Introduction
All organisms adapt to changes in their environment by adjustments in gene expression, and in all organisms, from Escherichia coli to man, the most important control point is at transcription initiation. All, that is, except those belonging to one very small family of early-branching eukaryotes, which seems to have completely lost the ability to regulate transcription by RNA polymerase II.
The organisms concerned are unicellular, spindle- like flagellates that flourish in the digestive systems of arthropods, in the blood, macrophages and brains of vertebrates from humans to lizards, and even in the sap of coconut palms and lemon trees. Many of them are able to multiply both in a vertebrate (or plant) and an invertebrate, which serves to transmit the parasites from one vertebrate (or plant) to the next. Adaptation to the two distinct environments, with different temperatures, nutrients and defences, requires major changes in gene expression. Yet this seems to be achieved in the total absence of any developmental regulation of RNA polymerase II; perhaps even without any specific polymerase II transcription initiation. This extraordinary state of affairs might be written off as an irrelevant evolutionary quirk (and, indeed, might have even gone unnoticed) if it were not for the fact that some of the organisms concerned, the trypanosomes and the leishmanias, kill millions of people every year (http://www.who.ch).
The leishmanias cause a spectrum of diseases ranging from self-resolving skin ulcers to lethal infection of the internal organs. One-and-a-half to two million people are newly infected every year in the tropics and southern Europe. Leishmania has an extracellular form in the gut of the vector, the sand-fly, but multiply as spherical aflagellate forms within the lysosomes of mammalian macrophages. Leishmania must be phagocytosed without activating the host macrophage and must combat oxidative, acidic and proteolytic stresses. The South American trypanosome Trypanosoma cruzi is transmitted in the faeces of reduviid bugs. Within mammals, it multiplies freely in the cytoplasm of a variety of cell types; an active uptake mechanism involving the recruitment of lysosomes to the site of attachment is critical. Approximately 17 million people are currently infected. The sleeping sickness trypanosome, Trypanosoma brucei, is never intracellular; one form lives in the gut of tsetse flies (the ‘insect form’) and the other, ‘bloodstream form’ is found in the blood and tissue fluids of the mammal, where it evades the immune response by antigenic variation. Sleeping sickness is fatal unless treated and most of the 300 000 people who are currently infected will die of the disease. Survival of these parasites within their arthropod vectors requires the ability to combat proteolysis and anti-microbial peptides via expression of various surface glycoproteins or glycolipids.
Gene organization and RNA processing
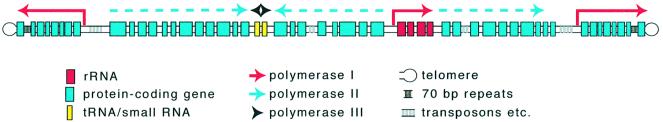
Even a cursory glance at the organization of Trypanosoma and Leishmania chromosomes is enough to reveal that something extremely unusual is going on (Figure 1; see parasite genome pages at http://www.tigr.org and http://www.ebi.ac.uk). The Leishmania major chromosome I is 285 kb long. About one third of the way along the chromosome, there is a 1.6 kb region lacking open reading frames (ORFs). To the left of this are 29 predicted protein-coding ORFs, all oriented towards the left telomere, and to the right, 50 ORFs oriented towards the right telomere (Myler et al., 1999; McDonagh et al., 2000). All are transcribed by RNA polymerase II (based on their sensitivity to α amanitin). Similar arrangements are apparent in other chromosomes from T.brucei and L.major, except that in longer chromosomes there are more changes in the direction of transcription, and polymerase I and III transcription units are present.
Fig. 1. Schematic diagram of a generic T.brucei chromosome. The diagram is meant only to show typical features. It does not illustrate a particular chromosome and is not to scale. Information is taken from Myler et al. (1999), personal communications, other references in the text and the websites listed below. Trypanosomes and leishmanias are diploid. Although homologous chromosomes can differ considerably in size, this is mostly due to variations in the numbers of tandemly repeated genes or non-coding repeated sequences, particularly sub-temomeric repeats. Haploid genome sizes range from 30 to 40 Mb with ∼10 000 genes, and most chromosomes are 0.3–10 Mb long. Trypanosoma brucei also has many smaller ‘intermediate’ and ‘minichromosomes’ that have repetitive regions, rRNA genes and VSG genes; at the end of some of the large T.brucei chromosomes there are VSG expression sites transcribed by RNA polymerase I. Current relevant websites are http://www.cbs.dtu.dk/services/GenomeAtlas/Eukaryotes/Leishmania/major/Friedlin/Lmajor_Friedlin_1.structureatlas.lin.html; http://www.sanger.ac.uk/Projects/L_major/; http://www.tigr.org/tdb/mdb/tbdb/ index.shtml; http://www.tigr.org/tdb/; http://parsun1.path.cam.ac.uk/; http://www.sanger.ac.uk/Projects/T_brucei/; http://www.ebi.ac.uk/parasites/ paratable.html#cruzi.
The arrangement of genes in trypanosomes and Leishmania (and in other related parasites from the same order, kinetoplastida) is reminiscent of that in bacterial operons, especially as protein coding regions are almost never interrupted by introns; the single exception so far is the gene encoding poly(A) polymerase (Mair et al., 2000a). However, upon closer scrutiny, the similarity vanishes as genes clustered in a common orientation do not share a common regulatory pattern. The results of many experiments suggest that genes that are next to each other and share a common orientation are co-transcribed. Individual mRNAs are cleaved from the precursor by a trans splicing reaction, which adds a capped RNA of ∼40 nucleotides (nt) called the ‘spliced leader’ (SL) to the 5′-end, and by polyadenylation at the 3′-end (Ullu et al., 1993; Matthews et al., 1994). 5′ trans splicing enables efficient production of translatable mRNAs by RNA polymerase I or even by introduced bacteriophage polymerases (see Irmer and Clayton, 2001).
