Abstract
The 2a (polymerase) protein of cucumber mosaic virus (CMV) was shown to be phosphorylated both in vivo and in vitro. In vitro assays using 2a protein mutants and tobacco protein kinases showed that the 2a protein has at least three phosphorylation sites, one of which is located within the N-terminal 126 amino acid region. This region is essential and sufficient for interaction with the CMV 1a protein. When phosphorylated in vitro, the 2a protein N-terminal region failed to interact with the 1a protein. Since the 1a–2a interaction is essential for the replication of CMV, this suggests that phosphorylation of the N-terminal region of the 2a protein negatively modulates the interaction in vivo, and may have a regulatory role acting directly in viral infection.
Keywords: phosphorylation/protein interaction/replicase/virus replication
Introduction
Protein phosphorylation and dephosphorylation, which regulate diverse cellular functions including gene expression in acute response to various extracellular stimuli (Karin and Hunter, 1995; Hardie, 1999), are among the most studied biochemical events in cell signaling. Distinct external stimuli elicit specific transcriptional responses by initiating intracellular signaling that mediates activation of unique protein kinases, which in turn phosphorylate distinct transcription factors (Hill and Treisman, 1995; Cao et al., 1997). Several protein kinases in the plant kingdom have been functionally identified. Mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases are conserved and are involved in resistance signal transduction to plant viruses and stress responses, respectively (Shuqun and Klessig, 1998; Petersen et al., 2000; Saijo et al., 2000; Frye et al., 2001; Yang et al., 2001). Other protein kinases such as Pto and Xa21 themselves have also been referred to as resistance (R) gene products (Song et al., 1995; Sessa et al., 2000).
Phosphorylation may also play a role in regulating cell-to-cell trafficking in plants (Lee and Lucas, 2001) as it does in animals (Lampe et al., 2000). It has been demonstrated that plant viral gene products, such as movement proteins of tobacco mosaic virus and tomato mosaic virus, are phosphorylated in vitro and in vivo (Haley et al., 1995; Karpova et al., 1999; Kawakami et al., 1999; Matsushita et al., 2000; Waigmann et al., 2000). The role of phosphorylation of these movement proteins appears to be to regulate the functions of the movement proteins, by either preventing cell-to-cell movement (Kawakami et al., 1999; Waigmann et al., 2000) or enabling translational dissociation of movement protein– RNA complexes (Karpova et al., 1999). Phosphorylation of the capsid protein of potato virus A downregulates its ability to bind RNA in vitro, suggesting that a plant mechanism regulates the formation of virus particles (Ivanov et al., 2001). Phosphorylation has also been identified as playing a role in either the stability or the function of plant viral-encoded RNA-dependent RNA polymerases (RdRp) or replicases. The viral-encoded component of the RdRp of turnip yellow mosaic virus has been reported to be phosphorylated in insect cells (Hericourt et al., 2000). This phosphorylation occurred at several threonine and serine residues, and was suggested to be a prelude to ubiquitylation and targeted turnover of this protein, which is known to accumulate only to low levels in plant cells. Potential phosphorylation of a viral-encoded protein also has been suggested as the basis for infection of a resistant host (cowpea) by a strain of cucumber mosaic virus (CMV; Kim and Palukaitis, 1997). Resistance in cowpea was shown to be due to nucleotide changes in the 2a gene encoding the viral polymerase subunit of the replicase. Specifically, substitution of a phenylalanine residue (in the 2a protein encoded by the restricted CMV strain Fny) to tyrosine (in the resistance breaking CMV strain B) and substitution of a nearby alanine residue (Fny-CMV) to serine (B-CMV) have been shown to enable Fny-CMV to infect cowpea (Kim and Palukaitis, 1997). Therefore, it was suggested that virus infectivity could be regulated by phosphorylation of the 2a protein. Here we investigated whether the 2a protein of CMV is phosphorylated in vivo and in vitro in order to ascertain whether phosphorylation of CMV-encoded, replicase-associated proteins has a role in the virus infection cycle.
CMV has a tripartite, positive-sense RNA genome of three RNAs designated as RNAs 1–3. RNA 3 encodes two proteins involved in the movement of the virus, while RNAs 1 and 2 each encode one protein involved in replication of the viral genome, designated 1a protein (110 kDa) and 2a protein (97 kDa), respectively (Palukaitis et al., 1992). A small (13 kDa) protein is also encoded by RNA 2, but is not involved in virus replication per se (Ding et al., 1996). The active CMV replicase consists of both 1a and 2a proteins, as well as one or more host factors. This RdRp participates in the synthesis of both double-stranded and single-stranded RNAs, and was isolated and purified from infected tobacco tissue (Hayes and Buck, 1990). The 2a protein has a conserved domain, which shares sequence motifs with many viral RdRp, including the Mg2+-binding GDD motif (Argos, 1988). The N-terminal half of the 1a protein shares common sequence motif with nsP1 protein of Sindbis virus, which has been shown to be involved in RNA capping (Mi and Stollar, 1991). On the other hand, the C-terminal half of the 1a protein has sequence similarity to many viral and cellular helicases (Gorbalenya et al., 1989). The interaction between proteins 1a and 2a has been well studied in brome mosaic virus (BMV), which is taxonomically closely related to CMV. Mutations in the 1a and/or 2a proteins of BMV have been shown to either abolish or markedly reduce RNA replication levels, due to interruption of interactions between the 1a protein and the 2a protein normally leading to the formation of a functional replicase complex (Kao et al., 1992; O’Reilly et al., 1995, 1998). Several lines of evidence indicate that heterologous combinations of 1a and 2a proteins of BMV (and the bromovirus cowpea chlorotic mottle virus) and CMV (and the cucumovirus tomato aspermy virus) fail to interact with each other, demonstrating viral species-specific interactions (Traynor and Ahlquist, 1990; Dinant et al., 1993; O’Reilly et al., 1997; Masuta et al., 1998). The N-terminal 115 amino acids of the BMV 2a protein and the helicase-like region of >50 kDa of the BMV 1a protein are both necessary and sufficient for the 1a–2a protein interaction in the yeast two-hybrid system (Kao and Ahlquist, 1992; O’Reilly et al., 1997).
Our findings show that the CMV 1a and 2a proteins interact in vitro and phosphorylation of the 2a protein inhibits this interaction. This suggests that phosphorylation has a regulatory role on the formation of the replicase complex in vivo. A model describing the effects of phosphorylation of the 2a protein on its functions in CMV infection is discussed.
Results
CMV 2a protein is phosphorylated in vivo and in vitro
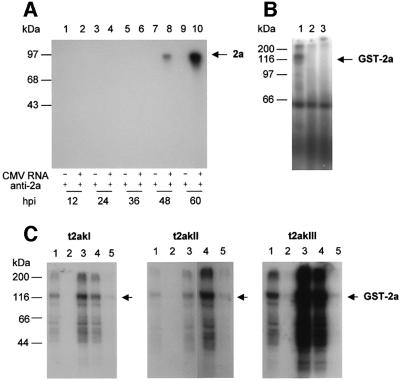
To examine whether the 2a protein was phosphorylated in vivo, tobacco protoplasts were infected by CMV, metabolically labeled with 32P-phosphate and the proteins were subjected to immunoprecipitation analysis using rabbit anti-2a polyclonal antibody. As shown in Figure 1A, phosphorylated 2a protein (97 kDa) accumulated in CMV-infected protoplasts between 48 and 60 h post-inoculation (h.p.i.). 32P-labeled 2a protein could not be detected before 48 h.p.i., although 2a protein accumulation was detected as early as 3 h.p.i. by western blot analysis (Gal-On et al., 1994; Kim and Palukaitis, 1997; data not shown). These data indicate that the 2a protein is a phosphoprotein, but phosphorylation appears to take place late during the replication cycle.
