Abstract
Saccharomyces cerevisiae Mgs1 protein, which possesses DNA-dependent ATPase and single strand DNA annealing activities, plays a role in maintaining genomic stability. We found that mgs1 is synthetic lethal with rad6 and exhibits a synergistic growth defect with rad18 and rad5, which are members of the RAD6 epistasis group important for tolerance of DNA damage during DNA replication. The mgs1 mutant is not sensitive to DNA-damaging agents, but the mgs1 rad5 double mutant has increased sensitivity to hydroxyurea and a greatly increased spontaneous mutation rate. Growth defects of mgs1 rad18 double mutants are suppressed by a mutation in SRS2, encoding a DNA helicase, or by overexpression of Rad52. More over, mgs1 mutation suppresses the temperature sensitivity of mutants in POL3, encoding DNA polymerase δ. mgs1 also suppresses the growth defect of a pol3 mutant caused by expression of Escherichia coli RuvC, a bacterial Holliday junction resolvase. These findings suggest that Mgs1 is essential for preventing genome instability caused by replication fork arrest in cells deficient in the RAD6 pathway and may modulate replication fork movement catalyzed by yeast polymerase δ.
Keywords: DNA replication/homologous recombination/MGS1/RAD6 epistasis group
Introduction
DNA-damaging agents cause DNA lesions that can block the progression of DNA polymerases (Friedberg et al., 1995). However, the DNA replication fork can also be hindered during normal vegetative growth in the absence of exogenous DNA-damaging agents. For example, the fork may encounter topological stress, tightly bound protein complexes or aberrant DNA structures. These obstacles can block fork progression leading to gaps or double strand breaks, which can be lethal lesions. Cells have developed mechanisms to enable them to survive such potentially lethal situations. Because these fork block sites do not contain damaged DNA on template DNA, it is likely that the stalled replication complexes are released and normal replication restored by an error-free mechanism. In contrast, translesion polymerases act at forks stalled due to DNA lesions and perform error-prone and error-free DNA repair (Woodgate, 1999; Kunz et al., 2000). Recently, there has been increasing evidence that the homologous recombination pathway plays an important role in the error-free process. DNA gaps and double strand breaks caused by replication fork arrest are excellent substrates for homologous recombination (Haber, 1999; Cox et al., 2000; Kowalczykowski, 2000; Kuzminov, 2001). Thus, DNA replication, repair and recombination systems are coordinated to prevent genome instability caused by DNA replication arrest.
In Saccharomyces cerevisiae, Rad6 and Rad18 play central roles in the post-replication repair (PRR) pathway which supports both error-free and error-prone DNA repair (Prakash et al., 1993; Friedberg et al., 1995; Xiao et al., 2000). Rad6 is a ubiquitin-conjugating enzyme (UBC) that forms a stable complex with Rad18, an ATPase that binds single-stranded DNA (Bailly et al., 1994, 1997). The RAD6 subpathway is the error-prone branch of PRR and it involves translesion polymerases that bypass replication-blocking lesions in a process known as translesion DNA synthesis (Liefshitz et al., 1998; Xiao et al., 2000). In contrast, Rad5 participates in an error-free subpathway of PRR (McDonald et al., 1997). Rad5 is a DNA-dependent ATPase that has homology to the SNF2/SWI2 family of helicases (Johnson et al., 1994; Pazin and Kadonaga, 1997); however, no helicase activity has been detected in Rad5 (Johnson et al., 1994). Rad5 associates with Rad18 and recruits the Mms2–Ubc13 complex to chromatin in response to DNA damage (Ulrich and Jentsch, 2000). Ubc13 is a UBC that forms a stable complex with a non-canonical UBC variant, Mms2 (Broomfield et al., 1998; Brusky et al., 2000). The cellular targets of Ubc13–Mms2 and the function of Rad5 remain unknown; however, it is likely that these proteins play an important role in error-free PRR.
The phenotype of rad6 and rad18 mutants includes hypersensitivity to radiation, slow growth and a high spontaneous mutation rate (Lawrence and Christensen, 1979; Prakash, 1981). This phenotype is suppressed by mutations in SRS2 (Aboussekhra et al., 1989; Schiestl et al., 1990), which encodes a DNA helicase with 3′ to 5′ polarity (Rong and Klein, 1993). Recently, it was shown that srs2 specifically suppresses mutations in the RAD5-dependent error-free branch of the RAD6 pathway (Ulrich, 2001). Suppression by srs2 requires homologous recombination, suggesting that Srs2 may channel lesions into the error-free Rad5-dependent PRR subpathway. This may prevent aborted recombination repair at stalled replication forks.
Saccharomyces cerevisiae MGS1 (maintenance of genome stability 1) encodes a protein with centrally located homology to the central region of Escherichia coli RuvB, a Holliday junction branch migration protein, and Rfc, the eukaryotic clamp loader protein (Hishida et al., 2001). The homologous region includes the Walker A and B, and sensor I and II motif sequences, which are characteristic of the AAA+ class ATPase family (Hishida et al., 2001). The MGS1 orthologues are highly conserved in prokaryotes and eukaryotes (Barre et al., 2001; Hishida et al., 2001; Kawabe et al., 2001). The S.cerevisiae Mgs1 protein possesses DNA-dependent ATPase and DNA annealing activities (Hishida et al., 2001). The mgs1Δ mutant has an increased rate of homologous recombination, which is elevated further when combined with a mutation in TOP3, a type IA topoisomerase (Hishida et al., 2001). Recently, it was shown that a mouse homologue of Mgs1, Whip1, interacts physically with the N-terminal portion of mouse Werner’s syndrome protein (mWrn) and co-localizes with mWrn in the nucleus (Kawabe et al., 2001). WRN encodes a member of RecQ helicase family, and mutants in WRN are tightly associated with the human hereditary disease Werner’s syndrome (Yu et al., 1996; Gray et al., 1997). Werner’s syndrome cells have an increased rate of sister chromatid exchange, slow replication and increased sensitivity to 4-nitroquinoline oxide and camptothecin (Shen and Loeb, 2000). These results suggest that Wrn protein is involved in maintaining genome stability during replication. Based on these results, we proposed that Mgs1 may play a role in maintaining proper DNA topology, which is required for genome stability during replication (Hishida et al., 2001).
To understand further the biological role of Mgs1, genetic interactions between MGS1 and S.cerevisiae DNA repair genes were characterized by screening for synthetic lethality in the presence of mgs1Δ. This screen identified rad18. The synthetic lethality of the mgs1 rad18 double mutant is suppressed by activation of homologous recombination. In addition, an mgs1 mutation partially suppresses the growth defect in a pol3 strain. These results suggest that Mgs1 interacts with the DNA replication machinery and plays a role in preventing genomic instability caused by replication fork arrest. This mechanism may provide an alternative pathway that functions when PRR and homologous recombination are impaired.
Results
Isolation of mutants that require Mgs1 for growth
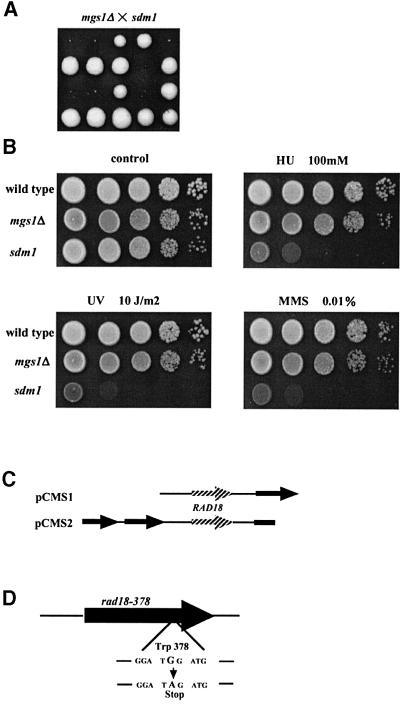
To isolate mutants that require Mgs1 for growth, 120 000 colonies were screened using a red/white colony-sectoring assay. One mutant was isolated and it was called synthetic growth defect with mgs1Δ (sdm1). Tetrad analysis revealed that the mgs1Δ sdm1 double mutant is unable to grow at 37°C, and forms very small colonies after 5 days at 30°C on rich agar plates (Figure 1A). The sdm1 single mutant is highly sensitive to hydroxyurea (HU) and methylmethane sulfonate (MMS), and extremely sensitive to UV light (Figure 1B). Two plasmids were isolated from a yeast genomic library that complement the MMS sensitivity of sdm1. These two plasmids also complement the HU and UV sensitivities of the sdm1 mutant and the growth defect of the mgs1Δ sdm1 double mutant (data not shown). Both complementing plasmids include a yeast genomic sequence corresponding to the open reading frame (ORF) of the RAD18 gene (Figure 1C). To determine whether the sdm1 mutation is allelic to RAD18, the RAD18 region in the sdm1 mutant was sequenced. A single G to A transition mutation was found at the second base of the 378th codon of RAD18 (TGG→TAG), substituting an amber nonsense codon for tryptophan (Figure 1D). Therefore, we named the sdm1 mutant allele rad18-378. mgs1Δ was combined with a rad18 null allele, and heterozygous diploid cells were subjected to tetrad analysis. The mgs1Δ rad18Δ double mutant did not grow at 37°C and formed extremely small colonies at 30°C (data not shown).
