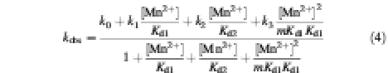Abstract
Interactions with divalent metal ions are essential for the folding and function of the catalytic RNA component of the tRNA processing enzyme ribonuclease P (RNase P RNA). However, the number and location of specific metal ion interactions in this large, highly structured RNA are poorly understood. Using atomic mutagenesis and quantitative analysis of thiophilic metal ion rescue we provide evidence for metal ion interactions at the pro-RP and pro-SP non-bridging phosphate oxygens at nucleotide A67 in the universally conserved helix P4. Moreover, second-site modifications within helix P4 and the adjacent single stranded region (J3/4) provide the first evidence for metal ion interactions with nucleotide base functional groups in RNase P RNA and reveal the presence of an additional metal ion important for catalytic function. Together, these data are consistent with a cluster of metal ion interactions in the P1–P4 multi-helix junction that defines the catalytic core of the RNase P ribozyme.
Keywords: metal ion interactions/phosphorothioate/ribonuclease P/ribozyme
Introduction
The closely packed conformations adopted by structural RNAs result in a high degree of unfavorable electrostatic repulsion that must be offset by interactions with positively charged monovalent and divalent ions (Misra and Draper, 1998). While RNA secondary and some tertiary structural elements can form in the presence of monovalent ions (Shelton et al., 2001; Wadkins et al., 2001), divalent ions are generally required by large structural RNAs to both fold and stabilize their active conformations. Metal ion interactions are also essential for catalytic function of the large RNA enzymes (ribozymes) derived from group I and group II introns, and RNase P RNA (Hanna and Doudna, 2000). Like one-third of all protein enzymes, these ribozymes are thought to act as metalloenzymes, where metal ions directly interact with the substrate phosphate to catalyze phosphodiester bond hydrolysis or transesterification reactions (Narlikar and Herschlag, 1997). The location of individual metal ion binding sites within the RNA catalysts themselves, however, are not well defined, and only a few interactions have been tied biochemically to a specific aspect of enzyme function.
An effective tool for analysis of divalent metal ion interactions in RNA involves the ability of phosphorothioate modifications to disrupt specific magnesium (Mg2+) interactions at non-bridging phosphate oxygens (Eckstein, 1985). This approach relies on the principle that sulfur is a very poor ligand for Mg2+ while softer metal ions, such as manganese (Mn2+) and cadmium (Cd2+), can accept both oxygen and sulfur in their inner coordination sphere. Thus, rescue of the deleterious effects of site-specific sulfur substitutions by inclusion of low concentrations of such ‘thiophilic’ metal ions provides strong evidence for direct metal ion coordination (Jaffe and Cohn, 1978). This method has the general limitations that (i) steric constraints imposed by the larger sulfur atom can alter the geometry of metal ion interactions to preclude thiophilic metal rescue (e.g. Brautigam et al., 1999), and (ii) the method cannot be applied to examine equally important outer sphere interactions with ribose and nucleotide base functional groups. Thiophilic metal ions can also bind to positions other than the site of interest and have indirect effects on structure and activity (Basu and Strobel, 1999). Moreover, it is generally assumed that phosphorothioate modifications do not significantly perturb the native structure or introduce de novo metal ion binding sites and, like other modification studies, there is always a possibility that the effect of metal ion rescue can be indirect. Despite these limitations, the use of phosphorothioate analogs has provided compelling evidence for sites of specific inner sphere metal ion coordination within the conserved structural domains, and at the reactive phosphates of all three of the large ribozymes (Christian et al., 2000; Gordon and Piccirilli, 2001; Shan et al., 2001 and references therein).
An important extension of the metal rescue method, developed by Herschlag, Piccirilli and colleagues, involves quantitative analysis of the dependence of reaction rate on the concentration of the rescuing metal ion(s) (Shan et al., 2001 and references therein). In this approach, the apparent affinity for a particular metal ion interaction can be measured, and compared with the effect of additional enzyme or substrate modifications that may alter its affinity. This type of analysis has allowed the delineation of individual metal ion interactions at the group I intron 5′ splice site (Shan et al., 1999), and provided evidence for individual metal ion ligands in the hammerhead (Peracchi et al., 1997) and the group II ribozyme (Gordon et al., 2000). In principle, quantitative analysis of metal rescue should be useful as a general method for the analysis of interactions within the complex structures of large ribozymes, when inner sphere coordination with non-bridging oxygens can be measured. However, successful application of this experimental approach requires detailed understanding of the reaction kinetics, knowledge of individual candidate functional groups within the hundreds of nucleotides that make up the RNA of interest, and the ability to analyze quantitatively an individual step in the ribozyme reaction.
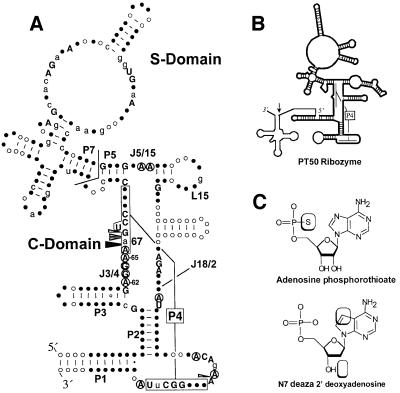
We adapted this approach to identify metal ion interactions in the catalytic RNA subunit of the essential bacterial tRNA processing enzyme ribonuclease P (RNase P RNA). RNase P RNA is a large, ∼400 nucleotide RNA that forms a specific complex with tRNA precursors in vitro, and catalyzes the divalent metal ion-dependent hydrolysis of a specific phosphodiester bond to generate the mature tRNA 5′ end (for a review see Altman and Kirsebom, 1999; Kurz and Fierke, 2000). RNase P enzymes are found in all three phylogenetic domains, and, with the possible exception of the activity identified in spinach chloroplasts (Thomas et al., 1995), all members of this class of enzymes characterized to date contain essential and homologous RNA subunits (Figure 1A). A reasonably detailed perspective of the RNA secondary structure and the role individual structural elements in RNase P function has been obtained by phylogenetic comparative analysis, chemical and enzymatic probing, and kinetic analysis of mutant and modified RNAs. The conserved core of RNase P RNA is composed of two domains: a substrate binding (S) domain that interacts with the T-stem and loop of tRNA, and a catalytic (C) domain that interacts with the pre-tRNA cleavage site at the base of the acceptor stem (Loria and Pan, 1996) (Figure 1A). While the C-domain presumably contains binding sites for catalytic metal ions, these sites have yet to be defined for RNase P RNA, and are generally not well understood for the large ribozymes. The identification of functional groups and elements of ribozyme structure that bind metal ions, especially those linked to transesterification or hydrolysis, remains a central goal of understanding RNA catalysts.