Fig. 1. Phosphorylation of CMV 2a protein in vivo and in vitro. (A) In vivo phosphorylation. Tobacco protoplasts were either not infected (lanes 1, 3, 5, 7 and 9) or infected with total CMV-As RNAs (lanes 2, 4, 6, 8 and 10) by electroporation. The protoplasts were incubated for various times in the presence of [32P]orthophosphate. At the time intervals indicated in hours post-inoculation (h.p.i.), protoplasts were harvested, lysed and proteins were immunoprecipitated using anti-2a antibody. The immunoprecipitated proteins were analyzed by SDS–PAGE and autoradiography. The position of the 2a protein is indicated by an arrow. (B) In vitro phosphorylation. Proteins were solubilized from tobacco membranes and fractionated by chromatography on Q-Sepharose. The bound fraction eluting with 1 M NaCl was used as a source of protein kinases in an in vitro phosphorylation reaction with [γ-32P]ATP, using GST–2a (lane 1), free GST (lane 2) or no added protein (lane 3) as a substrate. The reaction mixtures were analyzed by SDS–PAGE and autoradiography. (C) In vitro phosphorylation. Three tobacco kinases—t2akI (p60), t2akII (p55) and t2akIII (p35)—obtained by hydrophobic interaction chromatography on phenyl–Sepharose followed by anion-exchange chromatography on Q-Sepharose (lane 1), were further fractionated by affinity chromatography on heparin– Sepharose (lanes 2–5). After the flow-through (lane 2), the three protein kinases were each eluted stepwise from the heparin–Sepharose matrix: 0.1–0.6 M NaCl eluate (lane 3); 0.6 M NaCl eluate (lane 4); and 1.5 M eluate (lane 5). Equal volumes of column eluate were used for each assay, and equal volumes of reaction mixture were analyzed by SDS–PAGE and autoradiography. The position of the phosphorylated GST–2a protein is indicated by an arrow in (B) and (C). The positions of molecular weight standards are indicated on the left.
To assay for the in vitro phosphorylation of CMV 2a protein, isolated 2a protein expressed in Escherichia coli was used as a substrate for tobacco protein kinases. The 2a open reading frame (ORF) in RNA 2 of the As strain of CMV was expressed in E.coli (Guan and Dixon, 1991) as a glutathione S-transferase (GST) fusion protein. The GST–2a protein was purified further by glutathione– agarose chromatography and was used as a substrate for the phosphorylation reaction. To identify the protein kinase activity that phosphorylates the 2a protein, soluble extracts as well as membrane-bound extracts that had been solubilized were prepared separately from tobacco leaf tissues and further purified by absorption and affinity chromatography. Both the soluble and solubilized fractions were used in phosphorylation assays. The soluble fraction did not phosphorylate GST–2a protein (data not shown), whereas the solubilized fraction demonstrated phosphorylation of GST–2a protein (Figure 1B, lane 1). The solubilized fraction did not phosphorylate the GST control protein (Figure 1B, lane 2), although there was phosphorylation of an endogenous plant protein of ∼60 kDa (Figure 1B, lane 3). These data suggest that a tobacco membrane fraction contains one or more tobacco kinases capable of phosphorylating the CMV protein 2a. We designated the tobacco 2a protein kinases as t2ak.
The solubilized t2ak activities were subjected to further purification by chromatography on phenyl–Sepharose, Q-Sepharose and heparin–Sepharose, yielding three kinase activities of 60, 55 and 35 kDa, designated t2akI, t2akII and t2akIII, respectively (S.H.Kim, Y.P.Lee, J.B.Sohn and Y.I.Park, manuscript in preparation). The elution profiles from heparin–Sepharose showed that all three kinases were able to phosphorylate GST–2a protein in vitro (Figure 1C).
CMV 2a protein has at least three different phosphorylation sites
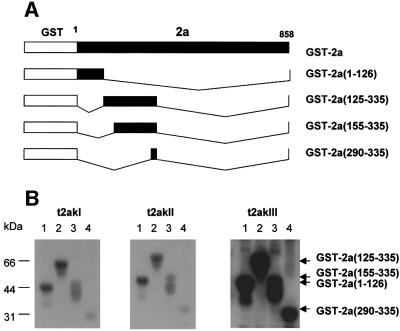
To determine whether there was more than one site of phosphorylation in the entire 2a protein of 858 amino acids, and whether these kinases showed differences in sequence targeting, several types of GST fusion proteins were prepared with truncated versions of the 2a protein (Figures 2A and 3A). These truncated GST–2a fusion proteins were assayed for phosphorylation with each of the three t2aks. As shown in Figure 2B (lane 1), the truncated GST–2a(1–126) fusion protein was phosphorylated by each of the three t2aks. This indicates that the N-terminal fragment of 126 amino acids is sufficient for phosphorylation of the 2a protein. However, the truncated GST–2a(125–335) fusion protein, which lacks the N-terminal fragment of 126 amino acids (Figure 2A), was also phosphorylated by the three t2aks (Figure 2B, lane 2). In the latter case, this phosphorylation domain was delimited to a fragment of 46 amino acids using more truncated fusion proteins, GST–2a(155–335) and GST–2a(290–335) (Figure 2A and B, lanes 3 and 4 in B). These results indicate that at least two phosphorylation sites are present in the N-terminal 335 amino acid region of the 2a protein.
Fig. 2. Localization of the N-terminal in vitro phosphorylation domains of the 2a protein. (A) Schematic representation showing the truncation mutants of the 2a protein fused to GST. Mutants were generated by deletions between restriction enzyme sites in the cDNA clone of CMV RNA 2. The name of the mutant and the amino acid sequences of the 2a protein remaining in the truncated construct are indicated on the right. (B) Phosphorylation analysis of the truncation mutant 2a proteins. The truncated GST–2a fusion proteins were incubated with [γ-32P]ATP and each of the three tobacco 2a kinases (t2akI, t2akII or t2akIII) obtained after affinity chromatography on heparin–Sepharose (eluates in buffer B containing 0.6, 1.5 and 0.6 M NaCl, respectively). Samples: GST–2a(1–126) (lane 1); GST–2a(125–335) (lane 2); GST–2a (155–335) (lane 3); and GST–2a(290–335) (lane 4). The reaction mixtures were analyzed by SDS–PAGE and autoradiography. The positions of molecular weight standards are indicated on the left. The relative positions of the phosphorylated proteins are indicated by arrows on the right.