Fig. 1. Identification of the sdm1 mutation. (A) Growth defect of the mgs1Δ sdm1 double mutant. Tetrad analysis of a cross between mgs1Δ and sdm1. All tiny colonies were mgs1Δ sdm1 double mutant. mgs1Δ and sdm1 mutations were confirmed by Leu+ prototropy and UV-sensitive phenotype, respectively. (B) Phenotypes of the sdm1 mutant with respect to growth and sensitivity to MMS, HU and UV. Wild-type, mgs1Δ and sdm1 cells were serially diluted and spotted onto YPAD plates containing 0.01% MMS or 100 mM HU. For UV sensitivity, plates on which cells were spotted were irradiated and incubated at 30°C for 3 days. (C) DNA inserts that complement the MMS sensitivity of the sdm1 mutant. (D) sdm1 mutation and wild-type sequence. Sequence analysis revealed that sdm1 is an allele of RAD18; a nonsense codon substitutes for tryptophan codon 378.
Relationship between MGS1 and genes in the RAD6 epistasis group
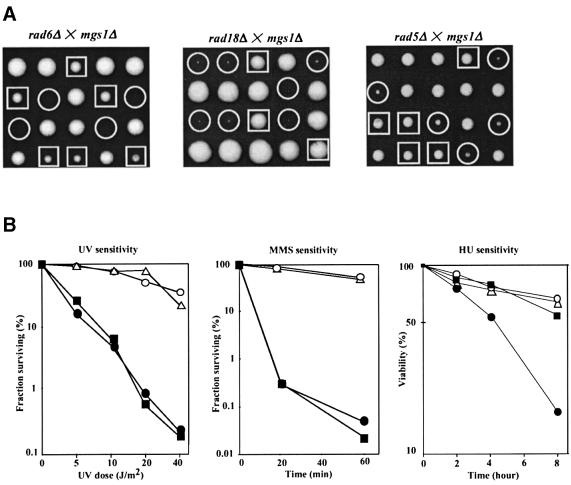
RAD18 is a member of the RAD6 epistasis group that contributes to DNA damage tolerance. Because MGS1 interacts with RAD18, mgs1Δ was also combined with mutants in other genes in the RAD6 pathway in a background with a wild-type RAD5 gene (see Materials and methods). (Experiments reported above were carried out in a rad5G535R background.) Tetrad analysis was carried out with diploid strains heterozygous for mgs1Δ and a deletion of rad6, rad18, rad5, rad30, rev1, rev3 or rev7, and the double mutants were examined for UV sensitivity and growth rate. The mgs1Δ mutation did not affect UV sensitivity of any mutants or the growth of rad30Δ, rev1Δ, rev3Δ or rev7Δ mutants (data not shown). However, an mgs1Δ rad6Δ double mutant did not grow at 26, 30 or 37°C (Figure 2A). The mgs1Δ rad18Δ double mutant was as defective in growth in this strain background as in the rad5G535R background (Figure 2A). A rad5Δ mutation also caused a synergistic growth defect in mgs1Δ, which was less severe than the growth defect in an mgs1Δ rad18Δ strain (Figure 2A). The mgs1Δ rad18Δ double mutant was very difficult to study, because it grows very poorly and was unstable, spontaneously producing fast growing colonies. Therefore, the phenotype and DNA repair characteristics of mgs1Δ mutants were studied in rad5Δ cells.
Fig. 2. Synergistic growth defect due to mgs1Δ and mutants belonging to the RAD6 epistasis group. (A) Tetrads of diploid cells formed by the crosses indicated were dissected and grown on YPAD at 30°C for 4 or 5 days. mgs1Δ was synthetic lethal with rad6Δ, and showed a strong synergistic growth defect with rad18Δ and rad5Δ. Squares indicate the rad6Δ, rad18Δ and rad5Δ mutants. Circles indicate the rad6Δ mgs1Δ, rad18Δ mgs1Δ and rad5Δ mgs1Δ double mutants. (B) UV, MMS and HU sensitivity of the mgs1Δ rad5Δ cells. UV, MMS and HU sensitivity was determined as described in Materials and methods. Wild type (open circles); mgs1 (open triangles); rad5 (filled squares); and rad5 mgs1 (filled circles).
mgs1Δ is no more sensitive than wild-type yeast to UV, MMS or HU; rad5Δ is more sensitive to UV and MMS than wild-type yeast, but it is no more sensitive to HU than wild type (Figure 2B). The rad5Δ mgs1Δ mutant has a similar level of sensitivity to UV and MMS as rad5Δ, but enhanced sensitivity to HU (Figure 2B). These results suggest that mgs1 mutants require Rad5, Rad6 and Rad18 for normal growth, but not other genes in the RAD6 epistasis group. The synergistic sensitivity of the rad5Δ mgs1Δ double mutant to HU, but not UV or MMS, suggests that Mgs1 plays a role in preventing HU-induced interference with DNA replication in the rad5 mutant.
Rad5 has been assigned to the error-free subpathway of PRR, suggesting that Mgs1 may play a redundant role in this process. This possibility was tested by measuring spontaneous reversion and recombination frequencies in an mgs1 mutant background. Intrachromosomal mitotic recombination between heteroallelic DNA sequences was measured using a lys2BA::URA3::lys2BG cassette on chromosome II (see Materials and methods). As shown in Table I, the rate of recombination increased slightly in mgs1Δ and rad5Δ strains. In the mgs1Δ rad5Δ double mutant, Lys+ recombination frequency increased significantly. The majority of Lys+ recombinants were Lys+Ura–, which result from intrachromatid popout or unequal sister chromatid exchange. In addition, the mgs1Δ rad5Δ double mutant had a 284- and 644-fold increase in the ade2-1 and trp1-1 reversion frequency, respectively (Table II). These results suggest that Mgs1 is functionally redundant with RAD5-dependent error-free PRR. Thus, MGS1 is likely to play a role in maintaining genomic stability during replication.
Table I. Rate of intrachromosomal recombination.
| Genotype | Lys+ Ura+ (relative rec.) | Lys+ Ura– (relative rec.) |
|---|---|---|
| MGS1 RAD5 | 1.6 × 10–5 (1.0) | 1.9 × 10–5 (1.0) |
| mgs1 RAD5 | 5.3 × 10–5 (3.3) | 10.8 × 10–5 (5.7) |
| MGS1 rad5 | 2.3 × 10–5 (1.4) | 8.6 × 10–5 (4.5) |
| mgs1 rad5 | 9.2 × 10–5 (5.7) | 50.2 × 10–5 (26.4) |
Recombination rates between the direct repeat sequences in lys2BA::URA3::lys2BG of the haploid strains were determined. Recombination rates were calculated according to the median method described by Lea and Coulson (1949).