Fig. 1. (A) Consensus secondary structure of bacterial RNase P RNA (Brown, 1999). Helices are designated P for paired region. Helix P4, referred to in the text, is boxed and connected by brackets. Elements of non-Watson–Crick structure joining helices are designated J for joining and numbered according to the helices that they connect. The bent line between P5 and P7 marks the boundary between substrate binding (S) and catalytic (C) domains. Nucleotides are numbered according to E.coli RNase P RNA. Upper case letters indicate nucleotide positions conserved among all known bacterial RNase P RNAs. Lower case letters indicate 80% conservation in nucleotide identity. Filled and open circles reflect 100 and 80% conservation in nucleotide position, respectively. Arrows indicate locations of phosphorothioate sensitivity (Harris and Pace, 1995). Filled arrows indicate position of thiophilic metal ion rescue (Christian et al., 2000). Circled nucleotides indicate positions important to catalysis identified by NAIM (Kazantsev and Pace, 1998; Kaye et al., 2002). (B) Schematic diagram of PT50 secondary structure. Dark lines represent Native RNase P RNA sequences and thin lines represent pre-tRNA and joining sequences. 5′ and 3′ ends are indicated by italics. Arrow indicates position of cleavage site. (C) Nucleotide analogs used in the current work. Boxes indicate positions of atomic substitutions.
Specific regions of the RNase P RNA that are involved in catalytic function have been identified by mutational and chemical modification, and crosslinking experiments. These studies showed that conserved sequences in P4, L15, J5/15 and J18/2 within the C-domain are in proximity to the pre-tRNA cleavage site, and contain chemical groups that are important for catalysis (Altman and Kirsebom, 1999; Kurz and Fierke, 2000 and references therein). In particular, site-specific functional group modification experiments identified non-bridging phosphate oxygens in helix P4 that, when substituted with sulfur, dramatically inhibit ribozyme activity (Figure 1A). Rescue of the phosphorothioate interference effects by Mn2+ and Cd2+ strongly argues that one role of helix P4 is to position divalent metal ions that are important for catalytic function (Christian et al., 2000). This hypothesis is supported by the observation that point mutations (C70U and U69 deletion in Escherichia coli RNase P RNA) in helix P4 and adjacent to the positions of phosphorothioate sensitivity enhances the use of Ca2+ as a catalytic metal ion (Frank and Pace, 1997). These experiments, however, do not distinguish between effects on ribozyme structure, substrate positioning or direct effects on active site function. Moreover, it is not known how many metal ions interact with the P4 metal binding site, or what other elements of structure contribute to the affinity of metal ions in the C-domain.
Further insight into the structural elements that may be involved in metal ion binding in E.coli RNase P comes from the recent identification of base functional groups important to catalysis by nucleotide analog interference mapping (NAIM) and site-specific modification (Kazantsev and Pace, 1998; Kaye et al., 2002). All of the base functional groups identified occur at highly conserved nucleotide positions within the ribozyme’s C-domain. Moreover, these functional groups cluster within the same discrete regions of helix P4, J5/15 and J18/2 associated with catalysis in previous structural studies noted above. Of particular interest is the sensitivity of the interference signal to Mg2+ concentration, and the finding that a significant number of these functional groups are located adjacent to the position of metal ion coordination in helix P4. The proximity of these catalytically important base functional groups to the site of metal ion coordination at A67, combined with the observed sensitivity of the interference signal to Mg2+ concentration, suggested that these moieties participate directly, or indirectly in metal ion interactions important for ribozyme function.
In the present analysis we utilized atomic mutagenesis and quantitative analysis of thiophilic metal ion rescue to characterize metal ion binding within helix P4, and to analyze the contribution of base functional groups to metal ion interactions within the ribozyme’s conserved catalytic core. This approach permitted the measurement of distinct metal ion affinities for both the pro-RP and pro-SP non-bridging phosphate oxygens at A67. Intriguingly, second-site modifications targeting base functional groups in P4 and J3/4 identified additional metal ligands at A65 and A66, and revealed the presence of a new metal ion bound in the universally conserved P1–P4 multi-helix junction that defines the conserved core of RNase P RNA. Taken together, these data are most consistent with a cluster of metal ion interactions in the P1–P4 multi-helix junction that is crucial to RNase P function.
Results
Effects of phosphorothioate modification and thiophilic metal ion rescue on ribozyme catalysis
To identify ligands for the metal ions bound within the catalytic core of the RNase P ribozyme, we examined the effect of individual functional group modifications on the binding of metal ions within helix P4. These experiments utilized self-cleaving E.coli RNase P ribozymes covalently linked to pre-tRNA substrate sequences (Frank et al., 1994). The tethered construct used in these experiments (PT50; Figure 1B) both accurately cleaves its pre-tRNA substrate and has a pH and metal ion dependence that is essentially identical to that of the native RNase P RNA (Christian et al., 2000). The PT50 ribozyme provides a convenient experimental means of introducing atomic substitutions adjacent to the site of metal coordination in helix P4 by oligonucleotide-directed ligation (Moore and Sharp, 1992; Figure 1B). Modified ribozymes are defined by the type and position of the incorporated atomic substitutions. RNase P RNAs containing an SP phosphorothioate at A67, for example, is referred to in the text as A67SP, while ribozymes containing additional dN7dA modification at A65 are referred to as dN7dA65,A67SP (Figure 1C).
