Abstract
The Williams Syndrome Transcription Factor (WSTF), the product of the WBSCR9 gene, is invariably deleted in the haploinsufficiency Williams–Beuren Syndrome. Along with the nucleosome-dependent ATPase ISWI, WSTF forms a novel chromatin remodeling complex, WICH (WSTF–ISWI chromatin remodeling complex), which is conserved in vertebrates. The WICH complex was purified to homogeneity from Xenopus egg extract and was found to contain only WSTF and ISWI. In mouse cells, WSTF interacts with the SNF2H isoform of ISWI. WSTF accumulates in pericentric heterochromatin coincident with the replication of these structures, suggesting a role for WSTF in the replication of heterochromatin. Such a role is supported by the in vitro activity of both the mouse and frog WICH complexes: they are involved in the assembly of regular spaced nucleosomal arrays. In contrast to the related ISWI-interacting protein ACF1/WCRF180, WSTF binds stably to mitotic chromosomes. As dysfunction of other chromatin remodeling factors often has severe effects on development, haploinsufficiency of WSTF may explain some of the phenotypes associated with this disease.
Keywords: chromosomes/DNA replication/nucleosome/WICH/WSTF
Introduction
Williams–Beuren syndrome is a developmental disorder characterized by congenital vascular and heart disease, particular facial features, growth deficiency, mental retardation and characteristic behavior patterns (Lenhoff et al., 1997; Francke, 1999). This disorder is caused by haploinsufficiency of ∼1.6 Mb of chromosome 7 (Francke, 1999). The deletion is uniform in size because it arises by inter- or intrachromosomal crossover events within regions of high sequence identity flanking the typical deletion. More than 15 genes reside in the deleted area and several of those may contribute to the complex multi-system clinical phenotype (Francke, 1999). One of the genes that invariably maps to the deletion region is WBSCR9, which encodes an ∼170 kDa protein called Williams syndrome transcription factor (WSTF) (Lu et al., 1998; Peoples et al., 1998). WSTF is expressed in many tissues and is subject to alternative splicing (Lu et al., 1998; Peoples et al., 1998).
WSTF belongs to a family of proteins with similar domain structure, the BAZ (Jones et al., 2000a), also referred to as the WAL family (Poot et al., 2000). WSTF is related in subdomain architecture to ACF1 and WCRF180, subunits of the chromatin remodeling factors ACF, WCRF and CHRAC (Ito et al., 1999; Bochar et al., 2000; Guschin et al., 2000; LeRoy et al., 2000; Poot et al., 2000; Eberharter et al., 2001). hACF1 (LeRoy et al., 2000; Poot et al., 2000), BAZ1A (Jones et al., 2000a) and WCRF180 (Bochar et al., 2000) are essentially identical (with only three amino acid differences between the sequences published by Poot et al. and Bochar et al., which may reflect polymorphism), and this protein is the human ortholog of the Drosophila ACF1 protein (Ito et al., 1999). hACF1 (WCRF180/BAZ1A) and WSTF share an N-terminal WAC (WSTF/ACF1/cbp146) domain (Ito et al., 1999), which is followed by a DDT domain (Doerks et al., 2001), BAZ motifs (Jones et al., 2000a), a WAKZ domain (Ito et al., 1999), a PHD finger and a C-terminal bromodomain (Haynes et al., 1992) (see Figure 1). The WAC domain, DDT domain, BAZ and WAKZ motifs had been identified by their conservation in various proteins; no functionality has been assigned to these domains and motifs. A difference between WSTF and ACF1/WCRF180 is highlighted by the fact that domain analysis tools (e.g. RPS-BLAST, www.ncbi.nlm. nih.gov/Structure/cdd/wrpsb.cgi) identify a FERM domain (Chishti et al., 1998) related sequence in WSTF (Figure 1), but not in ACF1, despite the fact that the significance of this domain in WSTF is unclear.
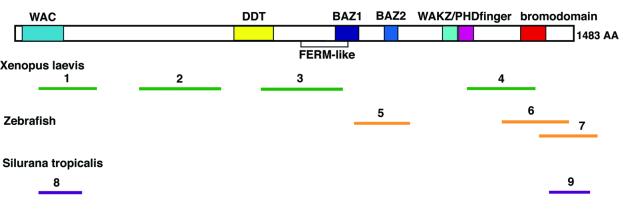
Fig. 1. WSTF is conserved in vertebrates. The domain structure of WSTF is shown above. Bars below show the matching positions of corresponding ESTs from X.laevis (1–4), zebrafish (D.rerio, 5–7) and S.tropicalis (8–9). Accession numbers: 1, BI447904; 2, BG264264; 3, BI447594; 4, BG345707; 5, AI794397; 6, AI436874; 7, AW466480; 8, BG514920; 9, BG515236.
The dynamic nature of eukaryotic chromatin requires special mechanisms to alter histone–DNA contacts. ATP-dependent chromatin remodeling factors use the energy gained from ATP hydrolysis to remodel nucleosomes (Kingston and Narlikar, 1999; Varga-Weisz, 2001). Most of these enzymes are multi-subunit complexes and all contain a SNF2 superfamily ATPase (Eisen et al., 1995). Four classes of ATP-dependent chromatin remodeling factors can be distinguished that contain either the SNF2, CHD/Mi-2, INO80 or ISWI ATPases (Kingston and Narlikar, 1999; Varga-Weisz, 2001). Three ISWI-containing complexes, nucleosome remodeling factor (NURF), CHRAC and ACF (Tsukiyama et al., 1995; Ito et al., 1997; Varga-Weisz et al., 1997), have been purified from Drosophila extracts and either disrupt or enhance the periodic organization of nucleosome arrays. Related ISWI-containing complexes have subsequently been purified from human cell extracts, Xenopus and budding yeast (LeRoy et al., 1998; Tsukiyama et al., 1999; Bochar et al., 2000; Guschin et al., 2000; LeRoy et al., 2000; Poot et al., 2000). ISWI complexes mobilize nucleosomes, resulting in alterations in the translational position of the histone octamer without displacement from DNA (Hamiche et al., 1999; Längst et al., 1999). Genetic analysis implicates ISWI in transcriptional regulation and maintenance of chromosome structure (Deuring et al., 2000; Goldmark et al., 2000).
Three BAZ/WAL proteins interact independently with ISWI to form chromatin remodeling factors: ACF1/WCRF180 in the ACF, WCRF and CHRAC complexes from fly, frog and human (Ito et al., 1999; Bochar et al., 2000; Guschin et al., 2000; LeRoy et al., 2000; Poot et al., 2000; Eberharter et al., 2001), TIP5 (also called BAZ2A and WALp3) in the NoRC complex (Strohner et al., 2001) and NURF301 in NURF (Xiao et al., 2001). Whereas ISWI alone has nucleosome remodeling activity, ACF1 enhances the ATP-dependent chromatin assembly activity of ISWI (Ito et al., 1999) and the efficiency of ISWI-catalyzed nucleosome sliding by an order of magnitude, and determines the directionality of this sliding (Guschin et al., 2000; Eberharter et al., 2001). The largest subunit of NURF, NURF301, shares structural motifs with ACF1 and also modulates the nucleosome mobilizing property of ISWI (Xiao et al., 2001).
Here we show that WSTF forms a nucleosome remodeling factor containing ISWI and that this factor is conserved across vertebrate phyla. Mammalian WSTF is targeted to pericentromeric heterochromatin during its replication and is distinguished from the related ACF1 protein by its ability to interact with mitotic chromatin. We suggest that WSTF is important for the dynamics of chromosome structure in vertebrates, which may explain some aspects of Williams–Beuren syndrome.
