Abstract
Ribosome recycling factor (RRF) together with elongation factor G (EF-G) disassembles the post- termination ribosomal complex. Inhibitors of translocation, thiostrepton, viomycin and aminoglycosides, inhibited the release of tRNA and mRNA from the post-termination complex. In contrast, fusidic acid and a GTP analog that fix EF-G to the ribosome, allowing one round of tRNA translocation, inhibited mRNA but not tRNA release from the complex. The release of tRNA is a prerequisite for mRNA release but partially takes place with EF-G alone. The data are consistent with the notion that RRF binds to the A-site and is translocated to the P-site, releasing deacylated tRNA from the P- and E-sites. The final step, the release of mRNA, is accompanied by the release of RRF and EF-G from the ribosome. With the model post-termination complex, 70S ribosomes were released from the post-termination complex by the RRF reaction and were then dissociated into subunits by IF3.
Keywords: antibiotics/elongation factor G/protein synthesis/RRF/translocation
Introduction
After the translation termination step in prokaryotes, ribosome recycling factor (RRF) and elongation factor G (EF-G) disassemble the post-termination ribosomal complex of ribosome, deacylated tRNA and mRNA (for reviews, see Janosi et al., 1996a,b; Kaji and Hirokawa, 2000; Kaji et al., 2001). The disassembly requires GTP hydrolysis (Hirashima and Kaji, 1972; Ogawa and Kaji, 1975). These conclusions were mostly derived from experiments using a model post-termination complex, puromycin-treated polysome. In this system, the release of ribosome from mRNA was measured by following the conversion of polysome to monosome (70S ribosome). The conclusions obtained with this model post-termination complex were confirmed by a system with a natural termination complex formed at the amber codon situated at the seventh codon of the coat cistron of phage R17 (Ogawa and Kaji, 1975).
The crystal structure of Thermotoga maritima RRF has been solved and was shown to be a near perfect mimic of tRNA. RRF has two domains, I and II, corresponding to the anticodon and acceptor arms of tRNA, respectively. On the basis of this structural similarity between tRNA and RRF, it was postulated that RRF might be translocated like tRNA on the ribosome during the disassembly of the post-termination complex (Selmer et al., 1999).
In this study, we examined the effects of various inhibitors on the release of tRNA and mRNA from the model post-termination complex by RRF and EF-G, and propose that RRF is not only a near perfect mimic of tRNA but also performs a functional mimicry on the ribosome. The release of tRNA partially took place with EF-G alone, but the release of mRNA was strictly dependent on both RRF and EF-G. The dissociation constant of RRF for the 50S subunit was ∼2 × 10–6 M. IF3 was shown to dissociate the 70S ribosomes released by RRF and EF-G from the model post-termination complex.
Results
Release of ribosome-bound tRNA by the concerted action of RRF and EF-G or by EF-G alone
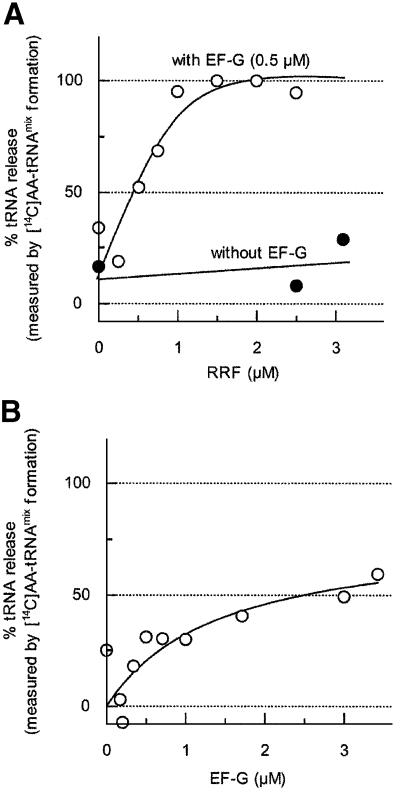
As shown in Figure 1, we examined the dose-dependent release of tRNA by RRF. It is clear from this figure that the release of tRNA took place in an RRF dose-dependent manner. More importantly, RRF and EF-G released all of the bound tRNA during the disassembly reaction. When the complex was incubated with 1.5 µM RRF and 0.5 µM EF-G, almost all of the ribosome-bound deacylated tRNA was released in 15 min. Most of the ribosomes in naturally occurring polysome isolated from growing Escherichia coli are known to have peptidyl-tRNA in the P-site and deacylated tRNA in the E-site (the A-site is empty) (Remme et al., 1989; Stark et al., 1997). During our preparation of polysome, tetracycline is present in the crude extract to ensure that the A-site is empty. In fact, puromycin removed 100% of the ribosome-bound peptidyl group from our polysome, indicating that all of the peptidyl-tRNA was on the P-site (Hirashima and Kaji, 1972). In other words, all of the ribosomes were in the post-translocation state with the A-site empty. From Figure 1, we conclude that RRF and EF-G release tRNA from both the P- and E-sites. This figure also shows another important point that RRF alone does not release tRNA.
Fig. 1. Release of deacylated tRNA from the model post-termination complex by RRF and EF-G. Release of deacylated tRNA from the model post-termination complex (1.0 A260 units) was examined as described in Materials and methods. (A) Release of tRNA by various concentrations of RRF in the presence (open circles) or absence (closed circles) of EF-G (0.5 µM). (B) Release of tRNA by various concentrations of EF-G alone. The percentage tRNA release relative to the total number of deacylated tRNAs bound to the model post-termination complex (1838 c.p.m., measured by the aminoacylation with [14C]amino acids mixture) was plotted against the amount of added RRF (A) or EF-G (B).
As shown in Table I, the release of deacylated tRNA from the model complex by RRF and EF-G took place with GMPPCP (a non-hydrolyzable GTP analog) almost as well as with GTP. This is understandable because a single round of tRNA translocation by EF-G occurs even in the presence of GMPPCP (Inoue-Yokosawa et al., 1974; Rodnina et al., 1997) and this will permit the release of tRNA by a single round of translocation of RRF. Although GMPPCP is known as a translocation inhibitor because it fixes EF-G on the ribosome (Kuriki et al., 1970), this action will not influence the release of tRNA by RRF that involves a single round of translocation. In contrast, as shown in Table I, GDP did not release tRNA. GDP has been reported not to promote normal translocation (Inoue-Yokosawa et al., 1974; Rodnina et al., 1997), although complete translocation, albeit slow, was observed with GDP under certain conditions (Rodnina et al., 1997). In this context, the data shown in Table I are consistent with the notion that RRF is translocated from the A- to the P-site by EF-G and releases the ribosome-bound tRNA by this movement.
Table I. Requirement for GTP or GTP analog for the release of tRNA from the model post-termination complex by RRF and EF-G.
| Nucleotides | [14C]AA-tRNAmix formeda (c.p.m.) |
|---|---|
| GTP | 1859.5 ± 132.0 |
| GDP | 330.2 ± 22.2 |
| GMPPCP | 1846.6 ± 315.5 |
| No nucleotides | 310.8 ± 141.9 |
| Total tRNA | 1756.0 ± 113.0 |
aRelease of tRNA from the model post-termination complex (1.5 A260 units) by RRF (1.5 µM) and EF-G (0.5 µM) was examined as in Figure 1A, except that 0.4 mM GTP, GDP, GMPPCP or no nucleotide was used. The average ± SEM are shown.
