Abstract
Phosphatidylinositol transfer protein α (PITPα) is a ubiquitous and highly conserved protein in multicellular eukaryotes that catalyzes the exchange of phospholipids between membranes in vitro and participates in cellular phospholipid metabolism, signal transduction and vesicular trafficking in vivo. Here we report the three-dimensional crystal structure of a phospholipid-free mouse PITPα at 2.0 Å resolution. The structure reveals an open conformation characterized by a channel running through the protein. The channel is created by opening the phospholipid-binding cavity on one side by displacement of the C-terminal region and a hydrophobic lipid exchange loop, and on the other side by flattening of the central β-sheet. The relaxed conformation is stabilized at the proposed membrane association site by hydrophobic interactions with a crystallographically related molecule, creating an intimate dimer. The observed open conformer is consistent with a membrane-bound state of PITP and suggests a mechanism for membrane anchoring and the presentation of phosphatidylinositol to kinases and phospholipases after its extraction from the membrane. Coordinates have been deposited in the Protein Data Bank (accession No. 1KCM).
Keywords: membrane association/phospholipid-binding protein/PITP
Introduction
Phosphatidylinositol transfer protein α (PITPα) belongs to a family of phospholipid-binding proteins, which is present in all mammalian tissues investigated to date (Wirtz, 1997). The protein is highly conserved (>98% sequence identity) among different mammalian species and shares extended sequence identity with its isoforms PITPβ (Wirtz, 1997) and retinal degeneration B protein β (RdgBβ) (Milligan et al., 1997). Other PITP domains are found in the recently identified human RdgBαs or N-terminal domain-interacting receptors (NIRs) (Lev et al., 1999). However, PITP has no sequence homology with its functional counterpart (Sec14p) in yeast (Sha et al., 1998). PITPα is a multifunctional protein with regulatory roles in intracellular lipid and vesicular trafficking and in lipid-mediated signal transduction pathways (Cunningham et al., 1995; Cockroft, 1999). In vitro, PITPα transfers phosphatidylinositol (PI) and phosphatidylcholine (PC) between membranes through the aqueous phase, exhibiting a 16-fold higher affinity for PI than for PC (van Paridon et al., 1987a), and thereby may regulate the phospholipid composition of the membranes. In vivo, an apparent key role for PITPα is the presentation of PI to PI-specific kinases (Cockroft, 1999) and phospholipases (Snoek et al., 1999), yielding metabolites essential for lipid signaling. Phosphorylation of PITPα by protein kinase C at the single site Ser166 is a prerequisite for its relocalization from the nucleus and the cytosol to the Golgi membrane (van Tiel et al., 2000), where it co-localizes with its isoform PITPβ. Little is known about the detailed way in which PITPs fulfil their functions in vivo. However, their biological importance has been demonstrated in mice carrying the vibrator mutation and in RdgB null Drosophila mutants. In the former case, a strong reduction in PITPα levels causes neurodegeneration (Hamilton et al., 1997), whereas in the latter the absence of RdgB causes light-induced retinal degeneration (Milligan et al., 1997).
Results and discussion
Structure determination
The structure of mouse PITPα was determined with molecular replacement, using PITPα from rat (Protein Data Bank accession No. 1FVZ) as the search motif. Recombinant mouse PITPα was crystallized using 11% (w/v) PEG 6000, 200 mM calcium acetate and 100 mM cacodylate pH 6.5. Prior to crystallization, the bound lipid, phosphatidylglycerol, was exchanged for di-brominated PC. The protein crystallized in the space group P3221 with unit cell dimensions a = b = 50.46 Å and c = 216.11 Å. One PITPα molecule is present in the asymmetric unit. The refined model consists of 256 amino acid residues (residues 2–257) and 216 water molecules, and has a crystallographic R-factor of 21.6% and an Rfree of 27.3% for data in the 30–2.0 Å resolution range. No density for lipid molecules was observed. Table I summarizes the statistics of the crystallographic data and the refinement. Coordinates and structure factors have been deposited in the Protein Data Bank with accession No. 1KCM.