Results
WSTF is a highly conserved protein in vertebrates
Searches of expressed sequence tag (EST) databases using the BLAST algorithm (Altschul et al., 1997) revealed that WSTF is highly conserved in mammals, with multiple ESTs from several organisms showing near sequence identity. In addition, many ESTs from amphibia (Xenopus laevis and Silurana tropicalis) and zebrafish (Danio rerio) share considerable sequence identity with human WSTF (Figure 1). Frog and fish ESTs more closely resemble human WSTF than human ACF1/WCRF180, and can be assigned as WSTF. This sequence conservation suggests a specific function for WSTF in vertebrates.
To study the localization and function of WSTF, we raised a rabbit polyclonal antiserum against an N-terminal peptide of human WSTF. The affinity-purified antibody detected a protein of ∼175 kDa in mouse NIH 3T3 (Figure 2A) and human (HeLa) nuclear extracts (Figures 2B and 3A). No cross-reactivity with mouse or human ACF1 (WCRF180) or with any protein in Drosophila embryo extract, which contains much ACF1, was observed (see Figure 2A and D). We concluded that this antiserum specifically recognizes the WSTF protein.
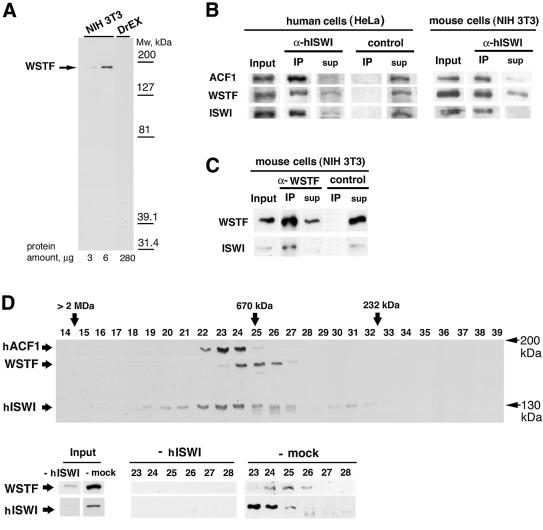
Fig. 2. WSTF and ISWI form a complex in mouse and human cells. (A) Western blot analysis of NIH 3T3 nuclear extract and Drosophila embryo extract using affinity-purified anti-WSTF antibody. (B) Co-immunoprecipitation of WSTF with affinity-purified anti-ISWI antibodies in HeLa cell nuclear extracts (left panel) and NIH 3T3 cell nuclear extracts (right panel). Input (12%); IP, immunoprecipitate (100%); sup, supernatant of immunoprecipitate. (C) Co-immunoprecipitation of ISWI with affinity-purified anti-WSTF antibodies. Input (4%); IP, immunoprecipitate (100%). Control immunoprecipitations in (B) and (C) were with the same amount of purified rabbit IgG. (D) Fractionation of WSTF, hACF1(WCRF180) and ISWI from crude HeLa nuclear extract by Superose-6 gel filtration chromatography. Upper panel: overlay of three separate western blots against WSTF, hACF1 (WCRF180) and hISWI of the fractions. Lower panel: fractionation of WSTF in HeLa nuclear extract immunodepleted with antibodies against hISWI (mock depletion was with pre-immune serum). Size standards were thyroglobulin (670 kDa) and catalase (232 kDa).
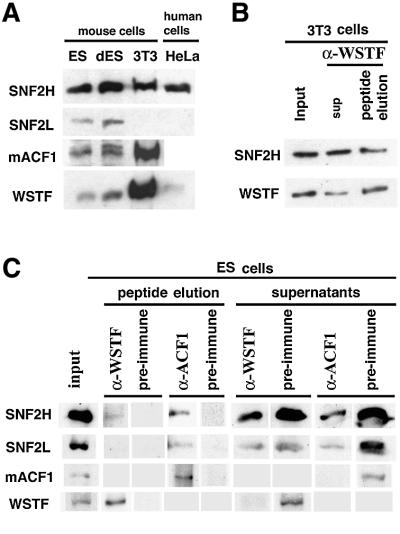
Fig. 3. WSTF interacts with the SNF2H isoform of ISWI. (A) Western blot of identical amounts of nuclear extract proteins from mouse ES cells, ES cells driven to differentiation (dES), NIH 3T3 and HeLa cells. (B) Western blot of affinity-purified mouse WSTF–ISWI complex: input, 10%; peptide eluate, 100%. (C) Immunoprecipitation of WSTF– and ACF1–ISWI complexes from ES cell nuclear extract. The complexes were eluted from the antibodies with the antigenic peptides and analyzed by western blots: input, 16%; peptide eluate, 100%.
WSTF forms a complex with ISWI
We used the anti-WSTF rabbit polyclonal antiserum in co-immunoprecipitation experiments to analyze the interaction of WSTF with human and mouse ISWI. An affinity-purified antiserum against human ISWI (α-hISWI), which recognizes both the SNF2H and SNF2L isoforms and co-immunoprecipitates hACF1 (Poot et al., 2000), efficiently depleted WSTF as well as hACF1 from nuclear extracts of HeLa and NIH 3T3 cells [Figure 2B, ‘input’ versus supernatant (‘sup’)]. Both WSTF and hACF1 were found in the ISWI immunoprecipitate (Figure 2B, ‘IP’). Conversely, the antiserum against WSTF (α-WSTF) precipitated both WSTF and ISWI (Figure 2C) from NIH 3T3 extracts. The WSTF–ISWI complex could be eluted from the immunocomplexes bound to protein A beads with the antigenic peptide (see below). We concluded that WSTF and ISWI form a complex in both human and mouse nuclei. The failure of the α-WSTF antiserum to co-immunoprecipitate hACF1 protein excluded the possibility of a ternary complex containing WSTF, hACF1 and ISWI (data not shown). We did not observe an interaction between WSTF and HuCHRAC-17, the histone-fold-containing subunit of HuCHRAC (Poot et al., 2000), or with topoisomerase II (data not shown).
We fractionated crude HeLa nuclear extract by gel filtration with Superose-6 and probed the fractions for the presence of WSTF, hACF1/WCRF180 and ISWI by western blot analysis (Figure 2D). WSTF fractionated in a single peak at ∼700 kDa. hACF1/WCRF180 ran in a slightly larger complex, consistent with earlier findings (Bochar et al., 2000; Poot et al., 2000). Most ISWI ran in a broad peak that includes both the hACF1/WCRF180- and WSTF-containing fractions. However, the majority of ISWI is found in the hACF1/WCRF180-containing fractions. To test if all the WSTF is in a complex with ISWI, we depleted the ISWI from crude HeLa nuclear extract with the anti-hISWI antiserum (Poot et al., 2000) and fractionated this depleted extract over the gel filtration column. The ISWI depletion not only removed most of the WSTF from the extract, but also the high molecular weight WSTF complex, whereas a mock depletion of the extract with the pre-immune serum had no effect on the fractionation of WSTF and ISWI (Figure 2D, ‘-mock’). We could further purify the ∼700 kDa WSTF–ISWI complex from both HeLa and NIH 3T3 extracts over several ion-exchange columns such as MonoQ, where the complex runs in one peak (data not shown). Taken together, the data strongly suggest that most of the WSTF is associated with ISWI in NIH 3T3 and HeLa cell extracts (although we cannot exclude that there is a minor fraction of WSTF in a complex that does not fractionate over the Superose-6). We attribute the fact that not all WSTF can be depleted with the antisera against ISWI to a weaker association of ISWI with WSTF as compared with hACF1, which can be depleted more efficiently (Figure 2B), and, therefore, to the presence of ‘free’ WSTF under some conditions.
