not dependent upon its ability to ubiquitylate p53 and that Tip60 is a direct target of the ubiquitin ligase activity of Mdm2 in living cells.
UV irradiation strongly induced Tip60 expression
Following DNA damage, the inhibitory effect of Mdm2 on p53 levels is relieved, resulting in the accumulation of active p53. Since our data indicate that Mdm2 also functions as a ubiquitin ligase for Tip60, we investigated the effect of UV irradiation on Tip60 expression. As expected, UV irradiation of Jurkat cells induced a slight increase in steady-state levels of p53 (data not shown). Strikingly, whereas Tip60 was undetectable in control cells, it was strongly induced by UV irradiation (Figure 7). Using cycloheximide experiments, we found that the accumulated Tip60 was stable, with a half-life of >3 h (data not shown). Although we could not measure the half-life of endogenous Tip60 in non-irradiated cells (see above), the half-life of exogenous Tip60 was ∼30 min (Figure 2B). Thus, this result strongly suggests that UV irradiation induced Tip60 stabilization. Consistent with a role of MDM2 in Tip60 degradation, we found that MDM2 levels decreased strongly after UV irradiation (Supple mentary figure 5), as shown previously by others (Blattner et al., 1999). Taken together, these results indicate that p53 and Tip60 levels both increase following DNA damage, and suggest a model in which p53 and Tip60 cooperate to induce the appropriate cellular response.
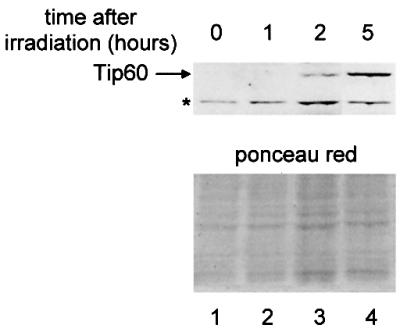
Fig. 7. Tip60 expression is induced by UV irradiation. Jurkat cells (2 × 106) were irradiated as indicated in Materials and methods and incubation was pursued for the period of time indicated. Cells were then collected, boiled in SDS loading buffer, and extracts were subjected to a western blot using the anti-Tip60 DT antibody (upper panel). The asterisk indicates a non-specific band. The membrane was also stained with Ponceau Red to check for loading differences (lower panel).
Discussion
Here, we show by various means (proteasome inhibition, half-life determination, ubiquitylation) that Tip60 is regulated by proteasome-mediated proteolysis. To our knowledge, this is the first demonstration of such a regulation for a HAT. Together with our unpublished data showing that the HAT activity of Tip60 is regulated through phosphorylation, our results indicate that the activity and intracellular levels of Tip60 are tightly controlled in cells.
The activity of HATs is crucial for the proper control of gene expression. Accordingly, many mechanisms regulating their activity have been described. This can occur by post-translational modifications, as has been shown for CREB-binding protein (CBP), which is activated via phosphorylation by cyclin E–cdk2 (Ait-Si-Ali et al., 1998). Also, phosphorylation of Tip60 results in an increase in the HAT activity of the enzyme (our unpublished observations). Another mechanism is through protein–protein interactions. This mechanism is widely used by viral transforming proteins. Binding of the viral protein E1A to the HAT CBP, p300 or pCAF has been shown to affect their HAT activity, although whether they are activated or repressed is still a matter of debate (Ait-Si-Ali et al., 1998; Reid et al., 1998; Chakravarti et al., 1999; Hamamori et al., 1999). Also, the HIV Tat protein represses Tip60 HAT activity through direct binding (Creaven et al., 1999). Recently, it has been shown that many cellular transcription factors similarly regulate the HAT activity of CBP/p300 through physical interactions (Hamamori et al., 1999; Chen et al., 2001; Soutoglou et al., 2001). Our data suggest that regulation of protein stability is an additional way of regulating HAT activity. The notion that it functions co-operatively with other mechanisms is an appealing, yet unproven, possibility.