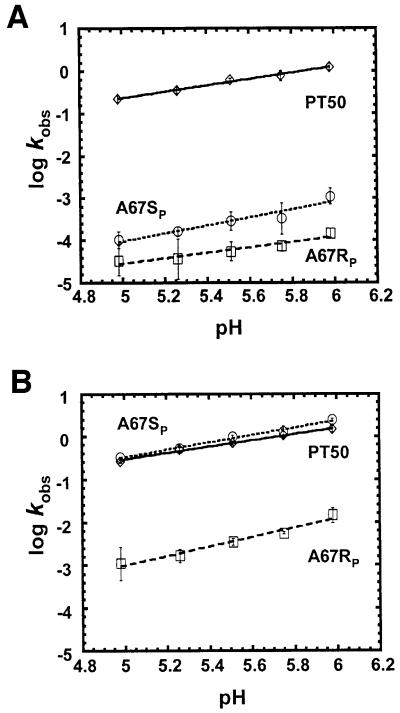
Because the current analysis of metal ion binding centered on thiophilic metal ion rescue of inhibition due to phosphorothioate substitution in helix P4, it was important to determine whether phosphorothioate substitution or thiophilic metal rescue resulted in changes in the rate-limiting step of the cleavage reaction, since such changes could complicate the interpretation of kinetic data (Shan and Herschlag, 2000). An important characteristic of the hydrolysis reaction characterized by RNase P is the dependence of the reaction rate on pH (Smith and Pace, 1993). Figure 2A shows that while phosphorothioate modification of either the pro-SP or pro-RP oxygens 5′ to A67 results in a decrease in the rate of catalysis by over three orders of magnitude, the dependence of the residual reaction rate on pH is maintained. The somewhat reduced pH dependence of A67RP relative to A67SP and PT50 appears to be due to the larger error associated with rates near the limit of detection (Figure 2A). The pH dependence of A67RP between 5.26 and 5.98 (0.80 ± 0.12) is indistinguishable from that observed for A67SP and PT50. Inclusion of 5 mM Mn2+ in the reaction results in a complete (∼5000-fold) rescue of the effect of A67SP phosphorothioate modification and partial, but significant (∼65-fold), rescue of the A67RP modification (Christian et al., 2000). In each case, the reactions retain pH sensitivities that show little to no change in dependence relative to magnesium alone (Figure 2B). These experiments therefore indicate that the modified ribozymes, in the presence and absence of rescuing metal ion, share the same rate-limiting step with each other and with the unmodified control.
Fig. 2. pH dependence of the reaction rates of phosphorothioate- modified ribozymes in the absence (A) and presence (B) of Mn2+. All reactions maintained a total concentration of 25 mM divalent metal ion: (A) 25 mM Mg2+, (B) 5 mM Mn2+, 20 mM Mg2+. Plots of log kobs versus pH are shown for the unmodified PT50 ribozyme (diamonds) and PT50 ribozymes that have either site-specific RP (squares) or SP (circles) phosphorothioate substitutions at A67. Data are fit to a linear equation and reflect at least three different trials. Slope of the pH dependence in the presence of 25 mM Mg2+ (A): 0.74 ± 0.04 for PT50, 0.95 ± 0.16 for A67SP, 0.63 ± 0.11 for A67RP (0.80 ± 0.12 between pH 5.26 and 5.98). Slope of the pH dependence in the presence of 5 mM Mn2+ and 20 mM Mg2+ (B): 0.73 ± 0.04 for PT50, 0.86 ± 0.06 for A67SP, 1.10 ± 0.11 for A67RP. All values contain an experimental error of 25% or less.
Quantitative analysis of metal ion interactions at A67
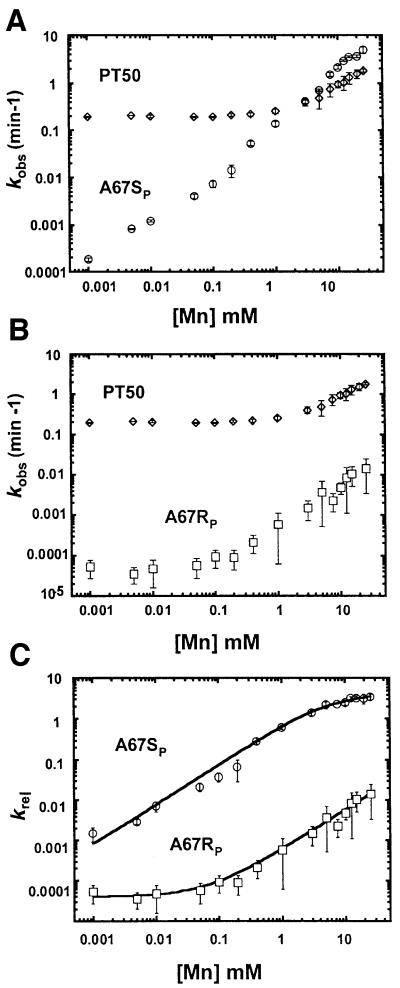
The apparent affinity for Mn2+ at the non-bridging phosphate oxygens of A67 was determined by examining the concentration dependence of the reaction rate on Mn2+ for ribozymes containing site-specific phosphorothioate modifications at either the pro-RP or pro-SP positions. Because PT50 shows a decrease in pH dependence of its reaction rate above 50 mM total divalent metal ion (Kaye et al., 2002), all reactions were carried out in a constant background of 10 mM Mg2+. This level of Mg2+ is in excess of that required for complete folding and catalytic activity of the ribozyme (see Pan et al., 1999 and references therein), but still allows a significant range of metal ion concentration in which to measure Mn2+ binding. In addition, the relative rate constant (krel) was examined to control for non-specific effects of increasing Mn2+ concentration on the reaction rate. Specifically, krel was derived from the observed rate of ribozymes containing a single sulfur substitution divided by the observed rate of the unmodified, oxygen-containing ribozyme (krel = ksobs/koobs). This ratio of reaction rates normalizes the contribution of all native functional groups in the molecule to the observed rate, allowing the measurement of the relative effect of an individual modification on cleavage as a function of thiophilic metal ion concentration. Subsequent fitting of relative rate values to models of single or multiple metal ion binding allows determination of the apparent metal ion affinity at the position of atomic substitution (see Materials and methods; Shan and Herschlag, 1999).
Figure 3 shows the measurement of the affinity of the rescuing metal for the A67RP and A67SP ribozymes. The data for Mn2+ rescue at each of the non-bridging phosphate oxygens at A67 fit well to a single binding isotherm. The apparent Mn2+ affinity (KMn, app) for the A67SP ribozyme was measured at 5.7 ± 0.8 mM. In contrast, the observed rate of A67RP RNA self-cleavage is not saturated at the highest concentration of Mn2+ tested (30 mM). As noted above, metal ion concentrations higher than 50 mM result in an apparent change in the rate-limiting step of the PT50 reaction (Kaye et al., 2002), precluding precise measurement of the affinity of the metal ion interaction with the A67RP modification. However, the binding isotherm of A67RP relative to A67SP is shifted to much higher concentrations of Mn2+ (Figure 3C). The KMn,app for the A67RP modification lies in a concentration range between the highest level of Mn2+ on the experimental curve and the theoretical concentration required for complete rescue of the phosphorothioate defect (>>15 mM), Mn2+ levels significantly higher than that required for binding at A67SP. The large difference in KMn, app for A67SP and A67RP is maintained even when background levels of Mg2+ are increased from 10 to 25 mM, but with a proportional loss in Mn2+ affinity (A67SP KMn, app ∼14 mM, A67RP KMn,app >>14 mM; data not shown). Taken together, these findings indicate the presence of two distinct metal ion ligands that may or may not involve the same metal ion, and that the binding of Mn2+ associated with A67SP and A67RP occurs at a position in the ribozyme normally occupied by Mg2+.