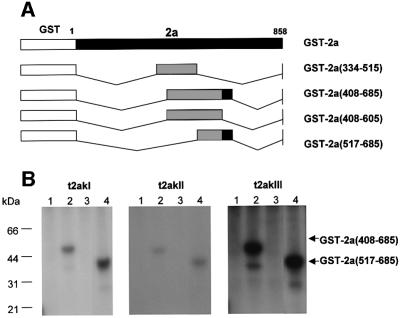
Fig. 3. Localization of the C-terminal in vitro phosphorylation domain of the 2a protein. (A) Schematic representation showing the truncation mutants of the 2a protein fused to GST. Mutants were generated by deletions between restriction enzyme sites in the cDNA clone of CMV RNA 2. The name of the mutant and the amino acid sequences of the 2a protein remaining in the truncated construct are indicated on the right. Regions phosphorylated are shown in black, while regions not phosphorylated are shown in gray. (B) Phosphorylation analysis of the truncation mutant 2a proteins. The truncated GST–2a fusion proteins were incubated with [γ-32P]ATP and each of the three tobacco 2a kinases (t2akI, t2akII or t2akIII) obtained after affinity chromatography on heparin–Sepharose. Samples: GST–2a(334–515) (lane 1); GST–2a (408–685) (lane 2); GST–2a(408–605) (lane 3); and GST–2a(517–685) (lane 4). The reaction mixtures were analyzed by SDS–PAGE and autoradiography. The positions of molecular weight standards are indicated on the left. The relative positions of the phosphorylated proteins are indicated by arrows on the right.
Deletion mutants also were produced by truncating both termini and various parts of the central conserved regions of the 2a protein (Figure 3A). When these truncated fusion proteins were assayed for phosphorylation (Figure 3B; lanes 1–4), only the 2a protein deletion mutants GST–2a(408–685) and GST–2a(517–685) (Figure 3A), which retained the highly conserved core region of the polymerase, i.e. the amino acid region from 605 to 685, were phosphorylated (Figure 3B, lanes 2 and 4). GST–2a(684–858), which contains the C-terminal fragment of 175 amino acids, was not phosphorylated (data not shown). Again, all three t2aks showed the same specificity (Figure 3B), although as observed before, t2akIII showed a higher activity. Therefore, taken together with the results of N-terminal truncated mutants (Figure 2), the 2a protein has at least three sites that can be phosphorylated independently of each other.
Phosphorylation of the 2a protein prevents the formation 1a–2a protein complexes
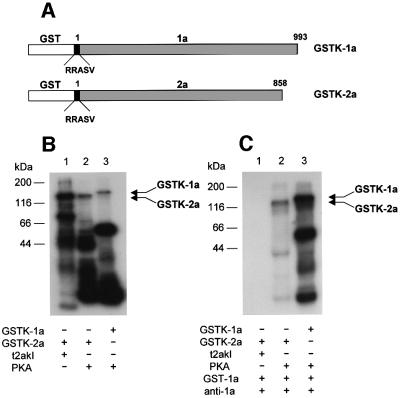
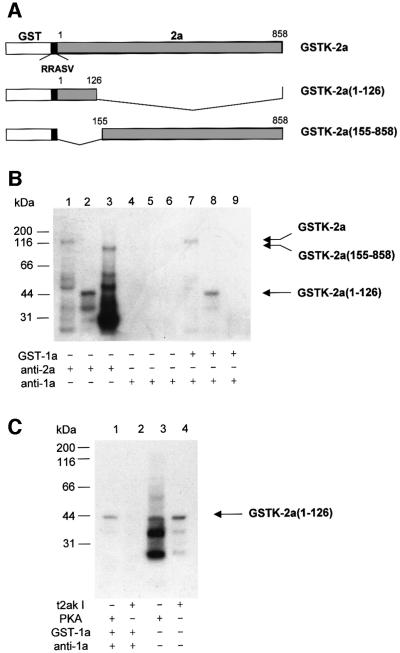
To examine whether the phosphorylation of the 2a protein modulates the interaction between 1a and 2a proteins, we used a co-immunoprecipitation assay with anti-1a antibody and various 32P-labeled 1a or 2a proteins. To obtain GST fusion proteins that were 32P-labeled at the C-terminus of GST rather than on the viral-encoded proteins, the ORFs encoding proteins 1a and 2a were fused to the 3′ end of the GST gene in the vector pGEX-KGK. These fusion proteins contained the pentapeptide sequence RRASV, which is a substrate for the catalytic subunit of cAMP-dependent protein kinase (PKA), introduced into the C-terminus of the GST. The 1a or 2a protein fused to GST containing the pentapeptide substrate sequence for PKA were designated GSTK-1a or GSTK-2a, respectively (Figure 4A). In the experiments below, GSTK-2a phosphorylated by PKA versus t2akI was compared for effects on the interaction with GST–1a. Although t2akIII showed higher levels of specific activity than t2akI (Figures 1C, 2B and 3B), it was thought that t2akIII might be a degradation product of t2akI (our unpublished observations); therefore, t2akI was used in the co-immunoprecipitation analyses.
Fig. 4. Phosphorylation inhibits the in vitro interaction of 1a and 2a proteins. (A) Schematic representation of the GSTK-1a and GSTK-2a fusion proteins. The pentapeptide substrate RRASV, which is a substrate for the catalytic subunit of cAMP-dependent protein kinase (PKA), was introduced into the 3′ end of the GST gene. (B) Phosphorylation analysis of the GSTK fusion proteins. GSTK-2a was phosphorylated in vitro using [γ-32P]ATP and either t2akI (lane 1) or PKA (lane 2), and GSTK-1a was phosphorylated in vitro using [γ-32P]ATP and PKA (lane 3). (C) Co-immunoprecipitation analysis of phosphorylated GSTK-2a and GSTK-1a fusions proteins. GSTK-2a, phosphorylated in vitro by either t2akI (lane 1) or PKA (lane 2), was mixed with unlabeled GST–1a, and incubated with anti-1a serum. GSTK-1a phosphorylated in vitro by PKA (lane 3), was mixed with carrier, unlabeled GST–1a and immunoprecipitated by incubation with anti-1a serum. The reaction mixtures in (B) and the immunoprecipitates formed in (C) were analyzed by SDS–PAGE and autoradiography. The positions of molecular weight standards are indicated on the left. The relative positions of the phosphorylated proteins are indicated by arrows on the right.
The GSTK-2a protein could be phosphorylated in its pentapeptide region by PKA (see below), while GST–1a or GST–2a, without the peptapeptide sequence specific for PKA, were not substrates for phosphorylation by PKA (data not shown). The GSTK-2a protein, phosphorylated at its pentapeptide region by PKA showed labeling of the full-length fusion protein, as well as of several other lower molecular weight proteins (Figure 4B, lane 2). The lower molecular weight proteins are probably derived from the 2a protein, as they are immunoprecipitated by anti-2a antibody (Figure 5B, lane 1). The pattern of labeling was not the same as observed when the GSTK-1a protein was used in place of GSTK-2a protein (Figure 4B, lane 3), or when t2akI was used in place of PKA to phosphorylate the GSTK-2a protein (Figure 4B, lane 1). After phosphorylation by PKA and addition of unlabeled GST–1a protein, the labeled GSTK-2a protein could be co-immunoprecipitated with anti-1a antibody (Figures 4C, lane 2, and 5B, lane 7), demonstrating the formation of 1a–2a protein complexes. Anti-1a antibody did not immunoprecipitate phosphorylated GSTK-2a protein in the absence of GSTK-1a (Figure 5B, lane 4). Thus, phosphorylation of the GST protein by PKA did not interfere with the ability of 1a protein and 2a protein to form a complex. In contrast, GSTK-2a phosphorylated by t2akI did not produce immunoprecipitable complexes after mixing with GSTK-1a and incubation with anti-1a antibody (Figure 4C, lane 1), indicating that the internally phosphorylated GSTK-2a protein was not able to form a complex with 1a protein. These results imply that formation of the replicase holoenzyme of CMV is inhibited by phosphorylation of the 2a protein.