Table II. Spontaneous reversion frequency at trp1-1 and ade2-1 alleles.
| Reversion frequency/107 viable cells (fold increase) at 30°C | ||
|---|---|---|
| Trp+ | Ade+ | |
| Wild type | 2.0 (×1) | 3.6 (×1) |
| mgs1 | 8.5 (×4.2) | 8.5 (×2.4) |
| rad5 | 13.3 (×6.7) | 13.3 (×3.7) |
|
mgs1 rad5 |
1288.2 (×644) |
1025.1 (×284) |
| Reversion frequency/107 viable cells (fold increase) at 25°C | ||
| Wild type | 1.2 (×1) | 1.5 (×1) |
| mgs1 | 5.9 (×4.9) | 4.3 (×2.8) |
| pol3-13 | 6.3 (×5.3) | 3.2 (×2.1) |
| mgs1 pol3-13 | 43.9 (×36.4) | 13.9 (×9.2) |
The reversion frequency was determined as described in Materials and methods. The data are the averages of five independent measurements.
Isolation of an mgs1 mutation that confers temperature sensitivity on rad18Δ
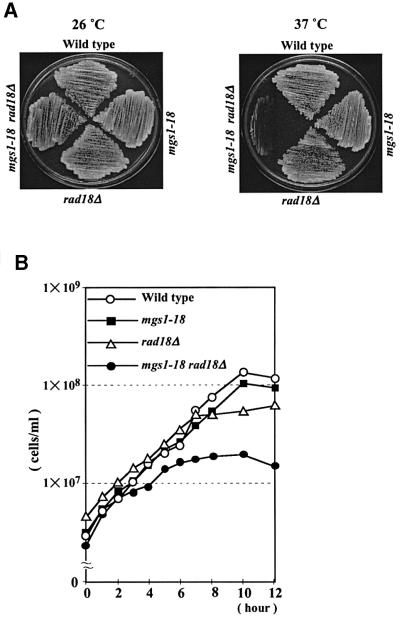
An mgs1 mutant, mgs1-18, was isolated that makes rad18Δ cells temperature sensitive for growth. In mgs1-18, both the 394th lysine codon (AAG) and the 411th glycine codon (GGG) are changed to that for arginine (AGG). Each of the two mutations, K394R and G411R, confer a modest degree of temperature sensitivity for growth on the rad18Δ strain (data not shown). As shown in Figure 3A, the mgs1-18 rad18Δ double mutant is severely defective in growth at 37°C (Figure 3A). The mgs1-18 rad18Δ cells were grown to early logarithmic phase (2–5 × 106 cells/ml) in liquid YPAD at 26°C. When the culture was shifted to 37°C, the double mutant ceased to grow after ∼5 h (Figure 3B).
Fig. 3. Temperature sensitivity of mgs1-18 rad18Δ. (A) Wild-type, mgs1-18, rad18Δ and mgs1-18 rad18Δ cells were streaked onto a YPAD plate and incubated at 30 (left) and 37°C (right) for 3 days. (B) Growth of mutant cells after temperature shift to 37°C. Wild-type, mgs1-18, rad18Δ and mgs1-18 rad18Δ cells were grown in liquid YPAD to early logarithmic phase (2–5 × 106 cells/ml) at 3°C, and then shifted to 37°C. Wild-type (open diamonds); mgs1-18 (filled squares); rad18Δ (open triangles); and mgs1-18 rad18Δ (filled circles).
Effect of mgs1 on cell cycle progression in a rad18 mutant
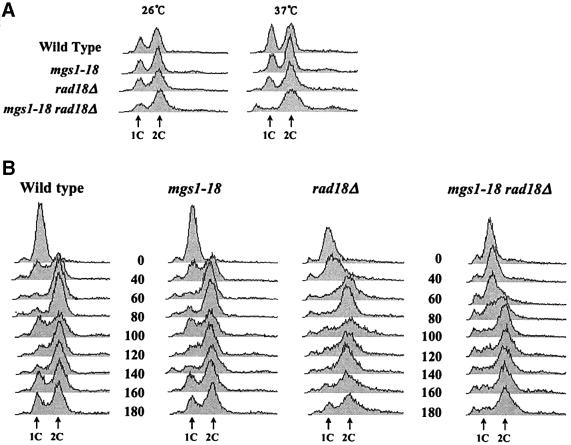
Cell cycle progression was analysed in wild-type, mgs1-18, rad18Δ and mgs1-18 rad18Δ cells using fluorescence-activated cell sorting (FACS) analysis to follow cellular DNA content. Asynchronous cultures were grown to early logarithmic phase at 26°C and shifted to 37°C for 6 h. As shown in Figure 4A, the cell cycle of wild-type, mgs1-18 and rad18Δ cells was unaffected by incubation at 37°C and had similar profiles. However, the mgs1-18 rad18Δ double mutant accumulated cells with 2C DNA content. Cells with <1C and >2C DNA content also accumulated. This is probably due to increased cell death, cellular debris or chromosome fragmentation. DNA synthesis was also examined in synchronized cells carrying mgs1 and/or rad18 mutations. As shown in Figure 4B, cells were synchronized with α-factor and then released and incubated at 37°C for the indicated times. The majority of wild-type, mgs1Δ and rad18Δ cells achieved 2C DNA content within 60 min of release from α-factor. In contrast, mgs1-18 rad18Δ cells achieved 2C DNA content 100 min after release from α-factor, indicating that mgs1-18 rad18 is delayed in S-phase progression.
Fig. 4. FACS analysis of mgs1-18 rad18Δ cells. (A) FACS analysis of the DNA content of asynchronized cells. Cells were incubated to early log phase and then shifted to 37°C for 6 h. (B) FACS analysis of the DNA content of synchronized cells. Cells were synchronized at G1 by treatment with α-factor at 26°C and then released at 37°C. Aliquots were taken at the indicated time points. DNA content was measured by FACS.
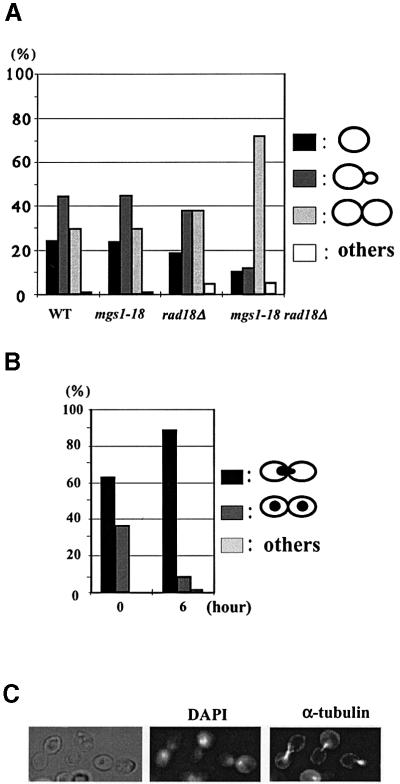
The morphology of the mgs1-18 rad18Δ cells was analysed 6 h after the shift to 37°C (Figure 5): ∼30% of wild type, mgs1-18 and rad18Δ cells were large budded (dumbbell-shaped), but >70% of mgs1-18 rad18Δ cells were large budded (Figure 5A). In addition, >90% of the large-budded mgs1-18 rad18Δ cells accumulated nuclei at the bud necks and had a short mitotic spindle; this morphology is characteristic of cells at late S/G2 (Figure 5B and C).
Fig. 5. Morphological analysis of mgs1-18 rad18Δ mutants grown at restrictive temperature. Wild-type, mgs1-18, rad18Δ and mgs1-18 rad18Δ cells were grown to early log phase at 26°C, and then grown at 37°C for 6 h. (A) Cells were harvested and the shapes were monitored under a microscope. (B) mgs1-18 rad18Δ double mutant cells (0 and 6 h) were stained with DAPI. At least 200 cells were examined. (C) The cells grown at 37°C for 6 h were harvested and fixed in formaldehyde. Nuclear and microtubular structures were visualized with DAPI and anti-tubulin antibodies, respectively.