Experiments with a variety of genes have shown that trans splicing and polyadenylation are inextricably coupled. Trans splicing signals are often U-rich polypyrimidine tracts, which precede AG acceptor sites. There are no dedicated polyadenylation signals. Instead, polyadenylation occurs a fixed distance (∼100–400 nt, depending on the species) upstream of the splice signal (LeBowitz et al., 1993; Matthews et al., 1994). Experiments in permeabilized cells have shown that splicing is both spacially and temporally coupled to trans splicing (Ullu et al., 1993). Thus one has to imagine stretches of chromosome of up to 150 kb being constitutively transcribed, with concurrent trans splicing and polyadenylation of the nascent chains.
Promoters and polymerases
Kinetoplastid genomes contain clear homologues of RNA polymerase core subunits. In most eukaryotes, the C-terminus of the largest polymerase II subunit consists of essential serine-rich heptapeptide repeats. The C-terminus of the corresponding kinetoplastid protein lacks the repeats but is nevertheless serine-rich, and the subunit is phosphorylated just as in other eukaryotes (see Gilinger and Bellofatto, 2001 for references). The gene encoding the core transcription factor TBP is in the Leishmania database, so other basal factors are probably also present.
Much effort has been expended in the search for transcriptional promoters in kinetoplastids. Polymerase I promoters were first identified in T.brucei, not only in the RRNA loci but also upstream of the genes encoding the major surface proteins of the bloodstream form (variant surface glycoprotein, VSG) and the insect form (the EP and GPEET procyclins). The promoters were characterized using nuclear run-on assays, primer extension, and transient transfection of plasmids containing the promoter followed by a trans splice acceptor signal and a reporter gene. The promoter structure resembles that of other eukaryotic rRNA promoters and the RRNA, VSG and EP promoters compete for common factors in an in vitro system (Laufer et al., 1999; Laufer and Günzl, 2001).
In kinetoplastids, RNA polymerase III transcribes most U RNAs in addition to tRNAs. Promoters have been identified and mapped in detail using an in vitro system and by transient transfection of plasmids designed for production of tagged RNAs. Transcription of U RNAs depends on the A and B boxes of an upstream tRNA (or tRNA-like) gene, while accurate initiation also requires a sequence at the beginning of the U RNA (Nakaar et al., 1997). The small nucleolar RNAs, in contrast, are transcribed as part of polycistronic polymerase II transcription units (Dunbar et al., 2000; Xu et al., 2001).
RNA polymerase II promoters for protein coding genes have proved elusive so far. Although there have been several reports of putative promoters, mapped by primer extension or transient transfection, none has been confirmed (McAndrew et al., 1998; Downey and Donelson, 1999). Experiments with transfected plasmids have demonstrated beyond any doubt that kinetoplastid RNA polymerase II can initiate in the absence of a specific kinetoplastid promoter. For example, in Leishmania and Leptomonas, it was possible to obtain expression of a marker from episomes containing nothing but the plasmid backbone and the marker gene preceded by a splice acceptor, which could be a synthetic polypyrimidine tract (Bellofatto et al., 1991; de Lafaille et al., 1992). Similarly, in T.brucei, a promoterless chloramphenicol acetyltransferase (CAT) reporter gene with pre-mRNA processing signals that was inserted in a gap between the U3 snRNA and 7SL RNA genes (which are both transcribed by polymerase III) was efficiently expressed, although the experiment was designed so that there was no selective pressure for CAT expression (Marchetti et al., 1998).
Despite these indications of totally random initiation, transcription from intact chromosomes (measured by run-on assays) is predominantly unidirectional. Directionality could either be conferred by promoters, or by transcriptional termination or attenuation, and there is evidence for both. The results of a study of episome transcription in Leishmania were most consistent with the idea that the strand preference for transcription was caused by termination (Wong et al., 1994). In contrast, results from the laboratory of P.Myler and K.Stuart support the existence of promoters (P.Myler, personal communication). Graded doses of UV irradiation, followed by nuclear run-on assays, have been used to analyse transcription on Leishmania chromosome I. The results indicate that the region between the oppositely-oriented transcription units (the coding strand inflexion point) is a site of bi-directional transcription initiation. Transcription of the coding strand was much lower than that of the non-coding strand, although it was probably slightly higher than the experimental background. Stably maintained episomes containing the relevant region expressed up to 10-fold more protein from reporter genes than analogous episomes lacking the region, although results were very variable. Similar nuclear run-on experiments with chromosome III also indicated that both promoters and terminators are present in regions where transcription changes direction and where a tRNA gene is found at the end of a polymerase II transcription unit. However, the different coding strand inflexion points do not show any obvious sequence similarity. Overall, all results so far are consistent with specific transcription from a few promoters per chromosome. In Leishmania, there may also be a background of low, non-specific and randomly initiated polymerase II transcription of the non-coding strand. In T.brucei, bi-directional transcription does occur (Liniger et al., 2001), but it is unlikely to be widespread as this organism has a constitutively active RNA interference system that destroys double-stranded RNAs (Djikeng et al., 2001).