Fig. 5. Phosphorylation inhibits the in vitro interaction of 1a protein with the N-terminal domain of the 2a protein. (A) Schematic representation showing the GSTK-2a fusion protein and truncation mutants of the 2a protein fused to GSTK. The two 2a protein mutants were generated by deletions between restriction enzyme sites in the cDNA clone of CMV RNA 2. The name of the mutant and the amino acid sequences of the 2a protein remaining in the truncated construct are indicated on the right. (B) Co-immunoprecipitation analysis of GSTK-2a and the truncation mutant 2a proteins. GSTK-2a and the truncated GSTK-2a fusion proteins were phosphorylated in vitro by PKA using [γ-32P]ATP. The labeled fusion proteins were incubated with either anti-2a serum (lanes 1–3) or anti-1a serum (lanes 4–6). Alternatively, GST–1a was mixed with the above labeled fusion proteins before anti-1a serum was added to the mixture (lanes 7–9). The immunoprecipitates were collected and analyzed by SDS–PAGE and autoradiography. Labeled fusion protein samples: GSTK-2a (lanes 1, 4 and 7); GSTK-2a(1–126) (lanes 2, 5 and 8); GSTK-2a(155–858) (lanes 3, 6 and 9). (C) Co- immunoprecipitation analysis of GST–1a and phosphorylated GSTK-2a(1–126) fusion proteins. GSTK-2a(1–126) was phosphorylated in vitro by either PKA (lanes 1 and 3) or t2akI (lanes 2 and 4). The labeled proteins were either analyzed directly by SDS–PAGE and autoradiography (lanes 3 and 4), or after sequential incubation with GST–1a and anti-1a serum, and collection of the immunoprecipitates (lanes 1 and 2). In (B) and (C), the positions of molecular weight standards are indicated on the left, and the relative positions of the phosphorylated proteins are indicated by arrows on the right.
The N-terminal 126 amino acid region of 2a protein is sufficient for interaction with 1a protein
Using the yeast two-hybrid system, it was shown that only the N-terminal 115 amino-acid region of the 2a protein of BMV is necessary and sufficient to form a complex with the helicase-like region of the 1a protein (O’Reilly et al., 1995). Therefore, to determine whether phosphorylation of the comparable region in the CMV 2a protein (amino acids 1–126; Figure 2) affected 1a–2a protein complex formation, it was first necessary to establish whether the N-terminal 126 amino acid region of the 2a protein of CMV could interact with the 1a protein.
The in vivo interaction between the CMV 1a and 2a proteins in the yeast two-hybrid system was first established. Yeast cells transformed with both pACT2/1a and pAS2-1/2a showed the induction of a high level of β-galactosidase activity (Table I), indicating that the 2a protein interacted specifically with the 1a protein in yeast cells. Moreover, the yeast strain harboring both pAS2-1/2a and pACT2 vectors was not able to grow in medium lacking Trp, Leu and His (data not shown). This indicates that the 2a protein alone is not able to function as a transcriptional activator. Yeast cells expressing a fusion of the GAL4 activation domain and the 1a protein (from pACT2/1a) and a truncated version of the 2a protein containing only the N-terminal 126 amino-acid region fused to GAL4 DNA binding domain [pBD/2a(1–126)] also induced β-galactosidase activity (Table I). This indicates that an interaction occurred between the 1a protein and the N-terminal 126 amino acids of the 2a protein, although not as strongly as observed between the 1a protein and the entire 2a protein (Table I). Nevertheless, the interaction led a higher activity than the positive controls: the SV40 T antigen and the human p53 protein (Table I). These results are consistent with the previous findings in the BMV system (O’Reilly et al., 1995), and indicate that only the N-terminal 126 amino acids of the 2a protein are necessary and sufficient for interaction with the 1a protein in vivo.
Table I. Summary of interactions between proteins expressed in the yeast two-hybrid system.
| Plasmid combinationsa | Appearance of colonies | Relative β-galactosidase-specific activityb |
|---|---|---|
| pACT2 alone | White | 0.4 |
| pACT2/1a alone | White | 0.1 |
| pAS2-1 alone | White | 0.1 |
| pAS2-1/2a alone | White | 0.3 |
| pACT2 and pAS2-1 | White | 0.3 |
| pACT2/1a and pAS2-1 | White | 0.3 |
| pACT2 and pAS2-1/2a | White | 0.3 |
| pACT2/1a and pAS2-1/2a | Blue | 55 |
| pBD/2a(1–126) alone | White | 0.04 |
| pACT2 and pBD/2a(1–126) | White | 0.2 |
| pACT2/1a and pBD/2a(1–126) | Blue | 7.3 |
| pACT2/p53 and pAS2-1/pSV40 | Blue | 3.2 |
| pACT2/pLaminC and pAS2-1/pSV40 | White | 0.01 |
aPlasmids are described in text and/or Materials and methods.
bSpecific activity is calculated in micromoles of O-nitrophenyl-galactosidase hydrolysed per minute per milligram of protein.
To establish in vitro interaction between the 1a protein and truncated 2a proteins, the GSTK-2a protein, as well as the truncation mutant proteins GSTK-2a(1–126) and GSTK-2a(155–858) (Figure 5A), were labeled with 32P by PKA and co-immunoprecipitated with unlabeled GST–1a protein (Figure 5B). The 32P-labeled GSTK-2a variants were successfully immunoprecipitated with anti-2a antibody (Figure 5B, lanes 1–3). Co-immunoprecipitation using anti-1a antibody showed that GST–1a was associated with both full-length GSTK-2a and GSTK-2a(1–126), but not with GSTK-2a(155–858) (Figure 5B, lanes 7–9). No immune complexes were formed without the addition of unlabeled GST–1a protein (Figure 5B, lanes 4–6). These results indicate that the N-terminal 126 amino acid region of the 2a protein interacts directly with the 1a protein, and the 2a protein sequences beyond amino acid 126 were neither required for the interaction, nor could they themselves form a complex with the 1a protein. The conclusions from the in vitro results (Figure 5B) are consistent with those from the in vivo results (Table I), although the latter results suggest that 2a protein sequences beyond amino acid 126 may act to stabilize the complex formed with the 1a protein.
Phosphorylation of the N-terminal region of 2a protein inhibits the interaction between 1a and 2a proteins
The 2a protein has at least three phosphorylation sites (Figures 2 and 3), and the N-terminal 126 amino acid region of the 2a protein contains at least one phosphorylation site (Figure 2). Therefore, to test whether the phosphorylation of the N-terminal region of the 2a protein could affect the interaction between 1a and 2a proteins, GSTK-2a(1–126) was phosphorylated by t2akI using [γ-32P]ATP (Figure 5C, lane 4) and then mixed with GST–1a protein to induce formation of 1a–2a protein complexes. Co-immunoprecipitation using anti-1a antibody showed that the GSTK-2a(1–126) phosphorylated by t2akI was not able to interact with the 1a protein (Figure 5C, lane 2). In contrast, GSTK-2a(1–126) phosphorylated by PKA (Figure 5B lane 3) did interact with the 1a protein to produce a complex that could be co-immunoprecipitated by anti-1a antibody (Figure 5C, lane 1). These data show that the phosphorylation of the N-terminal 126 amino-acid region of the 2a protein negatively affects the interaction between the 1a and 2a proteins.