Increased homologous recombination suppresses the growth defect of the mgs1 rad18 mutant
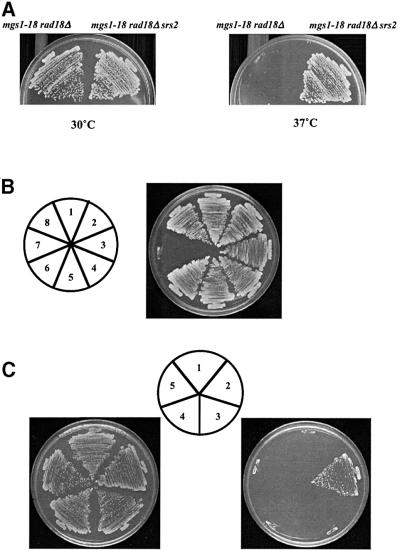
Prolonged incubation of mgs1-18 rad18Δ cells at the restrictive temperature led to spontaneous accumulation of faster growing cells or colonies. These spontaneous faster growing mutant cells were crossed with the wild-type strain and sporulated. Tetrad analysis revealed that an unlinked mutation was present in these cells that suppresses the UV sensitivity of rad18Δ cells. This single mutant displayed moderate UV sensitivity and suppressed the UV sensitivity of both mgs1-18 rad18Δ and rad18Δ mutants (data not shown). Previous studies showed that srs2 mutations suppress the UV sensitivity of both rad6 and rad18 mutants (Lawrence and Christensen, 1979; Aboussekhra et al., 1989; Schiestl et al., 1990). Thus, this suppressor mutant was crossed to an srs2Δ strain, sporulated and dissected. All 48 spores were moderately sensitive to UV (data not shown), suggesting that this suppressor mutation is tightly linked to SRS2. Sequence analysis of the SRS2 region in the suppressed strain revealed a frameshift at nucleotide 1297 of the SRS2 ORF, which truncates the Srs2 protein approximately two-thirds of the way from the translation start. srs2Δ also suppresses the growth defect of the mgs1-18 rad18Δ double mutant at the restrictive temperature (Figure 6A), as well as the poor growth phenotype of the mgs1Δ rad18Δ mutant (Figure 6B). The triple mutant grows at the same rate as wild-type yeast.
Fig. 6. Suppression of temperature sensitivity of mgs1-18 rad18. (A) srs2 mutation suppressed the temperature sensitivity for growth of the mgs1-18 rad18Δ mutant. mgs1-18 rad18Δ srs2 cells were streaked onto YPAD and incubated at 26 (left) or 37°C (right) for 3 days. (B) srs2Δ mutation suppresses the growth defect of the mgs1Δ rad18Δ mutant. Spore colonies resulting from diploid strain TO202 were streaked onto YPAD and incubated at 26°C: (1) wild-type; (2) mgs1Δ; (3) srs2Δ; (4) rad18Δ; (5) mgs1Δ srs2Δ; (6) srs2Δ rad18Δ; (7) mgs1Δ rad18Δ; (8) mgs1Δ rad18Δ srs2Δ. (C) Increased expression of Rad52p suppresses the growth defect of mgs1-18 rad18Δ. mgs1-18 rad18Δ cells were transformed with pRS426(1), pSC52 (2) and pSC51 (3). mgs1-18 rad18Δ rad51Δ cells were transformed with pRS426 (4) and pSC52 (5). All transformants were streaked onto SC –Ura and incubated at 26 (left) or 37°C (right) for 3 days.
Genetic interactions were also examined between mgs1 and other yeast recombination genes including Rad51 and Rad52, which play central roles in homologous recombination. mgs1-18 rad18Δ srs2Δ was lethal with rad52Δ and rad51Δ mutations at 37°C, respectively (data not shown). mgs1Δ rad18Δ srs2Δ was also lethal with rad52Δ and rad51Δ mutations, respectively (data not shown). These results suggest that loss of Srs2 function suppresses the synthetic lethality of the mgs1 rad18 strain by a mechanism that requires homologous recombination. This idea is consistent with the observation that overexpression of Rad52 suppresses the growth defect of mgs1-18 rad18Δ (Figure 6C). In contrast, overexpression of Rad51 does not suppress the lethality of the double mutant at restrictive temperature (Figure 6C). These results suggest that Rad52 is a limiting factor for the growth of the mgs1 rad18 double mutant. The role of homologous recombination for suppression of the growth defect was examined in an mgs1-18 rad18Δ rad51Δ triple mutant. In this strain background, overexpression of Rad52 did not suppress lethality (Figure 6C), suggesting that this suppression is dependent on homologous recombination function. These results clearly indicate that the lethal situation caused by the mgs1 rad18 double mutation is rescued by activation of homologous recombination function.
mgs1 suppresses the growth defect of DNA polymerase δ mutants
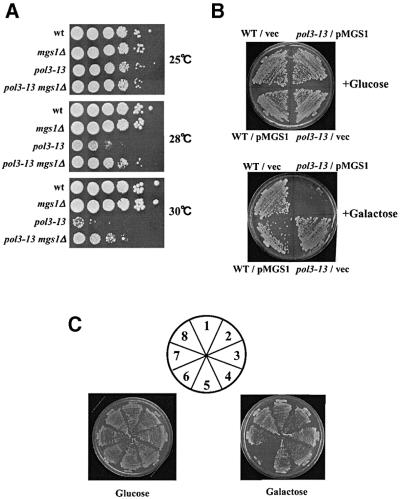
The above studies examine the genetic interactions between mgs1 and genes in the RAD6 epistasis group. The results suggest that Mgs1 is involved in normal DNA replication and an error-free PRR pathway. It is therefore possible that Mgs1 interacts with the DNA replication machinery in yeast. This idea was tested by crossing mgs1Δ with DNA replication mutants pol1-17, pol2-11 and pol3-13. These mutations are in the genes encoding DNA polymerase α, ε and δ, respectively. mgs1Δ did not alter the UV or MMS sensitivity or the growth rate of pol2-11 mutant. mgs1Δ slightly suppressed the temperature sensitivity of pol1-17 (data not shown). However, mgs1Δ clearly suppressed the growth defect of pol3-13 at 28 and 30°C (Figure 7A). In addition, overexpression of Mgs1 inhibited the growth of the pol3-13 mutant (Figure 7B). These results suggest that wild-type Mgs1 interferes with DNA replication in the pol3-13 mutant. pol3-13 is a DNA repair-deficient temperature-sensitive allele of pol3 with a cysteine to serine change in a conserved C-terminal residue (C1074S) (Giot et al., 1997). The growth of another pol3 allele, cdc2-3, which is temperature sensitive for growth but not DNA repair deficient, was also partially suppressed by mgs1Δ (data not shown). These results suggest that mgs1 mutation suppresses the DNA synthesis defect in pol3 mutants. However, mgs1Δ pol3-13 had a synergistic increase in mutation frequency (Table II), suggesting that Mgs1 prevents genomic instability resulting from a mutation in DNA polymerase δ.
Fig. 7. Mgs1 interacts genetically with DNA polymerase δ. (A) mgs1 mutation partially suppresses the growth defect caused by pol3-13 mutation. Wild-type, mgs1Δ, pol3-13 and mgs1Δ pol3-13 cells were incubated at 25°C. Cells were diluted and spotted onto YPAD plates. The plates were incubated at the indicated temperature for 3 days. (B) Overproduction of Mgs1 inhibits the growth of pol3-13. Wild-type or pol3-13 cells harbouring empty vector (vector) or plasmids with the galactose-inducible wild-type MGS1 gene (pMGS1) were plated onto SC galactose –Leu or SC glucose –Leu plates. Plates were incubated at 25°C for 3 days. (C) mgs1 suppresses the growth inhibition of pol3-13 cells caused by expression of RuvC. Vector, pSC135; pGA102, NLS-ruvC. (1) Wild-type/vector, (2) wild-type/pGA102, (3) mgs1Δ/vector, (4) mgs1Δ/pGA102, (5) pol3-13/vector, (6) pol3-13/pGA102, (7) mgs1Δ pol3-13/vector, (8) mgs1Δ pol3-13/pGA102.
