Abstract
The MinCDE proteins help to select cell division sites in normal cylindrical Escherichia coli by oscillating along the long axis, preventing unwanted polar divisions. To determine how the Min system might function in cells with multiple potential division planes, we investigated its role in a round-cell rodA mutant. Round cells lacking MinCDE were viable, but growth, morphology and positioning of cell division sites were abnormal relative to Min+ cells. In round cells with a long axis, such as those undergoing cell division, green fluorescent protein (GFP) fusions to MinD almost always oscillated parallel to the long axis. However, perfect spheres or irregularly shaped cells exhibited MinD movement to and from multiple sites on the cell surface. A MinE–GFP fusion exhibited similar behavior. These results indicate that the Min proteins can potentially localize anywhere in the cell but tend to move a certain maximum distance from their previous assembly site, thus favoring movement along the cell’s long axis. A new model for the spatial control of division planes by the Min system in round cells is proposed.
Keywords: bacteria/cell division/FtsZ/minCDE/RodA
Introduction
Bacterial cells such as those of Escherichia coli normally divide by binary fission, producing two daughter cells of equal size. Initiation of cell division is dependent on the formation of a polymeric ring structure, called the Z ring, which encircles the cell at the division plane (Bi and Lutkenhaus, 1991; Ma et al., 1996). The Z ring, which consists of a highly conserved tubulin-like protein (FtsZ), then recruits other cell division proteins to form a putative cell division protein machine that completes septum formation (Rothfield et al., 1999; Margolin, 2000).
The mechanism for the selection of the division plane between the daughter chromosomes is not well understood. Existing evidence suggests that both the nucleoid and the Min system play critical roles in the proper localization of the Z ring. The nucleoid appears to have an important negative influence on Z-ring formation, keeping Z rings away from regions of the cell that contain unsegregated nucleoids and only allowing Z-ring formation between segregating nucleoids at midcell (Yu and Margolin, 1999).
The Min system, which consists of the products of three genes in the min operon (minC, minD and minE), also has a negative role in Z-ring placement. MinC acts to inhibit FtsZ polymerization in vitro and Z-ring formation in vivo (Bi and Lutkenhaus, 1993; Hu et al., 1999; Pichoff and Lutkenhaus, 2001). MinC oscillates from pole to pole in E.coli, and this movement depends upon both MinD and MinE, which also oscillate back and forth and likely move together as a unit (Raskin and de Boer, 1997, 1999a,b; Hu and Lutkenhaus, 1999; Fu et al., 2001; Hale et al., 2001). MinC and MinD prevent Z-ring formation throughout the cell, whereas MinE suppresses this inhibition by MinC and MinD at the central division site, allowing medial cytokinesis to occur (de Boer et al., 1989, 1990, 1992; Bi and Lutkenhaus, 1993). MinD movement depends upon stimulation of its ATPase activity (de Boer et al., 1991) by MinE at the membrane (Hu and Lutkenhaus, 2001).
Complete deletion of the min operon (Δmin) leads to localization of Z rings at all nucleoid-free regions (Yu and Margolin, 1999), producing viable cells that divide at midcell but also large quantities of nucleoid-free minicells, the products of polar divisions (Adler et al., 1967). Because polar divisions occur at the expense of medial divisions and not in addition to them (Teather et al., 1974), Δmin mutant cells consist of minicells and short filaments. Nucleoid-mediated inhibition in Δmin cells is sufficient to ensure cell viability by blocking Z-ring formation at all non-polar sites except midcell (Yu and Margolin, 1999; Sun and Margolin, 2001; Yu et al., 2001).
In normal cylindrical E.coli cells, the division site corresponds to the location equidistant from the two cell poles. The Min system is presumably used to extend the inhibition of Z-ring formation from regions occupied by nucleoids to these nucleoid-free regions of the cell (Margolin, 2001). However, this working model for division site placement becomes much more complex in spherical cells. Spherical cells, in theory, do not have a defined middle, but instead possess an infinite number of potential division planes at the point of greatest cell diameter. Nevertheless, certain mutants of E.coli are able to grow and divide as spheres, producing two daughter spheres of similar size, indicating that cell division in the spheres is still precisely controlled. Interestingly, E.coli spheres are multinucleate and appear to divide in alternating perpendicular planes (Donachie et al., 1995; Begg and Donachie, 1998; Zaritsky et al., 1999; de Pedro et al., 2001; Pas et al., 2001), much like some other naturally spherical species (Westling-Haggstrom et al., 1977; Murray et al., 1983). Moreover, cell division is asymmetric in these spheres, as shown by FtsZ arcs (Addinall and Lutkenhaus, 1996; Pas et al., 2001) and cytokinetic furrows (Begg and Donachie, 1998) on one side of the cell during the early stages.
The importance of the Min system in toporegulation of the division site in bipolar, cylindrical E.coli prompted us first to test whether the Min system is important for the identification of proper division sites in round cells. We found that the Min proteins are important for selecting the proper division plane in round cells. In addition, we found that the pattern of Min protein movement in round cells depends on the cell geometry, with a bias toward movement along the cell’s long axis. These results have important implications for both the mechanism of Min protein movement and the regulation of division planes in cocci.
Results
The rodA Δmin mutant is viable but deficient in growth and division
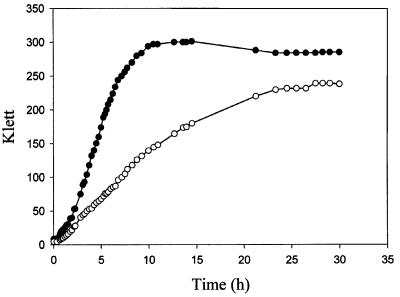
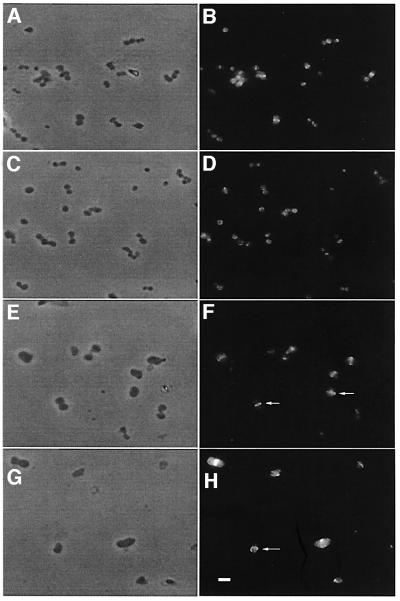
Strain KJB24 contains a rodA amber mutation resulting in spherical shape and an additional mutation allowing cells to be viable in rich media (Begg and Donachie, 1998). Strain WM1250, which is KJB24 containing a complete deletion of the minCDE locus, was able to form colonies but was clearly inhibited for growth relative to KJB24, with a growth rate about half that of the parent strain (Figure 1). Instead of producing short filaments and minicells like rod+ ΔminCDE mutants, the double mutant strain WM1250 produced a mixture of small and large cells with highly variable morphologies (Figure 2E and G). Individual KJB24 cells were smaller and more uniform in size (Figure 2A and C), with a mean area calculated from phase-contrast images of 1.50 ± 0.42 µm2, and ranged in size from 0.75 to 2.52 µm2. In contrast, individual WM1250 cells had a calculated mean area of 5.3 ± 1.9 µm2 and ranged in size from 2.2 to 9.3 µm2, except for the occasional minicell or bud (see below). This significant enlargement of WM1250 cells likely represents the round-cell equivalent of short filaments and is similar to the enlargement of KJB28, a rodA ftsA (ts) double mutant, at 42°C (data not shown). Unlike WM1250, however, most KJB28 cells at the non-permissive temperature had a very regular spherical morphology (data not shown).