Table I. Summary of data and refinement statistics.
| Data set statisticsa | |
| Resolution limits (Å) (outer shell) | 30–1.99 (2.06–1.99) |
| Space group | P3221 |
| Unit cell parameters (Å, °) | a = 50.46, b = 50.46,c = 216.11 |
| α = 90, β = 90, γ = 120 | |
| Mosaicity (°) | 0.62 |
| Oscillation range (°) | 1.0 |
| Total oscillation for data set (°) | 131 |
| Total No. of reflections | 231 585 |
| No. of unique reflections (outer shell) | 22 060 (2012) |
| Redundancy (outer shell) | 4.33 (2.70) |
| Rsym (%) (outer shell)b | 5.1 (17.1) |
| Completeness (%) (outer shell) | 96.7 (91.1) |
| I/σ(I) (outer shell) | 25.7 (7.3) |
| Refinement statistics | |
| Resolution range (Å) | 30–2.0 |
| Total No. of reflections | 20 780 |
| No. of protein residues | 256 |
| Total No. of non-hydrogen atoms | 2346 |
| No. of water molecules | 216 |
| Rwork (%)c | 21.6 |
| Rfree (%)d | 27.3 |
| R.m.s.d. bond lengths (Å) | 0.012 |
| R.m.s.d. bond angles (°) | 1.42 |
| Average B-factor (all protein atoms) (Å2) | 24.3 |
aNumbers in parentheses indicate the values in the last resolution shell.
bRsym = ∑|Ih – <Ih>|/∑Ih, where <Ih> is the average intensity over symmetry equivalents.
cRwork = ∑||Fobs| – |Fcalc||/∑|Fobs|
dRfree was calculated using a randomly selected 5.1% of the reflections.
Overall structure
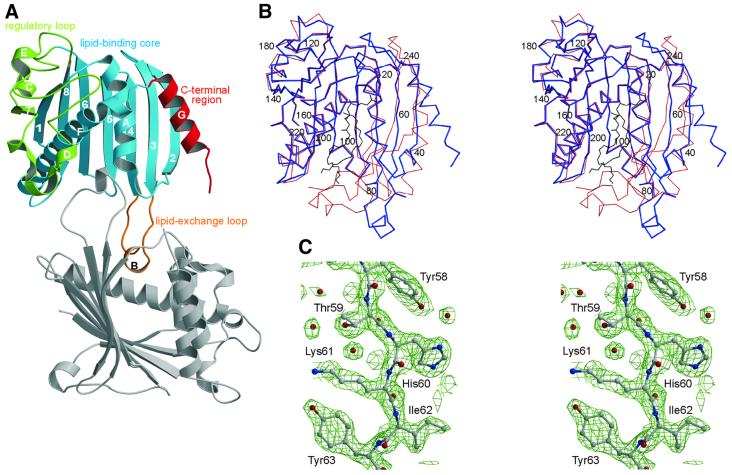
Here we present the X-ray structure of mouse PITPα (Mr 32 kDa, 271 amino acid residues) without bound phospholipid. The apo-PITPα structure reveals an open form, which is stabilized through dimerization in the crystal (see Figure 1A). In contrast, a recently determined structure of rat PITPα (Yoder et al., 2001), whose sequence differs only by the single conservative replacement of Ile167 for valine, shows a closed monomeric form (see Figure 1B) that fully encapsulates a PC molecule. This lipid-enclosed state of the protein most probably corresponds to the phospholipid transport intermediate of PITPα. As with the closed form, the main structural feature of apo-PITPα is a β-sheet structure made by strands 1–8 (Figure 1A). Helices A and F face the interior of the β-sheet and together with it define the lipid-binding core (residues 1–118 and 191–238), the dominant functional region of PITPα. Another three functional regions of the molecule can be defined: the lipid exchange loop (residues 65–83) which contains the small helix B and acts as a lid to the lipid-binding cavity, the regulatory loop (residues 119–190) and the C-terminal region (residues 239–271) containing helix G. The predominant structural differences between the closed and open state are: (i) a swing of ∼90° of the lipid exchange loop; (ii) a twist and flattening out of the peripheral β-strands 2, 3 and 4 of the lipid-binding core; and (iii) a 20° swing of the C-terminal helix G, accompanied by a partial unwinding of residues 254–257 and a disordering of the C-terminal tail (residues 258–271). All structural changes are located on one side of the PITPα molecule, with only minor changes in the regulatory loop and the major part of the lipid-binding core (Figure 1B). The consequence of the large structural rearrangements in PITPα is a widening of the lipid-binding cavity and opening on two sides of the cavity creating a channel. A small opening of ∼6 × 14 Å is found between the N-terminus of helix G and strands 2 and 3, and a large opening of 20 × 22 Å is located between the β-sheet and helices A and F (see Figure 1B). The small opening is of a hydrophilic nature and lies in the vicinity of the site for lipid head group binding. The large opening exposes a hydrophobic surface of ∼1300 Å2 formed by the lipid exchange loop and part of the lipid-binding core (see Figure 2A). In the crystal, the PITPα monomers are packed face-to-face, covering the hydrophobic areas, with residues 70–78 of the lipid exchange loops inserting into each other’s lipid-binding cavities. Thus, in the dimer, the lipid exchange loops substitute protein–lipid interactions as found in the closed form between the sn-1 acyl chain of PC and PITPα.