The mgs1 mutation alleviates the growth defect in a DNA polymerase δ mutant expressing E.coli RuvC
Previous studies demonstrated that Holliday junction-like structures form at an increased rate during replication in yeast mutants in DNA polymerase (Zou and Rothstein, 1997). This result implies that mutations in DNA polymerase δ cause the replication fork to stall, leading to the accumulation of Holliday-like structures. This also might indicate that mgs1 mutation suppresses the formation of Holliday junctions in pol3 mutants. This idea was tested by expressing E.coli Holliday junction-specific endonucleases RuvC and RusA in the mgs1Δ pol3-13 mutant. To ensure that the bacterial proteins entered the yeast nucleus, an SV40 nuclear localization signal (NLS) was added to the N-terminus of the RuvC and RusA ORFs, respectively. NLS-fused RuvC and RusA complemented the UV sensitivity of an E.coli ruvC mutant, indicating that these fusion proteins are functional in vivo in E.coli (data not shown). NLS-RuvC and NLS-RusA were expressed in yeast cells under the control of the GAL1 promoter. Although RusA was a useful tool for detecting the formation of Holliday junctions in fission yeast (Doe et al., 2000; Boddy et al., 2001), RusA expression severely inhibited cell growth in wild-type budding yeast (data not shown). Overexpression of RuvC had no effect on the growth of wild-type or the mgs1Δ strain, and it inhibited the growth of pol3-13 at the permissive temperature (Figure 7C), consistent with the accumulation of RuvC-cleavable Holliday junction structures in the pol3 mutant (Zou and Rothstein, 1997). These results suggest that Holliday junctions accumulated at replication forks in a pol3 mutant are cleaved by RuvC and that this process causes lethal DNA double strand breaks to accumulate. However, as shown in Figure 7C, mgs1 mutation suppressed this growth inhibition. Thus, a mutation in MGS1 may prevent the formation of Holliday junctions in a DNA polymerase δ mutant.
Discussion
Role of Mgs1 in spontaneously growing cells
This study characterizes genetic interactions between yeast mgs1 and other yeast DNA repair, recombination and replication genes. In a screen for mutants that are synthetically lethal with mgs1, an allele of rad18, rad18-378, was identified. Additional analysis of double mutants between mgs1Δ and genes in the RAD6 epistasis group showed that mgs1Δ causes a synergistic growth defect with rad5Δ and synthetic lethality with rad6Δ (Figure 2A). Increased homologous recombination promoted by srs2 mutation or Rad52 overexpression rescues the growth defect in mgs1-18 rad18Δ at the restrictive temperature (Figure 6). In addition, mgs1Δ partially improves the growth of cells with temperature-sensitive alleles of POL3 (Figure 7A). Although mgs1Δ causes a severe growth defect with rad5Δ, the rad5Δ mgs1Δ double mutant has a similar level of sensitivity to UV and MMS as the rad5Δ single mutant (Figure 2B). However, rad5Δ mgs1Δ has increased HU sensitivity and spontaneous mutation and recombination frequencies (Tables I and II). HU does not directly damage DNA, but inhibits DNA synthesis by depleting nucleotide pools, which leads to replication fork arrest. These results suggest that the growth defect of rad5Δ mgs1Δ is due to a defect in processing stalled replication forks, but not a defect in DNA repair. These results suggest that Mgs1 plays a pivotal role in preventing genome instability caused by replication fork arrest, which may be an alternative to the error-free subpathway of RAD6-dependent PRR.
mgs1-18 rad18Δ cells experience a delay in S-phase progression at the restrictive temperature, and are large budded with undivided nuclei and short spindles (Figures 4 and 5), indicating that these cells arrest at late S/G2 at the restrictive temperature. Similar characteristics have been observed in leaky DNA synthesis mutants such as pol1, rfc1 and rfc2 (Lucchini et al., 1990, 1988; Zou and Stillman, 1997; Noskov et al., 1998). These results indicate that completion of DNA replication is blocked in mgs1-18 rad18 cells, even though the bulk DNA content is doubled eventually. In addition, srs2 suppresses the severe growth defect of mgs1Δ rad18Δ and mgs1-18 rad18Δ (Figure 6A and B). srs2 also suppresses the slow growth and hypermutation phenotypes of rad5 mgs1 (data not shown). srs2 was first identified as a suppressor of the UV sensitivity of rad6 and rad18 mutants (Lawrence and Christensen, 1979; Aboussekhra et al., 1989). The suppression is specific for the RAD5-dependent error-free branch of the RAD6 pathway (Ulrich, 2001). Therefore, srs2 suppresses mgs1-18 rad18 by bypassing RAD6-dependent PRR.
Previous studies show that srs2 suppression of rad6 and rad18 depends on the RAD52 homologous recombination pathway, possibly by channelling aborted repair events into recombinational repair (Schiestl et al., 1990). How ever, the rad6 srs2 rad52 triple mutant, which is defective in both PRR and homologous recombination, is not lethal, indicating that there is an alternative pathway for reinitiating replication at an arrested replication fork. This study shows that the mgs1 rad18 srs2 triple mutation, which does not cause a growth defect, is a synthetic lethal with rad52Δ and rad51Δ mutations. In addition, overexpression of RAD52 rescues the lethality of mgs1-18 rad18Δ mutants by increasing the level of homologous recombination activity (Figure 6C). This result indicates that the RAD52 homologous recombination pathway is essential for the growth of the mgs1 rad18 mutant, possibly by restoring replication from arrested replication forks in a recombination-dependent manner. These results demonstrate that Mgs1 participates in a novel error-free pathway, which may play an essential role in restoring replication from arrested replication forks when RAD6-dependent PRR and homologous recombination pathways are impaired.
Functional interaction between Mgs1 and DNA polymerase δ
Previous studies show that mgs1 increases recombination between repeated sequences and causes a synergistic growth defect in a top3 (topoisomerase III) strain (Hishida et al., 2001). It was also shown previously that overproduction of Mgs1 makes cells sensitive to HU and the topoisomerase I inhibitor camptothecin (Hishida et al., 2001). These results suggest that Mgs1 influences DNA topology during replication and plays a role in DNA replication. This study shows that mgs1Δ suppresses the temperature sensitivity for growth of mutants of DNA polymerase δ (Figure 7A). mgs1Δ also synergistically increases the mutation frequency in a pol3 mutant (Table II). These results suggest that deficiency of Mgs1 allows replication fork progression in the presence of the mutant DNA polymerase δ at the expense of replication fidelity. Mgs1 may regulate the processivity and fidelity of replication catalysed by polymerase δ. Expression of RuvC specifically inhibits the growth of pol3-13 at the permissive temperature but does not affect the growth of wild-type and mgs1Δ cells (Figure 7C). This is consistent with the suggestion that RuvC-cleavable Holliday junction structures accumulate in a DNA polymerase δ mutant (Zou and Rothstein, 1997). mgs1Δ suppresses RuvC-mediated growth inhibition of pol3-13 mutants (Figure 7C), indicating that RuvC resolvase inhibits the growth of DNA polymerase δ mutants in an Mgs1-dependent manner. Mgs1 deficiency may suppress the accumulation of Holliday junctions, and wild-type Mgs1 may promote formation of these structures in a pol3 mutant. Mgs1 possesses DNA annealing activity and affects DNA topology (Hishida et al., 2001); thus, it is possible that Mgs1 is required for regression of the replication fork, which facilitates formation of a Holliday junction at a stalled replication fork. However, it has not been proven that the E.coli Holliday junction resolvases exclusively target Holliday junction structures in yeast (Doe et al., 2000; Boddy et al., 2001), and this hypothesis needs rigorous testing.