Fig. 1. Growth curves for KJB24 (rodA, filled circles) and WM1250 (rodA ΔminCDE, open circles) grown in LB medium.
Fig. 2. Morphology and FtsZ ring localization in (A–D) KJB24 (rodA) and (E–H) WM1250 (rodA ΔminCDE). Cells were grown to logarithmic phase, fixed, permeabilized and incubated with anti-FtsZ polyclonal antibody. Two representative fields of cells are shown for each strain, imaged either with phase contrast (A, C, E and G) or epifluorescence (B, D, F and H). All are at the same magnification. Arrows in (F) indicate cells with apparently normal medial FtsZ rings, and the arrow in (H) indicates an FtsZ spiral. Scale bar = 3 µm.
Despite the apparent inhibition of cell division, the overall viability of the rodA ΔminCDE double mutant indicated that there were sufficient chromosome-containing daughter cells produced in the population to form colonies. In support of this idea, we often observed mother–daughter cell pairs in which the daughter cell had the appearance of a bud coming off the mother cell. In many cases, DNA was clearly present in the adjoining bud (data not shown).
In Δmin mutants of cylindrical cells, DNA-less minicells comprise a high percentage of the total cells. However, in both min– and min+ versions of KJB24, <1–2% of cells lacked DNA staining. Therefore, unlike in cylindrical cells, the absence of Min proteins in rodA cells did not detectably increase the frequency of anucleate cells. The presence of DNA in most cells of this Δmin derivative is consistent with the multinucleate nature of round E.coli cells (Donachie et al., 1995; Zaritsky et al., 1999).
FtsZ structures in min+ versus min– spheres
To investigate whether the abnormal cellular morphologies and position of constrictions in WM1250 correlated with abnormal FtsZ localization and ring structure, we examined the localization of FtsZ in the double mutant by using indirect immunofluorescence staining with affinity-purified FtsZ antibody. In KJB24 (rodA) cells, FtsZ staining was typically found at the center of the spheres, and just one FtsZ ring or arc was present per cell (Figure 2B and D). Although it was difficult to determine whether every FtsZ structure represented a ring or an arc, at least some of these FtsZ structures appeared to be complete rings (data not shown). We also visualized green fluorescent protein (GFP) fusions to FtsZ expressed in living KJB24 cells and observed fluorescent rings indistinguishable from those seen with immunofluorescence (data not shown).
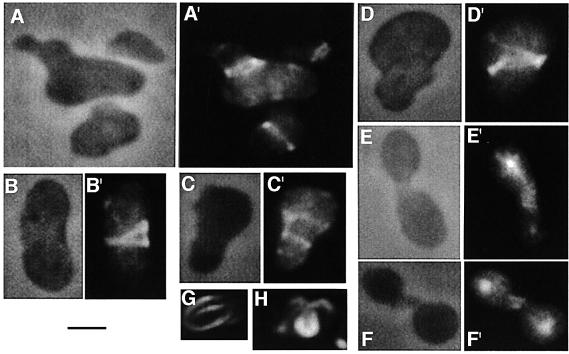
In WM1250, however, FtsZ localization and polymer structure were significantly more complex and correlated well with the abnormal cellular morphologies of this strain. Unlike KJB24, many WM1250 cells contained FtsZ spirals and/or multiple FtsZ bands in oblique planes (Figures 2H, arrow, and 3). FtsZ spirals or rings not perpendicular to the cell’s long axis are not normally observed in wild-type cells, although they are sometimes seen in cells with nucleoid defects (Sun et al., 1998; Yu and Margolin, 1999) or a phospholipid imbalance (Mileykovskaya et al., 1998), and often in the ftsZ26 mutant strain that makes spiral septa (Addinall and Lutkenhaus, 1996). The abnormal localization patterns of FtsZ in WM1250 suggest that the abnormal constrictions in this strain were directly caused by cell wall invagination along the aberrant FtsZ structures. This idea is similar to that previously proposed for the spiral septa formed by FtsZ26 (Addinall and Lutkenhaus, 1996). Moreover, elongated septa containing FtsZ were sometimes observed (Figure 3E, E′, F and F′); these may be a result of delayed disassembly of FtsZ, also observed under certain conditions in cylindrical cells lacking the Min proteins (Yu and Margolin, 2000). We ruled out the possibility that these defects were caused by an FtsZ deficiency, as the levels of FtsZ from rodA, rodA Δmin and wild-type cylindrical cells as normalized to total cell protein were comparable on immunoblots (data not shown).
Fig. 3. Details of abnormal FtsZ structures in WM1250. Cells were prepared as described in the legend to Figure 2. (A–F) Phase-contrast images of each cell or cells. (A′–F′) Corresponding immunofluorescence results with anti-FtsZ and Alexa 488 (as described in Materials and methods). (G and H) Three-dimensional immunofluorescence micrographs of FtsZ spirals as imaged by deconvolution. Scale bar = 2.5 µm.
Despite the many cases of abnormal localization of FtsZ in cells with aberrant morphology, about a quarter of the WM1250 cells were symmetrical like KJB24 cells and contained an apparently normal FtsZ ring at the cell center. For example, the WM1250 cells indicated with arrows in Figure 2F are clearly enlarged relative to the average KJB24 cell, yet they have apparently normal medial FtsZ rings that resemble FtsZ rings of KJB24 cells. Therefore, the loss of the Min system in round cells not only permits viability but also allows apparently normal medial placement of the FtsZ ring in a significant number of cells. This suggests that another factor, possibly nucleoid occlusion, must regulate division site placement in spheres to prevent FtsZ localization from being random in all cells in the absence of the Min system. These characteristics are very similar to the case of cylindrical Δmin mutant cells, many of which divide normally at midcell but which also assemble FtsZ rings in all nucleoid-free spaces (Yu and Margolin, 1999).
Back-and-forth oscillation of GFP–MinD in round cells with a long axis
To understand how the Min system might be involved in division plane specification, as well as to investigate how the Min proteins might move in more than one dimension, we monitored the movement of GFP–MinD in KJB24 cells immobilized in LB medium containing 1% agarose. This GFP–MinD construct was present either on a multicopy plasmid (pWM1255) or in single copy at the λ attachment site and contained the intact minE gene downstream in its normal context. The single-copy fusion construct WM1534 is essentially the same as a GFP–MinD fusion shown previously to be fully functional and to oscillate from pole to pole (Raskin and de Boer, 1999b). We verified that this fusion, expressed either in single copy or from a plasmid in cylindrical E.coli cells, resulted in typical pole-to-pole oscillation (data not shown).