Fig. 1. Crystal structure of apo-PITPα revealing an open conformation stabilized through dimerization. (A) With functional regions color-coded: regulatory loop (green), lipid-binding core (blue), C-terminal region (red) and the lipid exchange loop (orange); secondary structural elements are labeled numerically (β-strands) and with upper case roman letters (α-helices). The lipid-binding site, located in the core of the molecule, is partially occupied by the lipid exchange loop of the symmetry-related, dimeric partner molecule (gray). (B) Stereo representation of the Cα trace of the open (blue) and closed, PC-encapsulated (red) conformations showing the major structural rearrangements of the lipid exchange loop, the C-terminal region and β-strands 2, 3 and 4; the position of every 20th residue is indicated. The position of PC in the PC-encapsulated form is shown in black. (C) Stereo view of the 2Fo – Fc electron density map contoured at 1.5σ of residues 58–63 of β-strand 3 preceding the lipid exchange loop.
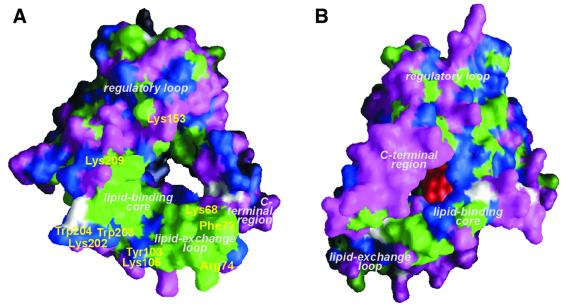
Fig. 2. Molecular surface representation of apo-PITPα showing the openings of the lipid-binding cavity. (A) Viewed from the side of membrane association showing exposed hydrophobic residues (green), charged residues (magenta), polar residues (blue) and glycine (white). The positions of residues thought to be involved in binding to the lipid layer at the interfacial region are indicated in yellow. (B) Viewed from the opposite side (using the same color code) with phosphorylinositol shown at its putative binding site in red.
Membrane association
We observe an open conformation of the PITPα monomer that is consistent with the biochemically defined characteristics of PITPα bound to a membrane (Tremblay et al., 1996, 1998; Voziyan et al., 1996). Dislodging and partial unfolding of the C-terminal region is consistent with susceptibility to proteolytic cleavage at Arg253 and Arg259 when PITPα is associated with membrane vesicles (Tremblay et al., 1996). Additionally, increased membrane affinity and a more relaxed conformation as compared with full-length PITP is observed for a C-terminally truncated mutant Δ260–271 (Voziyan et al., 1996; Tremblay et al., 1998), indicating that the C-terminal region may function to stabilize the closed and soluble form of PITPα (Yoder et al., 2001). Displacement of the C-terminal region and the lipid exchange loop, as observed in our structure, allows aromatic residues, Tyr103, Trp203 and Trp204, and positively charged residues, Lys105, Lys202 and Lys209, of the structurally invariant part of the lipid-binding core to contact the membrane and interact with the lipid interfacial region (Figure 2A). Additional interactions may come from Lys68, Phe72 and Arg74 of the lipid exchange loop and from Lys153 of helix D of the regulatory loop. At the same time, the lipid exchange loop may insert partially into the bilayer, anchoring the molecule to the membrane. Membrane insertion is consistent with a slight increase of surface pressure when PITPα is injected under a phospholipid monolayer spread at the air–water interface (Demel et al., 1977). These data, therefore, support the notion that the observed open structure of PITPα is consistent with the membrane-bound state of the protein. In addition to these data, it was reported recently that Cys95 can be chemically modified by N-ethylmaleimide provided the protein is associated with a membrane surface (Tremblay et al., 2001). The authors argued that this alkylating agent accesses Cys95 by way of the membrane. However, the observed open conformation indicates that Cys95, as part of the lipid-binding cavity, is close to the small hydrophilic hole, allowing access of the alkylating agent. Interestingly, Cys95 is modified in our crystal, as indicated by a residual electron density at 1.8 Å distance from the Sγ atom. The membrane association model proposed implies that a major part of the PC-binding site is intact, when PITPα is bound to the membrane. Extraction of a phospholipid from the membrane bilayer is probably facilitated by a low dielectric constant in the channel, reducing the bilayer stability at the site of interaction. Subsequent closing of the lipid-binding cavity by the lipid exchange loop and refolding of the C-terminus will dissociate the lipid–PITPα complex from the membrane.