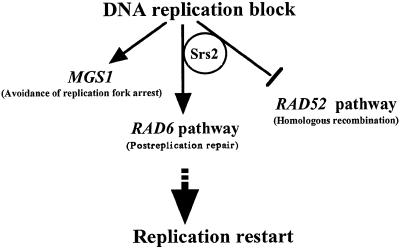
The MGS1 orthologues are highly conserved from bacteria to human (Barre et al., 2001; Hishida et al., 2001; Kawabe et al., 2001), suggesting that Mgs1 performs an important biological function. Evidence shown here indicates that the biological function of Mgs1 is related to DNA replication. If that is the case, it can be asked why a deficiency in Mgs1 does not cause lethality or sensitivity to UV, HU and MMS. When DNA replication forks proceed, they encounter endogenous DNA obstacles such as DNA topological stress, tightly bound proteins or specific DNA sequences and structures that inhibit DNA synthesis. Mgs1 may act to avoid fatal replication arrest caused by these obstacles. It is possible that Mgs1 is required to overcome replication blocks involving polymerase δ, which is redundant with the function of genes in the RAD6 epistasis group (Figure 8). In addition, in the absence of Mgs1 and Rad6, DNA replication can be restored at a blocked replication fork by homologous recombination only when recombination activity is induced or activated (i.e. Rad52 overexpression or srs2 mutation) (Figure 8). Mgs1 is essential for growth when PRR and homologous recombination pathways are impaired; this suggests that Mgs1 participates in an alternative pathway which restores replication at blocked replication forks involving DNA polymerase δ. The mouse orthologue of MGS1, Whip1, physically interacts with Werner syndrome protein (WRN), which belongs to the bacterial RecQ helicase family (Kawabe et al., 2001). Human WRN physically interacts with the DNA replication complex and stimulates DNA synthesis by polymerase δ (Lebel et al., 1999; Kamath-Loeb et al., 2000), implicating the WRN protein in restoring replication fork movement after DNA damage or topological stress. Hence, Mgs1 orthologues and RecQ helicases may act in a coordinated manner to regulate fork progression by DNA polymerase δ and may play a role in restoring replication at stalled forks.
Fig. 8. Proposed role of Mgs1 in resolving stalled replication forks. In rad6Δ cells, Mgs1 is essential for restoration of replication fork arrest. In the absence of Mgs1 and Rad6, homologous recombination proteins can relieve the block to replication and allow replication fork progression to resume. Mgs1 might facilitate fork progression catalysed by DNA polymerase δ in a RAD6 group-independent manner by rescuing the stalled replication forks. The T-bar sign indicates the processes suppressed by functions of the gene products.
In summary, Mgs1 may influence progression of DNA polymerase δ-dependent replication forks and play a role in preventing genomic instability during DNA replication. The present findings provide important clues to understanding the complex interplay between DNA replication, repair and recombination functions, especially at stalled replication forks.
Materials and methods
Strains and plasmids
Yeast strains are listed in Table III. The W303 strain and its derivatives carry a point mutation in the RAD5 gene, known as the rad5-535 allele (Fan et al., 1996). To avoid the possible effect of the mutation on the phenotypes we intended to study, we used strain W1588, in which the rad5-G535R allele of W303 has been replaced by the wild-type RAD5 gene. The strains derived from W1588, M31 (rad6), M35 (rad18) and C22 (rad5) were kindly provided by R.Woodgate (McDonald et al., 1997). Strains, pol1-17 and pol2-11, which are W303 derivatives, were gifts from A.Sugino. Strains C23-9B (cdc2-3) were kindly provided by L.Prakash. W23 was obtained after five back-crosses between C23-9B and the W1588-4B isogenic strain. All yeast manipulations were as described (Sherman and Hicks, 1986). 5-fluoro-orotic acid (5-FOA) was used to select Ura– cells as described (Rose and Hieter, 1990).
Table III. Saccharomyces cerevisiae strains used in this study.
| Strain | Genotype | Origin |
|---|---|---|
| W303-1A | MATa ade2-1 can1-100 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 rad5-G535R | Thomas and Rothstein (1989) |
| W303-1B | MATα ade2-1 can1-100 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 rad5-G535R | Thomas and Rothstein (1989) |
| TH102-1A | W303-1A, ade3::hisG | laboratory collection |
| TH102-1B | W303-1B, ade3::hisG | laboratory collection |
| TH103 | TH102-1A, mgs1::LEU2 | this study |
| TH104 | TH102-1B, rad18–378 | this study |
| TH105 | TH102-1A, mgs1::LEU2 rad18-378 | this study |
| TH106 | TH102-1A, mgs1::LEU2 rad18::TRP1 [pSL1(ARS/CEN ADE3 URA3 MGS1)] | this study |
| TD100 | TH103 × TH104 | this study |
| W1588-4A | W303-1A, RAD5 | McDonald et al. (1997) |
| W1588-4B | W303-1B, RAD5 | McDonald et al. (1997) |
| M31 | W1588-4B, rad6::LEU2 | McDonald et al. (1997) |
| M35 | W1588-4B, rad18::LEU2 | McDonald et al. (1997) |
| C22 | W1588-4B, rad5::HIS3 | McDonald et al. (1997) |
| TH201 | W1588-4A, mgs1::LEU2 | this study |
| TH202 | W1588-4A, mgs1::HIS3 | this study |
| TH203 | W1588-4A, mgs1-18 | this study |
| TH205 | W1588-4A, rad5::HIS3 mgs1::LEU2 | this study |
| TH210 | W1588-4A, rad18::TRP1 | this study |
| TH211 | W1588-4A, rad51::hisG-URA3-hisG | this study |
| TH212 | W1588-4A, rad52::TRP1 | this study |
| TH250 | W1588-4A, lys2BA::URA3lys2BG | this study |
| TH251 | W1588-4A, mgs1::LEU2, lys2BA::URA3lys2BG | this study |
| TH252 | W1588-4B, rad5::HIS3, lys2BA::URA3lys2BG | this study |
| TH253 | W1588-4A, rad5::HIS3 mgs1::LEU2, lys2BA::URA3lys2BG | this study |
| TH221 | W1588-4A, mgs1-18 rad18::TRP1 | this study |
| TH222 | W1588-4A, mgs1-18 rad18::TRP1 rad51::hisG | this study |
| TH223 | W1588-4A, mgs1-18 rad18::TRP1 srs2 | this study |
| TH230 | W1588-4A, mgs1-18 rad18::TRP1 srs2::HIS3 | this study |
| TH231 | W1588-4A, mgs1-18 rad18::LEU2 srs2::HIS3 rad52::TRP1 | this study |
| TH232 | W1588-4A, mgs1-18 rad18::TRP1 srs2::HIS3 rad51::hisG-URA3-hisG | this study |
| TH250 | W1588-4A, srs2::HIS3 | this study |
| TH251 | W1588-4A, srs2::HIS3 rad18::TRP1 | this study |
| TH252 | W1588-4A, mgs1::LEU2 srs2::HIS3 | this study |
| TH253 | W1588-4A, mgs1::LEU2 srs2::HIS3 rad18::TRP1 | this study |
| TH260 | W1588-4A, pol3-13.URA3 | this study |
| TH261 | W1588-4A, mgs1::LEU2 pol3-13.URA3 | this study |
| TD001 | W1588-4A × W1588-4A | this study |
| TD106 | TD001, MGS1/mgs1::HIS3 RAD6/rad6::LEU2 | this study |
| TD107 | TD001, MGS1/mgs1::LEU2 RAD5/rad5::HIS3 | this study |
| TD118 | TD001, MGS1/mgs1::LEU2 RAD18/rad18::TRP1 | this study |
| TO202 | TD001, MGS1/mgs1::LEU2 RAD18/rad18::TRP1 SRS2/srs2::HIS3 | this study |
To construct the mgs1::HIS3 allele, plasmid pDRH1 was constructed by replacing the AflII–XhoI fragment in the MGS1 coding region with the HIS3 gene. This plasmid was linearized by digestion with FspI and transformed into yeast to generate a strain with deletion of nucleotides from +234 to +1665 in the 1761 bp coding region of the MGS1 gene. The cassette to be used for construction of a rad18::TRP1 disrupted gene on the chromosome was made as follows. The TRP1 gene of Ydp-W (Berben et al., 1991) was amplified using the primers RAD18F (5′-ATGGACCACCAAATAACCACTGCAAGCGACTTCACGACTACTTCAATACCGGACGGCCAGTGAATTCCCGG) and RAD18R (5′-GCATCA GTTGAATCTTCGTTAGGCAAATTTCTTTCACCAGCAACTCTATCAAGCTAGCTTGGCTGCAGGT) to give a product that contains the TRP1 gene flanked by the 50 and 51 bp sequences which are identical to the N- or C-terminal parts of the RAD18 coding region, respectively. Plasmid srs2Δ::HIS3 was constructed by amplifying the coding region of SRS2 by PCR using the genomic DNA as template and replacing the XbaI fragment of the SRS2 coding region with a HIS3 cassette in pUC19. pol3-13.URA3 was constructed as follows. A 1500 bp PCR fragment containing the C-terminal part of POL3 with a flanking region was inserted into pRS306 (Sikorski and Hieter, 1989) and Cys1074 was altered to serine with PCR-mediated site-directed mutagenesis to produce pOL302. pOL302 was digested with Tth111I and integrated into the genome to produce the pol3-13 strain. Plasmid containing the rad51::hisG-URA3-hisG was kindly provided by A.Shinohara (Shinohara et al., 1992). Plasmid pSL1 was constructed by inserting the 3.9 kb HindIII–PstI fragment containing the MGS1 gene into HindIII–PstI-digested pDS1 vector (ADE3 URA3 CEN/ARS). Plasmid pH951 was constructed by inserting the 4.8 kb HindIII genomic fragment containing the MGS1 gene into the multicloning site of pUC19 vector. Plasmids pSC52 and pSC51 were constructed by subcloning the SalI fragment containing the RAD52 gene from Yrp7 (Adzuma et al., 1984) and the BamHI fragment containing the RAD51 gene from YEP-51 (Shinohara et al., 1992) into a vector, pRS426 (Stratagene), respectively. To construct the NLS-RuvC and NLS-RusA genes, ruvC and rusA coding regions amplified by PCR were fused in-frame to their 5′ end with SV40 NLS that have been added to the GAL4 activation domain sequence. NLS-ruvC and NLS-rusA were inserted into the galactose-inducible vector, p423GAL1 (Mumberg et al., 1994), to produce pGA102 or pGA153, respectively. The DNA sequences of the PCR-amplified fragments were confirmed by sequencing the appropriate regions.