In the round cells of WM1534, three basic classes of cells were examined for MinD movement: (i) those that had a cleavage furrow on one side, forming a kidney- or heart-shaped cell with a clear long axis perpendicular to the furrow; (ii) diplococci that were either in the last stages of cell division or were already separate daughter cells that had yet to completely separate; and (iii) single cells that appeared to be nearly spherical, sometimes larger than normal.
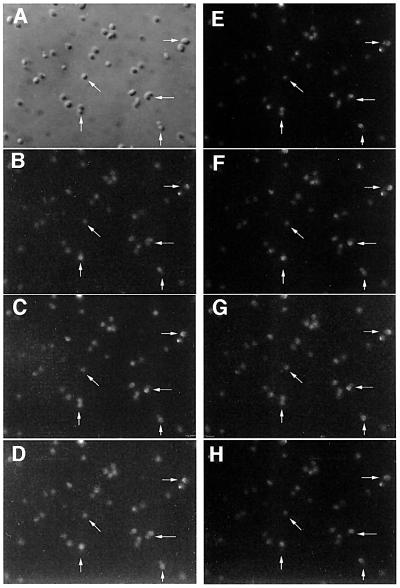
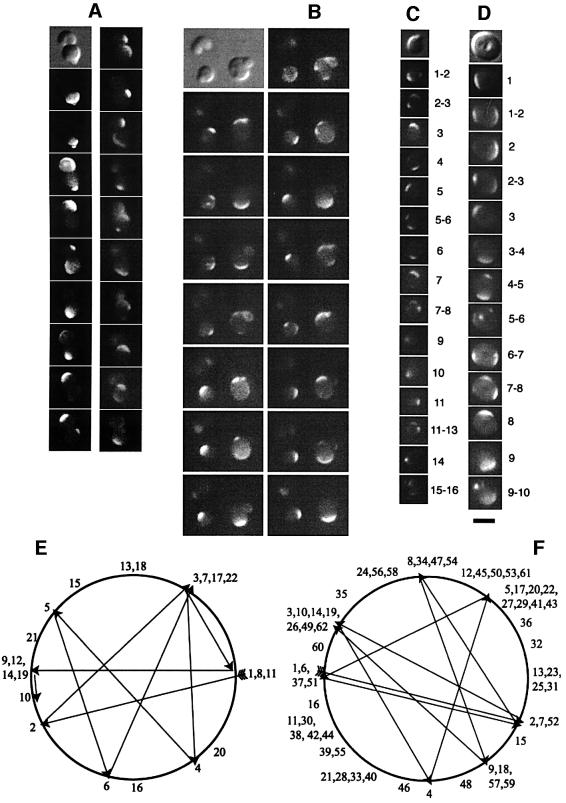
The direction of oscillation in the dividing cells in class (i) was almost exclusively in the direction of the long axis of the cell, perpendicular to the new cell division plane. We monitored 830 separate GFP–MinD migrations in time-lapse movies of a total of 29 class (i) cells. An individual migration was defined as the movement of fluorescence from one assembly point to another, usually occurring over a time course of 10–30 s. Of these 830 migrations, 825 were perpendicular to the developing division plane and parallel to the cell’s long axis. Therefore, MinD continued to assemble repeatedly in zones where it had assembled previously, similar to the pole-to-pole oscillation observed with cylindrical cells. A time-lapse series of a field of cells is shown in Figure 4; horizontal arrows indicate class (i) cells and the movement of GFP–MinD along the long axis. Figure 4 shows only a sub-region of the full field of cells actually observed and only a subset of the full time course. The full time-lapse dataset of this field of cells and others are available for viewing at our website (http://mmg.uth.tmc.edu/mmg2/wm/movie.html). The strong tendency of class (i) cells to exhibit GFP–MinD movement along the long axis is shown graphically in Figure 5A (filled circles).
Fig. 4. Time-lapse images of GFP–MinD movement in a field of rodA cells (WM1534). (A) Nomarski image of the field of cells. (B–H) Fluorescence images of the same field of cells at 10–20 s intervals. Indicated cells include class (i) cells with an asymmetric constriction with MinD oscillation perpendicular to the division plane (horizontal arrows); deeply constricted diplococci (class ii), apparently not fully septated with MinD oscillation perpendicular to the constriction (vertical arrows); and a nearly perfect sphere (class iii) with apparently random directionality of MinD movement (diagonal arrows).
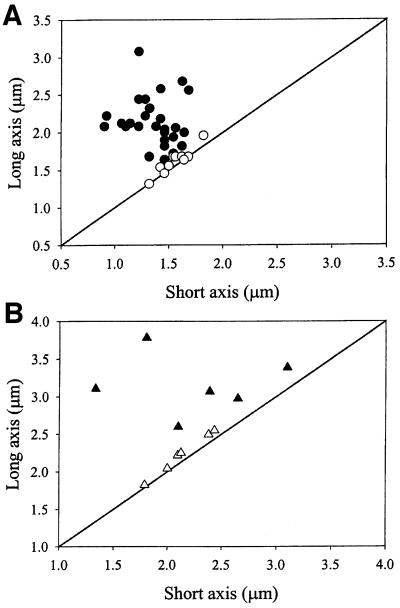
Fig. 5. Patterns of GFP–MinD and MinE–GFP movement correlate with cell geometry. (A) A total of 830 individual movements of GFP–MinD in 40 cells of WM1534 were monitored by time-lapse microscopy in fields similar to those shown in Figure 4, with a range of 14–41 movements per cell. Based on the measured dimensions of each of the 40 cells, the GFP–MinD movements were defined as either along the long axis, along the short axis or along multiple axes. Twenty-nine cells were defined as having predominantly long-axis migrations (filled circles), and the remaining 11 cells had movement to multiple sites (open circles). Of the 29 cells with regular long-axis migrations, three also exhibited occasional migrations along the short axis; this occurred twice out of 27 total migrations, once out of 26 total migrations and twice out of 38 total migrations, respectively, for each of these three cells. (B) A total of 29 MinE–GFP movements were measured in six cells with obvious long axes; all 29 movements were along the long axis (filled triangles). Another six cells were judged to have movement of MinE–GFP in multiple directions (open triangles). In both (A) and (B), the type of movement for each cell in the graph is shown superimposed upon the cell dimensions, with the diagonal line representing a perfectly round cell.
Because rodA mutant cells do not have conventional poles, the back-and-forth movement in class (i) pre-divisional cells suggests that geometric constraints, as opposed to poles per se, may influence MinD movement. However, as five of the 830 movements counted were clearly not along the long axis of the cell, it appears that MinD movement can occasionally deviate from its course. Moreover, of the movements along the long axis, GFP–MinD did not always assemble in exactly the same spot each time, but rather in a general location at the end of the cell (data not shown). This lack of specificity of assembly sites has important implications for the mechanism of movement, more tests of which are described below.