Figure 1B shows that tRNA is released partially in the presence of EF-G alone. Since the release of mRNA, measured by the conversion of puromycin-treated polysome to monosome, does not take place with EF-G alone (Hirashima and Kaji, 1973), the data presented in this figure show that the release of tRNA during the disassembly reaction is not tightly coupled with the release of mRNA from the ribosome. More importantly, it shows that the primary function of RRF and EF-G in the disassembly reaction is the release of mRNA, because the release of tRNA, though a prerequisite for the release of mRNA, can be achieved partially by EF-G without RRF.
Effect of EF-G inhibitors on the release of tRNA—evidence for a translocation-like movement of RRF
Our early work (Ishitsuka et al., 1970) as well as work from other laboratories (Lucas-Lenard and Haenni, 1969; Roufa et al., 1970; Inoue-Yokosawa et al., 1974; Spirin, 1985; Rodnina et al., 1997) established that the release of tRNA from the ribosome is an indication of the translocation of tRNA. The translocation of ribosome-bound tRNA by EF-G (Agrawal et al., 1998; Frank and Agrawal, 2000; Stark et al., 2000) can be detected by measuring the release of the ribosome-bound deacylated tRNA. As discussed in the Introduction and the Discussion, the near perfect structural mimicry of tRNA by RRF (Selmer et al., 1999) strongly suggests a functional mimicry of tRNA by RRF. As shown below, this expectation was met in that the release of tRNA by RRF and EF-G has every characteristic expected of the concept that RRF is translocated on the ribosome by EF-G.
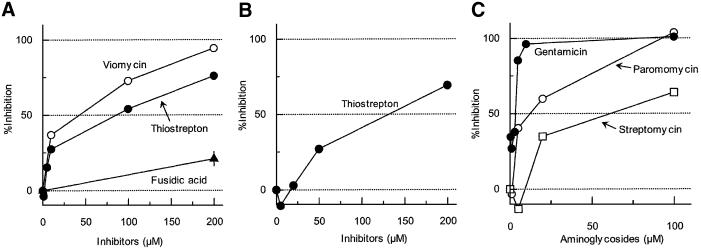
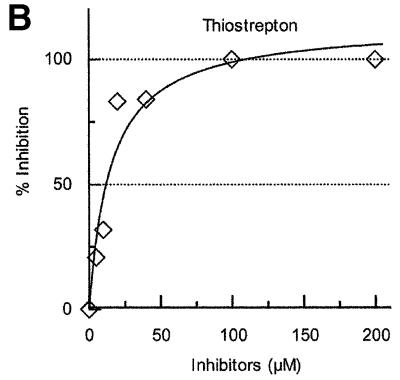
Thiostrepton is known to prevent translocation of tRNA by EF-G (Hausner et al., 1988; Rodnina et al., 1997, 1999). It has been reported that GTP hydrolysis is inhibited by this antibiotic (Cundliffe, 1990) by preventing the binding of EF-G/GTP to the ribosome (Bodley et al., 1970). In contrast, a recent report suggests that it actually permits the binding of EF-G and GTP hydrolysis but inhibits the release of produced inorganic phosphate and hence translocation (Rodnina et al., 1999). Thus, one can obtain a ribosomal complex with EF-G in the pre-translocation state fixed by thiostrepton (Stark et al., 2000). As shown in Figure 2A, thiostrepton inhibited the release of tRNA from the post-termination complex by RRF and EF-G. Approximately 90 µM of thiostrepton inhibited the tRNA release ∼50% (Figure 2A and Table II).
Fig. 2. Effects of antibiotics on the release of tRNA from the model post-termination complex. (A) Effects of EF-G/translocation inhibitors on tRNA release by RRF and EF-G. Effects of viomycin (open circles), thiostrepton (closed circles) and fusidic acid (closed triangles) on the release of tRNA by RRF (1.5 µM) and EF-G (0.5 µM) were examined as in Figure 1A. (B) Effect of thiostrepton on tRNA release by EF-G only. The effect of thiostrepton on the release of tRNA by EF-G (2.4 µM) alone was examined as in Figure 1B. (C) Effects of aminoglycosides on tRNA release by RRF and EF-G. The effects of paromomycin (open circles), gentamicin (closed circles) and streptomycin (open squares) on the release of tRNA by RRF (1.5 µM) and EF-G (0.5 µM) were examined as in Figure 1A.
Table II. Comparison of concentrations of inhibitors required for 50% inhibition of the release of tRNA and mRNA from the model post-termination complex by RRF and EF-G.
| Inhibitors | Concentration required for 50% inhibition |
|
|---|---|---|
| tRNA release | mRNA release | |
| Thiostrepton | 92.3 µM | 12.5 µM |
| Viomycin | 58.3 µM | 25.0 µM |
| Fusidic acid | No inhibition | 10.0 µMa |
| Paromomycin | 11.4 µM | 1.2 µM |
| Gentamicin | 2.1 µM | 0.5 µM |
| Streptomycin | 68.5 µM | 25.0 µMa |
| Tobramycin | Not tested | 3.7 µM |
| Neomycin | Not tested | 1.7 µM |
| G418 | Not tested | 63.0 µM |
| Kanamycin | Not tested | 20.4 µM |
aThese values were estimated from Hirashima and Kaji (1973).
Viomycin is also known to prevent translocation of tRNA (Modolell and Vazquez, 1977; Hausner et al., 1988), but the GTP hydrolysis is not inhibited by this antibiotic (Rodnina et al., 1997). Like thiostrepton, viomycin also inhibited the release of tRNA by RRF and EF-G (Figure 2A). The concentration of viomycin for 50% inhibition was estimated to be ∼60 µM (Table II).
Fusidic acid, an effective inhibitor of the elongation step (Pestka, 1968; Tanaka et al., 1968), is known to inhibit neither hydrolysis of EF-G-bound GTP (Bodley et al., 1969) nor translocation of aminoacyl-tRNA by EF-G (Inoue-Yokosawa et al., 1974). However, after the hydrolysis and the translocation, the release of EF-G is inhibited by this antibiotic (Bodley et al., 1969). Therefore, it permits a single round of tRNA translocation but prevents multiple rounds. As shown in Figure 2A, release of ribosome-bound tRNA by RRF and EF-G took place even with high concentrations of fusidic acid. This is what one would expect following the hypothesis that the release of tRNA by RRF and EF-G is due to a single round of a translocation-like movement of RRF by EF-G on the ribosome.