Tip60 is supposed to be present in mammalian cells within a large complex, containing at least 12 subunits, including the TRRAP p400 protein and two actin-related proteins (Ikura et al., 2000). The significance of this finding is highlighted by the fact that only the complex, but not purified Tip60, can efficiently acetylate nucleosomal substrates. In addition, the yeast NuA4 complex, which contains the TIP60-related ESA1 HAT, includes orthologues of TRRAP and actin-related proteins (Allard et al., 1999; Galarneau et al., 2000), suggesting that the Tip60 complex is conserved throughout evolution. Notably, purification of the Tip60-associated complex relied on overexpression of exogenous Tip60 (Ikura et al., 2000). Taken together with our results indicating that Tip60 is subjected to proteasome-mediated degradation, this fact emphasizes the importance of assessing whether Tip60 is really a stoichiometric component of the endogenous ‘Tip60 complex’. An interesting possibility is that Tip60 levels are very low in cells, and that some Tip60 complex would be free of Tip60 and thereby inactive. Consistent with this, a complex containing all subunits present in the Tip60 complex but devoid of Tip60 has recently been described (Fuchs et al., 2001). If this hypothesis is correct, regulating the amount or the activity of Tip60 would result in control of the activity of the whole Tip60 complex.
We found that Mdm2 binds to and induces the ubiquitylation of Tip60 in vitro (Figure 6A). In addition, we show that (i) Mdm2 induces the ubiquitylation of Tip60 (Figure 3B and C), (ii) Mdm2 binds Tip60 (Figure 4), (iii) physical binding is required for Mdm2 to induce Tip60 degradation (Figure 5) and (iv) degradation of Tip60 requires the ubiquitin ligase activity of Mdm2 (Figure 6B) and is independent of p53, the only substrate of Mdm2 described previously (Figure 6C). Taken together with the in vitro data, these data strongly indicate that Tip60 is a direct target of Mdm2 for ubiquitylation in living cells. Unfortunately, we were not able to detect full-length Tip60 in mouse cells with our antibodies, so we could not test whether Tip60 expression is deregulated in Mdm2–/–p53–/– MEFs. Nevertheless, one of the consequences of our findings is the fact that the same protein, Mdm2, regulates the stability of both p53 and Tip60. Tip60 has recently been demonstrated to be involved in DNA repair and apoptosis following irradiation (Ikura et al., 2000), two processes in which the p53 protein also plays a prominent role. Our results thus lead to the interesting hypothesis that Tip60 and p53 levels are co-regulated. Indeed, we observed that UV irradiation induced both p53 and Tip60 expression. Consequently, Tip60 and p53 could cooperate to mediate the appropriate cellular response to DNA damage. It was recently proposed that the Tip60 complex could be involved in the regulation of p53 activity (Nourani et al., 2001). Thus, an appealing hypothesis would be that the induction of a specific genetic programme following DNA damage involves the assembly on inducible promoters of a transcriptionally active complex relying on p53 and Tip60.
Aberrant accumulation of Mdm2 protein through amplification of the Mdm2-encoding gene is found in a significant number of human tumours (Momand et al., 1998). It is believed that one of the mechanisms by which overexpression of Mdm2 leads to cell transformation is through the degradation, and concomitant inactivation, of p53. Consistent with this hypothesis, the p53 gene is usually not mutated in tumours overexpressing Mdm2 (Freedman et al., 1999). However, a few tumours have been reported to harbour a mutated p53 gene and an amplified Mdm2 gene (Freedman et al., 1999). In addition, Mdm2 can induce tumour formation in a p53-independent manner (Jones et al., 1998), indicating that it very likely has other important targets. Indeed, Mdm2 binds directly to the E2F1 transcription factor (Martin et al., 1995), which is involved in cell cycle regulation and apoptosis, and E2F1 levels increase after UV irradiation (Blattner et al., 1999). However, it is presently not clear if Mdm2 is directly involved in the degradation of E2F1. Our results suggest that the ability of Mdm2 to bind to Tip60 and target it for degradation also contributes to its oncogenic capacity. Interestingly, it has recently been shown that Tip60 HAT activity is important for apoptosis (Ikura et al., 2000). It is thus tempting to speculate that some cells engaged in the process of malignant transformation could be rescued from Tip60-induced apoptosis by overexpressing Mdm2. If this hypothesis is correct, a possible consequence would be that the Tip60-encoding gene is the target of inactivating mutations in human tumours.