Fig. 3. Quantitative analysis of thiophilic metal ion rescue of RP and SP phosphorothioate modifications at A67. (A) A plot of kobs versus Mn2+ concentration is shown for the unmodified ribozyme (PT50; diamonds) and for ribozymes with a site-specific SP phosphorothioate modification at A67 (A67SP; circles). (B) Plot of kobs versus Mn2+ concentration for unmodified PT50 (PT50, diamonds) and for ribozymes with a site- specific RP phosphorothioate modification at A67 (A67RP; squares). Rate constants for all reactions were determined in a constant background of 10 mM Mg2+, 3 M NaCl, 50 mM PIPES pH 5.5 at 50°C. (C) Relative reaction rates (krel) for the pro-SP (A67SP; circles) or pro-RP (A67RP; squares) phosphate oxygen at A67 are plotted as a function of added Mn2+ and fit to an equation for binding of a single metal ion (Equation 1, Materials and methods). For A67SP, KMn, app = 5.7 ± 0.8 mM; for A67RP, KMn,app >>15 mM. Error bars reflect standard deviation of at least four experiments.
Evidence for additional metal ion interactions in helix P4 and the P1–P4 helix junction
To identify functional groups linked to metal ion interactions at A67, we examined nucleotide positions that were shown to be important to catalytic activity by NAIM and site-specific modification (Figure 1A; Kaye et al., 2002). In the current analysis, we targeted functional groups in J3/4 and helix P4 because of the prevalence of important functional groups in this region, as well as the obvious proximity to the A67 metal ion interactions. In particular, we focused on nucleotides that showed significant inhibition of observed reaction rate due to modification at the N7 position of purines, because of the association of this functional group with metal ion coordination in numerous structural studies (Feig and Uhlenbeck, 1999). Three such positions (A62, A65 and A66) are found in J3/4 and helix P4.
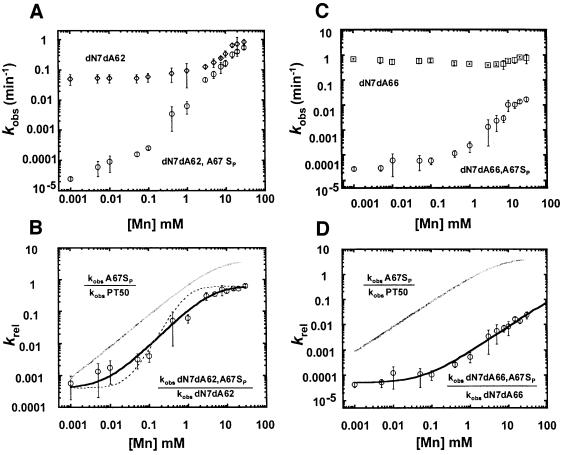
To examine whether the base functional groups analyzed above are linked to metal ion coordination at A67, we determined the effect of modifications at the N7 positions of A62, A65 and A66 on the apparent affinity for metal ions interacting with the pro-SP phosphate oxygen of A67. Because N7 deaza analogs are only available as deoxynucleotides, modified ribozymes were constructed with N7 dN7dA modifications at either A62, A65 or A66, in addition to an SP phosphorothioate modification 5′ to nucleotide A67 (A67SP; see Materials and methods). Previous studies with 2′deoxy alone at A62, A65 or A66 showed that 2′deoxy substitution does not appear to influence the effect of the N7 substitution (Kaye et al., 2002). In addition, dN7dA62, dN7dA62 or dN7dA62 modifications in combination with A67SP (e.g. dN7dA62,A67SP) retain log–linear dependence of reaction rate on pH and thus, like the phosphorothioate modifications alone, do not appear to result in a change in rate- limiting step (data not shown). The observed cleavage rates of ribozymes modified at two nucleotide positions (dN7dA62,A67SP, dN7dA65,A67SP or dN7dA66,A67SP) were determined over a range of Mn2+ concentrations, and compared with that of ribozymes lacking the phosphorothioate modification (dN7dA62, dN7dA65 or dN7dA66) (Figures 4A and C, and 5A). Analogous to the normalization of non-specific effects above, the relative rate constant (i.e. krel = kobs dN7dA62,A67SP/kobs dN7dA62) was plotted as a function of the rescuing metal ion concentration, allowing isolation of the rescuing metal interaction with the pro-SP phosphorothioate substitution at position A67 (Figures 4B and D, and 5B).
Fig. 4. Effect of second-site modification at A62 and A66 on metal binding affinity at A67. Observed reaction rates (kobs) for modified ribozymes dN7dA62 and dN7dA66 [diamonds (A) and squares (C), respectively] and dN7dA62,A67SP and dN7dA66,A67SP [circles (A) and (C), respectively] are plotted as a function of added Mn2+ as described in Figure 3. Relative reaction rates (krel) due to second-site dN7dA modification at A62 (B) or A66 (D) (circles) are plotted as a function of added Mn2+ and fit to an equation for binding of a single metal ion, as in Figure 3. For dN7dA62,A67SP, KMn, app = 4.8 ± 0.9 mM, nHill = 1.2 ± 0.2 (χ2 = 0.01). For dN7dA66,A67SP, KMn, app >>15 mM, nHill = 0.99 ± 0.01 (χ2 = 0.01). Gray curves in (B) and (D) reflect single metal ion binding curves of A67SP ribozymes for comparison. The dashed line in (B) shows theoretical fit for two metal ion binding (nHill = 2, χ2 = 0.22).