Isolation of sdm mutants
Strain TH103-1A (MATa mgs1::LEU2 ade2 ade3 leu2 trp1 ura3) was transformed with the plasmid pSL1 (MGS1 ADE3 URA3 CEN/ARS). Cells from one of the transformed clones were grown at 30°C in selective medium to logarithmic phase (1 × 107 cells/ml) and treated with N-methyl-N′-nitro-N-nitrosoguanidine (35 µg/ml) at 30°C for 30 min. This treatment reduced the cell viability to ∼10%. The cells were washed twice with TE buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA), plated onto YPD plates and incubated for 5 days at 30°C. About 120 000 colonies formed on YPD plates were screened. Non-sectored colonies were patched onto YPD plates, and replica-plated onto 5-FOA plates to confirm the dependence on plasmid pSL1 for viability. Mutants that did not grow or grew poorly on 5-FOA plates were crossed with the wild-type strain (TH102-1B) or mgs1Δ strain (TH103-1B) to determine whether the synthetic lethality was caused by a single locus mutation.
Sensitivity to UV light, HU and MMS
A quantitative assay for HU sensitivity was performed at 30°C in YPAD. Overnight cultures were inoculated into fresh YPAD at a 100-fold dilution and cells were allowed to grow at 30°C. HU was added to a final concentration of 100 mM and aliquots were taken at the indicated times. Cells were washed, diluted and plated on YPAD. The cell numbers were counted under a phase contrast microscope. Cell numbers versus colony-forming units of cells were quantified. A quantitative assay for MMS and UV sensitivity was performed as described by Interthal and Heyer (2000). Plates were incubated at 30°C for 3 days in the dark.
Reversion and recombination frequency
For determining reversion frequency of trp1-1 and ade2-1 alleles, strains were streaked out on YPAD plates and grown at 30 or 25°C as indicated. Five colonies for each strain were suspended in YPAD medium, grown for 12 h and plated at the appropriate dilution to determine the total cell number on YPAD plates, and the number of Trp+ or Ade+ revertants on synthetic complete plates plus 2% glucose (SC glucose) minus Trp, or SC glucose minus Ade plates, respectively. Plates were incubated in the dark for 5 days (–Trp plates) or 7 days (–Ade plates). Trp+ or Ade+ revertants per 107 survivors were calculated. Recombination rates were monitored in haploid strains containing lys2BA::URA3::lys2BG alleles. To construct lys2BA and lys2BG alleles, the LYS2 coding sequence was digested with either BamHI or BglII, blunt-ended and ligated. For determining recombination rates during mitosis, strains were streaked out on YPAD plates and grown at 30°C. Five colonies for each strain were scraped, suspended in YPAD medium, grown for 12 h and plated at appropriate dilutions to determine the total cell numbers on YPAD and the numbers of recombinants on SC glucose –Lys or SC glucose –Lys–Ura plates.
Isolation of mgs1 mutants that are temperature sensitive for growth with the rad18Δ mutation
Mutagenesis of the MGS1 gene was performed by PCR as described previously (Muhlrad et al., 1992; Umezu et al., 1998). PCR primers were designed to amplify the 2.4 kb region of pH951. The primers (1) 5′-GAACGGTTCTCATCAGAATTCAATATTGCG and (2) 5′-GCGTATCTATGCTCCTCAAGCCTGGCTCA are complementary to the sequence located 400 bp upstream from the initiation codon and 300 bp downstream from the termination codon of the MGS1 gene, respectively. Primer (1) was designed to introduce an EcoRI site. A BamHI site is located 260 bp downstream from the termination codon of the MGS1 ORF. The PCR product and vector pRS413 (Stratagene) were digested with EcoRI and BamHI. They were mixed in a 1:1 molar ratio and co-transformed into TH106 at 26°C. Transformants were selected on SC plates lacking histidine and then replica-plated onto 5-FOA plates to eliminate pSL1 using the plasmid shuffle technique (Boeke et al., 1987). To screen mgs1 mutants that were temperature sensitive for growth, colonies formed on a 5-FOA plate were patched onto a YPAD plate and incubated at 37°C. The mgs1 mutants that could not grow at 37°C in the rad18Δ mutant were selected. The 2.4 kb EcoRI–BamHI fragment containing the mgs1 mutant allele (mgs1-18) obtained by PCR mutagenesis was cloned into a yeast integration vector pRS306 to produce pMGS18. pMGS18 was linearized by digestion with MunI and integrated into the yeast genome. Strains that have left the mgs1-18 allele behind were selected using 5-FOA medium and temperature sensitivity phenotype.
Flow cytometry
To prepare asynchronized cells for analysis by flow cytometry, wild-type, mgs1-18, rad18Δ and mgs1-18 rad18Δ cells were grown in liquid YPAD to early logarithmic phase (2–5 × 106 cells/ml) at 26°C, and shifted to a non-permissive temperature, 37°C, for 6 h. Samples were fixed in 70% ethanol at 4°C. The fixed cells, washed and resuspended in 50 mM sodium citrate pH 7.5, were treated with RNase A (0.25 mg/ml) at 50°C for 1 h, and then with proteinase K (1 mg/ml) at 50°C for 1 h. Cells were stained with propidium iodide (16 µg/ml), and incubated for 1 h at 4°C. The DNA content of cells was analysed with a Becton-Dickinson FACScan flow cytometer. To prepare synchronized cells, cells were grown in liquid YPAD to early logarithmic phase (2–5 × 106 cells/ml) at 26°C and treated with α-factor (10 µg/ml) for 2 h. After confirming that cells were arrested at G1 phase under a microscope, cells were washed to remove α-factor and grown in YPAD at 37°C. The aliquots were taken at the indicated time points and processed for flow cytometry.
Morphological analysis
Wild-type, mgs1-18, rad18Δ and mgs1-18 rad18Δ cells were grown in liquid YPAD to early logarithmic phase (2–5 × 106 cells/ml) at 26°C, and then grown at 37°C for 6 h. Cells were harvested and stained with 4′,6-diamino-2-phenylindole (DAPI). Cell morphology (unbudded cell, small-budded cell or large-budded cell) was monitored under a microscope. DAPI and immunofluorescence staining were performed as previously described (Sherman and Hicks, 1986; Redding et al., 1991).