As shown in the preceding section, a significant amount of tRNA was released in the presence of EF-G alone. Figure 2B shows that this release of tRNA by EF-G alone was equally sensitive to the translocation inhibitor thiostrepton. Another inhibitor, viomycin, was also inhibitory to this release of tRNA by EF-G (data not shown). Therefore, the release of tRNA observed in Figures 1B and 2B can be regarded as a result of the translocation of mRNA on the ribosome. It should be mentioned here again that the model post-termination complex used here has both the P- and E-sites occupied but the A-site is empty. The data presented in Figures 1B and 2B therefore establish an important new concept that EF-G can release the P- and/or E-site-bound deacylated tRNA from the ribosome even if the A-site is empty. This is in sharp contrast to the well-accepted concept that the A-site has to be filled with tRNA for the release of tRNA by EF-G from the ribosome programmed with synthetic polynucleotides containing poly(U) (Ishitsuka et al., 1970; Holschuh et al., 1980).
Aminoglycosides inhibit tRNA release from the model post-termination complex
Our early work (Igarashi et al., 1969) as well as work from other laboratories (Cabanas et al., 1978; Misumi et al., 1978; Hausner et al., 1988) showed that streptomycin, one of the aminoglycosides, inhibits translocation. It has also been shown to inhibit disassembly of the model post-termination complex as measured by the release of ribosome from mRNA (Hirashima and Kaji, 1973). Streptomycin causes misreading during translation (Davies et al., 1964; Ruusala and Kurland, 1984). The misreading caused by aminoglycosides is at the step of binding of aminoacyl-tRNA to the programmed ribosome (Kaji and Kaji, 1965) due to the decreased dissociation rate of tRNA from the A-site (Karimi and Ehrenberg, 1994; Pape et al., 2000).
As shown in Figure 2C, 50% inhibition of the release of tRNA by RRF and EF-G was observed in the presence of 2 µM gentamicin or 11 µM paromomycin. It is noted that streptomycin was the weakest of all these aminoglycosides tested (∼70 µM was required for 50% inhibition) and this compares well with the equally weak inhibitory effect on the release of mRNA found in our early work (Hirashima and Kaji, 1973) (see Table II). These results also suggest that during the disassembly of the model post-termination complex, a translocation-like movement of RRF occurs on the ribosome, resulting in the release of tRNA from the ribosome.
Effect of EF-G inhibitors on the release of mRNA from ribosomes
In this section, we show the effect of various concentrations of inhibitors on the release of mRNA. The important conclusion derived from this series of experiments shown in Figure 3 is that the release of tRNA is a prerequisite for the release of mRNA that is the major function of RRF. As shown in Figure 3A, fusidic acid and GMPPCP, which did not influence the release of tRNA (see Figure 2A and Table I), inhibited the release of mRNA from the ribosome. Since both of these inhibitors are known to fix EF-G on the ribosome, these data strongly suggest that the release of EF-G from the ribosome is required for the release of mRNA by RRF and EF-G.
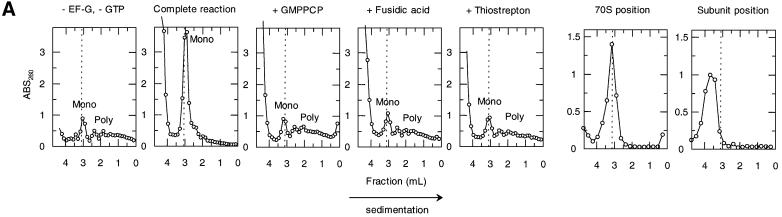
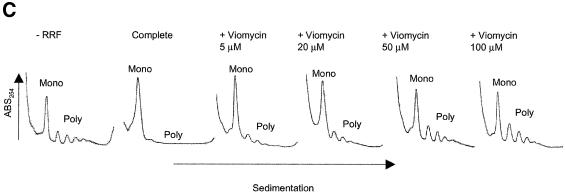
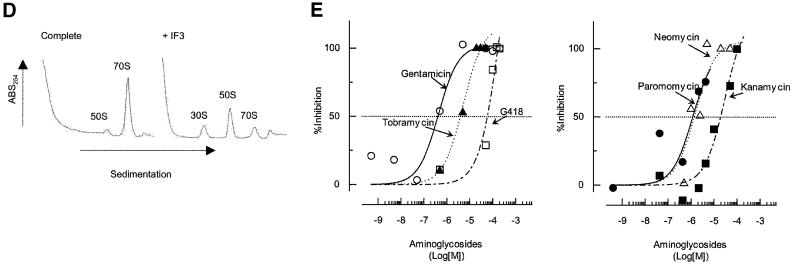
Fig. 3. Effects of antibiotics on the release of mRNA from ribosome by RRF and EF-G. (A) Effects of fusidic acid, GMPPCP (EF-G release inhibitors) and thiostrepton. The release of ribosome from the model substrate was examined by measuring the conversion of puromycin-treated polysome (marked as Poly) to monosome (marked as Mono) as described in Materials and methods except that 760 pmol of RRF and EF-G, and 3.2 A260 units of polysome were used. The ribosome sedimentation pattern was analyzed by 15–30% sucrose density gradient ultracentrifugation (Beckman SW50.1, 40 000 r.p.m., 75 min). Fractions were taken from the bottom of the tube and the A260 was measured. Sedimentation is from left to right. The first and second panel from the left show the control (minus EF-G) and complete reaction mixture, respectively. Note that in the control, the major portion of the ribosomes were polysomes, while the complete mixture showed that the polysomes were converted to the monosome. The effect of 370 µM GMPPCP (third panel from the left), 200 µM fusidic acid (fourth panel) and 20 µM thiostrepton (fifth panel) are shown. In the experiment where the effect of GMPPCP was tested, GTP was omitted. The positions of 70S ribosomes (6th panel) and subunits (7th panel) were determined by sedimentation of washed ribosome (2.0 A260 units) either in buffer R or in buffer R containing 8 mM EDTA, respectively. Note that the background level of these panels was completely flat, indicating that the polysomes observed in the first, third, fourth and fifth panels were significantly above the background. (B) Effect of thiostrepton. Effect of various amounts of thiostrepton on the release of mRNA from the model post-termination complex was examined as in (A). (C) Effect of various concentration of viomycin. Experimental conditions were the same as (A) except that A254 was measured continuously by an ISCO UA-6 spectrophotometer (ISCO). Sedimentation was from left to right. (D) Absence of ribosome subunit formation during the RRF reaction—effect of IF3. Left trace: the experimental conditions were identical to those of (C) except that the ultracentrifugation time was increased to 240 min. Right trace: the same as the left panel except that 100 µg of IF3 was added. (E) Effect of aminoglycosides. The conditions were the same as (B) except that an ISCO UA-6 spectrophotometer was used. Gentamicin (open circles), tobramycin (closed triangles), G418 (open squares), paromomycin (closed circles), neomycin (open triangles) and kanamycin (closed squares) were tested. In (B) and (E), 88% of polysomes were converted to monosome by RRF and EF-G without inhibitor, and this value was set as 100% conversion.