Diplococci [class (ii)] displayed two types of GFP–MinD patterns. The first was movement from one daughter sphere to the other, with the migration nearly always occurring along the longest axis of the combined cells (Figures 4, vertical arrows, 5A and 6A). This pattern is essentially the same as that observed for the class (i) cells, except that the constrictions are significantly deeper and more symmetrical. This pattern also indicates that the cytoplasms of the two daughter cells are continuous.
Fig. 6. Time courses of GFP–MinD movement in individual rodA Δmin (WM1558) cells belonging to the three morphological classes: (A) a class (ii) cell in the final stages of cytokinesis; (B) three cells, early in asymmetric division (top left, class i) and two other cells with buds; (C) a class (iii) cell with no obvious long axis (a perfect sphere) attached to, but apparently closed off from, a bud; and (D) another class (iii) cell. (A)–(D) are subsets of the actual time courses. The first panel is a Nomarski image of the cells, and the following panels represent subsequent time points, ranging from 20 to 60 s intervals, reading down. In (A) and (B), the time courses start on the left, read down, and then continue at the top of the right column of images. In (A), the right column of images was taken starting 5 min after the last image in the left column. Although the cytoplasms of the two cells appear joined in the first three panels, the separate oscillations observed thererafter suggest that the septum had recently closed. Each consecutive MinD assembly point in time courses (C) and (D) is defined by a number, and numbers corresponding to an assembly point or transition between points are shown to the right of each of the panels showing GFP–MinD fluorescence. (E and F) Diagrams of GFP–MinD movement over the entire time courses for cells shown in (C) and (D), respectively. The first nine GFP–MinD movements for each time course are indicated by arrows, and numbers are positioned at the approximate centers of GFP–MinD arcs. Scale bar = 3 µm.
The second pattern, which was less frequently observed, was separate oscillations in the two daughter cells, suggestive of a complete separation of the daughter cell cytoplasms by the septum. Figure 6A shows a rare example of two connected cells that initially displayed MinD movement from one cell to the other (first several panels), but which subsequently exhibited separate movements within each cell. The change in the pattern of MinD movement, from perpendicular to the growing septum to roughly parallel to it, suggests that the septum completed its closure during the time course. In other such diplococci examined, we observed movement parallel to the septum but also occasionally in other directions. These movements correlated with the shape of the daughter cells; if their long axes were parallel to the recently formed septum, the MinD movement was also parallel. However, we did not find enough of these cells that had clear movement to be able to make a firm conclusion.
GFP–MinD assembles at multiple sites in spheres lacking an obvious long axis
In spheres with no obvious long axis (class iii), MinD also moved, migrating repeatedly to a distal site. However, unlike in class (i) cells, the direction of movement usually drifted markedly over the course of several assembly– disassembly cycles. In 11 nearly spherical cells of WM1534 followed over hundreds of MinD movements, in the same batch of cells used for monitoring class (i) cells, MinD movement was more random (Figure 5A, open circles) than the back-and-forth movement of MinD in cells with a clear long axis (Figure 5A, filled circles). The average ratio of the long to short axis of cells with back-and-forth movement was 1.60 ± 0.39, compared with 1.04 ± 0.04 for cells with more random movement. Therefore, movement to multiple sites over time was almost exclusively observed in cells lacking an obvious long axis.
Figure 6C and D shows subsets of typical time courses for class (iii) cells, and the consecutive movements of MinD throughout the full time courses for these cells are tracked by number in Figure 6E and F, respectively, with the first nine movements shown with arrows. It is clear from these data that MinD usually assembles at a site distal from the most recent assembly site but that these sites are not always at the exact opposite side of the cell and drift markedly. Over a time course of several minutes, the movement of MinD to multiple assembly sites indicated that many positions on the cell surface, not just two fixed opposing points, were permissive for MinD assembly. It should be emphasized that because the cells were immobilized in agar, this movement to multiple points did not represent actual rotation of the cell itself; in any case, the type of movement observed is incompatible with simple rotation of the cell. These results were supported by many other examinations of MinD movement in spherical cells (Figures 4, slanted arrows, and 6B; data not shown).
GFP–MinD movement in round cells lacking the Min system
The pattern of GFP–MinD movement was similar in min+ and Δmin rodA (WM1558) cells. For example, pre-divisional cells of WM1558 containing a single, clear central constriction exhibited oscillation perpendicular to the plane of the constriction (Figure 6A and B, cell at top left). Likewise, non-dividing WM1558 cells with nearly spherical dimensions exhibited MinD assembly at a multitude of locations opposite from the previous assembly site (Figure 6B, bottom left cell, C and D; data not shown). Because buds are common in WM1558, we often observed migration of GFP–MinD into the bud if it was contiguous with the mother cell (Figure 6B, right cell). The results with both min+ and Δmin cells indicate that GFP–MinD can diffuse to multiple cellular locations and, in the absence of geometric constraints, can display a highly changeable localization pattern.
MinE–GFP movement in round cells
Because MinD and MinE probably co-migrate (Hale et al., 2001), we also monitored MinE–GFP movement in round cells by using a plasmid containing minD followed by minE–GFP under the control of the pBAD promoter. This MinE–GFP construct (pWM1079) is very similar to those described previously that are functional for MinE activity and exhibit a MinD-dependent dynamic localization cycle (Fu et al., 2001; Hale et al., 2001). We verified that this fusion was able to form MinE rings and oscillate from pole to pole in cylindrical E.coli cells (data not shown).
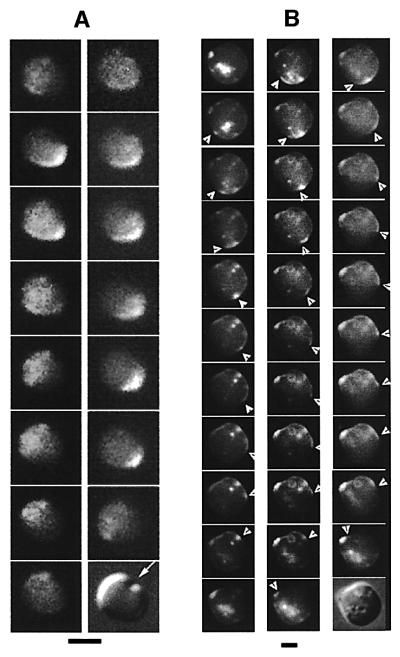
In rodA and Δmin rodA cells expressing this MinE–GFP fusion (WM1595), the general patterns for the three classes of cells were similar to what we observed for MinD movement. MinE–GFP nearly always oscillated back and forth along the long axis of class (i) and (ii) cells. (Figures 7A and 5B, filled triangles). In cells with no obvious long axis, MinE–GFP mostly moved in multiple directions (Figure 5B, open triangles). Unlike GFP–MinD, MinE–GFP tended to form large diffuse zones of fluorescence, analogous to those of MinE–GFP in cylindrical cells (Fu et al., 2001; Hale et al., 2001; data not shown), except that MinE rings were much less distinct in round cells. The diffuse nature of MinE localization in round cells made the migration patterns harder to detect, particularly in smaller cells. As a result, fewer cells exhibited clear patterns of MinE–GFP movement than with GFP–MinD, with the clearest patterns observed in larger cells (Figure 5B). Interestingly, in some very large symmetrical cells, we observed a circular sweeping motion of MinE–GFP along the cell surface (Figure 7B, arrowheads; data not shown) that may represent MinE following MinD around the cell at multiple sites.