In contrast to the protein-coding genes, the genes encoding the SL precursor RNA, SLRNA, have clearly identifiable promoters with a short concensus initiator element (Luo et al., 1999). Most evidence indicates that this transcription is by polymerase II: the SL has a 5′ cap structure (Mair et al., 2000b), and the results of inhibitor studies and immunodepletion experiments with an active in vitro extract both support polymerase II involvement (Gilinger and Bellofatto, 2001). Several SL-promoter-binding proteins have been identified (Wen et al., 2000; Matkin et al., 2001), but it is not known whether these proteins are also involved in transcription of mRNAs. Indeed, the SLRNA transcription complex normally terminates within poly(T) stretches (Sturm et al., 1999), which is reminiscent of polymerase III transcription. The same cannot possibly be true of the complex that transcribes protein-coding genes, as they are preceded by polypyrimidine trans splicing signals!
The nature of the polymerase II complex is still one of the major mysteries of kinetoplastid molecular biology. The absence of known specific polymerase II promoters for protein-coding genes has, in the past, been an insuperable impediment to the development of in vitro transcription systems, so promoter identification would be a major advance. The completion of the genome projects may tell us which basal transcription factors are present, so may provide some hints as to why polymerase II transcription in kinetoplastids is so unusual.
Models of developmental regulation: trypanosome surface proteins
The most extensively studied kinetoplastid genes are those encoding the major surface proteins of T.brucei. The VSGs form a dense surface layer that protects the bloodstream form from the humoral immune response and complement activation. Up to 1000 different VSG genes, pseudogenes and fragments are located within chromosomes, and between 20 and 40 VSG ‘expression sites’ are found at telomeres. In the expression sites (Figure 1), a single telomere-proximal VSG gene is preceded by up to 10 other genes, an RNA polymerase I promoter and a repetitive region that may transcriptionally insulate the expression site from the rest of the chromosome (Pays et al., 2001; Vanhamme et al., 2001). In bloodstream trypanosomes, a single expression site is fully transcriptionally active, so that each organism expresses only one VSG (Chaves et al., 1999). The remaining expression sites show very weak transcription that is restricted to the promoter-proximal region. In insect-form trypanosomes, only the weak transcription type is seen. Antigenic variation occurs by two different mechanisms. Gene rearrangements move VSG genes between expression sites, from chromosome internal to telomeric locations, and transcriptional switches result in activation of one expression site while another is silenced. The control of VSG transcription is still not understood, but all evidence so far points to epigenetic regulation mechanisms (Borst and Ulbert, 2001). Regulation via chromatin structure is one possibility (Horn, 2001). Remarkable new evidence indicates that the nucleus of each bloodstream trypanosome contains a single RNA polymerase I ‘VSG transcription factory’ that is separate from the nucleolus (Navarro and Gull, 2001). The nature of this site, and its control, are exciting topics for the future.
The major surface proteins of insect-form T.brucei consist mainly of EP or GPEET repeats and are undetectable in bloodstream forms. Each diploid parasite has eight to 10 EP and GPEET genes present in pairs or triplets, preceded by an RNA polymerase I promoter and followed downstream by a few unrelated co-transcribed genes. The regulation of the EP/GPEET proteins is important, because expression of an exposed invariant surface antigen in the bloodstream form would lead to immune destruction. Correspondingly, the genes are under multiple layers of control (Hotz et al., 1998). Their transcription is down-regulated ∼10-fold in bloodstream forms, most likely by a chromatin-mediated mechanism (Hotz et al., 1998). Any EP RNAs that are still produced in the bloodstream forms are rapidly degraded, giving another 10-fold regulation (Furger et al., 1997; Hotz et al., 1997), and the few surviving EP RNAs are extremely poorly translated. Together, these measures give 1000-fold regulation; it is not clear whether there are additional blocks to protein processing. The GPEET protein is produced only transiently during and after differentiation to the insect form, and the subsequent down-regulation is post-transcriptional (Vassella et al., 2000).
Although all kinetoplastids are capable of producing mRNAs using RNA polymerase I (as shown by reporter gene assays), the sleeping sickness trypanosomes are the only ones known to use this capacity. There are two known properties of polymerase I that may account for its use in producing surface proteins. One is the susceptibility to regulation, which is needed for antigenic variation. The other is the transcription rate; a chromosomally integrated reporter gene transcribed by polymerase I yields 10 times as much product as the identical gene transcribed by polymerase II (Biebinger et al., 1996). VSG constitutes 10% of the total surface protein of a bloodstream trypanosome, and VSG mRNA is correspondingly abundant. Yet all this RNA has to be produced from a single gene. Tubulin mRNAs, which are produced by RNA polymerase II, only achieve high abundance because there are ∼30 tubulin genes, and other abundant proteins in kinetoplastids are also encoded by tandemly repeated genes. The EP/GPEET genes may be transcribed by polymerase I because of the need to make large amounts of RNA and protein in a regulatable fashion. The insect stages of other kinetoplastids are coated either by glycolipids or by the products of very large families of surface glycoprotein genes.
Models of developmental regulation: polymerase II loci and mRNA degradation
At the time of writing, over 30 developmentally regulated polymerase II transcribed genes had been studied in kinetoplastids. No developmental regulation of polymerase II transcription, polyadenylation or trans splicing has been found. Instead, sequences in the 3′-untranslated regions (UTRs) determine the mRNA abundance by modulating RNA degradation (Zilberstein and Shapira, 1994; Charest et al., 1996; Coughlin et al., 2000; Quijada et al., 2000; Acosta-Serrano et al., 2001; Brittingham et al., 2001). The same regulatory sequences often also modulate translation efficiency.