Materials and methods
Vectors
PCDNA3 Tip60 wt and mutants were constructed by PCR and inserted into pCDNA3 HA, in-frame with the HA tag. PCMV 2N3T Tip60 and pCMV 2N3T Ku80 were constructed by inserting the corresponding cDNA (Ku80 cDNA was a kind gift from Dr C.Müller) into the pCMV 2N3T vector (Magnaghi-Jaulin et al., 1998), which expresses proteins with three HA tags and two nuclear localization signals at their N-terminus. PCMV 2N3T RbAp48 has been described in Nicolas et al. (2000). PSG5 His-ubiquitin and pSG5 HA-ubiquitin were kind gifts from Dr D.Bohmann. PCMV NeoBam Mdm2 expresses human Mdm2 and was a kind gift from Dr B.Vogelstein and was described in Martin et al. (1995). PGEX Mdm2 expresses mouse Mdm2 and was a kind gift from Dr J.Piette. PXJ Mdm2 (expressing mouse Mdm2) and pXJ Mdm2 C462A were kind gifts from Dr B.Wasylyk (Argentini et al., 2000). Details of constructions are available upon request. PCDNA3 Mdm2 Δ101 expressed human Mdm2 deleted for the first 101 amino acids from a CMV promoter. pGEX Mdm2 Δ417 expressed human Mdm2 deleted for the first 417 amino acids as a GST fusion protein in E.coli.
Cell culture and transfection
U2OS, SAOS2 and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with antibiotics and fetal calf serum (FCS) (10%). Transfections were performed by the calcium phosphate co-precipitation method. Jurkat cells were cultured in RPMI 1640, supplemented with antibiotics and FCS (10%).
Proteasome inhibition was achieved by treatment with ALLN (100 µM), MG132 (3 µM) or lactacystine (10 µM), as indicated in the figure legend. In some experiments, cells were treated with cycloheximide (30 µg/ml) to inhibit protein translation. UV irradiation (8 mJ/cm2) was performed using a UV crosslinker (Hoefer).
Antibodies
The anti-HA (12CA5) and anti-myc (9E10) antibodies were purchased from Roche Diagnostics, and the anti-Mdm2 antibody (SMP14) from Santa Cruz. We raised two independent polyclonal anti-Tip60 antibodies. For the first one (anti-Tip60DT), two rabbits were immunized with an N-terminal peptide (LPVLRRNQDNEDEWPC) from Tip60 (the immunization was performed by Eurogentec). The serum was collected, as well as pre-immune serum. For western blotting experiments, the antibody was further subjected to affinity purification on a column harbouring the peptide. For the other anti-Tip60 antibody (anti-Tip60 SK), two rabbits were immunized with a bacterially produced His-Tip60 1–211 fusion protein. Immune and pre-immune sera were collected. Both antibodies recognize exogenous HA-Tip60 from transfected cell extracts very efficiently in western blotting experiments as well as in immunoprecipitation experiments (data not shown).
Analysis of Tip60 steady-state levels
For endogenous Tip60, human cells (HeLa, Jurkat, U2OS or SAOS2) were lysed directly by boiling in Laemmli sample buffer and extracts were blotted using an anti-Tip60 antibody. For transfected Tip60, cells were lysed in 200 µl of luciferase lysis buffer (Promega). Luciferase activity was measured with a kit according to the manufacturer’s instructions (Promega). Sixty microlitres of 4× Laemmli sample buffer were then added to the cells to prepare total cell lysates. After boiling and centrifugation, a defined amount of lysate (standardized according to luciferase activity) was collected and was adjusted to 20 µl with 1× Laemmli sample buffer, before being subjected to western blot analysis using the anti-HA antibody.
Purification of His-tagged protein from transfected cells
Cells transfected with His-tagged ubiquitin as described in the figure legends were treated with 100 µM ALLN (Calbiochem) for 12 h before extraction. Twenty-four hours after transfection, cells were then collected in 1 ml of buffer A (6 M guanidium–HCl, 100 mM Na2HPO4/NaH2PO4 pH 8.0, 10 mM imidazole). Lysates were sonicated to reduce the viscosity. After centrifugation, supernatants were subjected to a preclearing step with 50 µl protein A–protein G beads (Sigma) for 15 min at room temperature. Extracts were then incubated with 50 µl of nickel-NTA–agarose beads (Qiagen) for 3 h at room temperature. Beads were then washed twice in buffer A, twice in buffer A diluted five times in 50 mM Tris pH 6.8, 20 mM imidazole, and twice in 50 mM Tris pH 6.8, 20 mM imidazole. Beads were then eluted twice with 30 µl SDS loading buffer supplemented with 200 mM imidazole, and supernatants were subjected to SDS–PAGE and western blotting.
Immunoprecipitations
For co-immunoprecipitation experiments from transfected cells, cells were treated with ALLN (100 µM) for 12 h and were collected 24 h after transfection in 500 µl of lysis buffer (300 mM NaCl, 50 mM Tris pH 8.0, 0.4% NP-40, 10 mM MgCl2, 2.5 mM CaCl2) supplemented with protease inhibitors (Complete mini EDTA-free, Roche Diagnostics), RNase 100 µg/ml and DNase (Coger). After centrifugation, total cell extracts were diluted in 500 µl of dilution buffer (50 mM Tris pH 8.0, 0.4% NP-40) and then subjected to immunoprecipitation as described (Nicolas et al., 2000).