Fig. 7. MinE–GFP movement in round cells. (A) Typical MinE–GFP movement along the long axis of a rodA (WM1467) cell, perpendicular to the new division plane. Time-lapse images are spaced 10–30 s apart, from top to bottom, ending with a Nomarski image of the cell at the bottom (the arrow indicates the cell division furrow). (B) Circular MinE–GFP movement in a large rodA Δmin cell (WM1595). Time-lapse images were taken at intervals of 15–60 s. The time course reads from top to bottom in each of the three columns, ending with a Nomarski image of the cell. The trailing edge of the circular, wave-like movement is indicated with arrowheads. Bright fluorescent spots inside the cell indicate MinE–GFP movement in and out of surface aberrations. These patterns are much more apparent in the movie of this time course (see our website: http://mmg.uth.tmc.edu/mmg2/wm/movie.html). Scale bars = 2 µm.
A model for control of the division plane in spherical cells by the Min system
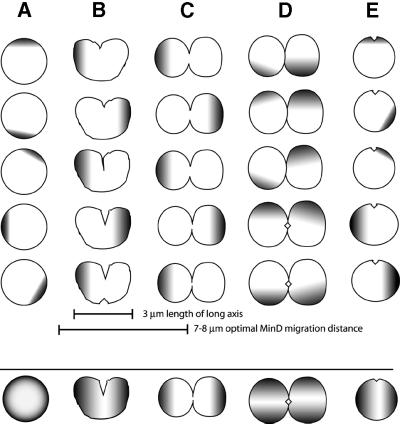
The importance of the Min system for proper cell division in round cells, as well as the patterns of movement observed in different cell shapes, prompted us to propose a model to explain how the Min proteins might regulate division plane specification in round cells that divide in alternating perpendicular planes in two dimensions. In E.coli rodA mutants, division constriction starts on one side of the cell. This asymmetric constriction produces two daughter cells that are not spherical, at least on solid media, but instead have a long axis parallel with the most recent division plane (Begg and Donachie, 1998). Because GFP–MinD stably oscillates in the same direction as this long axis (see Discussion), it is reasonable to suggest that MinD, and the division inhibitor MinC, should always oscillate in a direction parallel to the last division plane (Figure 8B–D). This would lead to the lowest time-averaged concentration of MinC (Meinhardt and de Boer, 2001) at precisely the location that would give rise to a division furrow: the medial perpendicular plane (Figure 8D, bottom cell). In cells with no obvious long axis, GFP–MinD assembles at all points on the cell surface (Figure 8A). As such symmetrical cells seem to be found only in liquid cultures and not on solid media, it remains to be seen if and how the division plane is eventually selected in these cells.
Fig. 8. Schematic of MinD movement in spherical cells, and a model to explain regulation of division planes in cocci. (A–D) Four general stages of growth and division, starting with a spherical cell (A), which then initiates an asymmetric invagination (B), which deepens and becomes an asymmetrical, circumferential constriction (C) and finally becomes two separate cells that are ready to divide in a plane perpendicular to the previous division (D). The five cells within each column above the large horizontal line are representative short-term time courses from top to bottom. The gradients represent the observed movement of MinD (and presumably that of MinC) typical of each stage in the division cycle. Typical long axis distances of round cells and optimal MinD migration distances (see text) are shown by scale bars. The cells below the horizontal line represent the predicted time-averaged location of the MinCD inhibitor in each cell type. It is proposed here that alternating perpendicular division planes in E.coli spherical cells either arise (D) or are stabilized subsequent to the initial formation of the cleavage furrow (E) because of the strong tendency of the MinCD division inhibitor complex to move along the long axis of the cell. This asymmetry is generated either by the previous division process itself or by the initiation of the new constriction, and is maintained and stabilized by the direction of MinCD oscillation.
Discussion
The problem of division plane placement in cocci is complex. Round cells of E.coli are multinucleate and divide centrally and in alternating perpendicular planes, making them tractable model systems for cocci. Evidence from the present study indicates that the Min system exerts spatial control of division site positioning in round cells. The abnormal pattern of FtsZ polymers, including spirals, multiple arcs and elongated septal structures, suggests that, as in cylindrical cells, the Min system normally blocks FtsZ assembly in specific regions of the cell. The Min system may be especially important in round cells because (i) one division plane must be selected from many possible planes and (ii) multiple nucleoid-free spaces probably exist (Donachie et al., 1995; Zaritsky et al., 1999) that could result in unwanted FtsZ assembly at multiple sites. The FtsZ spirals we observed may be examples of such hyper-assembly. In addition, the elongated septal structures containing FtsZ seen in some rodA min– cells (Figure 3F and F′) are consistent with the apparent requirement for Min proteins in proper disassembly of used FtsZ (Yu and Margolin, 2000).
Although FtsZ polymerizes abnormally in many rodA cells lacking the Min system, sometimes assembling into spirals or polymers that appear to be following different trajectories, it still assembles at discrete locations in a significant proportion of cells. This indicates that some other factors, such as nucleoid occlusion (Woldringh et al., 1991, 1994; Margolin, 2000), must exert negative control over FtsZ nucleation and assembly. The viability of the rodA Δmin double mutant suggests that, for the same reason that Δmin mutants of cylindrical E.coli are viable, a sufficient number of division events occurs between multiple nucleoids in these cells to generate viable daughters. The presence of DNA in buds of the double mutant is consistent with this notion and contrasts with the DNA-less minicells of min mutants of cylindrical E.coli. Work is in progress to overcome some of the technical limitations in visualizing nucleoids in spherical cells and to understand how the Min system and the nucleoid segregation apparatus function to position division planes in these cells.
What factors might cause MinD to migrate along the long axis of the cell versus the short axis? One possibility is that one or more components on the cell membrane, either protein or lipid, might act as favored assembly points for MinD. For example, cell poles have characteristics quite distinct from the rest of the cell wall, including lack of peptidoglycan turnover (de Pedro et al., 1997) and different phospholipid composition (Mileykovskaya and Dowhan, 2000; Koppelman et al., 2001). If MinD assembly favored such sites, it could explain assembly at poles with rapid diffusion between assembly sites. There is good evidence for rapid cytoplasmic diffusion in E.coli (Elowitz et al., 1999). MinD also assembles at non-polar sites in filamentous E.coli cells, but it could always be argued that these sites might also harbor distinct membrane domains.