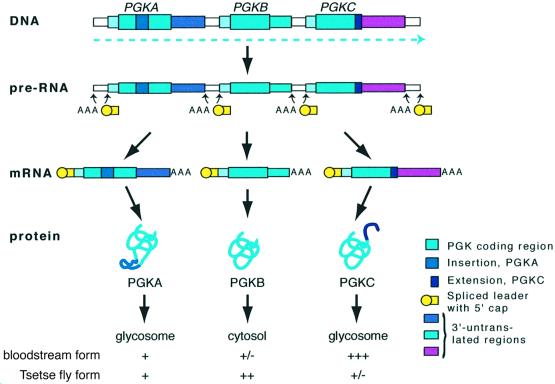
The phosphoglycerate kinase (PGK) gene cluster from chromosome I of T.brucei is a useful example. There are three genes, PGKA, PGKB and PGKC, with other unrelated predicted genes, with the same orientation located upstream and downstream (Figure 2). The PGKB enzyme is located in the cytosol, and is expressed exclusively in the tsetse fly form of T.brucei. PGKC, in contrast, is expressed almost exclusively in the bloodstream form, and is targeted to a specialized peroxisome-like glycolytic organelle, the glycosome. The developmental regulation of PGK expression is crucial as PGK activity in the cytosol of bloodstream forms inhibits cell growth (Blattner et al., 1998). PGKA is present at a low level throughout the life cycle. Early experiments involving both nuclear run-on assays and RT–PCR established that all three genes were co-transcribed by RNA polymerase II and that there was no developmental regulation of the transcription rate. The low expression of PGKA can be attributed to a poor trans splicing signal (Kapotas and Bellofatto, 1993), and the expression patterns of PGKB and PGKC are determined by their 3′-UTRs (Blattner and Clayton, 1995).
Fig. 2. The PGK locus of T.brucei, showing patterns of transcription, processing, translation and protein targeting in bloodstream forms and procyclic (tsetse fly) forms.
At present, one exception to the general picture is known. In the insect kinetoplastid Crithidia fasciculata, several mRNAs linked to nuclear and mitochondrial DNA replication begin to accumulate prior to S phase then decline rapidly, resulting in stepwise protein accumulation. The 5′-UTRs of these mRNAs contain an octamer consensus sequence, which is needed for cell-cycle regulation (Mahmood et al., 1999), and proteins that bind this sequence have been identified (Mahmood et al., 2001).
The general RNA degradation apparatus in trypanosomatids
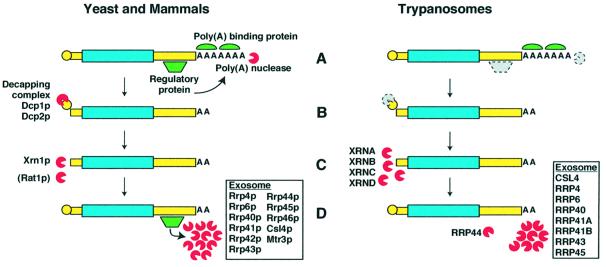
In yeast and mammalian cells, the major pathway of mRNA degradation is initiated by deadenylation (Figure 3; Caponigro and Parker, 1996; Mitchell and Tollervey, 2000, 2001; Shyu and Wilkinson, 2000). Trimming of the poly(A) tails to a short oligo(A) triggers mRNA decapping. The mRNAs are then degraded in the 5′→3′ direction by cytoplasmic exonucleases (such as the yeast Xrn1p), and in the 3′→5′ direction by a large exonuclease complex, the exosome (Butler, 2002). Regulatory proteins bind to specific motifs in the 3′-UTR and may, for example, stimulate deadenylation or exosome action. Regulated degradation can also involve endonuclease cleavage in the 3′-UTR (Caponigro and Parker, 1996).
Fig. 3. mRNA degradation in mammals and trypanosomes. Some of the known Saccharomyces cerevisiae participants are shown to the left; corresponding T.brucei proteins are on the right. Red pacmen are exonucleases. Where clear homologues are not yet established, the protein is illustrated in grey with a dotted border. Processes are: (A) deadenylation reducing the poly(A) tail to oligo(A); (B) decapping; (C) 5′→3′ degradation; (D) 3′→5′ degradation. For references, see the text.
Kinetoplastids clearly possess this general machinery. Poly(A) binding proteins have been identified from several species (Hotchkiss et al., 1999; Bates et al., 2000). We have found four T.brucei genes encoding proteins similar to the yeast Xrn1p and its nuclear homo logue Rat1p (H.Irmer, H.Salm and S.Freese, unpublished observations). Decapping activity has been demonstated in Leptomonas seymouri (V.Bellofatto, personal communication), and T.brucei has nuclear and cytoplasmic exosomes that are rather smaller than the yeast exosome (Estévez et al., 2001; Figure 3).
Regulated mRNA degradation: mechanisms and signals
Experiments concerning the mechanism of mRNA degradation in kinetoplastids have mainly been restricted to observations that protein synthesis inhibitors increase or decrease the abundance of particular mRNAs (Teixeira et al., 1995; Graham and Barry, 1996; Di Noia et al., 2000; Brittingham et al., 2001). The specificity of such effects is not clear. In yeast, cycloheximide inhibits mRNA decapping (Beelman and Parker, 1994; Jacobs-Anderson and Parker, 1998).
In most cases, it has not been possible to find short linear signals determining kinetoplastid mRNA half-lives, perhaps because the secondary structures of the mRNAs concerned are important. However, there are exceptions. The first elements to be identified were in the 3′-UTR of the VSG mRNAs of T.brucei (Berberof et al., 1995): they are an 8mer and 14mer, which stimulate bloodstream-form expression and suppress insect-form expression, and regulate both mRNA levels and translation. The rapid degradation of VSG mRNA may be important after VSG transcription is shut off, as bloodstream trypanosomes differentiate into insect forms.