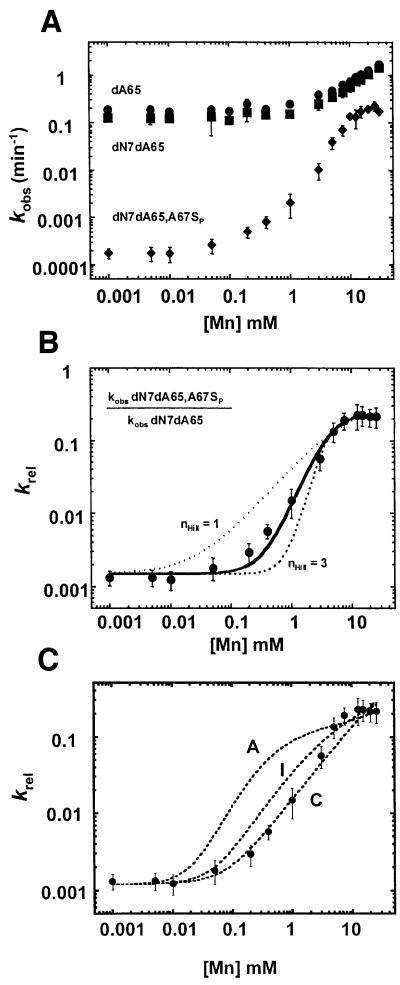
Fig. 5. Second-site modification at A65 reveals an additional metal ion interaction. (A) Observed reaction rates (kobs) for modified ribozymes dA65 (circles), dN7dA62 (squares) and dN7dA66,A67SP (diamonds) are plotted as a function of added Mn2+ as described in Figure 3. (B) Relative reaction rate (krel) is plotted as a function of added Mn2+ and fit to the Hill equation (Equation 3, Materials and methods). The solid line represents a fit with nHill = 2. The dotted lines represent fits with nHill = 1 and nHill = 3 as indicated. KMn, app = 4.4 ± 0.3 mM. (C) Fits of binding data in (B) to theoretical models for two metal ion binding (Scheme 1 and Equation 4). A, I and C reflect models for anti-cooperative, independent and cooperative binding (m values of 10, 1 and 0.1, respectively).
As shown in Figure 4, second-site dN7dA modifications at A62 and A66 both fit well to a single binding isotherm, as observed for the A67SP modification alone, but have different effects on the apparent metal ion affinity. The apparent affinity of Mn2+ for the A67SP ribozyme is essentially unchanged (4.8 ± 0.9 mM versus 5.7 ± 0.8 mM) when the N7 of A62 is replaced with carbon. We conclude from these data that although the dN7dA62 ribozyme has a measurable decrease in the rate of catalysis, this defect is not linked to direct metal ion interactions at A67 (Figure 4B). Second-site modification at A66, however, does influence the apparent affinity of the metal bound at A67 (Figure 4D). While the data still fit to a single binding isotherm, we do not observe saturation at the highest concentration of Mn2+ tested, similar to the behavior of the A67RP ribozyme (Figure 3). The KMn, app for the dN7dA66,A67SP ribozyme again lies in the concentration range between the highest Mn2+ concentration on the experimental curve and the theoretical concentration required for complete rescue of the phosphorothioate defect (>>15 mM), which is significantly higher than the Mn2+ levels required for binding to A67SP phosphorothioate modification alone (Figure 4D). The metal ion interaction at the A67 phosphate oxygen, therefore, is significantly weakened by modification of the nucleotide base at A66. This suggests that metal ion coordination to the A67 pro-SP oxygen is directly or indirectly linked to the N7 of A66. The simplest interpretation of this observation is that the A67 pro-SP oxygen and the N7 of A66 both form contacts to the same metal ion.
In contrast to the single binding isotherms observed in the relative rate constant for second site modifications at A62 and A66, the binding isotherm for second site dN7dA modification at A65 is more complex (Figure 5A and B). In this case the data fit the Hill equation with a curve that is consistent with the binding of at least two metal ions (nHill = 2) but not consistent with Hill values of three or larger. Importantly, krel in this experiment, like those above, examines metal ion binding to only the sulfur substituted non-bridging phosphate oxygen at A67 (krel = kobs dN7d65,A67SP/kobs dN7dA65). Thus, these data suggest that two metal ions either interact directly with the pro-SP non-bridging oxygen of A67, or are thermodynamically coupled to interactions involving this functional group.
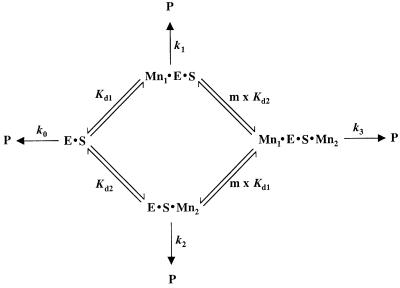
To explore the nature of metal ion binding at A67SP further, we examined theoretical models for the binding of two metal ions which give Mn2+ concentration dependencies that approximate the observed binding data. Scheme 1 reflects a general kinetic scheme for the binding of two metal ions required for activity (from Wang et al., 1999).
Scheme 1.
Using this framework, the binding of metal ions in the ground state can be modeled as either cooperative, anti-cooperative or independent (see Materials and methods). Given known values for kobs, [Mn2+] and the observed reaction rate in the absence of Mn2+, simulations were carried out over a range of assigned values of metal ion affinity and degree of cooperativity and plotted with the experimental data. We find that the metal ion dependence observed with second site modification at A65 is most consistent with models for cooperative binding of two metal ions with affinities that are either identical (∼2.5 mM) or distinct (∼0.05 and 5 mM) (Figure 5C).
Discussion
In the present study, we characterize catalytically important base functional groups within E.coli RNase P RNA, the proximity to phosphate oxygens linked to metal binding of which suggested that they may contribute to the affinity of essential metal ions. The metal ion affinity at non-bridging phosphate oxygens was determined to provide a basis of comparison for the effects of second-site nucleotide base modifications. The concentration dependence of Mn2+ rescue was found to be distinct for each of the pro-RP and pro-SP non-bridging phosphorothioate modified ribozymes at A67 (Figure 3). Although distinct metal ion affinities may imply the presence of two distinct metal ions, it is also possible that modification of different ligands of the same metal ion could have distinct effects on binding activity. The apparent differences in affinity may be due to changes in the steric or electrostatic environment engendered by phosphorothioate modification, as has been observed in protein enzymes (Brautigam and Steitz, 1998). However, the differences in binding affinity do not appear to be due to changes in rate-limiting step, since the pH dependence of the reaction rate is not significantly altered by phosphorothioate modification, in either the presence or absence of rescuing metal ion (Figure 2).
Some insight into the nature of metal ion coordination to an individual phosphate can be gained through the examination of high resolution RNA structures which provide a large number of metal sites for comparison. Using the Metalloprotein Database (Castagnetto et al., 2002) we surveyed 324 metal binding sites in crystal structures of 10 different RNAs [tRNAPhe, the group I intron P4–P6 domain, the hammerhead, hairpin and human delta l virus (HDV) ribozymes, 4.5S RNA, 5S RNA, the large rRNA subunit, the small rRNA subunit, and the HIV genomic dimerization site] as a representative sample of the available structural data (Cate et al., 1996; Scott et al., 1996; Correll et al., 1997; Ferre-D’Amare et al., 1998; Ennifar et al., 1999; Batey et al., 2000; Jovine et al., 2000; Klein et al., 2001; Ogle et al., 2001). We found only one occurrence of a single metal ion bound to both non-bridging of the same phosphate, in contrast to 13 examples of non-bridging oxygens of the same phosphate binding-independent metal ions (data not shown). Thus, while the current experiments do not distinguish between the same or distinct metal ions binding to the non-bridging oxygens at A67, we favor the latter interpretation given its prevalence in the existing structural database.