Another possibility, suggested by the present study, is that cell geometry imposes constraints on where MinD assembles and moves. If the total amount of MinD in a given cell cannot reassemble until it reaches a certain critical concentration of MinE and, therefore, a critical distance down a MinE gradient (Margolin, 2001; Rothfield et al., 2001), the prediction would be that MinD has the potential to assemble at any site on the cell surface, provided that it meets the above criteria. Our results show that over a time course in a rodA cell with a long axis, MinD moves the longest possible distance, with only rare forays to the lateral wall. The maximum and preferred distance that MinD moves in filamentous E.coli cells before reassembling appears to be ∼7–8 µm (Raskin and de Boer, 1999b; data not shown), more than twice the length of an average E.coli cylindrical or round-shaped cell. The poles of the average cylindrical cell prevent MinD from moving its preferred distance. In round-shaped or irregularly shaped cells, most of which are 1–3 µm in diameter on average (Figure 5), the longer axis will be heavily favored for MinD migration because it is closer to the preferred maximum migration distance. Our results are entirely consistent with this model. As more is learned about the molecular mechanism by which MinD moves and assembles, including whether it moves via the cytoplasm, membrane, or both, more precise calculations can be made concerning the surface area and volume of the spaces needed for MinD movement and assembly.
The time-averaged concentration of MinCD is predicted to be lowest at the center of a rodA cell with a long axis (Meinhardt and de Boer, 2001). Nevertheless, the occasional lateral movements suggest that MinD does not have to assemble at the ends of the long axis but can, in fact, assemble anywhere in the cell; assembly is heavily favored at the ends because, stochastically, this is usually the farthest point from the previous assembly site (and MinE), as discussed above. This ability to assemble anywhere only becomes obvious, however, in spherical cells with no obvious long axis; this characteristic is indicated in our model in Figure 8A. In these cells, over time, MinD can assemble at all locations, although movement is usually to one or more distal sites (Figure 6E and F). This tendency to migrate the longest distance possible from the previous assembly site may result from the need for MinD to move sufficiently far down the MinE gradient to allow new assembly.
We suggest that the movement of MinD, and therefore of the MinC inhibitor, perpendicular to the newest division plane can help to explain how E.coli cocci divide in alternating perpendicular planes. In the case of rodA cells growing on solid media, the daughter cells become heart- or kidney-shaped as a result of the deep medial constriction of the mother cell (Begg and Donachie, 1998). This causes them to develop a long axis parallel to the last division plane, which should then set up a MinCD gradient that favors midcell division in the perpendicular plane (Figure 8B–D). Additional support for the idea of perpendicular division planes in E.coli cocci comes from studies of the pattern of cell wall growth in round E.coli cells grown in broth (de Pedro et al., 2001). The striking morphological problems of Δmin rodA mutants are fully consistent with an important role of the Min system in spatial control of septation.
On the other hand, this model does not explain: (i) the presence of symmetrical class (iii) cells in the rodA population grown in broth; and (ii) the subpopulation of Δmin rodA mutant cells with apparently normal medial FtsZ rings. The presence of class (iii) cells can be explained if they are, in fact, defective products of division and will not be able to divide again. However, the second observation is harder to reconcile with a model that absolutely requires the Min system for identification of the proper division plane. Therefore, an alternative model for how the Min system controls division plane specification in spheres is needed. In this model, some other factor, such as nucleoid positioning, may control the initial choice of the division plane when the cell has no long axis (Figure 8E). The assembly of an FtsZ arc, and the process of invagination that follows, results in the formation of a long axis, which then would orient the Min oscillation, reinforcing and stabilizing the initial FtsZ arc and preventing FtsZ assembly elsewhere. In spherical cells undergoing artificial DNA replication downshifts and upshifts, constrictions are formed that are tilted relative to one another, resulting in butterfly shapes (Zaritsky et al., 1999). Perhaps Min oscillation is randomized in these enlarged cells and positioning of constriction sites depends more heavily on nucleoid positioning.
Intriguingly, homologs of MinCDE are present in several bacterial genera that are known to divide in alternating perpendicular planes, including Neisseria and Deinococcus. At least in Neisseria gonorrhoeae, the Min system appears to be required for proper selection of division sites, as it has been shown recently that inactivation of minC or minD results in abnormal cell division (Ramirez-Arcos et al., 2001; Szeto et al., 2001). These results are consistent with our work with round E.coli. However, Staphylococcus aureus, which also appears to divide in alternating perpendicular planes (Tzagoloff and Novick, 1977), does not contain recognizable homologs of the Min proteins. This observation makes it even more likely that positioning of the nucleoid must also play a role in division site selection, as it does in cylindrical E.coli cells.
Materials and methods
Construction of a rodA Δmin double mutant
All bacterial strains are derivatives of E.coli K12 and are listed in Tables I and II. Strain KJB24 contains a rodA (am) mutation, causing the cells to grow as spheres, and a Tn5 insertion that allows growth on rich media (Begg and Donachie, 1998). Because of this Tn5, KJB24 is already kanr, so the kanr ΔminCDE::aph allele of WM947 could not be moved into KJB24 directly. Instead, a tetracycline-resistance (tetr) marker linked to ΔminCDE was used. First, a fadR::Tn10 marker from strain CAG18497 was introduced by P1 transduction into WM947. The fadR::Tn10 confers tetr and is co-transducible with minCDE. Tetr kanr transductants containing both fadR::Tn10 and ΔminCDE::kan were isolated and purified, and one isolate (WM1192) was chosen as the donor to transduce ΔminCDE::kan with a linked tetr marker. A phage P1 lysate of WM1192 was used to transduce KJB24 to tetr. To ascertain which of these transductants were ΔminB rodA double mutants, we made P1 lysates from several transductants, which were used to transduce wild-type E.coli K12 (MG1655) to tetr. These transductants were then screened for kanr and examined microscopically for the production of minicells. One of several KJB24 strains that were able to transduce kanr and a minicell phenotype to MG1655 was chosen (WM1250) and defined as a rodA ΔminCDE mutant.