The 3′-UTRs of the T.brucei insect-form surface protein genes, EP and GPEET, provide the best-characterized example of regulatory motifs. These 3′-UTRs are dissimilar, apart from a 16mer stem–loop and U-rich 26mer. The 16mer enhances translation in insect forms by an unknown mechanism, but plays no role in developmental regulation (Furger et al., 1997). Deletions of, and point mutations in, the 26mer abolished or reduced developmental regulation (Hotz et al., 1997; Schürch et al., 1997). The 26mer is normally in a single-stranded conformation (Drozdz and Clayton, 1999). Examination of the PGKB 3′-UTR, which also mediates insect form-specific expression, revealed a similar U-rich sequence, which is predicted to be single-stranded. Specific deletion of this U-rich domain again abolished stage-specific regulation at both the RNA and protein levels (L.Quijada, C.Guerra-Giraldez and C.Clayton, manuscript in preparation). These regulatory sequences bear a strong resemblance to the destabilizing AU-rich elements (AREs) found in the 3′-UTRs of many mammalian mRNAs involved in cell growth and differentiation (Mitchell and Tollervey, 2000).
To study the mechanism of mRNA degradation, we have used CAT transgenes bearing different 3′-UTRs: the 3′-UTR of an unregulated actin (ACT) gene, those of the insect form-specific EP1 and PGKB genes, and mutant EP1 and PGKB 3′-UTRs, that lack ARE sequences, so give constitutive expression. The poly(A) tail length was not affected by the 3′-UTR (Irmer and Clayton, 2001; L.Quijada, C.Guerra-Giraldez and C.Clayton, manuscript in preparation). Degradation of CAT–ACT mRNA, and the mutated, constitutively expressed versions of the two regulated mRNAs, involved deadenylation, followed by degradation in both 5′→3′ and 3′→5′ directions (Irmer and Clayton, 2001). In contrast, in bloodstream forms, the EP1 26mer instability element caused rapid destruction of the EP1 3′-UTR. This may be a consequence of extremely rapid deadenylation or of endonuclease cleavage. The PGKB regulatory sequence stimulated deadenylation (L.Quijada, C.Guerra-Giraldez and C.Clayton, manuscript in preparation). Efforts to find proteins that regulate degradation of the EP mRNAs have failed so far.
The T.cruzi genes encoding mucin-like proteins are expressed predominantly in the vertebrate forms of the parasite. Here again, regulation is determined by an ARE (Di Noia et al., 2000). A protein that binds to the ARE has been identified. Overexpression of this protein in T.cruzi caused destabilization of mucin mRNA (D’Orso and Frasch, 2001). The sequence contains an RNA-binding domain and a glutamine-rich region. The genomes of T.cruzi, T.brucei and L.major contain several other genes encoding similar proteins. We do not know how many of these are involved in mRNA regulation, nor do we know whether the resemblance between the AU-rich elements in kinetoplastids and those in mammalian mRNAs is indicative of conservation of the degradation mechanism.
The finding of AREs in several regulated mRNAs suggests that particular expression patterns may correlate with specific 3′-UTR sequence motifs. Interestingly, a conserved regulatory sequence of ∼300 nt has recently been found in the 3′-UTRs of several Leishmania mRNAs that all show preferential expression in the mammalian intracellular forms (B.Papadopoulou, personal communication). In the future, DNA array results should reveal classes of RNAs that share particular regulation patterns, and possibly also regulatory motifs. A major task for the future will be the identification of the proteins that are responsible for regulation of different classes of mRNAs, and the elucidation of the mechanisms involved. It is clearly conceivable that some regulation is also mediated by small RNAs that initiate RNA interference.
Control of translation, protein processing and protein degradation
Translational control is probably extremely important in kinetoplastids, but has until now been rather neglected. Results from the hybridization of trypanosome and Leishmania gene arrays so far indicate that ∼1% of (detectable) mRNAs differ at least 2-fold in abundance between life-cycle stages (S.Diehl, F.Diehl, N.El-Sayed, C.Clayton and J.Hoheisel, manuscript in preparation; S.Beverley, personal communication). Corresponding analyses of the 1000 most abundant soluble proteins indicate that rather more, ∼3%, are developmentally regulated (K.Matthews and F.van Duersen, and B.Papadopoulou, personal communications). For many genes, the reported regulation of mRNA is not sufficient to explain the differences seen at the level of the protein product (e.g. Priest and Hajduk, 1994; Saas et al., 2000). Regulation of translation, protein sorting and degradation could all contribute.
Environmental signals and post-transcriptional regulation
We know nothing as yet about how changes in the environment of kinetoplastids cause changes in gene expression, although several triggers such as temperature shifts and pH changes have been defined (Zilberstein and Shapira, 1994; Matthews, 1999). Cyclic AMP is involved in trypanosome differentiation, but we do not understand how it works or how synthesis is controlled (Matthews, 1999). Protein kinases and other potential components of signal transduction pathways have been identified (Parsons and Ruben, 2000; Vassella et al., 2001), but very little is known about their functions. The connections between signal transduction and mRNA degradation are a mystery and are a major topic for future research. In fact, only a little more is known about such connections in mammalian cells, so it may be that results from the relatively uncomplicated and easily manipulated kinetoplastids will provide clues about regulation in more complex systems.