We next examined WSTF, ACF1 and the two isoforms of ISWI (SNF2L and SNF2H) (Okabe et al., 1992; Aihara et al., 1998; Lazzaro and Picketts, 2001) in nuclear extracts of different cell lines (Figure 3A). All extracts exhibited similar levels of SNF2H protein. SNF2L was observed only in the embryonic stem (ES) cell and differentiated embryonic stem cell extracts; we were not able to detect SNF2L in nuclear extracts of NIH 3T3 cells, which represent a more differentiated, fibroblastic cell type, nor in HeLa nuclear extract. The NIH 3T3 cell extracts exhibited dramatically higher levels of WSTF than the ES cells or HeLa extracts. The levels of ACF1 paralleled those of WSTF. Interestingly, the differentiated embryonic cell nuclear extracts contained an ACF1 isoform with slightly slower electrophoretic migration. This may represent a cell-type-specific splice variant of ACF1 as the ES cells differentiate into a very heterogeneous cell mixture. Because the NIH 3T3 cell nuclear extract is devoid of detectable SNF2L, we conclude that both WSTF and ACF1 interact with the SNF2H isoform of mammalian ISWI, as demonstrated previously for ACF1 in HeLa cell extracts (LeRoy et al., 2000; Poot et al., 2000). Indeed, the antiserum specific to the H-isoform of ISWI detected the SNF2H in affinity-purified WSTF– ISWI complex (Figure 3B), whereas SNF2L was not detectable in the extract or the affinity-purified complex (not shown). Therefore, WSTF and ACF1 may compete for the same pool of SNF2H in the nucleus.
We performed co-immunoprecipitation experiments with ES cell nuclear extract to find out whether WSTF and ACF1 interact with SNF2L in an extract that contains both ISWI isoforms (Figure 3C). The efficiency of ISWI co-precipitation with the antibodies against WSTF and ACF1 was low in this extract, probably because there is little WSTF and ACF1 in the ES extract to start with. We eluted the proteins from the antibody complexes bound to protein A with the antigenic peptides to reduce background. An antibody against mouse ACF1 precipitated SNF2H and some SNF2L together with the ACF1. However, we could detect only SNF2H in the immunoprecipitate of the antibody against WSTF. Although we cannot exclude the possibility that there is some interaction of SNF2L with WSTF, all experiments together indicate that SNF2H is the predominant isoform of ISWI with which WSTF interacts.
A two-subunit WSTF–ISWI complex from Xenopus egg extract
Previously, four discrete ISWI complexes, ISWI-A, -B, -C and -D, have been purified from Xenopus egg extracts, and ISWI-C was shown to contain the Xenopus homolog of ACF1 (Guschin et al., 2000). We examined the remaining ISWI complexes for WSTF with the WSTF antiserum. The antiserum strongly recognized the 200 kDa subunit of the ISWI-B complex (Figure 4A and B) and failed to react with any components of the ISWI-A and ISWI-D complexes (data not shown). The sequence [nine amino acids (aa)] of one tryptic peptide, derived from ‘p200’ obtained by Edman degradation, and five sequences obtained by mass spectrometry (13–16 aa) all matched sequences from WSTF ESTs. We therefore conclude that ‘p200’ is the Xenopus WSTF homolog. Furthermore, this WSTF–ISWI complex consists of only two subunits. A more careful analysis of the size of the WSTF from Xenopus indicated that it is actually smaller than 200 kDa, at ∼180 kDa.
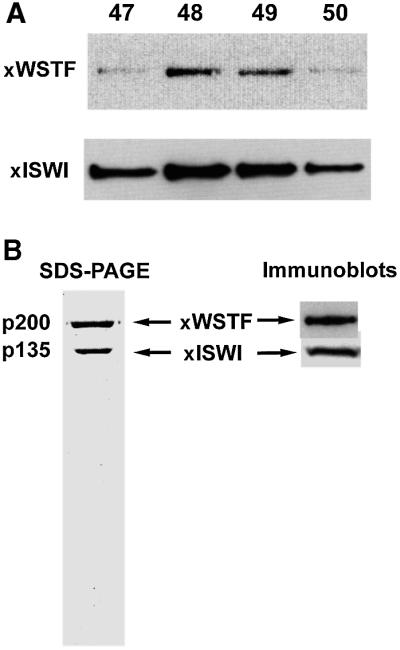
Fig. 4. WSTF forms a complex with ISWI in Xenopus egg extracts. (A) Western blot of co-purification of Xenopus WSTF (anti-human WSTF antibodies) and Xenopus ISWI (anti-Xenopus ISWI antibodies) over MonoS chromatography during ISWI-B purification. This is the penultimate step in the purification scheme. (B) SDS–PAGE of the final purified complex showing the two polypeptides. Immunoblot shows the antibody reactivity with the respective antisera.
The WSTF–ISWI complex is a chromatin remodeling factor
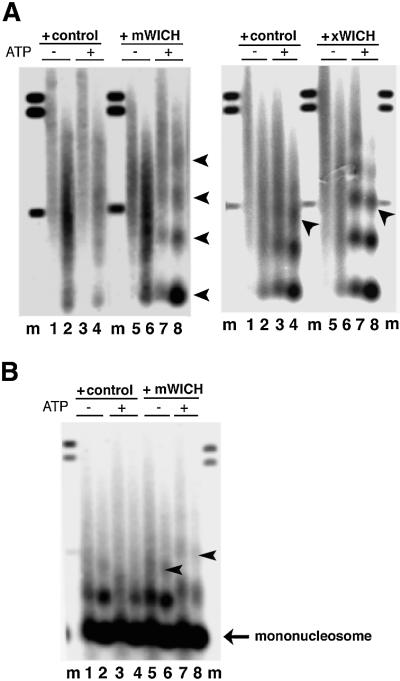
We examined the WSTF–ISWI complexes for nucleosome remodeling activities. The mouse complex was tested following immuno-affinity purification and peptide elution. A property of several ISWI complexes is the ability to create regular nucleosomal arrays from irregular chromatin in vitro (Varga-Weisz et al., 1997; Tsukiyama et al., 1999; Guschin et al., 2000; LeRoy et al., 2000; Poot et al., 2000; Längst and Becker, 2001). To test whether the WSTF–ISWI complexes share this activity we assembled a chromatin-like structure on plasmid DNA with a Drosophila chromatin assembly extract in the absence of ATP, and stripped endogenous nucleosome spacing activities with a sarcosyl wash. This template is characterized by the absence of a regular nucleosome repeat structure when assayed by partial micrococcal nuclease digest (Varga-Weisz et al., 1997). Addition of mock eluate in the presence of ATP did not improve the regularity of the chromatin structure (Figure 5A, left panel, lanes 1–4). However, the mouse WSTF–ISWI complex reconfigures the chromatin to a regularly spaced nucleosome array in an ATP-dependent fashion (Figure 5A, left panel, lanes 5–8). The same activity was exhibited by the purified frog complex (Figure 5A, right panel). There is some residual spacing activity in the sarcosyl-stripped chromatin in the presence of ATP (Figure 5A, right panel, buffer control, lanes 1 and 2 versus 3 and 4); however, the frog complex significantly improved the regularity of the nucleosomal array in an ATP-dependent manner (lanes 5 and 6 versus 7 and 8). The frog complex also increased the internucleosomal spacing (repeat length): the DNA protected by three nucleosomes (trinucleosome) runs below the 491 bp marker in the control experiment (Figure 5A, right panel, lane 4, arrow), whereas it runs with this marker in the chromatin that was remodeled by the frog WSTF–ISWI complex (lanes 7 and 8, arrow).