Other methods
Overproduction of Mgs1 and NLS-RuvC in yeast cells was performed as described previously (Hishida et al., 2001).
Acknowledgments
Acknowledgements
We thank R.Woodgate, J.P.McDonald, L.Prakash and A.Sugino for strains, and A.Shinohara for plasmids. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, and by a grant from the Human Frontier Science Program to H.S.
References
- Aboussekhra A., Chanet,R., Zgaga,Z., Cassier-Chauvat,C., Heude,M. and Fabre,F. (1989) RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res., 17, 7211–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzuma K., Ogawa,T. and Ogawa,H. (1984) Primary structure of the RAD52 gene in Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V., Lamb,J., Sung,P., Prakash,S. and Prakash,L. (1994) Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev., 8, 811–820. [DOI] [PubMed] [Google Scholar]
- Bailly V., Lauder,S., Prakash,S. and Prakash,L. (1997) Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding and ATP hydrolytic activities. J. Biol. Chem., 272, 23360–23365. [DOI] [PubMed] [Google Scholar]
- Barre F.X., Soballe,B., Michel,B., Aroyo,M., Robertson,M. and Sherratt,D. (2001) Circles: the replication–recombination–chromo some segregation connection. Proc. Natl Acad. Sci. USA, 98, 8189–8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berben G., Dumont,J., Gilliquet,V., Bolle,P.A. and Hilger,F. (1991) The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast, 7, 475–477. [DOI] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard,P.H., McDonald,W.H., Shanahan,P., Yates,J.R.,3rd and Russell,P. (2001) Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell, 107, 537–548. [DOI] [PubMed] [Google Scholar]
- Boeke J.D., Trueheart,J., Natsoulis,G. and Fink,G.R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol., 154, 164–175. [DOI] [PubMed] [Google Scholar]
- Broomfield S., Chow,B.L. and Xiao,W. (1998) MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl Acad. Sci. USA, 95, 5678–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusky J., Zhu,Y. and Xiao,W. (2000) UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet., 37, 168–174. [DOI] [PubMed] [Google Scholar]
- Cox M.M., Goodman,M.F., Kreuzer,K.N., Sherratt,D.J., Sandler,S.J. and Marians,K.J. (2000) The importance of repairing stalled replication forks. Nature, 404, 37–41. [DOI] [PubMed] [Google Scholar]
- Doe C.L., Dixon,J., Osman,F. and Whitby,M.C. (2000) Partial suppression of the fission yeast rqh1(–) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J., 19, 2751–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H.Y., Cheng,K.K. and Klein,H.L. (1996) Mutations in the RNA polymerase II transcription machinery suppress the hyper recombination mutant hpr1Δ of Saccharomyces cerevisiae. Genetics, 142, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Giot L., Chanet,R., Simon,M., Facca,C. and Faye,G. (1997) Involvement of the yeast DNA polymerase δ in DNA repair in vivo. Genetics, 146, 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M.D., Shen,J.C., Kamath-Loeb,A.S., Blank,A., Sopher,B.L., Martin,G.M., Oshima,J. and Loeb,L.A. (1997) The Werner syndrome protein is a DNA helicase. Nature Genet., 17, 100–103. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (1999) DNA recombination: the replication connection. Trends Biochem. Sci., 24, 271–275. [DOI] [PubMed] [Google Scholar]
- Hishida T., Iwasaki,H., Ohno,T., Morishita,T. and Shinagawa,H. (2001) A yeast gene, MGS1, encoding a DNA-dependent AAA+ ATPase is required to maintain genome stability. Proc. Natl Acad. Sci. USA, 98, 8283–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H. and Heyer,W.D. (2000) MUS81 encodes a novel helix–hairpin–helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet., 263, 812–827. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1994) Yeast DNA repair protein RAD5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J. Biol. Chem., 269, 28259–28262. [PubMed] [Google Scholar]
- Kamath-Loeb A.S., Johansson,E., Burgers,P.M. and Loeb,L.A. (2000) Functional interaction between the Werner syndrome protein and DNA polymerase δ. Proc. Natl Acad. Sci. USA, 97, 4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y. et al. (2001) A novel protein interacts with the Werner’s syndrome gene product physically and functionally. J. Biol. Chem., 276, 20364–20369. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S.C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci., 25, 156–165. [DOI] [PubMed] [Google Scholar]
- Kunz B.A., Straffon,A.F. and Vonarx,E.J. (2000) DNA damage-induced mutation: tolerance via translesion synthesis. Mutat. Res., 451, 169–185. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. (2001) DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl Acad. Sci. USA, 98, 8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.W. and Christensen,R.B. (1979) Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol., 139, 866–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D.E. and Coulson,C.A. (1949) The distribution of the numbers of mutants in bacterial populations. J. Genet., 49, 264–285. [DOI] [PubMed] [Google Scholar]
- Lebel M., Spillare,E.A., Harris,C.C. and Leder,P. (1999) The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem., 274, 37795–37799. [DOI] [PubMed] [Google Scholar]
- Liefshitz B., Steinlauf,R., Friedl,A., Eckardt-Schupp,F. and Kupiec,M. (1998) Genetic interactions between mutants of the ‘error-prone’ repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res., 407, 135–145. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Mazza,C., Scacheri,E. and Plevani,P. (1988) Genetic mapping of the Saccharomyces cerevisiae DNA polymerase I gene and characterization of a pol1 temperature-sensitive mutant altered in DNA primase–polymerase complex stability. Mol. Gen. Genet., 212, 459–465. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Falconi,M.M., Pizzagalli,A., Aguilera,A., Klein,H.L. and Plevani,P. (1990) Nucleotide sequence and characterization of temperature-sensitive pol1 mutants of Saccharomyces cerevisiae. Gene, 90, 99–104. [DOI] [PubMed] [Google Scholar]
- McDonald J.P., Levine,A.S. and Woodgate,R. (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Hunter,R. and Parker,R. (1992) A rapid method for localized mutagenesis of yeast genes. Yeast, 8, 79–82. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov V.N., Araki,H. and Sugino,A. (1998) The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces. Mol. Cell. Biol., 18, 4914–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1997) SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein–DNA interactions? Cell, 88, 737–740. [DOI] [PubMed] [Google Scholar]
- Redding K., Holcomb,C. and Fuller,R.S. (1991) Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J. Cell Biol., 113, 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L. (1981) Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet., 184, 471–478. [DOI] [PubMed] [Google Scholar]
- Prakash S., Sung,P. and Prakash,L. (1993) DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet., 27, 33–70. [DOI] [PubMed] [Google Scholar]
- Rong L. and Klein,H.L. (1993) Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem., 268, 1252–1259. [PubMed] [Google Scholar]
- Rose M.D.F.W. and Hieter,P. (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schiestl R.H., Prakash,S. and Prakash,L. (1990) The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics, 124, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J.C. and Loeb,L.A. (2000) The Werner syndrome gene: the molecular basis of RecQ helicase-deficiency diseases. Trends Genet., 16, 213–220. [DOI] [PubMed] [Google Scholar]
- Sherman F.G.F. and Hicks,J. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S.cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein,R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell, 56, 619–630. [DOI] [PubMed] [Google Scholar]
- Ulrich H.D. (2001) The srs2 suppressor of UV sensitivity acts specifically on the RAD5- and MMS2-dependent branch of the RAD6 pathway. Nucleic Acids Res., 29, 3487–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H.D. and Jentsch,S. (2000) Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J., 19, 3388–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K., Sugawara,N., Chen,C., Haber,J.E. and Kolodner,R.D. (1998) Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics, 148, 989–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R. (1999) A plethora of lesion-replicating DNA polymerases. Genes Dev., 13, 2191–2195. [DOI] [PubMed] [Google Scholar]
- Xiao W., Chow,B.L., Broomfield,S. and Hanna,M. (2000) The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics, 155, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.E. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- Zou H. and Rothstein,R. (1997) Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell, 90, 87–96. [DOI] [PubMed] [Google Scholar]
- Zou L.J.M. and Stillman,B. (1997) CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol. Cell. Biol., 17, 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]