For co-immunoprecipitation of endogenous proteins, 400 µl of HeLa nuclear extracts (Computer Cell Culture Center) were diluted in 400 µl of washing buffer (150 mM NaCl, 50 mM Tris pH 8.0, 0.4% NP-40, 5 mM MgCl2, 1.25 mM CaCl2) and treated as described above.
GST pull-downs
For GST pull-down experiments, recombinant GST–Mdm2 and GST were produced in E.coli, and purified according to Nicolas et al. (2000). The amount of recombinant proteins was assessed by SDS–PAGE followed by Coomassie Blue staining. 35S-labelled Tip60 was produced by in vitro transcription–translation (TNT, Promega) from the pCDNA3 HA-Tip60 vector, using T7 polymerase. GST pull-down assays were performed in washing buffer for 2 h at room temperature with ∼1 µg of GST fusion proteins. After three washes, beads were resuspended in 1× SDS loading buffer and were subjected to SDS–PAGE, followed by fluorography (Amplify; Amersham).
In vitro ubiquitylation assays
For Mdm2-mediated ubiquitylation, 2 µl of rabbit reticulocyte lysate-translated 35S-labelled Tip60 were incubated in 40 µl of 25 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM dithiothreitol (DTT), 2 mM ATP and 2 mM MgCl2, in the presence of bacterially expressed GST–Mdm2 or GST–Mdm2 Δ417 (500 ng) supplemented with 50 ng E1, 50 ng UbcH5 and 6 µg ubiquitin (Sigma) where indicated. After incubation at 30°C for 2 h, reaction products were analysed by SDS–PAGE followed by fluorography.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank E.Nicolas and M.Callige for helpful discussions, Drs J.Piette, D.Bohmann, C.Müller and B.Wasylyk for advice and materials, and Drs L.Vandel and M.Grigoriev for critical reading of the manuscript. This work was supported by a grant to D.T. from La Ligue Nationale Contre le Cancer as an ‘équipe labellisée’, by Sidaction (Sidaction—Ensemble Contre Le Sida postdoctoral fellowship to C.L. and contract 99-1249/23026-01-00/A010-1 to S.K.), and by the German–Israeli Foundation for Scientific Research and Development (M.S.). G.L. is a recipient of a studentship from the French Ministry of Science (MENR).
References
- Ait-Si-Ali S. et al. (1998) Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature, 396, 184–186. [DOI] [PubMed] [Google Scholar]
- Akhtar A. and Becker,P.B. (2001) The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition. EMBO rep., 2, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard S., Utley,R.T., Savard,J., Clarke,A., Grant,P., Brandl,C.J., Pillus,L., Workman,J.L. and Cote,J. (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J., 18, 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argentini M., Barboule,N. and Wasylyk,B. (2000) The contribution of the RING finger domain of MDM2 to cell cycle progression. Oncogene, 19, 3849–3857. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Blattner C., Sparks,A. and Lane,D. (1999) Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol. Cell. Biol., 19, 3704–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady M.E., Ozanne,D.M., Gaughan,L., Waite,I., Cook,S., Neal,D.E. and Robson,C.N. (1999) Tip60 is a nuclear hormone receptor coactivator. J. Biol. Chem., 274, 17599–17604. [DOI] [PubMed] [Google Scholar]
- Chakravarti D., Ogryzko,V., Kao,H.Y., Nash,A., Chen,H., Nakatani,Y. and Evans,R.M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell, 96, 393–403. [DOI] [PubMed] [Google Scholar]
- Chen C.J., Deng,Z., Kim,A.Y., Blobel,G.A. and Lieberman,P.M. (2001) Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol., 21, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creaven M., Hans,F., Mutskov,V., Col,E., Caron,C., Dimitrov,S. and Khochbin,S. (1999) Control of the histone-acetyltransferase activity of Tip60 by the HIV-1 transactivator protein, Tat. Biochemistry, 38, 8826–8830. [DOI] [PubMed] [Google Scholar]
- Freedman D.A., Wu,L. and Levine,A.J. (1999) Functions of the MDM2 oncoprotein. Cell Mol. Life Sci., 55, 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M., Gerber,J., Drapkin,R., Sif,S., Ikura,T., Ogryzko,V., Lane,W.S., Nakatani,Y. and Livingston,D.M. (2001) The p400 complex is an essential E1A transformation target. Cell, 106, 297–307. [DOI] [PubMed] [Google Scholar]
- Galarneau L. et al. (2000) Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell, 5, 927–937. [DOI] [PubMed] [Google Scholar]