Fig. 5. The WSTF–ISWI complex is a chromatin remodeling factor, WICH. (A) Left panel: the mouse WSTF–ISWI complex reconfigures irregular chromatin into a regular nucleosomal array. Eluates from the pre-immune serum (+control) or anti-WSTF antiserum (+WICH) were added to sarcosyl-stripped chromatin, assembled in Drosophila embryo extracts without ATP. Addition of 1 mM ATP was as indicated. Micrococcal nuclease digestion was for 30 s (lanes 1, 3, 5 and 7) or 60 s (lanes 2, 4, 6 and 8). Lanes labeled ‘m’ contain size marker DNA fragments: 0.49, 1.1 and 1.2 kbp. Arrows indicate mono-, di-, tri- and tetranucleosome DNA fragments (from the bottom upwards). Right panel: same experiment as in the left panel, but with the purified Xenopus WICH (300 ng MonoS fraction, purification buffer as control). Arrows indicate the position of the trinucleosome DNA fragment. (B) WICH mobilizes nucleosomes. Nucleosomes were assembled with polyglutamic acid as carrier on plasmid DNA. This chromatin was incubated with mock eluate (+control) or the immunopurified WSTF complex (+mWICH). Addition of 1 mM ATP was as indicated above the panels. Micrococcal nuclease digestion was for 30 s (lanes 1, 3, 5 and 7) or 60 s (2, 4, 6 and 8). Lanes labeled ‘m’ contain size marker DNA fragments: 0.49, 1.1 and 1.2 kbp. Arrows indicate a change of internucleosomal repeat length.
To assess the ability of the WSTF–ISWI complex to mobilize nucleosomes, we assembled a chromatin-like structure on plasmid DNA with purified HeLa histones and polyglutamic acid as carrier. This type of artificial chromatin is characterized by a non-physiological ‘close-packed’ nucleosomal spacing. Purified ACF complex from Drosophila and Xenopus has been shown previously to reposition nucleosomes in a salt and ATP-dependent manner to a spacing with greater internucleosomal distances (Ito et al., 1997; Guschin et al., 2000). We found that addition of the affinity-purified mouse WSTF–ISWI complex also caused a repositioning of the nucleosomes to a greater spacing in an ATP-dependent manner (Figure 5B, compare lane 5 with 7, and 6 with 8). These findings indicate that the interaction of WSTF with ISWI results in a nucleosome remodeling factor that can mobilize and ‘space’ nucleosomes. We named this complex the WSTF–ISWI chromatin remodeling complex, or WICH.
WSTF is targeted to pericentromeric heterochromatin during its replication
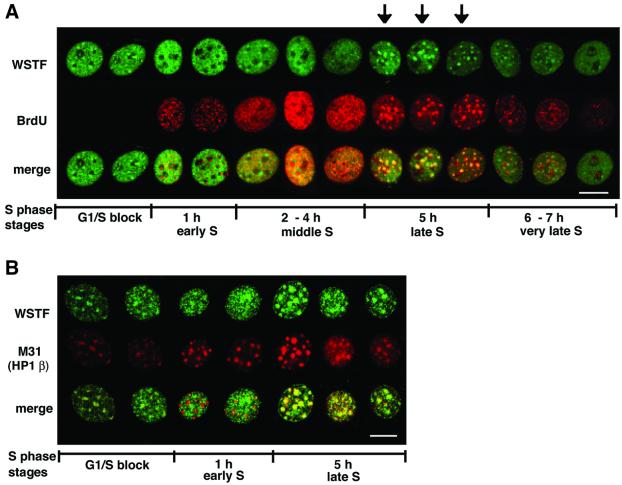
We next determined the localization of WSTF in mouse cells using indirect immunofluorescence and confocal microscopy. The affinity-purified α-WSTF antibody predominantly stained the nuclei of NIH 3T3 cells (Figure 6). In addition, ∼40–50% of the nuclei of an asynchronous log-phase culture exhibited foci of staining over a more even background. These foci were similar in number, size and distribution to the pericentromeric heterochromatin foci. We determined whether the WSTF foci are localized to pericentromeric heterochromatin by staining the cells for both WSTF and M31 (also called HP1β), a marker for pericentromeric heterochromatin (Wreggett et al., 1994). Indeed, the WSTF foci co-localized with the M31 foci (Figure 6) and, therefore, WSTF targets pericentromeric heterochromatin in at least a subset of the cell population.
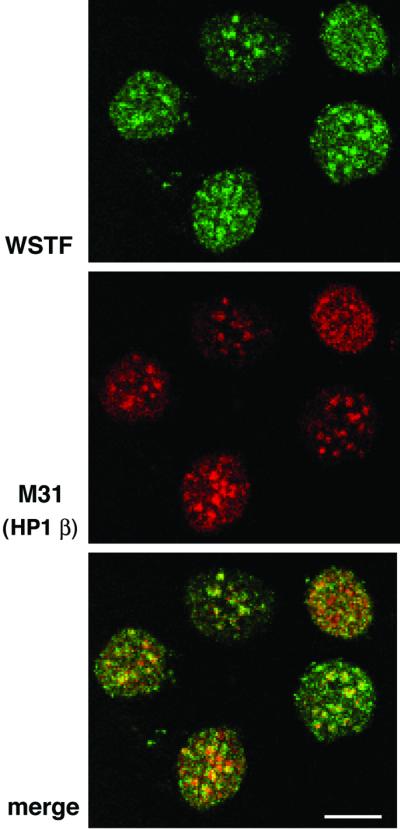
Fig. 6. WSTF co-localizes with M31 (mouse HP1β), a marker protein for mouse pericentromeric heterochromatin. NIH 3T3 cells were fixed and stained with affinity-purified anti-WSTF antibodies (top, FITC, green) and rat monoclonal antibodies against M31 (middle, Texas Red). Lower panel: merged image (yellow). Bar = 10 µm.
To understand the restriction of pericentromeric heterochromatin localization of WSTF to a subset of cells, we analyzed this localization pattern through the replication phase in NIH 3T3 cells. Levels of WSTF were constant and its association with ISWI was maintained throughout S phase (data not shown). To demarcate cells passing through S phase, we used the nucleotide analog bromodeoxyuridine (BrdU), which is incorporated into newly synthesized DNA and can be visualized with anti-BrdU antibodies. Cells were arrested at the G1/S boundary using an inhibitor of DNA polymerase α, aphidicolin, and then released into S phase by washing with fresh medium. S phase in NIH 3T3 cells usually takes 7–8 h to complete and occurs in stages (early, middle, late and very late S phase), which are characterized by specific patterns of BrdU incorporation (Fox et al., 1991; O’Keefe et al., 1992). Cells blocked at the G1/S boundary showed WSTF foci over a granular nuclear staining; however, the foci were rather weak compared with the general nuclear staining in these cells (Figure 7A, ‘G1/S block’). One hour after release from the G1/S block, overall WSTF staining became more intense and even, and it did not show pronounced foci but was excluded from the pericentromeric heterochromatin (see below; Figure 7B, ‘1 h’). There was no co-staining of WSTF with the BrdU incorporation sites. Two to 4 h after release, BrdU incorporation was seen in a granular pattern throughout the nucleus. At this time the WSTF staining became more uneven and there was an extensive overlap of the WSTF staining and the BrdU incorporation staining (Figure 7A, ‘2–4 h’). At ∼5 h into S phase the cells replicated the pericentromeric heterochromatin, which was visualized by the incorporation of the BrdU into large foci (Fox et al., 1991; O’Keefe et al., 1992). At this time, the WSTF staining was clearly concentrated into large foci, which co-localized with the BrdU foci (Figure 7A, ‘5 h’). At very late S stage, 6–7 h after release from the chemical block, BrdU incorporation again became more granular and was particularly apparent around the perimeter of the nucleus. At that point, WSTF staining persisted in the pericentromeric heterochromatin; however, the foci were not as intense as at the 5 h time point (Figure 7A, ‘6–7 h’). At this time, there was little co-localization of BrdU incorporation foci and WSTF staining.