Acknowledgments
Acknowledgements
This review cites only a small (usually recent) sample of the literature. I apologize to the many colleagues whose work was not cited due to space constraints. I am indebted to Debra Peattie for her hospitality while drafting this review and to Pedro Huertas for the use of his computer (although he didn’t know about it at the time). I thank Vivian Bellofatto, Steve Beverley, Keith Matthews, Peter Myler, Marc Ouellette and Barbara Papadopoulou for communicating unpublished results, often in considerable detail. Kinetoplastid genome sequences and information about chromosome structure came from the sequencing consortia, particularly from Al Ivens, Neil Hall and Matt Berriman at the Sanger Centre (Hinxton, Cambridge, UK), F.van Duersen and Najib El-Sayed, TIGR (Rockville, MD). I thank Antonio Estévez, Christina Guerra, Daniel Nilsson and Piet Borst for reading and correcting the manuscript. Work in my laboratory cited here was supported by EMBO, the Deutsche Akademische Austauschdienst and the Deutsche Forschungs gemeinschaft.
References
- Acosta-Serrano A., Almeida,I.C., Freitas-Junior,L.H., Yoshida,N. and Schenkman,S. (2001) The mucin-like glycoprotein super-family of Trypanosoma cruzi: structure and biological roles. Mol. Biochem. Parasitol., 114, 143–150. [DOI] [PubMed] [Google Scholar]
- Bates E.J., Knuepfer,E. and Smith,D.F. (2000) Poly(A)-binding protein I of Leishmania: functional analysis and localisation in trypanosomatid parasites. Nucleic Acids Res., 28, 1211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C.A. and Parker,R. (1994) Differential effects of translational inhibition in cis and trans on the decay of the unstable MFA2 mRNA. J. Biol. Chem., 269, 9687–9692. [PubMed] [Google Scholar]
- Bellofatto V., Torres-Munoz,J.E. and Cross,G.A.M. (1991) Stable trans formation of Leptomonas seymouri by circular extrachromosomal elements. Proc. Natl Acad. Sci. USA, 88, 6711–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berberof M., Vanhamme,L., Tebabi,P., Pays,A., Jefferies,D., Welburn,S. and Pays,E. (1995) The 3′-terminal region of the mRNAs for VSG and procyclin can confer stage specificity to gene expression in T. brucei. EMBO J., 14, 2925–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebinger S., Rettenmaier,S., Flaspohler,J., Hartmann,C., Peña-Diaz,J., Wirtz,L.E., Hotz,H.R., Barry,J.D. and Clayton,C.E. (1996) The PARP promoter of Trypanosoma brucei is developmentally regulated in a chromosomal context. Nucleic Acids Res., 24, 1202–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner J. and Clayton,C.E. (1995) The 3′-untranslated regions from the Trypanosoma brucei phosphoglycerate kinase genes mediate developmental regulation. Gene, 162, 153–156. [DOI] [PubMed] [Google Scholar]
- Blattner J., Helfert,S., Michels,P. and Clayton,C.E. (1998) Com partmentation of phosphoglycerate kinase in Trypanosoma brucei plays a critical role in parasite energy metabolism. Proc. Natl Acad. Sci. USA, 95, 11596–11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P. and Ulbert,S. (2001) Control of VSG gene expression sites. Mol. Biochem. Parasitol., 114, 17–27. [DOI] [PubMed] [Google Scholar]
- Brittingham A., Miller,M.A., Donelson,J.E. and Wilson,M.E. (2001) Regulation of GP63 mRNA stability in promastigotes of virulent and attenuated Leishmania chagasi. Mol. Biochem. Parasitol., 112, 51–59. [DOI] [PubMed] [Google Scholar]
- Butler J.S. (2002) The yin yang of the exosome. Trends Cell Biol., 12, 90–96. [DOI] [PubMed] [Google Scholar]
- Caponigro G. and Parker,R. (1996) Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest H., Zhang,W.-W. and Matlashewski,G. (1996) The develop mental expression of Leishmania donovani A2 amastigote-specific genes is post-transcriptionally mediated and involves elements in the 3′-untranslated region. J. Biol. Chem., 271, 17081–17090. [DOI] [PubMed] [Google Scholar]
- Chaves I., Rudenko,G., Dirks-Mulder,A., Cross,M. and Borst,P. (1999) Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J., 18, 4846–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin B.C., Teixeira,S.M., Kirchhoff,L.V. and Donelson,J.E. (2000) Amastin mRNA abundance in Trypanosoma cruzi is controlled by a 3′-untranslated region position-dependent cis-element and an untrans lated region-binding protein. J. Biol. Chem., 275, 12051–12060. [DOI] [PubMed] [Google Scholar]
- de Lafaille M.A.C., Laban,A. and Wirth,D.F. (1992) Gene expression in Leishmania: analysis of essential 5′ DNA sequences. Proc. Natl Acad. Sci. USA, 89, 2703–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J.M., D’Orso,I., Sánchez,D.O. and Frasch,A.C.C. (2000) AU-rich elements in the 3′-untranslated region of a new mucin-type gene family of Trypanosoma cruzi confers mRNA instability and modulates translation efficiency. J. Biol. Chem., 275, 10218–10227. [DOI] [PubMed] [Google Scholar]
- Djikeng A., Shi,H., Tschudi,C. and Ullu,E. (2001) RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA, 7, 1522–1530. [PMC free article] [PubMed] [Google Scholar]
- D’Orso I. and Frasch,A.C.C. (2001) TcUBP-1, a developmentally regulated U-rich RNA-binding protein involved in selective mRNA destabilization in trypanosomes. J. Biol. Chem., 276, 34801–34809. [DOI] [PubMed] [Google Scholar]