Discussion
The work presented here shows that the core polymerase protein of CMV replicase, protein 2a, is phosphorylated in vivo as well as in vitro. In vivo phosphorylation occurs late during infection in protoplasts, at 48 and 60 h.p.i. (Figure 1A). Previous studies have indicated that the 2a protein can be detected in protoplasts as early as 3 h.p.i. and in infected plants between 24 and 36 h.p.i. (Gal-On et al., 1994, 1995; Kim and Palukaitis, 1997). Viral replication is usually detected at 3 or 6 h.p.i. in protoplasts and between 24 and 36 h.p.i. in leaves (Gal-On et al., 1994, 1995; Hellwald and Palukaitis, 1995). The kinetics of phosphorylation indicated that it occurred after much of the viral replication had already taken place. Thus, phosphorylation does not appear to be involved in activating the polymerase in vivo. This is in contrast to the situation with DNA-dependent RNA polymerase II, in which phosphorylation is associated with the transition of polymerase function from initiation to elongation (Payne et al., 1989).
The 2a protein could be phosphorylated in vitro by three different protein kinases solubilized from tobacco membranes (Figures 1B, 2 and 3). The in vitro sites of phosphorylation were delimited to three domains, bordered by amino acids 1–126, 290–335 and 605–685 (Figures 2 and 3). All three domains contain multiple Ser, Thr and Tyr residues, as possible sites of phosphorylation. Phosphorylation domain I, containing amino acids 1–126, was shown here to be sufficient for interaction with the 1a protein, both in vivo (Table I) and in vitro (Figures 4 and 5). Phosphorylation domain II, bordered by amino acids 290–335, represents the beginning of the central region of the polymerase, which is highly conserved in amino acid sequence between CMV strains (Rizzo and Palukaitis, 1988). However, the effects of phosphorylation of this domain remain unknown. Phosphorylation domain III, bordered by amino acids 605–685, includes the highly conserved GDD motif (amino acids 609–611), which is present in all viral RdRp (Poch et al., 1989). This domain also contains the two amino acid positions previously demonstrated to be involved in the activation of a host defense response in cowpea plants infected by Fny-CMV (Kim and Palukaitis, 1997), amino acids 631 and 641. The 2a protein encoded by both As-CMV and Fny-CMV contains Phe and Ala at positions 631 and 641, while the 2a protein encoded by B-CMV contains Tyr and Ser, respectively, at these two positions. However, no difference in the extent of phosphorylation in vitro of the fusion proteins containing phosphorylation domain III [GST–2a(408–685)] was observed for this segment of the 2a protein from As-CMV and Fny-CMV versus B-CMV (data not shown). Thus, it seems unlikely that the t2aks described here are associated with any effect involving phosphorylation of domain III on the hypersensitive response in cowpea (Kim and Palukaitis, 1997).
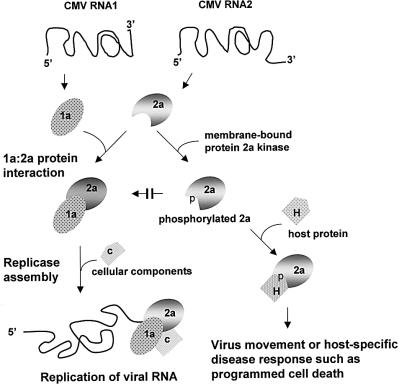
In vitro phosphorylation of the 2a protein was demonstrated to interfere with the ability of the 1a protein to interact with the 2a protein (Figure 4). Moreover, phosphorylation of the N-terminal domain consisting of amino acids 1–126 also interfered with the ability of the 1a protein to interact with this region of the 2a protein (Figure 5). As in vivo phosphorylation of the 2a protein cannot be detected during the exponential phase of virus replication in protoplasts (compare Figure 1 with Gal-On et al., 1994, 1995; Kim and Palukaitis, 1997), what is the significance of the observations concerning in vitro phosphorylation on replicase formation in vivo? We suggest that there may be two effects. These are presented in the diagram in Figure 6 and described below.
Fig. 6. A model proposing the effect of phosphorylation on the functions of the 2a protein. Top: the 1a and 2a proteins are translated from CMV RNAs 1 and 2, respectively. Left path: the 1a protein is membrane localized. The 1a and 2a proteins are capable of direct interaction. Interaction of the 2a protein with the 1a protein leads to its localization on tonoplast membranes. In association with one or more cellular components and viral RNA, the 1a–2a protein complex forms the active CMV replicase, which binds to the ‘+’ strand of the viral RNA and begins the replication cycle. Right path: phosphorylation of the 2a protein occurs at three different domains by various membrane-associated kinases. Phosphorylation in the N-terminal domain of the 2a protein prevents its interaction with the 1a protein, and thus inhibits the formation of new replicase complexes. The N-terminally phosphorylated 2a protein is now available for interaction with other host factors, including those involved in either hindering or promoting virus movement, or host-specific disease responses, such as programmed cell death. The extent of phosphorylation in other domains may influence or limit these additional proposed interactions.
It is know that rate of replication of CMV RNA in transfected protoplasts and inoculated leaves is reduced at the same time as the accumulation the 2a protein either reaches a plateau or begins a decline (Gal-On et al., 1994, 1995; Hellwald and Palukaitis, 1995; Kim and Palukaitis, 1997). This coincides with the maximum level of activity of the CMV RdRp in the inoculated leaves (Jaspars et al., 1985; Hellwald and Palukaitis, 1995). Thus, in susceptible host plants, phosphorylation of the 2a protein does not occur at the beginning of infection, preventing the interaction of the 1a and 2a proteins to form the CMV replicase. It is not known whether such phosphorylation may occur more rapidly in non-host species, preventing the formation of the viral replicase. Certainly, the data here show that the kinases already pre-exist in the susceptible host tobacco, and do not have to be induced by viral infection. It is unlikely that early during infection the 2a protein is protected from phosphorylation by interaction with the 1a protein, since at all times examined there appears to be a molar excess of 2a protein over 1a protein (Gal-On et al., 1994, 1995; Hellwald and Palukaitis, 1995). However, the 2a protein could be protected early during infection by association with host protein(s). Thus, we propose that the change in the rate of CMV RNA replication and level of replicase coincide with the phosphorylation of newly synthesized 2a protein. This phosphorylation of 2a protein would then prevent the formation of new 1a–2a protein complexes (Figure 6), leading to either a plateau or reduction in replicase activity, depending on the rate of turnover of 1a and 2a proteins. Phosphorylation would also lead to the presence of free 2a protein, either leading to its degradation or making it available for other functions. Interestingly, there is a pool of 2a protein free in the cytoplasm of 3-day-infected tobacco plants, not associated with 1a protein, which is all membrane associated (Gal-On et al., 2000). While it has not been shown that this 2a protein is phosphorylated, we suggest that it may be so, and in the model proposed in Figure 6 we speculate that this pool of 2a protein may have another function besides replication. One of these functions may be in activating host defense responses (Kim and Palukaitis, 1997; Canto and Palukaitis, 1999). Another function of phosphorylation of the 2a protein may be associated with virus movement.