Fig. 7. WSTF is targeted to pericentromeric heterochromatin during its replication. (A) NIH 3T3 cells were synchronized at the G1/S border, released into S phase, fixed at the indicated times after release and stained for WSTF (top, FITC, green) and BrdU (middle, Texas Red) incorporation. The merged image (yellow) is shown at the bottom. Arrows indicate cells with WSTF foci at late S phase. (B) WSTF (top, green) and M31 (HP1β, middle, red) staining through S phase. Bottom panel: merged image (yellow). Bar = 10 µm.
The most intriguing result from this time course experiment was the accumulation of WSTF in the foci of replicating heterochromatin. To better understand the relationship between pericentromeric heterochromatin and WSTF through S phase, we stained the cells at different points through S phase with antibodies against the pericentromeric heterochromatin protein M31, also called HP1β (Wreggett et al., 1994), together with antibodies against WSTF. WSTF is somewhat enriched in these pericentromeric foci before the release of the G1/S block (Figure 7B,‘G1/S block’) and it is also found throughout the nucleus. One hour after release from the G1/S block, WSTF is distributed more evenly in the nucleus in a granular pattern. Co-localization analysis with M31 shows that WSTF is actually excluded from the pericentromeric heterochromatin at this point (Figure 7B, ‘1 h’). Five hours into S phase, when cells replicate the pericentromeric heterochromatin, WSTF is clearly enriched in the M31-stained foci. These data document a dynamic association between WSTF and pericentromeric heterochromatin. WSTF co-localizes with DNA replication foci at middle to late S phase (2–5 h after G1/S). Outside of this time, there is no close link between WSTF localization and DNA replication foci. These data indicate a close relationship between the replication of the pericentromeric heterochromatin and WSTF accumulation at these structures in S phase. ACF1 is targeted to replicating heterochromatin in NIH 3T3 cells too; however, unlike WSTF, it remained in these structures into G2 phase (N.Collins, C.García-Jiménez, G.Dellaire and P.Varga-Weisz, submitted).
The N-terminal ∼350 amino acids of hACF1, including the WAC domain, have been shown to target a marker protein to pericentromeric heterochromatin in mouse cells (Tate et al., 1998), indicating a role for the WAC domain in heterochromatin targeting. However, we did not find targeting of the N-terminal 400 amino acids, including the WAC domain of WSTF fused to green fluorescent protein to heterochromatin, although the fusion protein is nuclear (I.Kukimoto and P.Varga-Weisz, unpublished data). Further studies are necessary to reveal the mechanism of heterochromatin targeting of WSTF.
Stable association of WSTF with mitotic chromosomes distinguishes it from ACF1/WCRF180
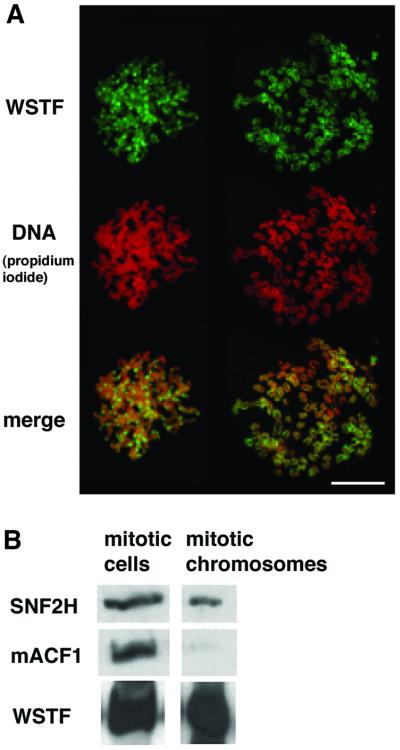
Immunostaining of fixed cells and spread chromosomes indicated that WSTF is associated with metaphase chromosomes. In less condensed chromosomes, such as in the earlier stages of chromosome condensation, enrichment of WSTF at the pericentromeric heterochromatin is apparent (Figure 8A, compare left and right sets of chromosomes). We have not been able to detect ACF1 (WCRF180) on mitotic chromosomes in fixed cells with indirect immunofluorescence experiments using antisera against both the N- and C-terminal peptides of ACF1 (N.Collins and P.Varga-Weisz, unpublished observations). To verify this clear distinction between WSTF and ACF1, we probed samples from whole mitotic cells and from isolated mitotic chromosomes for the presence of WSTF, ACF1 and ISWI. We observed a clear depletion of ACF1 from the mitotic chromosomes, which is in strong contrast to the retention of WSTF (Figure 8B).
Fig. 8. WSTF binds mitotic chromosomes. (A) Anti-WSTF staining (FITC, green) of spread mitotic chromosomes from NIH 3T3 cells, and propidium iodide staining for DNA (red, middle). Bottom: merged image (yellow). Bar = 10 µm. (B) Western blot analysis of whole mitotic NIH 3T3 cells and purified mitotic chromosomes. Each sample contains the same amount of chromatin.
Discussion
WSTF–ISWI complex, a novel nucleosome remodeling factor
Here we demonstrate that WSTF, the product of a gene deleted in Williams–Beuren syndrome, forms a novel chromatin remodeling complex with the ATPase ISWI. This complex mobilizes nucleosomes and reconfigures irregular chromatin to a regular nucleosome array structure.
According to its domain structure, WSTF belongs to the BAZ/WAL family of proteins, which also includes ACF1, WCRF180 and TIP5 (BAZ2A, WALp3) (Bochar et al., 2000; Jones et al., 2000a; Poot et al., 2000; Strohner et al., 2001). All these proteins have now been shown to interact with ISWI to form NURF. In addition, WSTF is similar to the p301 subunit of the ISWI containing NURF in a subdomain arrangement over a large proportion of the protein, and is also related to the human BPTF (Jones et al., 2000b; Xiao et al., 2001). One common motif that WSTF shares with ACF1 and p301 of NURF is the WAC domain. Interestingly, this domain is also conserved in an ISWI-interacting protein in yeast, which may indicate that this domain is important for the nucleosome remodeling mechanism (Gelbart et al., 2001). ACF1, TIP5 and p301 substantially enhance the nucleosome remodeling activity of the ISWI ATPase, and alter its nature and substrate specificity (Ito et al., 1999; Eberharter et al., 2001; Gelbart et al., 2001; Strohner et al., 2001; Xiao et al., 2001). The interaction of ISWI with several distinct subunits that impart different functionality to the resulting complex points to a uniquely ‘promiscuous’ role of the ISWI ATPase as compared with the other nucleosome remodeling ATPases.
Several ISWI complexes with BAZ/WAL proteins are found in the same tissues, e.g. the frog egg contains an ACF/CHRAC homolog, a WSTF–ISWI complex and at least one further ISWI complex (Guschin et al., 2000; our data). In addition, mouse NIH 3T3 cells contain both ACF1 and WSTF complexes with the ISWI isoform SNF2H. In vitro, the WSTF–ISWI complex exhibits essentially identical activity to purified ACF and CHRAC: it mobilizes nucleosomes and reconfigures irregular chromatin to a regular nucleosomal array structure. These findings lead to many questions: how is the ISWI allocated between these complexes? What is their relation to each other? Do they perform redundant or opposing roles? However, the conservation of WSTF in vertebrates points to a unique function of WSTF compared with ACF1. Using cell biological means, we observed a clear distinction between ACF1 and WSTF: whereas we have been unable to detect ACF1 on mitotic chromosomes, WSTF clearly stains condensed chromosomes, which may point to a unique role for WSTF during mitosis.