Putative inositol-binding site
The specificity of PI over PC could not be explained from the phosphorylcholine-binding site in the closed PC–PITPα structure. Based on biochemical data, van Paridon et al. (1987b) have suggested the existence of separate head group-binding sites, one highly specific for the phosphorylinositol head group and one less specific for the phosphorylcholine head group. In contrast, Tremblay et al. (2001) present a model in which the phosphorylinositol moiety fits within the observed site for phosphorylcholine binding. In our open structure, a twist of the peripheral β-strands 2, 3 and 4, flattening the concave β-sheet, yields an opening of the lipid-binding cavity close to the polar head group-binding site opposite to the membrane association site. This opening is located between residues 48 and 56 of β-strands 2 and 3, respectively, and residue 240 of the loop between helices F and G. We have used SuperStar (Verdonk et al., 1999) to locate possible binding sites for the phosphorylinositol moiety in the open conformation, using propensity maps for hydroxyl and carbonyl probes. Besides the known phosphate-binding site (formed by residues Gln22, Thr97, Thr114 and Lys195) as observed in the PC–PITPα complex, we predict a second adjacent site for phosphate moieties. This novel putative phosphate-binding site is formed primarily by Gln22 and Thr97 and is shifted 4.2 Å away from the phosphate of PC in PC–PITPα towards the channel opening. Furthermore, a cluster of peaks with high hydroxyl propensity was observed close to Thr59 and Lys61 (β-strand 3) and Glu86 and Asn90 (β-strand 4), which may indicate positions of hydroxyls of the inositol moiety. In comparison with the closed PC–PITPα structure, Thr59 is displaced by 4.3 Å (Cα–Cα distance) from the choline-binding site and becomes part of the putative inositol-binding site in the open structure. This binding site has residues Gln22, Thr59, Glu86, Asn90 and Thr97 in common with the phosphorylinositol-binding site proposed by Tremblay et al. (2001). However, due to conformational differences, the predicted position of phosphorylinositol is shifted by 4 Å, suggesting two separate sites for phosphorylinositol and phosphorylcholine binding in the open conformation. In support of this, mutagenesis data indicated that Ser25, Thr59 and Glu248 were essential for transferring PI, but not for PC (Alb et al., 1995). In both models of phosphorylinositol binding, Thr59 is involved in hydrogen bonding to the inositol moiety. Glu248 of the C-terminal helix G is part of the rim of the small hydrophilic hole, favoring the phosphorylinositol-binding site in the open structure. Ser25 is located in helix A oriented towards helix F and not towards the lipid-binding site, and thus may have an indirect effect on lipid specificity. When the phosphorylinositol moiety is placed at the alternative site in the open structure, the hydroxyl groups at positions 3, 4 and 5 are positioned in front of the hydrophilic hole of the lipid-binding channel (Figure 2B). Hence, by binding close to the opening near the polar head group (possibly by interacting with residues 160–190 of the regulatory loop), PI-specific enzymes may possibly have direct access to the protein-bound inositol moiety. Thus, the open conformation of PITPα supports the hypothesis that an alternative binding site specific for the phosphorylinositol moiety of PI exists. This site could be of significance in PI signaling. The precise binding mode must be verified experimentally, e.g. by structure determination of PITPα in complex with PI.