As shown in Figure 3B and C, the translocation inhibitors thiostrepton and viomycin also inhibited the release of mRNA. Concentrations of thiostrepton and viomycin required to give 50% inhibition of mRNA release were estimated to be ∼12.5 and 25.0 µM, respectively. This is less than those required for the inhibition of tRNA release (summarized in Table II). Table II shows that, without any exception, concentrations required to inhibit tRNA release are higher than those that inhibit mRNA release. These results show that the release of tRNA appears to be a necessary but not sufficient condition for the release of mRNA. The extreme left panel of Figure 3A and the extreme left panel of Figure 3C establish that neither RRF nor EF-G alone release mRNA from the post-termination complex, respectively. In the second panels (from the left) of Figure 3A and C, very few if any ribosomal subunits were observed. These panels show that disassembly is complete, yet there is no peak at the position where the subunits are sedimented (see the extreme right panel of Figure 3A for the positions of subunits). This point was strengthened further in the next experiment shown in Figure 3D.
In Figure 3D, the same experiments as in Figure 3A and C were performed but the centrifugation time was increased to show that there were no significant amounts of subunits present after the RRF reaction (left panel). Furthermore, when IF3 was added to the RRF reaction mixture, most of the released 70S ribosomes were dissociated into their subunits (right panel). We conclude that in our system the major products, if not all, of the RRF reaction are in the form of 70S ribosomes that can be dissociated into their subunits by IF3. It appears therefore that the ‘recycled ribosome’ is dissociated by IF3 into subunits, which are ready to engage in a new round of translation as suggested in an early work (Subramanian et al., 1968).
Effect of aminoglycosides on the release of mRNA from ribosomes
Figure 3E shows the effect of various concentrations of aminoglycosides on the release of mRNA from the model post-termination complex. As shown in this figure, all aminoglycosides tested showed inhibitory effects on the release of mRNA from the ribosome. Since all of them inhibit translocation, these data, together with their effect on the release of tRNA, further support the notion that RRF is translocated on the post-termination complex by EF-G. Gentamicin is an effective inhibitor of this reaction, with 50% inhibition at ∼0.5 µM. This is comparable with but less than the concentration required for 50% inhibition of tRNA release (2 µM) (Figure 2C and Table II). Likewise, the effectiveness of paromomycin on the release of tRNA (50% inhibition at 11 µM) was similar but less than that on the release of mRNA (50% inhibition at 1 µM). These data are consistent with the concept that the release of tRNA precedes the release of mRNA in the disassembly of the model post-termination complex. It is noted in Table II and Figure 3E that other aminoglycosides, tobramycin, kanamycin, G148 and neomycin, are all inhibitory to the release of mRNA, supporting the notion that aminoglycosides in general are inhibitory to the action of RRF and EF-G. This result further supports the notion that the disassembly catalyzed by RRF involves a translocation-like movement of RRF.
Determination of the binding constant of RRF to the 50S ribosomal subunits
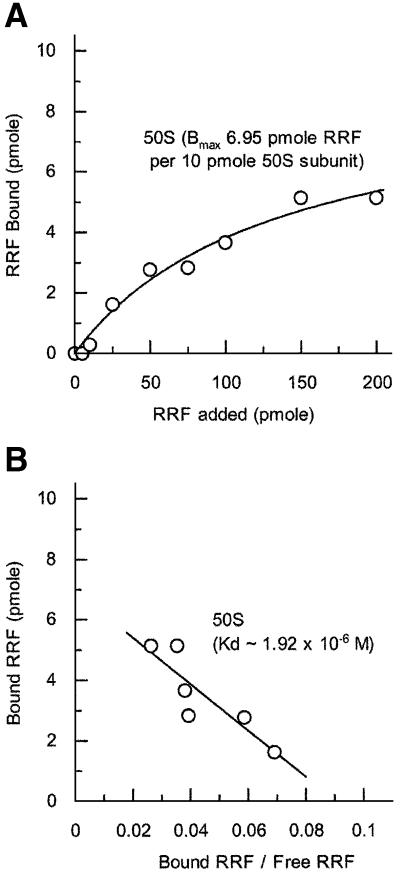
As noted in the Discussion, it is important to measure the binding constant of RRF to the subunits of the ribosome in order to explain the observation that the ribosome may be dissociated into subunits during the RRF reaction on the post-termination complex of mRNA with high affinity for the ribosome (Karimi et al., 1999). In the experiments described in Figure 4, various amounts of RRF were mixed with the 50S subunits and the bound RRF was determined (Figure 4A). The dissociation constant of RRF from 50S ribosomes was estimated to be 1.92 × 10–6 M (Figure 4B). Since the amount of bound RRF was measured after isolation of the RRF–ribosome complex by the microcon technique, the off rate of bound RRF must be much slower than the on rate. In a similar way, the complex of aminoacyl-tRNA with the programmed ribosome can be isolated through sucrose gradients (Kaji and Kaji, 1963) or by the millipore filter technique (Nirenberg and Leder, 1964). Therefore, the dissociation constant calculated above does not represent the true dissociation constant. Rather, the value obtained in this fashion represents a value proportionally related to the true dissociation constant. In fact, the dissociation constant of tRNA from the tRNA–ribosome complex measured after isolation of the complex was comparable to the value obtained without isolation of the complex (Lill et al., 1986). It is important to point out that the binding of RRF to 30S subunits was too low to calculate a dissociation constant (data not shown). This is in contrast to the report that RRF binds to 30S subunits as detected by the BIACORE technique, which also detected binding of RRF to 50S subunits (Ishino et al., 2000). We refrained from using this technique because our preliminary results suggested that modification of RRF often changes the behavior of the native RRF with regard to its interaction with the ribosome.
Fig. 4. Binding of RRF to the 50S ribosomal subunit. (A) The binding of RRF (at the indicated amounts) to 10 pmol of the 50S ribosomal subunit was examined as described in Materials and methods. Approximate Kd values were determined from the Scatchard plot (B) of the data shown in (A).
Discussion
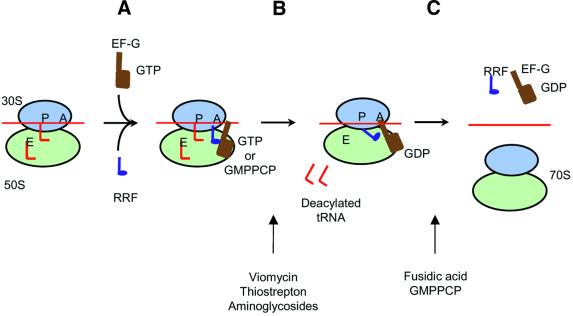
In this study, we examined the effects of various inhibitors on the disassembly of the post-termination complex by RRF and EF-G to reveal the molecular process of the disassembly reaction. Based on these data, we propose the possible action of RRF for the disassembly of a typical post-termination complex as shown in Figure 5.
Fig. 5. Model for disassembly of the post-termination complex by EF-G and RRF. (A) RRF binds to the A-site of the post-termination complex. EF-G binds to the factor-binding site. (B) RRF is translocated (or makes a certain movement, see Discussion) by EF-G, resulting in the release of tRNA from the complex. Two tRNAs, one each from the P- and E-sites, are presumed to be released in this step. This step is sensitive to thiostrepton, viomycin and aminoglycosides. (C) The next step is the release of mRNA, RRF and EF-G. EF-G release is required for the release of mRNA from the ribosome, hence fusidic acid and GMPPCP inhibit this step.