Given a distinct metal ion interaction the affinity of which we could accurately measure (A67SP), we asked whether secondary modification at nucleotide base functional groups identified by NAIM alter the affinity of metal ion interactions at A67. We found that base modification at A62 showed little effect on metal ion binding at A67 and is likely to interact with other elements within the C-domain (Figure 6A). In contrast, dN7dA modifications at A65 and A66 produce large effects on the observed metal ion binding properties of the pro-SP non-bridging oxygen at A67. Second-site dN7dA modification of A66 led to a significant weakening of metal ion affinity to the A67SP phosphorothioate modification, while dN7dA modification of A65 revealed the presence of an additional metal ion associated with the A67 pro-SP oxygen (Figure 6A).
Fig. 6. Summary of proposed metal ion interactions. (A) Schematic of possible metal ion interactions proposed in the current work. Functional groups proposed to be directly or indirectly linked to metal ion interactions at A67 are colored red. Functional groups not linked to metal ion interactions at A67 are colored blue. (B) Consensus secondary structure of the RNase P ribozyme modified from Figure 1 to highlight positions of functional groups analyzed in the current work, shown using the color scheme in (A), in addition to positions of strong phosphorothioate effects that could not be rescued in the presence of thiophilic metal ions, shown in yellow (Christian et al., 2000). Small filled circles indicate portion of molecule shown in (C). (C) Position of functional groups in the current three-dimensional model of the RNase P ribozyme (Massire et al., 1998). Stereo view shows universally conserved four helix junction displaying functional groups and color scheme used in (A) and (B). Large spheres indicate position of strong phosphorothioate effects. Small spheres indicate positions of N7 functional groups tested in the current work.
Interestingly, the presence of the additional metal ion is only ‘revealed’ by the dN7dA modification at position A65. One possible explanation for this effect is that while the A67 phosphorothioate modification disrupts the binding of both of these metal ions (e.g. MA2+ and MB2+; Figure 6A), the presence of an additional interaction specific to only one of the metal ions (MB2+) may help to stabilize the binding of the second metal ion relative to the first. In this scenario, MB2+ becomes revealed when its affinity is further reduced due to modification of the N7 position of A65, and subsequently becomes more dependent on interaction with the non-bridging phosphate oxygen at A67. Consistent with this hypothesis, the binding of two metal ions with significantly different affinities can be fit to the observed Mn2+ binding dependence of dN7dA65,A67SP ribozymes using a general kinetic scheme for two metal binding (Figure 5C; Wang et al., 1999). The association of N7 at A65 with a position of tight metal ion binding is also suggested by the observation that this functional group can only be detected by modification interference at low levels of divalent metal ion (1–2 mM), concentrations significantly below the apparent metal ion affinities measured in these experiments. Because the log–linear dependence of the reaction rate is maintained over the divalent metal ion concentration range from 1.25 to 40 mM, the loss of modification interference at the N7 of A65 at higher levels of divalent metal ion (>2 mM) does not appear to be due to a change in rate-limiting step of the reaction, but is more consistent with the saturation of metal binding linked to this functional group. The absence of a deaza modification at A65 may thus ensure that MB2+ is essentially saturated over the range of added Mn2+ that we measure for single metal ion binding of MA2+ in the context of the A67 pro-SP phosphorothioate modification alone.
It is important to note that the current experiments do not exclude the possibility that MB2+ is the same as that potentially associated with A67RP (MN2+), since metal ion binding at these positions was measured in different structural backgrounds (dA65,A67SP and wild type, respectively). The simplest interpretation, however, is that the large difference in apparent Mn2+ affinity at A65 and A67RP (<2 versus >15 mM, respectively) is indicative of distinct metal ions. The experimental data also do not distinguish between a direct interaction of MB2+ with A67SP, and an indirect thermodynamic linkage through additional interactions involving this functional group. In fact, recent functional group modification data suggests that A65 may form a contact with a second universally conserved nucleotide (A361), which occurs immediately adjacent to the 3′ strand of helix P4 and is thus likely to be in close contact with A66 and A67. The interaction of two distinct divalent metal ions with a single non-bridging phosphate oxygen nevertheless remains an intriguing possibility, as there is precedent for this motif in theoretical and tested models of metal ion binding to the Tetrahymena group I intron scissile phosphate and in recent crystal structures of the small and large subunits of the ribosome (Klein et al., 2001; Ogle et al., 2001; Shan et al., 2001).
The position of MA2+, MB2+ (and possibly MN2+) within helix P4 and the P1–P4 multi-helix junction suggests that an important aspect of their function lies in determining the ribozyme’s active conformation (Figure 6B and C). These metal ions appear to be adjacent to, and may help to stabilize, a sharp bend predicted in the backbone structure that occurs between helices P3 and P4, as well as being positioned within or adjacent to highly conserved single-stranded regions whose precise juxtaposition is likely to comprise a central element of the enzyme’s active site (Chen et al., 1998; Massire et al., 1998). Although monovalent ion conditions differ, the binding of two metal ions characterized here (MA2+ and MB2+) occurs roughly over the concentration range found to be required for the folding of the active RNase P ribozyme in both E.coli and Bacillus subtilis ([Mg2+]1/2 ∼2–3 mM) as assayed by hydroxyl radical cleavage, oligonucleotide binding, spectroscopy and catalytic activity (see Pan et al., 1999 and references therein). The range of metal ion affinities is also similar to that of catalytic metal ion interactions within the group I ribozyme (KMn,app ∼0.06–13 mM; Shan et al., 1999) and perhaps (in the case MN2+) the binding of a single metal ion linked to the transition state for pre-tRNA cleavage in the native B.subtilis ribozyme (KdMg ∼36 mM; Beebe et al., 1996). Owing to the nature of the current experiments and the lack of a detailed structural understanding of the ribozymes catalytic core, it is not possible to predict the nature or the extent to which MA2+ or MB2+ (or MN2+) provides a local or global structural role in determining ribozyme activity. Nevertheless, the observation that the phosphorothioate modifications examined here do not alter the pH dependence of the reaction supports the view that a change to a different rate-limiting step, such as folding, has not been engendered by disrupting these metal ion interactions, and that we are examining the effect of metal ion binding to the native and active form of the ribozyme. However, misfolding without an observed change in pH dependence is still possible if the unfolded, inactive conformation is in rapid equilibrium with the folded, active state.