Table I. Strains.
| Strain | Relevant genotype | Source/reference |
|---|---|---|
| MG1655 | wild-type K12 strain | laboratory collection |
| TG1 | F′ traD36 lacIqZΔM15 proA+B+/supE Δ(hsdM-mcrB)5 (rk-mk+McrB-) thi Δ(lac-proAB) | laboratory collection |
| W3110 | wild-type K12 strain | laboratory collection |
| JM109 | F′, traD36, proAB+, lacIqZΔM15, recA1, endA1, gyrA96, thi, hsdR17 (rk–,mk+), supE44, Δ(lac-proAB), relA1 | laboratory collection |
| WM947 | ΔminCDE::aph in MG1655 | Yu and Margolin (1999) |
| WM1192 | fadR::Tn10 in WM947 | this work |
| KJB24 | rodA (am), kanr, thyA in W3110 (Sup0) | Begg and Donachie (1998) |
| KJB28 | ftsA12 (ts) leu::Tn10 in KJB24 | Begg and Donachie (1998) |
| WM1250 | fadR::Tn10, ΔminCDE::aph in KJB24 | this work |
| WM1264 | Ptrc90-GFP-minD-minE from pWM1255 inserted in chromosome at attB, in W3110 | this work |
| WM1255 | pWM1255 in TG1 | this work |
| WM1079 | pWM1079 in JM109 | this work |
| WM1467 | pWM1079 in KJB24 | this work |
| WM1595 | pWM1079 in WM1250 | this work |
| WM1534 | KJB24 containing chromosomal GFP-minD-minE from WM1264 | this work |
| WM1558 | WM1534 containing fadR::Tn10, ΔminCDE::aph from WM1192 | this work |
Table II. Plasmids.
| Plasmid | Description | Source/reference |
|---|---|---|
| pBAD33 | pACYC184 (p15a) derivative containing the araBAD promoter | Guzman et al. (1995) |
| pDSW207 | pBR322 derivative containing Ptrc90 and GFP lacking a stop codon | Weiss et al. (1999) |
| pWM1079 | ParaBAD-minD-minE-GFP in pBAD33 | Sun and Margolin (2001) |
| pWM1191 | Plac-GFP-minD-minE in pBC (SK+) | this work |
| pWM1255 | Ptrc90-GFP-minD-minE in pDSW207 | this work |
Construction of fusions of GFP to MinD and MinE in round cells
A single-copy GFP–minD fusion containing the minE gene immediately downstream in order to allow MinD movement (Raskin and de Boer, 1999b) was constructed as follows. First, minDE was PCR amplified from E.coli genomic DNA using primers MinD6 (5′-TTGAATTCGCACGCATTATTGTTG, second codon of minD underlined) and MinE3 (5′-CTGGTACCTTATTTCAGCTCTTC, reverse complement of the minE stop codon underlined). The PCR product was cleaved with EcoRI and KpnI, and cloned between the EcoRI and KpnI sites of pWM678, which contains GFP without a stop codon inserted at the BamHI site of pBC SK(+) such that GFP is in the same reading frame as lacZ. This plasmid (pWM1191) was then cleaved with NcoI and KpnI to release a fragment containing the downstream portion of GFP fused to minD plus the minE gene, and this fragment was cloned into NcoI–KpnI-cleaved pDSW207 (Weiss et al., 1999). This plasmid (pWM1255) expresses GFP–MinD, as well as MinE from the pTrc207 promoter, and cells containing pWM1255 exhibited normal GFP–MinD oscillation in the absence of induction. However, to control the expression of the fusion protein more precisely, it was moved to the chromosome using the λ InCh system (Boyd et al., 2000) making strain WM1264. The ability of the single-copy fusion in WM1264 to oscillate was confirmed, and this fusion was then transferred to KJB24 or WM1250 by phage P1 transduction, selecting for Apr. These strains were designated WM1534 or WM1558, respectively.
To monitor MinE–GFP movement in spherical cells, we introduced plasmid pWM1079 into KJB24 and WM1250 to make WM1467 and WM1595, respectively. This plasmid contains minDE sequences upstream of GFP in plasmid pBAD33, with minE lacking its stop codon, under the control of the pBAD promoter (Sun and Margolin, 2001).
Growth conditions, measurements and cell measurements
All the E.coli strains were grown in LB medium, consisting of 10 g of tryptone, 5 g of yeast extract and 5 g of NaCl per liter. In some cases, the KJB24 derivatives were grown in M9 salts + 0.2% glucose + 1 mM MgSO4 + 5% casamino acids. Thymine was added to LB medium to support growth of KJB24 and its derivatives, which are thy–. No obvious difference in Min protein oscillation or cell morphology was noticed with growth in minimal versus rich media. For Min protein oscillation experiments, cells were generally grown to exponential phase at 30°C. For GFP–MinD experiments with the single-copy fusion, a saturated culture was diluted at least 100-fold into fresh medium containing 10–20 µM isopropyl-β-d-thiogalactopyranoside (IPTG) and grown for several hours before examination. For experiments with the plasmid-borne copy of GFP–MinD (pWM1255), no IPTG was added. For MinE–GFP measurements, cells were treated similarly, except that 0.001–0.01% l-arabinose was added to the medium. In both cases, induction levels were kept as low as possible to minimize perturbations to cellular physiology, yet high enough to produce fluorescence detectable by the camera (see below). Optical densities of cultures were measured with a Klett–Summerson meter, and the data were then analyzed and plotted with SigmaPlot (Jandel Scientific). Cell size distributions were measured by combining several phase-contrast images, like those shown in Figure 2, and analyzing particle sizes with ImageJ freeware. Care was taken to analyze only individual cells and not clumps of cells.
Detection of FtsZ in vivo
Immunofluorescence microscopy using affinity-purified anti-FtsZ polyclonal antibody conjugated to Alexa 488 (Molecular Probes, Eugene, OR) was performed essentially as described previously (Yu and Margolin, 1999). Deconvolution of FtsZ immunofluorescence images was performed with a Deltavision system as described previously (Sun and Margolin, 1998).
Monitoring GFP fusion localization in live cells
For microscopic examination, 3 µl of cell culture were mixed with an equal volume of molten growth medium containing 2% low-melt agarose, dropped onto a glass microscope slide and quickly covered with an 18-mm-square cover glass. As long as the agar was freshly made and was mixed well with the cells, all cells were completely immobilized on the slide. GFP fluorescence and Nomarski brightfield images were observed with a 100× oil immersion (N.A. 1.3) objective on an Olympus BX60 microscope fitted with a GFP filter cube and captured with a light-sensitive Photometrics CoolSnap fx cooled CCD camera driven by QED image capturing software. Time-lapse images were generally taken every 10–15 s, usually at exposures of 1–4 s each and sometimes with 2 × 2 binning to improve sensitivity, and saved as PICT files on a Macintosh G4 computer. At least one brightfield image per movie was also obtained with Nomarski optics. Because of the three-dimensional nature of the cells, care was taken to keep them in the same focal plane for the entire time course. To compile the time-lapse images, movies were made from these files with iMOVIE 2.2 software. To quantitate protein oscillations, cells of the three morphological classes were indicated in brightfield images, and then the movement of each fluorescent assembly point was scored for each of these cells. The data were compiled with Microsoft Excel and plotted with SigmaPlot.
Acknowledgments
Acknowledgements
We thank Ken Begg for sending us KJB24 and KJB28 and David Weiss for pD5W207. This work was supported by a grant from the National Institutes of Health (1-R01-GM61074-01).