In the first step (step A), RRF binds to the A-site of the post-termination complex. In support of this concept, the RRF reaction is inhibited, though weakly, by tetracycline, an A-site-specific inhibitor (Hirashima and Kaji, 1973; Kaji and Hirokawa, 2000). In addition, RRF competes with the peptide release factor, which presumably binds to the A-site (Pavlov et al., 1997). At this step, EF-G binds to the factor binding site of the ribosome.
Next (step B), we propose that RRF goes through a ‘translocation-like’ motion to occupy the P-site with the help of EF-G, resulting in the release of deacylated tRNA bound at the P- and E-sites [our model post-termination complex contains tRNA on the P- and E-sites (Remme et al., 1989; Stark et al., 1997) but the A-site is empty (Hirashima and Kaji, 1973)]. In this figure, we depict that the release of the P-site-bound tRNA by RRF is performed while a portion of RRF still keeps the A-site occupied. The rationale for this concept is that it is important for RRF to keep aminoacyl-tRNA away from the A-site during the disassembly reaction. Failure to do so would make disassembly very difficult.
Alternatively, the protection of the A-site against the incoming EF-Tu–aminoacyl-tRNA complex may be achieved in two different ways. First, EF-G occupies the factor-binding site until the complete disassembly occurs. Since the factor-binding site of the 70S ribosome is shared by both EF-G and EF-Tu (Moazed et al., 1988; Agrawal et al., 2000), this would prevent the A-site from being occupied by aminoacyl-tRNA. Another possibility is that the release of the tRNA is followed rapidly by the release of mRNA, so that there is no opportunity for the unwanted incoming aminoacyl-tRNA to occupy the A-site vacated by RRF. It should be pointed out that in Figure 5, RRF is not shown to occupy the E-site. This is based on our studies on the binding of RRF in relation to that of deacylated tRNA to the E-site (G.Hirokawa, M.C.Kiel, A.Muto, K.Igarashi and A.Kaji, in preparation).
Data consistent with the notion that RRF is translocated on the ribosome are as follows. (i) Release of mRNA from the model post-termination complex is accompanied by the release of tRNA (Figures 1 and 2). Our work (Ishitsuka et al., 1970) and that of others (Lucas-Lenard and Haenni, 1969; Roufa et al., 1970; Inoue-Yokosawa et al., 1974; Spirin, 1985; Rodnina et al., 1997) established that translocation can be observed by following the release of tRNA from the ribosome. (ii) Disassembly of the model post-termination complex takes place only when two factors, EF-G and RRF, are present simultaneously (Hirashima and Kaji, 1973). (iii) Inhibitors of translocation such as thiostrepton, viomycin and aminoglycosides inhibit the release of tRNA (Figure 2 and Table II) and mRNA (Figure 3 and Table II). (iv) Fusidic acid or GMPPCP do not inhibit the release of tRNA (Figure 2A and Table I), whereas they inhibit the release of mRNA (Figure 3A). This is because fusidic acid and GMPPCP allow for one round of translocation (Inoue-Yokosawa et al., 1974; Rodnina et al., 1997). Multiple rounds of translocation are inhibited by these agents because they fix EF-G on the ribosome (Bodley et al., 1969; Kuriki et al., 1970).
The release of tRNA must take place prior to the release of mRNA for three reasons. First, EF-G alone caused partial release of tRNA (Figure 1B) while the release of mRNA from the ribosome is strictly dependent on the presence of both EF-G and RRF (Figure 3). Secondly, every inhibitor that inhibits the release of tRNA also inhibits the release of mRNA at similar but lower concentrations (Table II). Thirdly, no inhibitor was found that stops tRNA release but does not stop mRNA release.
The last step of the disassembly is the release of mRNA as shown in Figure 5. We propose that EF-G must be released from the ribosome in order for the release of mRNA to occur. We arrive at this conclusion because fusidic acid and GMPPCP, which inhibit the release of EF-G from the ribosome, inhibit the release of mRNA (Figure 3A). In this last step, or in the earlier ‘translocation-like’ step, movement of domain I relative to domain II of RRF may take place. The relative movement of domains I and II of RRF was first suggested by the observation that the contact area of domains I and II is unusually small (Selmer et al., 1999). This notion was supported by our recent determination of the solution structure of RRF by NMR (Yoshida et al., 2001) as well as structural determination of other RRFs (Kim et al., 2000; Toyoda et al., 2000), which confirmed the structure determined by Selmer et al. (1999). It is possible that a specific interaction of RRF with EF-G (Rao and Varshney, 2001) may cause this intramolecular movement, resulting in the release of mRNA, tRNA and, finally, RRF from the ribosome. Since mRNA is tacked between the subunits (Shatsky et al., 1991; Noller et al., 2000; Yusupov et al., 2001) around the neck of the 30S subunit (Frank et al., 2000; Yusupova et al., 2001), the release of mRNA must require the concerted structural changes of the ribosome by EF-G (Frank and Agrawal, 2000) in addition to the flexing of the two domains of RRF. As shown in Figure 5, RRF is removed from the ribosome, probably by EF-G, during this process (M.C.Kiel and A.Kaji, unpublished observation).
We should point out that the above steps described in Figure 5 represent a typical mode of disassembly of a typical post-termination complex. These steps may not occur with certain types of mRNA. One example of such an mRNA is the short synthetic mRNA with a strong Shine–Dalgarno (SD) sequence and homopolynucleotides near the termination codon (Karimi et al., 1999). With such an mRNA, neither tRNA nor mRNA is released from the ribosomes. Instead, the ribosomes are dissociated into their subunits during the RRF reaction. In the mRNA used by Karimi et al. (1999), the strong SD sequence and homopolymer are close enough to the termination codon (Steitz, 1969; Yusupova et al., 2001) that the ribosome of the post-termination complex is influenced by these mRNA sequences. We propose that these two elements in the mRNA confer high affinity for the ribosome and make the P-site-bound tRNA difficult to be released by RRF and EF-G. As discussed above, the release of tRNA is a necessary requirement for the release of mRNA. Therefore, under these unusual conditions, mRNA and tRNA remain on the ribosome.
There are several observations that support the notion that homopolymers in the mRNA make the affinity of the mRNA for the ribosome unusually high. First, it is well known that one does not need any soluble factors to synthesize polypeptides in vitro under the direction of homopolymers (Pestka, 1969; Gavrilova and Spirin, 1974), while factors are necessary for the binding of natural mRNA to the ribosome. Secondly, as shown in Figure 1B, EF-G alone releases ∼50% of the bound tRNA from our model post-termination complex, which has deacylated tRNA at the P- and E-sites but no tRNA at the A-site. In contrast to these natural mRNAs, the release of tRNA by EF-G from the homopolymer ribosomal complex with deacylated tRNA is dependent on the presence of an A-site-bound tRNA (Ishitsuka et al., 1970; Holschuh et al., 1980). Thirdly, the ribosome can slide along natural mRNA in vivo for as far as 45 nucleotides without making polypeptides (Table 5 of Janosi et al., 1998). No such sliding has been reported with an in vitro system using synthetic homopolymers. Fourthly, RRF and EF-G do not release deacylated tRNA from the complex of homopolymer, ribosome and tRNA (M.C.Kiel and A.Kaji, unpublished observation).