Concluding remarks
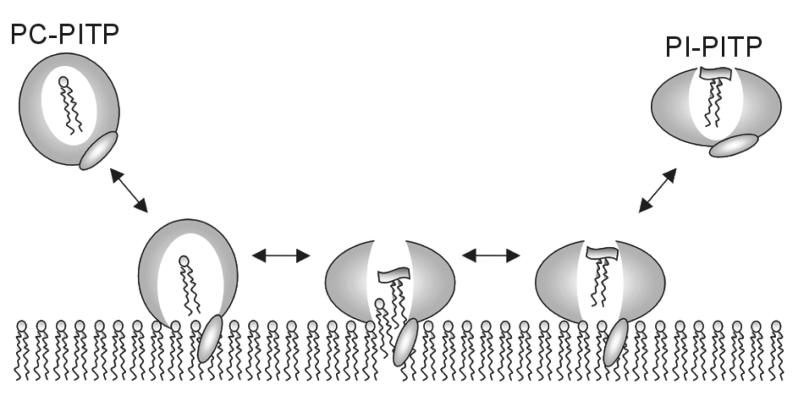
PITPα is considered to be unique among the lipid transfer proteins; besides its lipid transfer function, it may present PI to kinases (Cockroft, 1999) and phospholipases (Snoek et al., 1999) in signal transduction. A central β-sheet and α-helices form the lipid-binding cavity with access regulated by a lid, similar to other lipid-binding or sterol carrier proteins including PITP’s functional homolog, Sec14p from yeast (Sha et al., 1998), and PITP’s structural homolog, the cholesterol-binding START domain (Tsujishita and Hurley, 2000). This is the first time that a widely open channel, as seen in the structure of apo-PITPα (Figure 2A), has been observed in lipid-binding proteins. PITPα differs from its isoform PITPβ predominantly by non-conservative amino acid substitutions in the regulatory region, explaining the difference in the physiological roles of the two isoforms. The PITP domains in RdgBs are more distantly related, showing many non-conservative amino acid replacements throughout all structural regions except in the region close to the proposed PI-binding site. The open conformation presented here provides for the first time a structural basis for the association of PITP with the membrane and the mechanism by which it may extract a phospholipid molecule (Figure 3). Moreover, the structure of PITPα suggests that the inositol moiety of PI in its binding site is accessible to PI-modifying enzymes.
Fig. 3. Schematic representation of the proposed mechanism of phospholipid exchange by PITPα at the membrane interface. PITPα carrying a PC molecule has a closed conformation in solution. Upon binding to a membrane, the structure relaxes into an open conformation exposing a channel for phospholipid binding, which may accommodate PI in exchange for a PC molecule. Upon release from the membrane, PITPα carrying a PI molecule has a conformation closed at the site of membrane association and open at the polar head group region.
Materials and methods
Crystallization and data collection
Crystals of recombinant mouse PITPα, without the Met1 initiator, were grown at 4°C using the hanging drop vapor diffusion method by combining 4 µl of 14 mg/ml protein in 10 mM Tris–HCl pH 7.2 and 10 mM β-mercaptoethanol with 4 µl of a solution containing 11% (w/v) PEG 6000, 200 mM calcium acetate and 100 mM cacodylate pH 6.5, and equilibrated against a reservoir containing 500 µl of the same solution. Prior to crystallization, the protein sample was incubated with 16:0-18:0(6-7diBr) phosphatidylcholine (diBrPC) vesicles to replace the endogenous phosphatidylglycerol with diBrPC for multiwavelength anomalous dispersion (MAD) phasing using bromine. The crystal grew to maximum crystal dimensions of ∼0.9 × 0.3 × 0.2 mm and was cryoprotected in a solution of mother liquor with 34% (v/v) ethylene glycol and flash cooled in liquid nitrogen prior to data collection. The 2.0 Å resolution data were collected at 100 K and λ = 0.9322 Å at the ID-14 EH4 beamline at the European Synchrotron Radiation Facility (ESRF) in Grenoble. Data were indexed using DENZO merged with SCALEPACK (Otwinowski and Minor, 1997) and processed further using Truncate from the CCP4 suite (CCP4, 1994). The crystals belong to the trigonal space group P3221, with unit cell dimensions a = b = 50.46 Å, c = 216.11 Å, α = β = 90°, γ = 120°, and contain one monomer per asymmetric unit and a calculated solvent content of 50% (v/v). A summary of the data collection and final processing statistics are provided in Table I.