A number of resistance genes in plants contain kinase domains (Baker et al., 1997). Specific MAPKs are activated by plant responses to pathogens (Yang et al., 2001; Zhang and Klessig, 2001). Plant defense responses are also altered by MAPKs (Petersen et al., 2000; Frye et al., 2001; Zhang and Klessig, 2001). Thus, phosphorylation plays an important role in defense signaling. The observed phosphorylation of the 2a protein either may be the result of activation of a defense response, or itself may result in activation or negative regulation of defense responses. Previous work has shown that the 2a protein is the target of a resistance mechanism that prevents virus movement into vascular tissue (Hellwald and Palukaitis, 1995; Wintermantel et al., 1997). This resistance mechanism also involves the transgenic expression of either the entire 2a protein, or the N-terminal 590 amino acids of the 858 amino acid 2a protein (Wintermantel and Zaitlin, 2000). In addition, sequences in the phosphorylation domain III have already been identified as interacting with another defense response in cowpea plants, restricting virus movement (Kim and Palukaitis, 1997). Indeed, the phosphorylation of the 2a protein may also be necessary for the change in function from a replicase subunit to a factor promoting movement. Thus, phosphorylation of particular sequences in the 2a protein may promote interactions with other host proteins, leading to either the furtherance or the restriction of CMV infection, depending on the host. Regulation of these different functions may depend on the extent to which phosphorylation occurs within the three identified domains. Determination of the exact sites and extent of phosphorylation of the CMV 2a protein in vivo may establish whether the changes in function correlate with various phosphorylation reactions.
Materials and methods
Plant materials and infection
Tobacco plants (Nicotiana tabacum cv. Xanthi-nc) were mechanically inoculated with CMV strain As, a Korean isolate (obtained from the Plant Virus GenBank, Seoul Women’s University, Seoul, Korea), in 0.1 M sodium phosphate pH 7.0, using Celite 545 as an abrasive and rubbing onto leaves of 2-month-old plants. Plants were grown for a further 15–30 days before harvest. For preparation of proteins, tobacco plants were grown in the greenhouse under the natural light condition. Leaves of 2-month-old tobacco plants were harvested for preparation of protein extract.
cDNA cloning of CMV 1a and 2a genes
One microgram of the As-CMV genomic RNA, in a 20 µl reaction mixture volume, was combined with 10 pmol of an oligonucleotide complementary to the 3′ end 10 nucleotides of all CMV RNAs and containing PstI, SacI and XbaI restriction sites (5′-GAGAGGATCCTGCAGAGCTCTAGACCGCGGACGCGTGGTCTCCTT-3′). The RNAs and primer, in a mixture containing 50 mM Tris–HCl pH 8.3, 10 mM MgCl2, 10 mM β-mercaptoethanol, 100 mM KCl, 20 U of RNase inhibitor, and 15 U of AMV reverse transcriptase (US Biochemicals), were incubated at 42.5°C for 1 h. A 5 µl portion of first strand cDNA mixture was then subjected to the PCR with 0.475 µM of the above primer and 0.5 µM of 5′ primers specific to either the 1a gene (5′-GAGACCCGGGGCATATGGCGACGTCCTCGGTTCAAC-3′) or the 2a gene (5′-GAGACCCGGGGCATATGGCTTTCCCGCCCCC-3′), respectively, in a total volume of 50 µl. The reaction mixture also contained 10 mM Tris–HCl pH 8.8, 50 mM KCl, 1.5 mM MgCl2, 200 µM dNTP and 1.25 U of Taq polymerase. The thermocycling reaction was programmed as follows: 95°C for 1 min, 98°C for 20 s and 68°C for 5 min (25 cycles); and 72°C for 30 min. The amplified DNAs were digested with either SmaI and SacI, or SmaI and XbaI, and inserted into pGEX-KG (Guan and Dixon, 1991) to generate plasmids pKG1a and pKG2a, respectively, using standard procedures (Sambrook et al., 1989). Alternatively, the amplified DNAs were digested with SmaI and PstI and cloned into pQE30 (Qiagen).
Preparation of antibodies and recombinant proteins
The anti-1a and anti-2a antibodies were raised in rabbits against His6-tagged 1a and 2a proteins of As-CMV, purified from E.coli strain XL1-Blue containing full-length 1a and 2a ORFs in pQE30 (Qiagen), respectively. The antibodies were purified from crude serum by chromatography on protein A–Sepharose. To obtain soluble forms of the 1a and 2a proteins, the 1a and 2a ORFs were cloned into either pGEX-KG (Guan and Dixon, 1991) or pGEX-KGK. pGEX-KGK was generated from pGEX-KG by the introduction of a DNA fragment encoding the pentapeptide sequence (RRASV), which is a substrate for PKA, into the 3′ end of the GST gene. The GST-fused 1a and 2a proteins then were expressed in E.coli strain BL21 and purified by glutathione–agarose (Sigma) chromatography (Guan and Dixon, 1991).
Labeling of GSTK fusion proteins
One hundred nanograms of GSTK fusion proteins purified from E.coli were incubated in either 20 or 40 µl of kinase buffer [20 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM dithiothreitol (DTT), 12.5 mM MgCl2], including 1 µCi/µl [γ-32P]ATP and 1 U/µl PKA, at 4°C for 30 min (Blanar and Rutter, 1992). The reactions were terminated by the addition of sodium phosphate pH 8.0, sodium pyrophosphate and EDTA, to final concentrations of 10 mM each. Unincorporated, free [γ-32P]ATP was removed by chromatography through a Sephadex G-25 spin column equilibrated in 20 mM Tris–HCl pH 8.0, 1 mM EDTA, 100 mM NaCl, 0.1% NP-40 and 1 mM phenylmethylsulfonyl fluoride (PMSF).
Preparation and analysis of tobacco protoplast
Protoplasts were prepared from tobacco leaves and inoculated with CMV RNAs as described previously (Gal-On et al., 1994, 1995). The protoplasts were washed and then incubated with constant light in incubation medium containing 250 µCi/ml of [32P]orthophosphoric acid (6000 Ci/mmol; DuPont-NEN Research Products, Boston, MA) for either 12, 24, 36, 48 or 60 h. The metabolic labeling was stopped by rinsing the protoplasts with incubation medium and the 32P-labeled protoplasts were stored at –80°C. Total proteins were extracted by vortexing the protoplasts in extraction buffer [20 mM Tris–HCl pH 8.0, 10 mM EDTA, 10 mM EGTA, 25 mM NaF, 10 mM benzamidine, 10 mM sodium orthovanadate, 0.3% (v/v) 2-mercaptoethanol and 1 mM PMSF] containing 1% Triton X-100, intermittently for 1 h on ice. The extracts were centrifuged at 15 000 g for 20 min, and chromatographed through Sephadex G-25 spin columns equilibrated with immunoprecipitation buffer [50 mM Tris–HCl pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.02% NaN3]. Purified rabbit anti-2a antibody was added to 20 µg/ml, and immune complexes were precipitated by co-incubation with protein A–Sepharose. The precipitates were collected by centrifugation, were rinsed with 1000 vols of immunoprecipitation buffer and were analyzed by electrophoresis on a 7% SDS–polyacrylamide gel (Schägger and von Jagow, 1987). The gel was exposed to X-ray film between two sheets of intensifying screen at –70°C for 1 h.
Preparation of plant protein extracts
One hundred grams of tobacco leaf tissue were ground in liquid nitrogen with a mortar and pestle. The resulting powder was stirred in 2.5 vols of extraction buffer (see above), before centrifugation at 40 000 g for 20 min (Ti50.2; Beckman). The supernatant containing soluble proteins was removed. The pellet was resuspended in 200 ml of extraction buffer containing 2 M KCl and stirred on ice for 1 h. The resulting extracts containing solubilized membrane proteins were clarified by centrifugation.