Turning our attention to the strong SD sequence, it is well accepted that this sequence is designed to bind to the ribosome. In support of the notion that the SD sequence makes it harder for the ribosome to slide along mRNA, we mention here two observations. First, the ‘in vivo sliding of the ribosome’ mentioned above is effectively stopped by the presence of an SD sequence and an AUG (Figure 7 of Janosi et al., 1998). Secondly, the release of tRNA by EF-G from the natural initiation complex with a strong SD sequence requires tRNA on the A-site (Roufa et al., 1970). This is in contrast to the EF-G-dependent release of tRNA from the puromycin-treated polysome having the A-site empty (Figure 1B). In other words, mRNA of the naturally occurring polysome represents an average typical mRNA that is more easily moved by EF-G without an A-site-bound tRNA. This concept that the conditions required for the movement of the ribosome-bound mRNA or tRNA by EF-G are dependent on the mRNA sequence is of general importance for understanding the ribosome slippage observed in vivo (Janosi et al., 1998). We should emphasize, however, that the function of EF-G remains the same during the slippage of the ribosome; it is still an enzyme promoting ‘translocation’ of the ribosome-bound mRNA.
Regarding the subunit dissociation during the disassembly reaction, we focus attention on the fact that RRF has a significant affinity for the 50S subunit (Figure 4). Based on this information, we postulate that RRF anchors itself on the 50S subunit and ‘lifts’ mRNA using its ‘footing’ on the 50S subunit. If the 30S subunit is tightly bound to mRNA due to the presence of tRNA, this action would ‘lift’ the 30S subunit together with mRNA, resulting in subunit dissociation because RRF has a footing on the 50S subunit. This has indeed been observed with high affinity mRNA (Karimi et al., 1999).
Another example of the case where the scheme shown in Figure 5 may not apply is the unusual situation where the termination codon is shared with the initiation codon as UAAUG. One such example is the behavior of the ribosome in translating RNA phage GA coat and lysis protein genes. In this system, the upstream ribosome is responsible exclusively for the translation of the downstream open reading frame (lysis gene). It appears that at this junction, 25% of released ribosomes are re-captured precisely at the AUG codon through the help of RRF. RRF is essential for correct initiation from the AUG codon (Inokuchi et al., 2000). A similar situation may exist at the junction between tryptophan synthetase A and B (Oppenheim and Yanofsky, 1980).
Materials and methods
Release of tRNA from the model post-termination complex
Release of tRNA from the model post-termination complex was examined as described in Hirashima and Kaji (1973) with modifications as follows. Polysome (0.6–1.8 A260 units) isolated from tetracycline-treated E.coli Q13 cells (Hirashima and Kaji, 1972) was incubated with 275 µM puromycin, RRF, EF-G and 0.4 mM GTP in 550 µl of buffer R [10 mM Tris–HCl, pH 7.4, 8.2 mM MgSO4, 80 mM NH4Cl, 0.14 mM dithiothreitol (DTT)] at 30°C for 15 min. The released tRNA was separated from the ribosomal complex either by nitrocellulose membrane (Millipore, φ13 mm) or by Microcon 100 filtration (Millipore, 330 g). The ribosome-bound filter was washed once more by 550 µl of buffer R and the wash was combined with the first eluate. Released tRNA thus obtained was concentrated either by Microcon 10 (Millipore, 14 000 g) or by ethanol precipitation with 20 µg of salmon testis DNA as a carrier, and the concentrated tRNA was aminoacylated with 0.15 µCi of [14C]amino acid mixture (Amersham, CFB25) according to Momose and Kaji (1965). The cold (4°C) trichloroacetic acid-insoluble radioactivity thus formed was regarded as a mixture of [14C]aminoacyl-tRNA ([14C]AA-tRNAmix) formed from deacylated tRNA released from the model post-termination complex.
To obtain the value of total tRNA bound to the model post-termination complex, polysome was incubated with 275 µM puromycin in buffer R at 30°C for 8 min, and further incubated in the presence of 18.5 mM EDTA for 7 min. Released tRNA was separated and [14C]aminoacylated as described above except that the filter was washed with a buffer (10 mM Tris–HCl, pH 7.4, 80 mM NH4Cl, 0.14 mM DTT). The exact amount of released tRNA could not be estimated, because neither the precise specific activity of commercially available [14C]amino acids mixture nor the purity of the commercial deacylated tRNA mixture was known.
Release of mRNA from the model post-termination complex
Release of mRNA from puromycin-treated polysome was examined as described in Hirashima and Kaji (1973). Briefly, polysome (0.5–1 A260 unit) (Hirashima and Kaji, 1972) was incubated with 1 µg of RRF, 108 µg of EF-G, 0.37 mM GTP and 275 µM puromycin in buffer R at 30°C for 15 min. The sedimentation profile of the ribosomes and the remaining polysomes in sucrose density gradient centrifugation (15–30% sucrose in buffer R; Beckman SW50.1 or Hitachi p55ST-2 rotor, 40 000 r.p.m., 75 min, 4°C) was analyzed either by A254 measurement with an ISCO UA-6 spectrometer or by A260 measurement of a manually fractionated sample. The conversion was expressed by measuring the area corresponding to the 70S ribosome and calculating the percentage of the total amount of ribosomes including residual polysomes before and after the RRF reaction. Where indicated, IF3 as prepared according to Shimizu et al. (2001) was added.
Binding of RRF to ribosomes
Washed ribosomes were prepared from MRE 600 cells as described in Kaji et al. (1966). Ribosomal subunits were prepared as described in Spedding (1990). RRF was incubated with ribosome in 40 µl of buffer BD [20 mM Tris–HCl, pH 7.4, 10 mM Mg(OAc)2, 25 mM KCl, 1 mM DTT] at 30°C for 10 min. The mixture was applied onto Microcon 100 (Millipore) and centrifuged for 5 min at 3000 g. The micro-spin column was washed with 40 µl of the same buffer and centrifuged again for 12 min at 3000 g. The washing was repeated again. Then, 40 µl of the buffer was added on the filter and the column was inverted after 1 min incubation at room temperature. The ribosome-bound RRF was recovered by centrifugation (1 min, 3000 g) of the inverted column. The ribosome-bound RRF was detected by western blot with rabbit antiserum against E.coli RRF (1:15 000 dilution). Bound RRF was quantified from the blotting of known amounts of RRF standards.