- Downey N. and Donelson,J.E. (1999) Search for promoters for the GARP and rRNA genes of Trypanosma congolense. Mol. Biochem. Parasitol., 104, 25–38. [DOI] [PubMed] [Google Scholar]
- Drozdz M. and Clayton,C.E. (1999) Structure of a regulatory 3′-untranslated region from Trypanosoma brucei. RNA, 5, 1632–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar D.A., Chen,A.A., Wormsley,S. and Baserga,S.J. (2000) The genes for small nucleolar RNAs in Trypanosoma brucei are organized in clusters and are transcribed as a polycistronic RNA. Nucleic Acids Res., 28, 2855–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez A.M., Kemp,T. and Clayton,C. (2001) The exosome of Trypanosoma brucei. EMBO J., 20, 3831–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger A., Schürch,N., Kurath,U. and Roditi,I. (1997) Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol. Cell. Biol., 17, 4372–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilinger G. and Bellofatto,V. (2001) Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res., 29, 1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham S.V. and Barry,J.D. (1996) Polysomal, procyclin mRNAs accumulate in bloodstream forms of monomorphic and pleomorphic trypanosomes treated with protein synthesis inhibitors. Mol. Biochem. Parasitol., 80, 179–192. [DOI] [PubMed] [Google Scholar]
- Horn D. (2001) Nuclear gene transcription and chromatin in Trypanosoma brucei. Int. J. Parasitol., 31, 1157–1165. [DOI] [PubMed] [Google Scholar]
- Hotchkiss T.L., Nerantzakis,G.E., Dills,S.C., Shang,L. and Read,L.K. (1999) Trypanosoma brucei poly(A) binding protein I cDNA cloning, expression and binding to 5′ untranslated region sequence elements. Mol. Biochem. Parasitol., 98, 117–129. [DOI] [PubMed] [Google Scholar]
- Hotz H.-R., Hartmann,C., Huober,K., Hug,M. and Clayton,C.E. (1997) Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a trypanosome surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res., 25, 3017–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz H.-R., Biebinger,S., Flaspohler,J. and Clayton,C.E. (1998) PARP gene expression: regulation at many levels. Mol. Biochem. Parasitol., 91, 131–143. [DOI] [PubMed] [Google Scholar]
- Irmer H. and Clayton,C.E. (2001) Degradation of the unstable EP1 mRNA in Trypanosoma brucei involves initial destruction of the 3′-untranslated region. Nucleic Acids Res., 29, 4707–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-Anderson J.S. and Parker,R.P. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapotas N. and Bellofatto,V. (1993) Differential response to RNA trans-splicing signals within the phosphoglycerate kinase gene cluster in Trypanosoma brucei. Nucleic Acids Res., 21, 4067–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer G. and Günzl,A. (2001) In vitro competition analysis of procyclin gene and variant surface glycoprotein gene expression site transcription in Trypanosoma brucei. Mol. Biochem. Parasitol., 113, 55–65. [DOI] [PubMed] [Google Scholar]
- Laufer G., Schaaf,G., Bollgonn,S. and Günzl,A. (1999) In vitro analysis of α-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol. Cell. Biol., 19, 5466–5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBowitz J.H., Smith,H.Q., Rusche,L. and Beverley,S.M. (1993) Coupling of poly(A) site selection and trans splicing in Leishmania. Genes Dev., 7, 996–1007. [DOI] [PubMed] [Google Scholar]
- Liniger M., Bodenmüller,K., Pays,E., Gallati,S. and Roditi,I. (2001) Overlapping sense and antisense transcription units in Trypanosoma brucei. Mol. Microbiol., 40, 869–878. [DOI] [PubMed] [Google Scholar]
- Luo H., Gilinger,G., Mukherjee,D. and Bellofatto,V. (1999) Transcription initiation at the TATA-less spliced leader RNA gene promoter requires at least two DNA-bindng proteins and a tripartite architecture that includes an initiator element. J. Biol. Chem., 274, 31947–31954. [DOI] [PubMed] [Google Scholar]
- Mahmood R., Hines,J.C. and Ray,D.S. (1999) Identification of cis and trans elements involved in the cell cycle regulation of multiple genes in Crithidia fasciculata. Mol. Cell. Biol., 19, 6174–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood R., Mittra,B., Hines,J.C. and Ray,D.S. (2001) Charac terization of the Crithidia fasciculata mRNA cycling sequence binding proteins. Mol. Cell. Biol., 21, 4453–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair G. et al. (2000a) A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA, 6, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair G., Ullu,E. and Tschudi,C. (2000b) Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem., 275, 28994–28999. [DOI] [PubMed] [Google Scholar]
- Marchetti M.A., Tschudi,C., Silva,E. and Ullu,E. (1998) Physical and transcriptional analysis of the Trypanosoma brucei genome reveals a typical eucaryotic arrangement with a close interspersion of RNA polymerase II- and III-transcribed genes. Nucleic Acids Res., 26, 3591–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkin A., Das,A. and Bellofatto,V. (2001) The Leptomonas seymouri spliced leader RNA promoter requires a novel transcription factor. Int. J. Parasitol., 31, 545–549. [DOI] [PubMed] [Google Scholar]