References
- Addinall S.G. and Lutkenhaus,J. (1996) FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol., 22, 231–237. [DOI] [PubMed] [Google Scholar]
- Adler H.I., Fisher,W., Cohen,A. and Hardigree,A. (1967) Miniature E.coli cells deficient in DNA. Proc. Natl Acad. Sci. USA, 57, 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K.J. and Donachie,W.D. (1998) Division planes alternate in spherical cells of Escherichia coli. J. Bacteriol., 180, 2564–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. and Lutkenhaus,J. (1991) FtsZ ring structure associated with division in Escherichia coli. Nature, 354, 161–164. [DOI] [PubMed] [Google Scholar]
- Bi E. and Lutkenhaus,J. (1993) Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol., 175, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D., Weiss,D.S., Chen,J.C. and Beckwith,J. (2000) Towards single-copy gene expression systems making gene cloning physiologically relevant: λ InCh, a simple Escherichia coli plasmid–chromosome shuttle system. J. Bacteriol., 182, 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A.J., Crossley,R.E. and Rothfield,L.I. (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine the proper placement of the division site in Escherichia coli. Cell, 56, 641–649. [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E. and Rothfield,L.I. (1990) Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc. Natl Acad. Sci. USA, 87, 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A.J., Crossley,R.E., Hand,A.R. and Rothfield,L.I. (1991) The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J., 10, 4371–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E. and Rothfield,L.I. (1992) Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol., 174, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro M.A., Quintela,J.C., Holtje,J.-V. and Schwarz,H. (1997) Murein segregation in Escherichia coli. J. Bacteriol., 179, 2823–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro M.A., Donachie,W.D., Holtje,J.V. and Schwarz,H. (2001) Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol., 183, 4115–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie W.D., Addinall,S. and Begg,K. (1995) Cell shape and chromosome partition in prokaryotes or, why E.coli is rod-shaped and haploid. BioEssays, 17, 569–576. [DOI] [PubMed] [Google Scholar]
- Elowitz M.B., Surette,M.G., Wolf,P.E., Stock,J.B. and Leibler,S. (1999) Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol., 181, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Shih,Y.L., Zhang,Y. and Rothfield,L.I. (2001) The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl Acad. Sci. USA, 98, 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L.M., Belin,D., Carson,M.J. and Beckwith,J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C.A., Meinhardt,H. and de Boer,P.A. (2001) Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J., 20, 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. and Lutkenhaus,J. (1999) Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol., 34, 82–90. [DOI] [PubMed] [Google Scholar]
- Hu Z. and Lutkenhaus,J. (2001) Topological regulation of cell division in E.coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell, 7, 1337–1343. [DOI] [PubMed] [Google Scholar]
- Hu Z., Mukherjee,A., Pichoff,S. and Lutkenhaus,J. (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl Acad. Sci. USA, 96, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppelman C.M., Den Blaauwen,T., Duursma,M.C., Heeren,R.M. and Nanninga,N. (2001) Escherichia coli minicell membranes are enriched in cardiolipin. J. Bacteriol., 183, 6144–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Ehrhardt,D.W. and Margolin,W. (1996) Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl Acad. Sci. USA, 93, 12998–13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. (2000) Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev., 24, 531–548. [DOI] [PubMed] [Google Scholar]
- Margolin W. (2001) Bacterial cell division: a moving MinE sweeper boggles the MinD. Curr. Biol., 11, R395–R398. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. and de Boer,P.A.J. (2001) Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl Acad. Sci. USA, 98, 14202–14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E. and Dowhan,W. (2000) Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol., 182, 1172–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E., Sun,Q., Margolin,W. and Dowhan,W. (1998) Localization and function of cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J. Bacteriol., 180, 4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray R.G., Hall,M. and Thompson,B.G. (1983) Cell division in Deinococcus radiodurans and a method for displaying septa. Can. J. Microbiol., 29, 1412–1423. [DOI] [PubMed] [Google Scholar]
- Pas E., Einav,M., Woldringh,C.L. and Zaritsky,A. (2001) Perpendicular planes of FtsZ arcs in spheroidal Escherichia coli cells. Biochimie, 83, 121–124. [DOI] [PubMed] [Google Scholar]
- Pichoff S. and Lutkenhaus,J. (2001) Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol., 183, 6630–6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Arcos S., Szeto,J., Beveridge,T.J., Victor,C., Francis,F. and Dillon,J.R. (2001) Deletion of the cell-division inhibitor MinC results in lysis of Neisseria gonorrhoeae. Microbiology, 147, 225–237. [DOI] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A. (1997) The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E.coli. Cell, 91, 685–694. [DOI] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A. (1999a) MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol., 181, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A. (1999b) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA, 96, 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L., Justice,S. and Garcia-Lara,J. (1999) Bacterial cell division. Annu. Rev. Genet., 33, 423–448. [DOI] [PubMed] [Google Scholar]
- Rothfield L.I., Shih,Y.-L. and King,G. (2001) Polar explorers: membrane proteins that determine division site placement. Cell, 106, 13–16. [DOI] [PubMed] [Google Scholar]
- Sun Q. and Margolin,W. (1998) FtsZ dynamics during the cell division cycle of live Escherichia coli. J. Bacteriol., 180, 2050–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q. and Margolin,W. (2001) Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J. Bacteriol., 183, 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Yu,X.-C. and Margolin,W. (1998) Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol. Microbiol., 29, 491–504. [DOI] [PubMed] [Google Scholar]
- Szeto J., Ramirez-Arcos,S., Raymond,C., Hicks,L.D., Kay,C.M. and Dillon,J.A. (2001) Gonococcal MinD affects cell division in Neisseria gonorrhoeae and Escherichia coli and exhibits a novel self-interaction. J. Bacteriol., 183, 6253–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R.M., Collins,J.F. and Donachie,W.D. (1974) Quantal behavior of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J. Bacteriol., 118, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff H. and Novick,R. (1977) Geometry of cell division in Staphylococcus aureus. J. Bacteriol., 129, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D.S., Chen,J.C., Ghigo,J.M., Boyd,D. and Beckwith,J. (1999) Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol., 181, 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westling-Haggstrom B., Elmros,T., Normark,S. and Winblad,B. (1977) Growth pattern and cell division in Neisseria gonorrhoeae. J. Bacteriol., 129, 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh C.L., Mulder,E., Huls,P.G. and Vischer,N. (1991) Topo regulation of bacterial division according to the nucleoid occlusion model. Res. Microbiol., 142, 309–320. [DOI] [PubMed] [Google Scholar]
- Woldringh C.L., Zaritsky,A. and Grover,N.B. (1994) Nucleoid partitioning and the division plane of Escherichia coli. J. Bacteriol., 176, 6030–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.-C. and Margolin,W. (1999) FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol., 32, 315–326. [DOI] [PubMed] [Google Scholar]
- Yu X.-C. and Margolin,W. (2000) Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J. Bacteriol., 182, 6203–6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Sun,Q. and Margolin,W. (2001) FtsZ rings in mukB mutants with or without the Min system. Biochimie, 83, 125–129. [DOI] [PubMed] [Google Scholar]
- Zaritsky A., Woldringh,C.L., Fishov,I., Vischer,N.O. and Einav,M. (1999) Varying division planes of secondary constrictions in spheroidal Escherichia coli cells. Microbiology, 145, 1015–1022. [DOI] [PubMed] [Google Scholar]