Acknowledgments
Acknowledgements
We thank Dr Takuya Ueda, University of Tokyo for providing the expression plasmid encoding the His-tagged IF3 gene, Dr Sidney Pestka, Robert Wood Johnson Medical School for providing the ribosomal subunits, and Dr Akikazu Hirashima, Yakult Pharmaceutical Inc. Co. and Dr Yoshio Inokuchi, Teikyo University for helpful discussions. This work was partially supported by NIH grant 1-R01-GH-60429-01A1 (to A.K.) and by Nippon Paint Research fund (to H.K.).
References
- Agrawal R.K., Penczek,P., Grassucci,R.A. and Frank,J. (1998) Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl Acad. Sci. USA, 95, 6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Heagle,A.B. and Frank,J. (2000) Studies of elongation factor G-dependent tRNA translocation by three-dimensional cryo-electron microscopy. In Garret,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 53–62.
- Bodley J.W., Zieve,F.J., Lin,L. and Zieve,S.T. (1969) Formation of the ribosome–G factor–GDP complex in the presence of fusidic acid. Biochem. Biophys. Res. Commun., 37, 437–443. [DOI] [PubMed] [Google Scholar]
- Bodley J.W., Lin,L. and Highland,J.H. (1970) Studies on translocation. VI. Thiostrepton prevents the formation of a ribosome–G factor– guanine nucleotide complex. Biochem. Biophys. Res. Commun., 41, 1406–1411. [DOI] [PubMed] [Google Scholar]
- Cabanas M.J., Vazquez,D. and Modolell,J. (1978) Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem. Biophys. Res. Commun., 83, 991–997. [DOI] [PubMed] [Google Scholar]
- Cundliffe E. (1990) Recognition sites for antibiotics within rRNA. In Hille,W.E., Dahlberg,A., Garett,R.A., Moore,P.B., Schlessinger,D. and Warner,J.R. (eds), The Ribosome: Structure, Function and Evolution. American Society for Microbiology Press, Washington, DC, pp. 479–490.
- Davies J., Gilbert,W. and Gorini,L. (1964) Streptomycin, suppression and the code. Proc. Natl Acad. Sci. USA, 51, 883–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. and Agrawal,R.K. (2000) A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature, 406, 318–322. [DOI] [PubMed] [Google Scholar]
- Frank J., Penczek,P., Grassucci,R.A., Heagle,A.B., Spahn,C.M.T. and Agrawal,R.K. (2000) Cryo-electron microscopy of the translational apparatus: experimental evidence for the paths of mRNA, tRNA and the polypeptide chain. In Garret,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 45–51.
- Gavrilova L.P. and Spirin,A.S. (1974) Non-enzymatic translation. Methods Enzymol., 30, 452–462. [PubMed] [Google Scholar]
- Hausner T.P., Geigenmuller,U. and Nierhaus,K.H. (1988) The allosteric three-site model for the ribosomal elongation cycle. New insights into the inhibition mechanisms of aminoglycosides, thiostrepton and viomycin. J. Biol. Chem., 263, 13103–13111. [PubMed] [Google Scholar]
- Hirashima A. and Kaji,A. (1972) Factor-dependent release of ribosomes from messenger RNA. Requirement for two heat-stable factors. J. Mol. Biol., 65, 43–58. [DOI] [PubMed] [Google Scholar]
- Hirashima A. and Kaji,A. (1973) Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J. Biol. Chem., 248, 7580–7587. [PubMed] [Google Scholar]
- Holschuh K., Bonin,J. and Gassen,H.G. (1980) Mechanism of translocation: effect of cognate transfer ribonucleic acids on the binding of AUGUn to 70S ribosomes. Biochemistry, 19, 5857–5864. [DOI] [PubMed] [Google Scholar]
- Igarashi K., Ishitsuka,H. and Kaji,A. (1969) Comparative studies on the mechanism of action of lincomycin, streptomycin and erythromycin. Biochem. Biophys. Res. Commun., 37, 499–504. [DOI] [PubMed] [Google Scholar]
- Inokuchi Y., Hirashima,A., Sekine,Y., Janosi,L. and Kaji,A. (2000) Role of ribosome recycling factor (RRF) in translational coupling. EMBO J., 19, 3788–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue-Yokosawa N., Ishikawa,C. and Kaziro,Y. (1974) The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J. Biol. Chem., 249, 4321–4323. [PubMed] [Google Scholar]
- Ishino T. et al. (2000) Interaction of ribosome recycling factor and elongation factor EF-G with E.coli ribosomes studied by the surface plasmon resonance technique. Genes Cells, 5, 953–963. [DOI] [PubMed] [Google Scholar]
- Ishitsuka H., Kuriki,Y. and Kaji,A. (1970) Release of transfer ribonucleic acid from ribosomes. A G factor and guanosine triphosphate-dependent reaction. J. Biol. Chem., 245, 3346–3351. [PubMed] [Google Scholar]
- Janosi L., Hara,H., Zhang,S. and Kaji,A. (1996a) Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv. Biophys., 32, 121–201. [DOI] [PubMed] [Google Scholar]
- Janosi L., Ricker,R. and Kaji,A. (1996b) Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie, 78, 959–969. [DOI] [PubMed] [Google Scholar]
- Janosi L. et al. (1998) Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J., 17, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji A. and Hirokawa,G. (2000) Ribosome-recycling factor: an essential factor for protein synthesis. In Garret,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds.), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 527–539.
- Kaji A. and Kaji,H. (1963) Specific interaction of soluble RNA with polyribonucleic acid induced polysomes. Biochem. Biophys. Res. Commun., 13, 186–192. [Google Scholar]
- Kaji A., Kiel,M., Hirokawa,G., Muto,A., Inokuchi,Y. and Kaji,H. (2001) The fourth step of protein synthesis: disassembly of the post-termination complex is catalyzed by elongation factor G and ribosome recycling factor, RRF, a near perfect mimic of tRNA. Cold Spring Harb. Symp. Quant. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Kaji H. and Kaji,A. (1965) Specific binding of sRNA to ribosomes: effect of streptomycin. Proc. Natl Acad. Sci. USA, 54, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H., Suzuka,I. and Kaji,A. (1966) Binding of specific sRNA to template ribosome complex: effect of proteolytic enzymes. J. Mol. Biol., 18, 219–234. [DOI] [PubMed] [Google Scholar]
- Karimi R. and Ehrenberg,M. (1994) Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur. J. Biochem., 226, 355–360. [DOI] [PubMed] [Google Scholar]
- Karimi R., Pavlov,M.Y., Buckingham,R.H. and Ehrenberg,M. (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell, 3, 601–609. [DOI] [PubMed] [Google Scholar]
- Kim K.K., Min,K. and Suh,S.W. (2000) Crystal structure of the ribosome recycling factor from Escherichia coli. EMBO J., 19, 2362–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriki Y., Inoue,N. and Kaziro,Y. (1970) Formation of a complex between GTP, G factor and ribosomes as an intermediate of ribosome-dependent GTPase reaction. Biochim. Biophys. Acta, 224, 487–497. [DOI] [PubMed] [Google Scholar]
- Lill R., Robertson,J.M. and Wintermeyer,W. (1986) Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry, 25, 3245–3255. [DOI] [PubMed] [Google Scholar]
- Lucas-Lenard J. and Haenni,A.L. (1969) Release of transfer RNA during peptide chain elongation. Proc. Natl Acad. Sci. USA, 63, 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi M., Nishimura,T., Komai,T. and Tanaka,N. (1978) Interaction of kanamycin and related antibiotics with the large subunit of ribosomes and the inhibition of translocation. Biochem. Biophys. Res. Commun., 84, 358–365. [DOI] [PubMed] [Google Scholar]
- Moazed D., Robertson,J.M. and Noller,H.F. (1988) Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature, 334, 362–364. [DOI] [PubMed] [Google Scholar]
- Modolell J. and Vazquez. D. (1977) The inhibition of ribosomal translocation by viomycin. Eur. J. Biochem., 81, 491–497. [DOI] [PubMed] [Google Scholar]
- Momose K. and Kaji,A. (1965) Sedimentation studies on aminoacyl-sRNA synthetase and activation of aminoacyl-sRNA transfer factor. Arch. Biochem. Biophys., 111, 245–252. [DOI] [PubMed] [Google Scholar]
- Nirenberg M. and Leder,P. (1964) RNA codewords and protein synthesis: the effect of trinucleotides upon the binding of sRNA to ribosomes. Science, 145, 1399–1407. [DOI] [PubMed] [Google Scholar]
- Noller H.F. et al. (2000) Studies on the structure and function of ribosomes by combined use of chemical probing and X-ray crystallography. In Garret,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington DC, pp. 129–150.