It has been hypothesized that the role of the metal ions located in helix P4 is linked to catalysis (Harris and Pace, 1995; Christian et al., 2000). However, short or long range crosslinking from the pre-tRNA cleavage site does not detect helix P4 or J3/4, where we observe metal ion coordination (see Christian and Harris, 1999 and references therein). Although short and long range crosslinking studies have given consistently similar results, the absence of a crosslink between the cleavage site and helix P4 may be due in part to the fact that long range crosslinking was done in the context mature tRNA, which lacks functional groups known to be important in binding and catalysis. Nevertheless, while other physical constraints place helix P4 within 10–15 Å of the cleavage site in current three-dimensional models of the ribozyme, there is insufficient structural information to directly link positions of metal ion coordination within helix P4 and the pre-tRNA cleavage site. Moreover, it is also important to point out that the catalytic role of the metal ions identified in helix P4 can be indirect, simply requiring their presence for catalysis to occur using other metals or RNA functional groups.
However, there are a number of observations that are more consistent with a direct role of functional groups and metal ions identified in helix P4 in catalytic function. First, the effects of phosphorothioate substitution at A67, G68 and A352 are two to four orders of magnitude larger than modification of other functional groups in the ribozyme, and occur under all experimental conditions tested (Christian et al., 2000). Secondly, the positions of metal ion coordination described here occur at highly or universally conserved nucleotides within the ribozyme’s catalytic core, and adjacent to nucleotide bases (U69 and C70) that influence the utilization of Ca2+ as a catalytic metal ion (Frank and Pace, 1997). The only other functional group modifications that inhibit ribozyme function at a magnitude similar to those identified in P4 are 2′deoxy and phosphorothioate substitutions at the cleavage site itself (Smith and Pace, 1993; Warnecke et al., 1996). Thirdly, deletion of the universally conserved bulged uridine at position 69 leads to both the unique loss of metal-dependent cleavage at positions A66 and A67, and a change in the number and affinity of metal ions required for catalysis (N.M.Kaye, N.H.Zahler, E.L.Christian and M.E.Harris, in preparation). Interestingly, similar effects on the number of metal ions required for catalysis were observed for 2′deoxy substitution at the pre-tRNA cleavage site (Smith and Pace, 1993). These observations suggest an important functional link between chemical groups and metal ions identified in helix P4 and the pre-tRNA cleavage site and thus provide compelling candidates for elements that may define a portion of the ribozyme’s active site.
Materials and methods
RNA synthesis and ribozyme reactions
The synthesis of wild-type and site-specific mutants of PT50 RNase P RNA has been described previously (Christian et al., 2000). Enzymatic reactions were performed as described below, but varied with respect to pH and divalent ion conditions. Four microliters of transcribed or ligated RNA (2 nM) dissolved in 0.1% NP-40 were mixed with 32 µl of 1.25× reaction buffer (3.75 M NaCl, 62.5 mM PIPES, pH 5.5 at 50°C) and annealed in a PTC-100 programmable thermal controller (MJ Research; 85°C for 1 min, cooled to 65°C at 0.2°C/s, maintained at 65°C for 5 min, cooled to 50°C at 0.25°C/s and incubated for 3.0 h at 50°C). Reactions were then initiated by adding 4 µl of 100 mM MgCl2. The final reaction conditions were 0.2 nM RNA, 10 mM MgCl2, 3 M NaCl, 50 mM PIPES pH 5.5, 50°C. MgCl2 was added by rapid pipetting for 5 s. Aliquots (2 µl) were removed at various times and quenched by mixing with formamide gel loading buffer containing at least a 2:1 excess of EDTA relative to the total divalent metal ion concentration (typically 80 mM EDTA, 98% formamide). Products were loaded directly onto 6% polyacrylamide (8 M urea) gels and the products visualized by PhosphorImager analysis (Molecular Dynamics). Experiments where Mn2+ concentrations were varied were performed as described above, but with corresponding changes in the concentration of MnCl2 used to start the reaction. Similarly, reactions at various pHs were as described but with reaction buffer pH adjusted accordingly.
Analysis of reaction kinetics
The pseudo first-order reaction rate constant (kobs) was determined by fitting plots of the fraction of RNA cleaved as a function of time to an equation for a single exponential:
where [P] is the amount of product formed at time t, [S]total is the initial amount of tethered ribozyme added to the reaction, A represents the maximal extent of the reaction and B is the amplitude of the exponential. In cases where modifications produced large rate defects, the rate of product accumulation begins to be augmented to a limited extent by a small fraction (5–20%) of very slowly cleaving ribozymes that is present in both wild-type and mutant preparations of ligated RNAs. These data were better fit by an equation for a double exponential increase as follows:
where k1 represents the larger first-order rate constant, k2 represents the smaller rate constant and C is the amplitude of the exponential for the small rate constant.
To control for non-specific effects on ribozyme structure due to changes in divalent ion concentration, the relative reaction rate (krel) was calculated from the ratio of the observed reaction rates (kobs) of molecules that differ by a single atomic substitution. In phosphorothioate substitution for example, krel is equal to the observed rate with and without the sulfur substitution (e.g. krel = kobss/kobso). Plots of krel versus added Mn2+ concentration ([Mn2+]) were fit with a non-linear form of the Hill equation for cooperative binding:
where n is the Hill coefficient that gauges the cooperativity of Mn2+ binding (n = 1 represents no cooperativity) and KMn represents the Mn2+ concentration required to attain half the maximal rate (kmax) of self-cleavage at saturating Mn2+ levels.
Theoretical models to distinguish between independent, cooperative and anti-cooperative interactions of metal ions in the observed binding data utilized Scheme 1 (see Results), which describes a general kinetic scheme for the binding of two metal ions, and Equation 4, which describes the predicted dependence of the observed self-cleavage rate on Mn2+ concentration (Wang et al., 1999).
Independent binding is indicated by m values equal to 1, while cooperative and anticooperative binding are indicated by m values <1 or >1, respectively. Simulations were carried out over a range of m values (0.01–100), as well as with assigned values for Kd1 and Kd2 (0.001–10 and 0.1–100, respectively) using Equation 4 and plotted (KaleidaGraph, Synergy Software) with the experimental data to assess fit to models for metal ion binding.
Acknowledgments
Acknowledgements
We thank Drs Joseph Piccirilli, Dan Herschlag, Alexander Krauchuk and Mark Caprara for helpful discussion and careful review of the manuscript. This work was supported by NIH grant GM56740 to M.E.H. N.M.K. is supported by NIH training grant GM08056.
References
- Altman S. and Kirsebom,L. (1999) Ribonuclease P. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 351–380.