Potential functions of WSTF in heterochromatin during DNA replication
Pericentromeric heterochromatin is a highly condensed structure around the centromere, which is particularly large and easily visualized in mouse NIH 3T3 cells. It contains few genes and is a transcriptionally repressive environment (Wallrath, 1998). This structure has recently been shown to be essential for chromosome stability (Wallrath, 1998; Taddei et al., 2001) and is a model to study the establishment, assembly and inheritance of a specific, repressed chromosome structure. Pericentromeric heterochromatin is characterized by hypermethylation of the DNA and hypoacetylation of histones, and these modifications are involved in the inheritance of this structure. We find that WSTF accumulates in pericentromeric heterochromatin during its replication. The cell-cycle-regulated targeting to this domain is best explained by a role of WSTF during the replication of heterochromatin. Two potential roles that are not mutually exclusive can be envisioned for WSTF during heterochromatin replication: it may facilitate DNA replication through the heterochromatin or may have a role in the assembly of heterochromatin post-replication. Because of its condensed nature, heterochromatin is a daunting obstacle for all incoming proteins. Dedicated mechanisms may exist to permit the establishment of active origins of replication in heterochromatin and to allow fork progression in this structure. An essential role in the establishment of an active origin of replication in the context of chromatin has been demonstrated in vitro for CHRAC, which contains the WSTF-related ACF1 (Alexiadis et al., 1998). The role of the WSTF–ISWI complex in establishment of a regular nucleosomal array from irregular chromatin is consistent with a function in chromatin assembly. Therefore, the complex may have a role in the assembly of heterochromatin, which is characterized by a very regular nucleosomal array structure (Wallrath and Elgin, 1995; Cryderman et al., 1999). Elucidation of the role of WSTF in S phase progression in vivo will require the creation of cells where WSTF function is deleted.
Mitotic association of WSTF
Whereas we find that most ACF1 is excluded from mitotic chromatin, WSTF is stably associated with metaphase chromosomes. This behavior also distinguishes WSTF from other known ATP-dependent nucleosome remodeling factor subunits, such as hbrm and BRG1, which are excluded from the condensed chromosomes during mitosis (Muchardt et al., 1996). At the early stages of chromosome condensation there is an apparent enrichment of WSTF in the pericentromeric chromatin of the chromosomes, which may simply reflect greater DNA concentration at these sites. However, unlike the situation during S phase, WSTF stains the chromosomes all over. Future studies will reveal whether WSTF has a role in chromosome condensation or decondensation, or in facilitating specific modifications of histones that occur during mitosis, such as histone H3 phosphorylation.
Haploinsufficiency of WSTF may explain aspects of Williams–Beuren syndrome
The in vitro activity of the WSTF–ISWI complex, as well as the localization of WSTF to condensed chromatin in vivo, suggests a global, structural role for this protein. Depletion of WSTF, as would occur in Williams–Beuren syndrome, may therefore have a noticeable effect. Furthermore, WSTF will compete for ISWI binding with other BAZ/WAL proteins such as ACF1, and depletion of WSTF may upset a tight balance between these proteins in their interaction with ISWI. The targeting of WSTF to pericentromeric heterochromatin during its replication suggests a function of WSTF in the replication of this condensed chromatin and/or the assembly of heterochromatin post-replication. Since pericentromeric heterochromatin has been shown to have functions in chromosome stability, depletion of WSTF may affect cell survival and cause chromosome loss or breakage. Mutations in several chromatin remodeling factors have been implicated in developmental abnormalities and mental retardation. For example, mutations within the PHD finger of the ATRX protein, which is homologous to the nucleosome remodeling ATPase SNF2, causes an X-linked developmental disorder and mental retardation syndrome (Picketts et al., 1996). Remarkably, the ATR-X gene also targets pericentromeric heterochromatin (McDowell et al., 1999). Another structural chromatin protein that is linked to heterochromatin, and mutation of which causes a mental retardation syndrome, is the methyl CpG binding protein MeCP2 (Amir et al., 1999). Thus, it is not unreasonable to propose that the identification of WSTF as a chromatin remodeling factor may suggest a role in the complex Williams–Beuren syndrome.
Materials and methods
Antibodies
Rabbit polyclonal anti-peptide antisera were constructed against the N-terminal peptide of human/mouse WSTF APLLGRKPFPLVKPLPGEE, against the C-terminal peptide of mouse ACF1 SPSTVDQVSTPLAAKKSRI, against peptide STQKRKMDGAPDGRGRKK of human/mouse SNF2H, against peptide KRATKTPMVKFSAFS of human SNF2L, and all peptides were coupled to keyhole limpet hemocyanin. Anti-hACF1 and anti-hISWI have been described previously (Poot et al., 2000). Monoclonal rat α-M31 antibodies were a kind gift from Dr Prim Singh (The Roslin Institute, Edinburgh). Antibody affinity purification with CNBr-activated Sepharose 4B (Amersham Pharmacia), concentration and further purification by (NH4)2SO4 precipitation was performed according to a standard protocol (Harlow and Lane, 1988).
Cell lines, tissue culture and BrdU labeling
Cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with pyruvate, 4500 mg/l glucose (Gibco-BRL) and 100 U/ml penicillin/streptavidin mixture (Gibco-BRL) at 37°C, 5–8% CO2. The NIH 3T3 medium contained 10% fetal bovine serum (FBS). ES cells (XX PGK 12.1; Norris et al., 1994) were grown in flasks coated with 0.1% gelatin in media containing 20% FBS, 2 mM glutamine, 1× non-essential amino acids (Gibco-BRL), 0.5 mM β-mercaptoethanol and 104 U/ml leukemia inhibitory factor (LIF) (Sheardown et al., 1997). Cells had been driven to differentiation by withdrawal of LIF for 9 days (Sheardown et al., 1997). BrdU labeling was performed by the addition of 10 µM BrdU (Sigma) to the medium for 10 min.
Immunofluorescence
All procedures were performed at the room temperature in phosphate-buffered saline (PBS). Incubation time with antibodies was 20 min. NIH 3T3 fibroblasts grown on coverslips were washed twice with PBS, fixed with 1% formaldehyde for 10 min, incubated with 0.5% Triton X-100 for 10 min and then blocked with 10% FBS for 1 h. After incubation in 10% FBS with affinity-purified α-WSTF antibodies (dilution 1:150) or with a mixture of antibodies (α-WSTF, dilution 1:150, and α–M31, dilution 1:100), cells were washed three times for 5 min each time with PBS, incubated with fluorescein isothiocyanate (FITC)-conjugated α-rabbit antibodies (dilution 1:100; Sigma) and finally washed three times for 5 min each time with PBS. For M31 staining, the last incubation step was repeated with Texas Red-conjugated α-rat antibodies (dilution 1:50; Vector). For BrdU staining, cells were fixed further with 2% formaldehyde for 10 min, incubated with 50 mM glycine for 10 min, treated with 0.1% Triton X-100 in 4 M HCl for 10 min and washed with PBS. Staining with primary mouse α-BrdU antibodies (dilution 1:100; Pharmingen) and secondary Texas Red-conjugated α-mouse (dilution 1:100; Vector) antibodies was as above. Slides were mounted in Vectashield (Vector) and immunofluorescence was monitored by confocal microscopy (MRC-600 instrument; Bio-Rad software).
Cell synchronization
For synchronization of cells at the G1/S border, the cells were maintained at 100% confluence for 1–2 days and then they were re-seeded to 60–80% confluence in media containing 3 µg/ml aphidicolin (Sigma) for 18 h.
Mitotic cell synchronization was by 0.05 µg/ml nocadazole (Sigma) for 18–20 h. Mitotic cells were collected by flask shaking, washed with PBS and resuspended in 75 mM KCl. Chromosome spread was obtained by applying of mitotic cells suspension to Cyto-Tek chambers and then spinning onto polylysine-coated slides (BDH) in a Cyto-Tek centrifuge (Sakura, Tokyo) at 2000 r.p.m. for 10 min. Fixation and immunostaining of chromosome spreads was as described previously.