- Ogawa K. and Kaji,A. (1975) Requirement for ribosome-releasing factor for the release of ribosomes at the termination codon. Eur. J. Biochem., 58, 411–419. [DOI] [PubMed] [Google Scholar]
- Oppenheim D.S. and Yanofsky,C. (1980) Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics, 95, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T., Wintermeyer,W. and Rodnina,M.V. (2000) Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nature Struct. Biol., 7, 104–107. [DOI] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D.V., Heurgue-Hamard,V., Buckingham,R.H. and Ehrenberg,M. (1997) Release factor RF3 abolishes competition between release factor RF1 and ribosome recycling factor (RRF) for a ribosome binding site. J. Mol. Biol., 273, 389–401. [DOI] [PubMed] [Google Scholar]
- Pestka S. (1968) Studies on the formation of transfer ribonucleic acid–ribosome complexes. V. On the function of a soluble transfer factor in protein synthesis. Proc. Natl Acad. Sci. USA, 61, 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. (1969) Studies on the formation of transfer ribonucleic acid–ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J. Biol. Chem., 244, 1533–1539. [PubMed] [Google Scholar]
- Rao A.R. and Varshney,U. (2001) Specific interaction between the ribosome recycling factor and the elongation factor G from Mycobacterium tuberculosis mediates peptidyl-tRNA release and ribosome recycling in Escherichia coli. EMBO J., 20, 2977–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remme J., Margus,T., Villems,R. and Nierhaus,K.H. (1989) The third ribosomal tRNA-binding site, the E site, is occupied in native polysomes. Eur. J. Biochem., 183, 281–284. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh,A., Katunin,V.I. and Wintermeyer,W. (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature, 385, 37–41. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh,A., Matassova,N.B., Katunin,V.I., Semenkov,Y.P. and Wintermeyer,W. (1999) Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl Acad. Sci. USA, 96, 9586–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roufa D.J., Skogerson,L.E. and Leder,P. (1970) Translation of phage Qβ mRNA: a test of the two-site model for ribosomal function. Nature, 227, 567–570. [DOI] [PubMed] [Google Scholar]
- Ruusala T. and Kurland,C.G. (1984) Streptomycin preferentially perturbs ribosomal proofreading. Mol. Gen. Genet., 198, 100–104. [DOI] [PubMed] [Google Scholar]
- Selmer M., Al-Karadaghi,S., Hirokawa,G., Kaji,A. and Liljas,A. (1999) Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science, 286, 2349–2352. [DOI] [PubMed] [Google Scholar]
- Shatsky I.N., Bakin,A.V., Bogdanov,A.A. and Vasiliev,V.D. (1991) How does the mRNA pass through the ribosome? Biochimie, 73, 937–945. [DOI] [PubMed] [Google Scholar]
- Shimizu Y., Inoue,A., Tomari,Y., Suzuki,T., Yokogawa,T., Nishikawa,K. and Ueda,T. (2001) Cell-free translation reconstituted with purified components. Nature Biotechnol., 19, 751–755. [DOI] [PubMed] [Google Scholar]
- Spedding G. (1990) Isolation and analysis of ribosomes from prokaryotes, eukaryotes and organelles. In Spedding,G. (ed.), Ribosomes and Protein Synthesis: A Practical Approach. Oxford University Press, New York, pp. 1–27.
- Spirin A.S. (1985) Ribosomal translocation: facts and models. Prog. Nucleic Acid Res. Mol. Biol., 32, 75–114. [DOI] [PubMed] [Google Scholar]
- Stark H., Orlova,E.V., Rinke-Appel,J., Junke,N., Mueller,F., Rodnina,M., Wintermeyer,W., Brimacombe,R. and van Heel,M. (1997) Arrange ment of tRNAs in pre- and post-translocational ribosomes revealed by electron cryomicroscopy. Cell, 88, 19–28. [DOI] [PubMed] [Google Scholar]
- Stark H., Rodnina,M.V., Wieden,H.J., van Heel,M. and Wintermeyer,W. (2000) Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell, 100, 301–309. [DOI] [PubMed] [Google Scholar]
- Steitz J.A. (1969) Nucleotide sequences of the ribosomal binding sites of bacteriophage R17 RNA. Cold Spring Harbor Symp. Quant. Biol., 34, 621–630. [DOI] [PubMed] [Google Scholar]
- Subramanian A.R., Ron,E.Z. and Davis,B.D. (1968) A factor required for ribosome dissociation in Escherichia coli. Proc. Natl Acad. Sci. USA, 61, 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Kinoshita,T. and Masukawa,H. (1968) Mechanism of protein synthesis inhibition by fusidic acid and related antibiotics. Biochem. Biophys. Res. Commun., 30, 278–283. [DOI] [PubMed] [Google Scholar]
- Toyoda T., Tin,O.F., Ito,K., Fujiwara,T., Kumasaka,T., Yamamoto,M., Garber,M.B. and Nakamura,Y. (2000) Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch. RNA, 6, 1432–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T. et al. (2001) Solution structure of the ribosome recycling factor from Aquifex aeolicus. Biochemistry, 40, 2387–2396. [DOI] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova,G.Z., Baucom,A., Lieberman,K., Earnest,T.N., Cate,J.H. and Noller,H.F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science, 292, 883–896. [DOI] [PubMed] [Google Scholar]
- Yusupova G.Z., Yusupov,M.M., Cate,J.H. and Noller,H.F. (2001) The path of messenger RNA through the ribosome. Cell, 106, 233–241. [DOI] [PubMed] [Google Scholar]