Phenyl–Sepharose high-performance hydrophobic interaction chromatography
Two hundred milliliters of membrane fraction were applied onto a 20 ml phenyl–Sepharose High Performance column (1.5 × 14 cm; Pharmacia, Piscataway, NJ) that had been equilibrated with extraction buffer, at a flow-rate of 1 ml/min. Unbound proteins were washed with 10 column vols of elution buffer A [20 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM benzamidine HCl, 1 mM PMSF] containing 2 M KCl at the same flow rate. Bound proteins were eluted with a reverse gradient of KCl from 2 to 0 M in 200 ml of elution buffer A, collecting every 5 ml fraction. Elution was completed with sequential flow of 60 ml elution buffer A and 60 ml 60% (v/v) ethylene glycol in elution buffer A.
Q-Sepharose high performance anion-exchange chromatography
The phenyl–Sepharose pool was loaded onto a 10 ml Q-Sepharose High Performance column (1.6 × 6 cm) equilibrated with elution buffer A at a flow rate of 1 ml/min. The column was washed with 10 column vols of elution buffer A containing 0.1 M KCl, and the bound proteins were eluted with a linear gradient of 0.1–0.5 M KCl in elution buffer A.
Heparin–Sepharose affinity chromatography
The Q-Sepharose pool was loaded onto a 5 ml heparin–Sepharose Fast Flow column (1.5 × 3.5 cm) equilibrated with elution buffer B [50 mM MES pH 7.3, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM benzamidine HCl, 1 mM PMSF] at a flow rate of 0.5 ml/min. The column was washed with 10 column vols of elution buffer B containing 0.1 M NaCl, and the bound proteins were eluted stepwise with two column vols of 0.6, 1 and 1.5 M NaCl, all in elution buffer B.
Phosphorylation assay
One hundred nanograms of GST-fused, As-CMV viral proteins were incubated in phosphorylation buffer [20 mM HEPES–NaOH pH 7.4, 12 mM MgCl2, 1 mM DTT] containing 2 µCi of [γ-32P]ATP (3000–6000 Ci/mmol; DuPont-NEN Research Products) in a 20 µl reaction volume. The reaction was initiated by adding 20 U of t2ak(s) purified from tobacco plant and incubation at 25°C for 15 min. (one t2ak unit equals one PKA unit, i.e. one unit transfers 1 pmol of [γ-32P]ATP to hydroxylated protein in 30 min at 4°C.) The reaction was terminated by boiling in SDS sample buffer for 2 min, and was analyzed by electrophoresis on 8% SDS–polyacrylamide gels and autoradiography.
Co-immunoprecipitation analysis
GSTK-2a was phosphorylated either by t2ak (Q-Sepharose or heparin–Sepharose pool) or PKA, as described above. The phosphorylated GST–2a was chromatographed through Sephadex G-25 equilibrated with 20 mM Tris–HCl pH 8.0, 1 mM EDTA, 100 mM NaCl, 1 mM DTT and 0.1% NP-40, before an equal amount of GST–1a was added. After 1 h, rabbit anti-1a antibody was added to the mixture, which was then incubated at 4°C for 1 h. The immune complex was precipitated by co-incubation with 20 µl of protein A–Sepharose (Pharmacia) for 1 h. The protein A–Sepharose precipitates then were collected by centrifugation, washed with several changes of the above buffer, and analyzed on 7% SDS–polyacrylamide gels. After electrophoresis, the gels were dried and exposed to X-ray film for 8 h at –80°C.
Yeast two-hybrid analysis
The gene encoding the CMV 1a protein was fused to the gene encoding the GAL4 activation domain in the plasmid pACT2. The genes encoding the CMV 2a and 2a(1–126) proteins were fused to the genes encoding the GAL4 DNA-binding domain in the plasmids pAS2-1 and pBD-GAL4, respectively. Competent cells of Saccharomyces cerevisiae strain Y190 were prepared and transformed by the use of the lithium acetate method of Gietz et al. (1992). Yeast transformants were plated on solid SD/–Leu/–Trp/–His/+50 mM 3AT plates. After 7 days, yeast colonies were transferred onto Whatman No. 5 filter paper. Snap-frozen filters were soaked for 1 h in a buffer containing 50 mM sodium phosphate pH 7.0, 10 mM KCl, 1 mM MgCl2, 35 mM 2-mercaptoethanol and 15 µg/ml of X-gal. The presence of an interaction was identified by the appearance of blue colonies (Iwabuchi et al., 1993; Li and Fields, 1993). Quantitative assays of β-galactosidase-specific activities were determined by the method described by O’Reilly et al. (1995). Genes encoding the SV40 large T antigen (pSV40) and the human p53 were inserted into pAS2-1 and pACT2, respectively, and used as positive interaction controls. The human lamin C gene (pLamin C) was cloned downstream of the GAL4AD in the plasmid pACT2, and was used as a non-interactive control.
Acknowledgments
Acknowledgements
The authors thank Mr Young Pyo Lee for excellent technical assistance. This work was supported by Basic Research Grant from Korea Science and Engineering Foundation (KOSEF) to Y.I.P. P.P. is supported by a grant-in-aid from the Scottish Executive Environment and Rural Affairs Department.
References
- Argos P. (1988) A sequence motif in many polymerases. Nucleic Acids Res., 16, 9909–9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B., Zambryski,P., Staskawicz,B. and Dinesh-Kumar,S.P. (1997) Signaling in plant–microbe interactions. Science, 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Blanar M.A. and Rutter,W.J. (1992) Interaction cloning: identification of a helix–loop–helix zipper protein that interact with cFos. Science, 256, 1014–1018. [DOI] [PubMed] [Google Scholar]
- Canto T. and Palukaitis,P. (1999) The hypersensitive response to cucumber mosaic virus in Chenopodium amaranticolor requires movement outside the initially infected cell. Virology, 265, 74–82. [DOI] [PubMed] [Google Scholar]
- Cao H., Glazebrook,J., Clarke,J.D., Volko,S. and Dong,X. (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell, 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Dinant S., Janda,M., Kroner,P.A. and Ahlquist,P. (1993) Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J. Virol., 67, 7181–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.W., Shi,B.J., Li,W.X. and Symons,R.H. (1996) An interspecies hybrid RNA virus is significantly more virulent than either parental virus. Proc. Natl Acad. Sci. USA, 93, 7470–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C.A., Tang,D. and Innes,R.W. (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl Acad. Sci. USA, 98, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-On A., Kaplan,I., Roossinck,M.J. and Palukaitis,P. (1994) The kinetics of infection of zucchini squash by cucumber mosaic virus indicates a function for RNA 1 in virus movement. Virology, 205, 280–289. [DOI] [PubMed] [Google Scholar]
- Gal-On A., Kaplan,I. and Palukaitis,P. (1995) Differential effects of satellite RNA on the accumulation of cucumber mosaic virus RNAs and their encoded proteins in tobacco vs. zucchini squash with two strains of helper virus. Virology, 208, 58–66. [DOI] [PubMed] [Google Scholar]
- Gal-On A., Canto,T. and Palukaitis,P. (2000) Characterization of genetically modified cucumber mosaic virus expressing histidine-tagged 1a and 2a proteins. Arch. Virol., 145, 37–50. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin,E.V., Donchenko,A.P. and Blinov,V.M. (1989) Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res., 17, 4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K.L. and Dixon,J.E. (1991) Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione-S-transferase. Anal. Biochem., 192, 262–267. [DOI] [PubMed] [Google Scholar]
- Haley A., Hunter,T., Kiberstis,P. and Zimmern,D. (1995) Multiple serine phosphorylation sites on the 30 kDa TMV cell-to-cell movement protein synthesized in tobacco protoplasts. Plant J., 8, 715–724. [DOI] [PubMed] [Google Scholar]
- Hardie D.G. (1999) Plant protein serine/threonine kinases: classification and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol., 50, 97–131. [DOI] [PubMed] [Google Scholar]