Structure determination and refinement
Despite detection of bromine in the crystal by fluorescence, no bromine could be located in Patterson maps. Using the recently determined structure of rat PITPα (Yoder et al., 2001) as a search model, we solved the structure by molecular replacement with the CCP4 program AmoRe (Navaza, 1994). Nearly 80% of the initial model could be built automatically using the ARP/wARP package (Perrakis et al., 1999). From the improved electron density map, it was possible to model the remaining part of the structure. However, no density for C-terminal residues 258–271 was observed, indicating flexibility of the C-terminus. Residues Trp203 and Trp204 showed poor side-chain density. Additional density at 1.8 Å distance from the Sγ of Cys95 was observed, indicating a chemical modification. Manual adjustments of the model were carried out with the program O (Jones et al., 1991). The program REFMAC5 (Winn et al., 2001) with application of the TLS option was used for subsequent refinements. Model quality was checked using PROCHECK (Laskowski et al., 1993) and WHATIF (Vriend, 1990). Analysis of the Ramachandran plot shows that 93% of the residues are in the most favored regions, 6% in additional allowed regions and 2% in generously allowed regions. Domain–domain contacts were calculated with LIGPLOT (Wallace et al., 1995). Figures were prepared using MOLSCRIPT (Kraulis, 1991), BOBSCRIPT (Esnouf, 1999) and GRASP (Honig and Nicholls, 1995).
Acknowledgments
Acknowledgements
We thank B.Bouma and D.R.Boer for assistance, M.D.Yoder for supplying the coordinates of rat PITPα, and the staff at the ESRF synchrotron in Grenoble, in particular R.B.G.Ravelli, for support in data collection.
References
- Alb J.G. Jr, Gedvilaite,A., Cartee,R.T., Skinner,H.B. and Bankaitis,V.A. (1995) Mutant rat phosphatidylinositol/phosphatidylcholine transfer proteins specifically defective in phosphatidylinositol transfer: implications for the regulation of phospholipid transfer activity. Proc. Natl Acad. Sci. USA, 92, 8826–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockroft S. (1999) Mammalian phosphatidylinositol transfer proteins: emerging roles in signal transduction and vesicular traffic. Chem. Phys. Lipids, 98, 23–33. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 . (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cunningham E., Thomas,G.M.H., Ball,A., Hiles,I. and Cockroft,S. (1995) Phosphatidylinositol transfer protein dictates the rate of inositol triphosphate production by promoting the synthesis of PIP2. Curr. Biol., 5, 775–783. [DOI] [PubMed] [Google Scholar]
- Demel R.A., Kalsbeek,R., Wirtz,K.W.A. and van Deenen,L.L.M. (1977) The protein-mediated net transfer of phosphatidylinositol in model systems. Biochim. Biophys. Acta, 466, 10–22. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to Molscript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Hamilton B.A. et al. (1997) The vibrator mutation causes neurodegeneration via reduced expression of PITPα: positional complementation cloning and extragenic suppression. Neuron, 18, 711–722. [DOI] [PubMed] [Google Scholar]
- Honig B. and Nicholls,A. (1995) Classical electrostatics in biology and chemistry. Science, 268, 1144–1149. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lev S., Hernandez,J., Martinez,R., Chen,A., Plowman,G. and Schlessinger,J. (1999) Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol. Cell. Biol., 19, 2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscovitch M. and Cantley,L.C. (1995) Signal transduction and membrane traffic: the PITP/phosphoinositide connection. Cell, 81, 659–662. [DOI] [PubMed] [Google Scholar]