For western blot analysis, nocadazole-arrested mitotic cells were washed with PBS containing 0.5 mM MgCl2 and 0.5 mM CaCl2 (PBS+), then boiled in SDS loading buffer. For mitotic chromosomes, isolation cells were resuspended in 75 mM KCl and pipetted with a 2 ml syringe through a thin needle (25G5/8, 0.5 × 16) on ice, 20–30 times. The chromosome suspension was overlaid on 30% sucrose in a PBS+ cushion and centrifuged at 2000 g for 10 min at 4°C. The pelleted chromosomes were resuspended in PBS+ and then boiled in SDS loading buffer.
Immunoprecipitations and WSTF–ISWI complex purification
Nuclear extracts were prepared according to Dignam et al. (1983). The nuclear extract buffer (buffer C) comprised 20 mM HEPES pH 7.9, 420 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 20% glycerol, 1% Triton X-100, 1 mM DTT, 0.2 mM PMSF and 1× Complete protease inhibitor (Roche). HeLa nuclear extract (100 µl, 3 mg/ml), NIH 3T3 nuclear extract (100 µl, 0.6 mg/ml) or ES nuclear extract (100 µl, 5 mg/ml) was incubated with 20–50 µl of affinity-purified antibodies (0.8–1.6 mg/ml) for 2 h on ice. Protein A–Sepharose CL-4B beads (Pharmacia) were washed twice with IP 400 (20 mM HEPES pH 7.9, 400 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 20% glycerol, 0.1% NP-40, 1 mM DTT) and once with IP 400 containing 1 mg/ml ovalbumin. Fifty microliters of 50% slurry beads were added to 150 µl of the extract/antibody mixture and incubated at 4°C for 1.5 h. Beads were washed twice with IP 400, once with IP 150 (as IP 400, but including 150 M KCl), and were then boiled in SDS loading buffer. For peptide elution, beads were incubated with the antigenic peptides at 0.5 mg/ml in IP 400 for 1 h at 30°C.
Purification of WICH complex
Immunopurification of the WSTF–ISWI complex was with NIH 3T3 cell nuclear extract and α-WSTF antiserum. Protein A–Sepharose CL-4B beads (250–300 µl; prepared as described above) were incubated with 0.5–0.75 ml of antiserum for 1.5 h at 4°C. Beads were washed three times in PBS containing 1 mM DTT, three times in IP 400 and once in IP 230 (as IP 400 but containing 230 mM KCl). Nuclear extract (0.6–5 mg/ml) was diluted with IP 0 (as IP400 but containing no KCl) to obtain a final NaCl concentration of 230 mM. Six hundred microliters of nuclear extract were incubated with antibodies bound to beads for 2 h at 4°C. Beads were then washed three times with IP 230 and once with IP 150. WICH complex was eluted from the beads by incubation for 1 h at 30°C in IP 400 containing 0.5 mg/ml N-terminal WSTF peptide.
The ISWI-B complex was purified from Xenopus egg extract as in Guschin et al. (2000). Protein sequence determination and mass spectrometry were performed by the Protein/DNA Technology Center (Rockefeller University).
Fractionation of HeLa nuclear extract by Superose-6 chromatography and immunodepletion of HeLa nuclear extract was as described previously (Poot et al., 2000). Drosophila embryo extract was prepared as described by Varga-Weisz et al. (1999).
Chromatin assays and micrococcal nuclease digestion
The nucleosome spacing assay using pBluescript plasmid DNA was performed as described previously (Varga-Weisz et al., 1999). Remodeling reaction was for 1 h at 30 and 27°C for mouse and Xenopus WICH, respectively. For assembly of chromatin with polyglutamic acid (PGA), the histone–PGA complexes (40 µg HeLa histones, 20 µg PGA in 800 µl 20 mM Tris–HCl pH 7) were spun at 20 000 g for 5 min at 4°C and mixed with DNA (pBluescript) at a final concentration of 0.9 mg/ml in 20 mM Tris–HCl pH 7, 5 mM KCl, 0.5 mM NaCl, 0.5% glycerol, 0.1% polyvinyl alcohol and 0.1% polyethylene glycol. The histone/PGA/DNA mixture was incubated at 37°C for 3 h, and MgCl2 was added to 5 mM. Chromatin assays were performed with 15 µl of the PGA-assembled chromatin to which the following was added: 15 µl of A50 buffer (20 mM HEPES pH 7.6, 50 M KCl, 5 mM NaCl, 5% glycerol, 1% polyvinyl alcohol, Sigma P-8136, 1% polyethylene glycol, Sigma P-2139), 1 mM ATP and 8 µl of immunopurified WSTF–ISWI complex or mock eluate in IP 400 containing the elution peptide. The reaction was carried out at 30°C for 90 min, followed by incubation at 37°C for 30 min. Micrococcal nuclease digestion, DNA purification and analysis were as described above.
Acknowledgments
Acknowledgements
We thank Dr Raymond Poot for antisera and immunodepleted extracts, Dr Prim Singh (Roslin Institute, Edinburgh) for anti-M31 antiserum, Drs Jacqueline Mermoud and Neil Brockdorff (MRC Clinical Sciences Center, London) for XX PGK 12.1 cells and protocols, Drs Nadine Collins, Custodia García-Jiménez and Raymond Poot for suggestions and protocols, and Drs Gill Elliot and Rob Cross for help with microscopy. Thanks also to Drs Iwao Kukimoto and Enrique Castaño for critical reading of the manuscript. Protein sequence determination and mass spectrometry were performed by the Protein/DNA Technology Center of the Rockefeller University. This work was funded by Marie Curie Cancer Care (L.B. and P.V.W.).
Note added in proof
A study by MacCallum et al. [MacCallum,D.E., Losada,A., Kobayashi,R. and Hirano,T. (2002) ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol. Biol. Cell, 13, 25-39], describing Xenopus WSTF–ISWI complex, was published during the revision of this manuscript.