- Hayes R.J. and Buck,K.W. (1990) Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell, 63, 363–368. [DOI] [PubMed] [Google Scholar]
- Hellwald K.-H. and Palukaitis,P. (1995) Viral RNA as a potential target for two independent mechanisms of replicase-mediated resistance against cucumber mosaic virus. Cell, 83, 937–946. [DOI] [PubMed] [Google Scholar]
- Hericourt F., Blanc,S., Redeker,V. and Jupin,I. (2000) Evidence for phosphorylation and ubiquitinylation of the turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus-insect cell system. Biochem. J., 349, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C.S. and Treisman,R. (1995) Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell, 80, 199–211. [DOI] [PubMed] [Google Scholar]
- Ivanov K.I., Puustinen,P., Merits,A., Saarma,M. and Mäkinen,K. (2001) Phosphorylation down-regulates the RNA binding function of the coat protein of potato virus A. J. Biol. Chem., 276, 13530–13540. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K., Li,B., Bartels,P. and Fields,S. (1993) Use of the 2-hybrid system to identify the domain of p53 involved in oligomerization. Oncogene, 8, 1693–1696. [PubMed] [Google Scholar]
- Jaspars E.M.J., Gill,D.S. and Symons,R.H. (1985) Viral RNA synthesis by a particulate fraction from cucumber seedlings infected with cucumber mosaic virus. Virology, 144, 410–425. [DOI] [PubMed] [Google Scholar]
- Kao C.C. and Ahlquist,P. (1992) Identification of the domains required for interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J. Virol., 66, 7293–7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C.C., Quadt,R., Hershberger,R.P. and Ahlquist,P. (1992) Brome mosaic virus RNA replication proteins 1a and 2a form a complex in vitro. J. Virol., 66, 6322–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. and Hunter,T. (1995) Transcriptional control by protein phosphorylation: signal transmission from cell surface to the nucleus. Curr. Biol., 5, 747–757. [DOI] [PubMed] [Google Scholar]
- Karpova O.V., Rodionova,N.P., Ivanov,K.I., Kozlovsky,S.V., Dorokhov,Y.L. and Atabekov,J.G. (1999) Phosphorylation of tobacco mosaic virus movement protein abolishes its translation repressing ability. Virology, 261, 20–24. [DOI] [PubMed] [Google Scholar]
- Kawakami S., Padgett,H.S., Hosokawa,D., Okada,Y., Beachy,R.N. and Watanabe,Y. (1999) Phosphorylation and/or presence of serine 37 in the movement protein of tomato mosaic tobamovirus is essential for intracellular localization and stability in vivo. J. Virol., 73, 6831–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.-H. and Palukaitis,P. (1997) The plant defense response to cucumber mosaic virus in cowpea is elicited by the viral polymerase gene and affects virus accumulation in single cells. EMBO J., 16, 4060–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe P.D., TenBroek,E.M., Burt,J.M., Kurata,W.E., Johnson,R.G. and Lau,A.F. (2000) Phosphorylation of connexin43 on serine368 by protein kinase C regulates gap junctional communication. J. Cell Biol., 149, 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y. and Lucas,W.J. (2001) Phosphorylation of viral movement proteins — regulation of cell-to-cell trafficking. Trends Microbiol., 9, 5–8. [DOI] [PubMed] [Google Scholar]
- Li B. and Fields,S. (1993) Identification of mutation in p53 that affect its binding to SV40 T antigen by using yeast two-hybrid system. FASEB J., 7, 957–963. [DOI] [PubMed] [Google Scholar]
- Masuta C., Ueda,S., Suzuki,M. and Uyeda,I. (1998) Evolution of a quadripartite hybrid virus by interspecific exchange and recombination between replicase components of two related tripartite RNA viruses. Proc. Natl Acad. Sci. USA, 95, 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita Y., Hanazawa,K., Yoshioka,K., Oguchi,T., Kawakami,S., Watanabe,Y., Nishiguchi,M. and Nyunoya,H. (2000) In vitro phosphorylation of the movement protein of tomato mosaic tobamovirus by a cellular kinase. J. Gen. Virol., 81, 2095–2102. [DOI] [PubMed] [Google Scholar]
- Mi S. and Stollar,V. (1991) Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology, 184, 423–427. [DOI] [PubMed] [Google Scholar]
- O’Reilly E.K, Tang,N., Ahlquist,P. and Kao,C.C. (1995) Biochemical and genetic analyses of the interaction between the helicase-like and polymerase-like proteins of the brome mosaic virus. Virology, 214, 59–71. [DOI] [PubMed] [Google Scholar]
- O’Reilly E.K., Paul,J.D. and Kao,C.C. (1997) Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J. Virol., 71, 7526–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly E.K., Wang,Z., French,R. and Kao,C.C (1998) Interactions between the structural domains of the RNA replication proteins of plant-infecting RNA viruses. J. Virol., 72, 7160–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palukaitis P., Roossinck,M.J., Dietzgen,R.G. and Francki,R.I.B. (1992) Cucumber mosaic virus. Adv. Virus Res., 41, 281–348. [DOI] [PubMed] [Google Scholar]
- Payne J.M., Laybourn,P.J. and Dahmus,M.E. (1989) The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIA. J. Biol. Chem., 264, 19621–19629. [PubMed] [Google Scholar]
- Petersen M. et al. (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell, 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Poch O., Sauvaget,I., Delarue,M. and Tordo,N. (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding elments. EMBO J., 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo T.M. and Palukaitis,P. (1988) Nucleotide sequence and evolutionary relationships of cucumber mosaic virus (CMV) strains: CMV RNA 2. J. Gen. Virol., 69, 1777–1787. [DOI] [PubMed] [Google Scholar]
- Saijo Y., Hata,S., Kyozuka,J., Shimamoto,K. and Izui,K. (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J., 23, 319–327. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schägger H. and von Jagow,G. (1987) Tricine sodium dodecyl sulfate polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 kDa to 100 kDa. Anal. Biochem., 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Sessa G., D’Ascenzo,M. and Martin,G.B. (2000) Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J., 19, 2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuqun Z. and Klessig,D.F. (1998) Resistance gene N-mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl Acad. Sci. USA, 95, 7433–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.Y. et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science, 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Traynor P. and Ahlquist,P. (1990) Use of bromovirus RNA2 hybrids to map cis- and trans-acting functions in a conserved RNA replication gene. J. Virol., 64, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E., Chen,M.-H., Bachmaier,R., Ghoshroy,S. and Citovsky,V. (2000) Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J., 19, 4875–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermantel W.M. and Zaitlin,M. (2000) Transgene translatability increases the effectiveness of replicase-mediated resistance to cucumber mosaic virus. J. Gen. Virol., 81, 587–595. [DOI] [PubMed] [Google Scholar]
- Wintermantel W.M., Banerjee,N., Oliver,J.C., Paolillo,D.J. and Zaitlin,M. (1997) Cucumber mosaic virus is restricted from entering minor veins in transgenic tobacco exhibiting replicase-mediated resistance. Virology, 231, 248–257. [DOI] [PubMed] [Google Scholar]
- Yang K.-Y., Liu,Y. and Zhang,S. (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl Acad. Sci. USA, 98, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. and Klessig,D.F. (2001) MAPK cascades in plant defense signaling. Trends Plant Sci., 6, 520–527. [DOI] [PubMed] [Google Scholar]