- Milligan S.C., Alb,J.G.,Jr, Elagina,R.B., Bankaitis,V.A. and Hyde,D.R. (1997) The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol., 139, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. (1994) AmoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Sha B., Phillips,S.E., Bankaitis,V.A. and Luo,M. (1998) Crystal structure of the Saccharomyces cerevisae phosphatidylinositol-transfer protein. Nature, 391, 506–510. [DOI] [PubMed] [Google Scholar]
- Snoek G.T., Berrie,C.P., Geijtenbeek,T.B., van der Helm,H.A., Cadée,J.A., Iurisci,C., Corda,D. and Wirtz,K.W.A. (1999) Overexpression of phosphatidylinositol transfer protein α in NIH3T3 cells activates a phospholipase A. J. Biol. Chem., 274, 35393–35399. [DOI] [PubMed] [Google Scholar]
- Tremblay J.M., Helmkamp,G.M.,Jr and Yarbrough,L.R. (1996) Limited proteolysis of rat phosphatidylinositol transfer protein by trypsin cleaves the C-terminus, enhances binding to lipid vesicles, and reduces phospholipid transfer activity. J. Biol. Chem., 271, 21075–21080. [DOI] [PubMed] [Google Scholar]
- Tremblay J.M., Voziyan,P.A., Helmkamp,G.M.,Jr and Yarbrough,L.R. (1998) The C-terminus of phosphatidylinositol transfer protein modulates membrane interactions and transfer activity but not phospholipid binding. Biochim. Biophys. Acta, 1389, 91–100. [PubMed] [Google Scholar]
- Tremblay J.M., Li,H., Yarbrough,L.R. and Helmkamp,G.M.,Jr (2001) Modifications of cysteine residues in the solution and membrane-associated conformations of phosphatidylinositol transfer protein have differential effects on lipid transfer activity. Biochemistry, 40, 9151–9158. [DOI] [PubMed] [Google Scholar]
- Tsujishita Y. and Hurley,J.H. (2000) Structure and lipid transport mechanism of a StAR-related domain. Nature Struct. Biol., 7, 408–414. [DOI] [PubMed] [Google Scholar]
- van Paridon P.A., Gadella,T.W.J., Somerharju,P.J. and Wirtz,K.W.A. (1987a) On the relationship between the dual specificity of the bovine brain phosphatidylinositol transfer protein and membrane phopsphatidylinositol levels. Biochim. Biophys. Acta, 903, 68–77. [DOI] [PubMed] [Google Scholar]
- van Paridon P.A., Visser,A.J.W.G. and Wirtz,K.W.A. (1987b) Binding of phospholipids to the phosphatidylinositol transfer protein from bovine brain as studied by steady-state and time-resolved fluorescence spectroscopy. Biochim. Biophys. Acta, 898, 172–180. [DOI] [PubMed] [Google Scholar]
- van Tiel C.M., Westerman,J., Paasman,M., Wirtz,K.W.A. and Snoek,G.T. (2000) The protein kinase C-dependent phosphorylation of serine 166 is controlled by the phospholipid species bound to the phosphatidylinositol transfer protein α. J. Biol. Chem., 275, 21532–21538. [DOI] [PubMed] [Google Scholar]
- Verdonk M.L., Cole,J.C. and Taylor,R. (1999) SuperStar: a knowledge-based approach for identifying interaction sites in proteins. J. Mol. Biol., 289, 1093–1108. [DOI] [PubMed] [Google Scholar]
- Voziyan P.A., Tremblay,J.M., Yarbrough,L.R. and Helmkamp,G.M.,Jr (1996) Truncations of the C-terminus have different effects on the conformation and activity of phosphatidylinositol transfer protein. Biochemistry, 35, 12526–12531. [DOI] [PubMed] [Google Scholar]
- Vriend G. (1990) WHAT IF: a molecular modelling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Isupov,M.N. and Murshudov,G.N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D, 57, 122–133. [DOI] [PubMed] [Google Scholar]
- Wirtz K.W.A. (1997) Phospholipid transfer proteins revisited. Biochem. J., 324, 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder M.D, Thomas,L.M., Tremblay,J.M., Oliver,R.L., Yarbrough,L.R. and Helmkamp,G.M.,Jr (2001) Structure of a multifunctional protein. Mammalian phosphatidylinositol transfer protein complexed with phosphatidylcholine. J. Biol. Chem., 276, 9246–9252. [DOI] [PubMed] [Google Scholar]