- Hamamori Y., Sartorelli,V., Ogryzko,V., Puri,P.L., Wu,H.Y., Wang,J.Y., Nakatani,Y. and Kedes,L. (1999) Regulation of histone acetyl transferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell, 96, 405–413. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya,R., Kazaz,A. and Oren,M. (1997) Mdm2 promotes the rapid degradation of p53. Nature, 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Hlubek F., Lohberg,C., Meiler,J., Jung,A., Kirchner,T. and Brabletz,T. (2001) Tip60 is a cell-type-specific transcriptional regulator. J. Biochem. (Tokyo), 129, 635–641. [DOI] [PubMed] [Google Scholar]
- Honda R., Tanaka,H. and Yasuda,H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett., 420, 25–27. [DOI] [PubMed] [Google Scholar]
- Ikura T., Ogryzko,V.V., Grigoriev,M., Groisman,R., Wang,J., Horikoshi,M., Scully,R., Qin,J. and Nakatani,Y. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell, 102, 463–473. [DOI] [PubMed] [Google Scholar]
- Jacobson R.H., Ladurner,A.G., King,D.S. and Tjian,R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science, 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- Jones S.N., Hancock,A.R., Vogel,H., Donehower,L.A. and Bradley,A. (1998) Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl Acad. Sci. USA, 95, 15608–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamine J., Elangovan,B., Subramanian,T., Coleman,D. and Chinnadurai,G. (1996) Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology, 216, 357–366. [DOI] [PubMed] [Google Scholar]
- Kubbutat M.H., Jones,S.N. and Vousden,K.H. (1997) Regulation of p53 stability by Mdm2. Nature, 387, 299–303. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carrol,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Groisman,R., Naguibneva,I., Robin,P., Lorain,S., Le Villain,J.P., Troalen,F., Trouche,D. and Harel-Bellan,A. (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature, 391, 601–605. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. and Roth,S.Y. (2001) Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev., 11, 155–161. [DOI] [PubMed] [Google Scholar]
- Martin K., Trouche,D., Hagemeier,C., Sorensen,T.S., La Thangue,N.B. and Kouzarides,T. (1995) Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature, 375, 691–694. [DOI] [PubMed] [Google Scholar]
- Momand J., Jung,D., Wilczynski,S. and Niland,J. (1998) The MDM2 gene amplification database. Nucleic Acids Res., 26, 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E., Morales,V., Magnaghi-Jaulin,L., Harel-Bellan,A., Richard-Foy,H. and Trouche,D. (2000) RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J. Biol. Chem., 275, 9797–9804. [DOI] [PubMed] [Google Scholar]
- Nourani A., Doyon,Y., Utley,R.T., Allard,S., Lane,W.S. and Cote,J. (2001) Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol., 21, 7629–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Q. and Pereira-Smith,O.M. (2000) Identification of an alternatively spliced form of the Tat interactive protein (Tip60), Tip60(β). Gene, 258, 141–146. [DOI] [PubMed] [Google Scholar]
- Reid J.L., Bannister,A.J., Zegerman,P., Martinez-Balbas,M.A. and Kouzarides,T. (1998) E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J., 17, 4469–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewack G.F., Ellis,T.W. and Hansen,U. (2001) Binding of TATA binding protein to a naturally positioned nucleosome is facilitated by histone acetylation. Mol. Cell. Biol., 21, 1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T. (2000) The p53 tumor suppressor gene: from molecular biology to clinical investigation. Ann. N.Y. Acad. Sci., 910, 121–139. [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Viollet,B., Vaxillaire,M., Yaniv,M., Pontoglio,M. and Talianidis,I. (2001) Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J., 20, 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E. and Berger,S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Turner B.M. (2000) Histone acetylation and an epigenetic code. BioEssays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- Yuan Z.M. et al. (1999) Role for p300 in stabilization of p53 in the response to DNA damage. J. Biol. Chem., 274, 1883–1886. [DOI] [PubMed] [Google Scholar]
- Zhou B.B. and Elledge,S.J. (2000) The DNA damage response: putting checkpoints in perspective. Nature, 408, 433–439. [DOI] [PubMed] [Google Scholar]