- Matthews K.R. (1999) Developments in the differentiation of Trypanosoma brucei. Parasitol. Today, 15, 76–80. [DOI] [PubMed] [Google Scholar]
- Matthews K.R., Tschudi,C. and Ullu,E. (1994) A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev., 8, 491–501. [DOI] [PubMed] [Google Scholar]
- McAndrew M., Graham,S., Hartmann,C. and Clayton,C.E. (1998) Testing promoter activity in the trypanosome genome: unexpected insights into RNA polymerase II initiation. Exp. Parasitol., 90, 65–76. [DOI] [PubMed] [Google Scholar]
- McDonagh P.D., Myler,P.J. and Stuart,K. (2000) The unusual gene organization of Leishmania major chromosome 1 may reflect novel transcription processes. Nucleic Acids Res., 28, 2800–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000) mRNA stability in eucaryotes. Curr. Opin. Genet. Dev., 10, 193–198. [DOI] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2001) mRNA turnover. Curr. Opin. Cell Biol., 13, 320–325. [DOI] [PubMed] [Google Scholar]
- Myler P.J. et al. (1999) Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl Acad. Sci. USA, 96, 2902–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaar V., Günzl,A., Ullu,E. and Tschudi,C. (1997) Structure of the Trypanosoma brucei U6 snRNA gene promoter. Mol. Biochem. Parasitol., 88, 13–23. [DOI] [PubMed] [Google Scholar]
- Navarro M. and Gull,K. (2001) A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature, 414, 759–763. [DOI] [PubMed] [Google Scholar]
- Parsons M. and Ruben,L. (2000) Pathways involved in environmental sensing in trypanosomatids. Parasitol. Today, 16, 56–62. [DOI] [PubMed] [Google Scholar]
- Pays E., Lips,S., Nolan,D., Vanhamme,L. and Perez-Morga,D. (2001) The VSG expression sites of Trypanosoma brucei: multipurpose tools for the adaptation of the parasite to mammalian hosts. Mol. Biochem. Parasitol., 114, 1–16. [DOI] [PubMed] [Google Scholar]
- Priest J.W. and Hajduk,S.L. (1994) Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J. Bioenerg. Biomembr., 26, 179–191. [DOI] [PubMed] [Google Scholar]
- Quijada L., Soto,M., Alonso,C. and Requena,J.M. (2000) Identification of a putative regulatory element in the 3′-untranslated region that controls expression of HSP70 in Leishmania infantum. Mol. Biochem. Parasitol., 110, 79–91. [DOI] [PubMed] [Google Scholar]
- Saas J., Ziegelbauer,K., von Haeseler,A., Fast,B. and Boshart,M. (2000) A developmentally regulated aconitase related to iron-regulatory protein-1 is localized in the cytoplasm and in the mitochondrion of Trypanosoma brucei. J. Biol. Chem., 275, 2745–2755. [DOI] [PubMed] [Google Scholar]
- Schürch N., Furger,A., Kurath,U. and Roditi,I. (1997) Contribution of the procyclin 3′ untranslated region and coding region to the regulation of expression in bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol., 89, 109–121. [DOI] [PubMed] [Google Scholar]
- Shyu A.-B. and Wilkinson,M.F. (2000) The double lives of shuttling mRNA binding proteins. Cell, 102, 135–138. [DOI] [PubMed] [Google Scholar]
- Sturm N.R., Yu,M.C. and Campbell,D.A. (1999) Transcription termination and 3′-end processing of the spliced leader RNA in kinetoplastids. Mol. Cell. Biol., 19, 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira S.M.R., Kirchhoff,L.V. and Donelson,J.E. (1995) Post-transcriptional elements regulating expression of mRNAs from the amastin/tuzin gene cluster of Trypanosoma cruzi. J. Biol. Chem., 270, 22586–22594. [DOI] [PubMed] [Google Scholar]
- Ullu E., Matthews,K.R. and Tschudi,C. (1993) Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol. Cell. Biol., 13, 720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L., Pays,E., McCulloch,R. and Barry,J.D. (2001) An update on antigenic variation in African trypanosomes. Trends Parasitol., 17, 338–343. [DOI] [PubMed] [Google Scholar]
- Vassella E., Den Abbeele,J., Butikofer,P., Renggli,C.K., Furger,A., Brun,R. and Roditi,I. (2000) A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev., 14, 615–626. [PMC free article] [PubMed] [Google Scholar]
- Vassella E., Kramer,R., Turner,C.M., Wankell,M., Modes,C., van den Bogaard,M. and Boshart,M. (2001) Deletion of a novel protein kinase with PX and FYVE-related domains increases the rate of differentiation of Trypanosoma brucei. Mol. Microbiol., 41, 33–46. [DOI] [PubMed] [Google Scholar]
- Wen L.-M., Xu,P., Benegal,G., Carvalho,M.R.M. and Buck,G.A. (2000) PPB1, a putative spliced leader RNA gene transcription factor in Trypanosoma cruzi. Mol. Biochem. Parasitol., 110, 207–221. [DOI] [PubMed] [Google Scholar]
- Wong A.K., Curotto de Lafaille,C. and Wirth,D.F. (1994) Identification of a cis-acting gene regulatory element from the lemdr locus of Leishmania enriettii. J. Biol. Chem., 269, 26497–26502. [PubMed] [Google Scholar]
- Xu Y., Liu,L., Lopez-Estrano,C. and Michaeli,S. (2001) Expression studies on clustered trypanosomatid box C/D small nucleolar RNAs. J. Biol. Chem., 276, 14289–14298. [DOI] [PubMed] [Google Scholar]
- Zilberstein D. and Shapira,M. (1994) The role of pH and temperature in the development of Leishmania parasites. Annu. Rev. Microbiol., 48, 449–470. [DOI] [PubMed] [Google Scholar]