References
- Aihara T., Miyoshi,Y., Koyama,K., Suzuki,M., Takahashi,E., Monden,M. and Nakamura,Y. (1998) Cloning and mapping of SMARCA5 encoding hSNF2H, a novel human homologue of Drosophila ISWI. Cytogenet. Cell Genet., 81, 191–193. [DOI] [PubMed] [Google Scholar]
- Alexiadis V., Varga-Weisz,P.D., Becker,P.B. and Gruss,C. (1998) In vitro chromatin remodeling by chromatin accessibility complex (CHRAC) at the SV40 origin of DNA replication. EMBO J., 17, 3428–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir R.E., Van den Veyver,I.B., Wan,M., Tran,C.Q., Francke,U. and Zoghbi,H.Y. (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genet., 23, 185–188. [DOI] [PubMed] [Google Scholar]
- Bochar D.A., Savard,J., Wang,W., Lafleur,D.W., Moore,P., Cote,J. and Shiekhattar,R. (2000) A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl Acad. Sci. USA, 97, 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishti A.H. et al. (1998) The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends Biochem. Sci., 8, 281–282. [DOI] [PubMed] [Google Scholar]
- Cryderman D.E., Tang,H., Bell,C., Gilmour,D.S. and Wallrath,L.L. (1999) Heterochromatic silencing of Drosophila heat shock genes acts at the level of promoter potentiation. Nucleic Acids Res., 27, 3364–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R. et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Dignam J., Martin,P., Shastry,B. and Roeder,R. (1983) Eukaryotic gene transcription with purified components. Methods Enzymol., 101, 582–598. [DOI] [PubMed] [Google Scholar]
- Doerks T., Copley,R. and Bork,P. (2001) DDT—a novel domain in different transcription and chromosome remodeling factors. Trends Biochem. Sci., 26, 145–146. [DOI] [PubMed] [Google Scholar]
- Eberharter A., Ferrari,S., Langst,G., Straub,T., Imhof,A., Varga-Weisz,P., Wilm,M. and Becker,P.B. (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodeling. EMBO J., 20, 3781–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Sweder,K.S. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M.H., Arndt-Jovin,D.J., Jovin,T.M., Baumann,P.H. and Robert-Nicoud,M. (1991) Spatial and temporal distribution of DNA replication sites localized by immunofluorescence and confocal microscopy in mouse fibroblasts. J. Cell Sci., 99, 247–253. [DOI] [PubMed] [Google Scholar]
- Francke U. (1999) Williams–Beuren syndrome: genes and mechanisms. Hum. Mol. Genet., 8, 1947–1954. [DOI] [PubMed] [Google Scholar]
- Gelbart M.E., Rechsteiner,T., Richmond,T.J. and Tsukiyama,T. (2001) Interactions of Isw2 chromatin remodelling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol., 21, 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark J.P., Fazzio,T.G., Estep,P.W., Church,G.M. and Tsukiyama,T. (2000) The Isw2 chromatin remodelling complex represses early meiotic genes upon recruitment by Ume6p. Cell, 103, 423–433. [DOI] [PubMed] [Google Scholar]
- Guschin D., Geiman,T.M., Kikyo,N., Tremethick,D.J., Wolffe,A.P. and Wade,P.A. (2000) Multiple ISWI ATPase complexes from Xenopus laevis. Functional conservation of an ACF/CHRAC homolog. J. Biol. Chem., 275, 35248–35255. [DOI] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos,R., Gdula,D.A. and Wu,C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell, 97, 833–842. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Haynes S.R., Dollard,C., Winston,F., Beck,S., Trowsdale,J. and Dawid,I.B. (1992) The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res., 20, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Bulger,M., Pazin,M.J., Kobayashi,R. and Kadonaga,J.T. (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell, 90, 145–155. [DOI] [PubMed] [Google Scholar]
- Ito T., Levenstein,M.E., Fyodorov,D.V., Kutach,A.K., Kobayashi,R. and Kadonaga,J.T. (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev., 13, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.H., Hamana,N., Nezu,J. and Shimane,M. (2000a) A novel family of bromodomain genes. Genomics, 63, 40–45. [DOI] [PubMed] [Google Scholar]
- Jones M.H., Hamana,N. and Shimane,M. (2000b) Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics, 63, 35–39. [DOI] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Längst G. and Becker,P. (2001) Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J. Cell Sci., 114, 2561–2568. [DOI] [PubMed] [Google Scholar]
- Längst G., Bonte,E.J., Corona,D.F. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- Lazzaro M.A. and Picketts,D.J. (2001) Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem., 77, 1145–1156. [DOI] [PubMed] [Google Scholar]
- Lenhoff H.M., Wang,P., Greenberg,F. and Bellugi,U. (1997) Williams syndrome and the brain. Sci. Am., 277, 68–73. [DOI] [PubMed] [Google Scholar]
- LeRoy G., Orphanides,G., Lane,W.S. and Reinberg,D. (1998) Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science, 282, 1900–1904. [DOI] [PubMed] [Google Scholar]
- LeRoy G., Loyola,A., Lane,W.S. and Reinberg,D. (2000) Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem., 275, 14787–14790. [DOI] [PubMed] [Google Scholar]
- Lu X., Meng,X., Morris,C.A. and Keating,M.T. (1998) A novel human gene, WSTF, is deleted in Williams syndrome. Genomics, 54, 241–249. [DOI] [PubMed] [Google Scholar]
- McDowell T.L. et al. (1999) Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl Acad. Sci. USA, 96, 13983–13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C., Reyes,J.C., Bourachot,B., Leguoy,E. and Yaniv,M. (1996) The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J., 15, 3394–3402. [PMC free article] [PubMed] [Google Scholar]
- Norris D.P., Patel,D., Kay,G.F., Penny,G.D., Brockdorff,N., Sheardown,S.A. and Rastan,S. (1994) Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell, 77, 41–51. [DOI] [PubMed] [Google Scholar]
- Okabe I., Bailey,L.C., Attree,O., Srinivasan,S., Perkel,J.M., Laurent, B.C., Carlson,M., Nelson,D.L. and Nussbaum,R L. (1992) Cloning of human and bovine homologs of SNF2/SWI2: a global activator of transcription in yeast S. cerevisiae. Nucleic Acids Res., 20, 4649–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe R.T., Henderson,S.C. and Spector,D.L. (1992) Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol., 116, 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples R.J., Cisco,M.J., Kaplan,P. and Francke,U. (1998) Identification of the WBSCR9 gene, encoding a novel transcriptional regulator, in the Williams–Beuren syndrome deletion at 7q11.23. Cytogenet. Cell Genet., 82, 238–246. [DOI] [PubMed] [Google Scholar]
- Picketts D.J., Higgs,D.R., Bachoo,S., Blake,D.J., Quarrell,O.W. and Gibbons,R.J. (1996) ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum. Mol. Genet., 5, 1899–1907. [DOI] [PubMed] [Google Scholar]
- Poot R.A., Dellaire,G., Hulsmann,B.B., Grimaldi,M.A., Corona,D.F., Becker,P.B., Bickmore,W.A. and Varga-Weisz,P.D. (2000) HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J., 19, 3377–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheardown S.A., Newall,A.E., Norris,D.P., Rastan,S. and Brockdorff,N. (1997) Regulatory elements in the minimal promoter region of the mouse Xist gene. Gene, 203, 159–168. [DOI] [PubMed] [Google Scholar]
- Strohner R., Nemeth,A., Jansa,P., Hofmann-Rohrer,U., Santoro,R., Langst,G. and Grummt,I. (2001) NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J., 20, 4892–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Maison,C., Roche,D. and Almouzni,G. (2001) Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nature Cell Biol., 3, 114–120. [DOI] [PubMed] [Google Scholar]
- Tate P., Lee,M., Tweedie,S., Skarnes,W.C. and Bickmore,W.A. (1998) Capturing novel mouse genes encoding chromosomal and other nuclear proteins. J. Cell Sci., 111, 2575–2585. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Daniel,C., Tamkun,J. and Wu,C. (1995) ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kD subunit of the nucleosome remodeling factor. Cell, 83, 1021–1026. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Palmer,J., Landel,C.C., Shiloach,J. and Wu,C. (1999) Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev., 13, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P.D. (2001) ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene, 20, 3076–3085. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Wilm,M., Bonte,E., Dumas,K., Mann,M. and Becker,P.B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature, 388, 598–602. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Bonte,E.J. and Becker,P.B. (1999) Analysis of modulators of chromatin structure in Drosophila. Methods Enzymol., 304, 742–757. [DOI] [PubMed] [Google Scholar]
- Wallrath L.L. (1998) Unfolding the mysteries of heterochromatin. Curr. Opin. Genet. Dev., 8, 147–153. [DOI] [PubMed] [Google Scholar]
- Wallrath L.L. and Elgin,S.C. (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev., 9, 1263–1277. [DOI] [PubMed] [Google Scholar]
- Wreggett K.A., Hill,F., James,P.S., Hutchings,A., Butcher,G.W. and Singh,P.B. (1994) A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet. Cell. Genet., 66, 99–103. [DOI] [PubMed] [Google Scholar]
- Xiao H., Sandaltzopoulos,R., Wang,H., Hamiche,A., Ranallo,R., Lee,K., Fu,D. and Wu,C. (2001) Dual functions of largest nurf subunit nurf301 in nucleosome sliding and transcription factor interactions. Mol. Cell., 8, 531–543. [DOI] [PubMed] [Google Scholar]