- Basu S. and Strobel,S.A. (1999) Thiophilic metal ion rescue of phosphorothioate interference within the Tetrahymena ribozyme P4–P6 domain. RNA, 5, 1399–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- Beebe J.A., Kurz,J.C. and Fierke,C.A. (1996) Magnesium ions are required by Bacillus subtilis ribonuclease P RNA for both binding and cleaving precursor tRNAAsp. Biochemistry, 35, 10493–10505. [DOI] [PubMed] [Google Scholar]
- Brautigam C.A. and Steitz,T.A. (1998) Structural principles for the inhibition of the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J. Mol. Biol., 277, 363–377. [DOI] [PubMed] [Google Scholar]
- Brautigam C.A., Sun,S., Piccirilli,J.A. and Steitz,T.A. (1999) Struc tures of normal single-stranded DNA and deoxyribo-3′-S-phosphorothiolates bound to the 3′–5′ exonucleolytic active site of DNA polymerase I from Escherichia coli. Biochemistry, 38, 696–704. [DOI] [PubMed] [Google Scholar]
- Brown J.W. (1999) The Ribonuclease P Database. Nucleic Acids Res., 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnetto J.M., Hennessy,S.W., Roberts,V.A., Getzoff,E.D., Tainer, J.A. and Pique,M.E. (2002) MDB: the metalloprotein database and browser at The Scripps Research Institute. Nucleic Acids Res., 30, 379–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- Chen J.L., Nolan,J.M., Harris,M.E. and Pace,N.R. (1998) Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. EMBO J., 17, 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian E.L. and Harris,M.E. (1999) The track of the pre-tRNA 5′ leader in the ribonuclease P ribozyme–substrate complex. Biochemistry, 38, 12629–12638. [DOI] [PubMed] [Google Scholar]
- Christian E.L., Kaye,N.M. and Harris,M.E. (2000) Helix P4 is a divalent metal ion binding site in the conserved core of the ribonuclease P ribozyme. RNA, 6, 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll C.C., Freeborn,B., Moore,P.B. and Steitz,T.A. (1997) Metals, motifs and recognition in the crystal structure of a 5S rRNA domain. Cell, 91, 705–712. [DOI] [PubMed] [Google Scholar]
- Eckstein F. (1985) Nucleoside phosphorothioates. Annu. Rev. Biochem., 54, 367–402. [DOI] [PubMed] [Google Scholar]
- Ennifar E., Yusupov,M., Walter,P., Marquet,R., Ehresmann,B., Ehresmann,C. and Dumas,P. (1999) The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Structure Fold Des., 7, 1439–1449. [DOI] [PubMed] [Google Scholar]
- Feig A.L. and Uhlenbeck,O.C. (1999) The role of metal ions in RNA biochemistry. In Gesteland,R.F., Cech,T.R. and Atkins,J.F., (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 287–319.
- Ferre-D’Amare A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- Frank D.N. and Pace,N.R. (1997) In vitro selection for altered divalent metal specificity in the RNase P RNA. Proc. Natl Acad. Sci. USA, 94, 14355–14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.N., Harris,M.E. and Pace,N.R. (1994) Rational design of self-cleaving pre-tRNA-ribonuclease P RNA conjugates. Biochemistry, 33, 10800–10808.8075082 [Google Scholar]
- Gordon P.M. and Piccirilli,J.A. (2001) Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nature Struct. Biol., 8, 893–898. [DOI] [PubMed] [Google Scholar]
- Gordon P.M., Sontheimer,E.J. and Piccirilli,J.A. (2000) Kinetic characterization of the second step of group II intron splicing: role of metal ions and the cleavage site 2′-OH in catalysis. Biochemistry, 39, 12939–12952. [DOI] [PubMed] [Google Scholar]
- Hanna R. and Doudna,J.A. (2000) Metal ions in ribozyme folding and catalysis. Curr. Opin. Chem. Biol., 4, 166–170. [DOI] [PubMed] [Google Scholar]
- Harris M.E. and Pace,N.R. (1995) Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA, 1, 210–218. [PMC free article] [PubMed] [Google Scholar]
- Jaffe E.K. and Cohn,M. (1978) Divalent cation-dependent stereo specificity of adenosine 5′-O-(2-thiotriphosphate) in the hexokinase and pyruvate kinase reactions. The absolute stereochemistry of the diastereoisomers of adenosine 5′-O-(2-thiotriphosphate). J. Biol. Chem., 253, 4823–4825. [PubMed] [Google Scholar]
- Jovine L., Djordjevic,S. and Rhodes,D. (2000) The crystal structure of yeast phenylalanine tRNA at 2.0 Å resolution: cleavage by Mg2+ in 15-year old crystals. J. Mol. Biol., 301, 401–414. [DOI] [PubMed] [Google Scholar]
- Kaye N.M., Curistian,E.L. and Harris,M.E. (2002) NAIM and site-specific functional group modification analysis of RNase P RNA: magnesium dependent structure within the conserved P1–P4 multihelix junction contributes to catalysis. Biochemistry, in press. [DOI] [PubMed] [Google Scholar]
- Kazantsev A.V. and Pace,N.R. (1998) Identification by modification-interference of purine N-7 and ribose 2′-OH groups critical for catalysis by bacterial ribonuclease P. RNA, 4, 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D.J., Schmeing,T.M., Moore,P.B. and Steitz,T.A. (2001) The kink-turn: a new RNA secondary structure motif. EMBO J., 20, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz J.C. and Fierke,C.A. (2000) Ribonuclease P: a ribonucleoprotein enzyme. Curr. Opin. Chem. Biol., 4, 553–558. [DOI] [PubMed] [Google Scholar]
- Loria A. and Pan,T. (1996) Domain structure of the ribozyme from eubacterial ribonuclease P. RNA, 2, 551–563. [PMC free article] [PubMed] [Google Scholar]
- Massire C., Jaeger,L. and Westhof,E. (1998) Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol., 279, 773–793. [DOI] [PubMed] [Google Scholar]
- Misra V.K. and Draper,D.E. (1998) On the role of magnesium ions in RNA stability. Biopolymers, 48, 113–135. [DOI] [PubMed] [Google Scholar]
- Moore M.J. and Sharp,P.A. (1992) Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science, 256, 992–997. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J. and Herschlag,D. (1997) Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes. Annu. Rev. Biochem., 66, 19–59. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen,D.E., Clemons,W.M.,Jr, Tarry,M.J., Carter,A.P. and Ramakrishnan,V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science, 292, 897–902. [DOI] [PubMed] [Google Scholar]
- Pan T., Fang,X. and Sosnick,T. (1999) Pathway modulation, circular permutation and rapid RNA folding under kinetic control. J. Mol. Biol., 286, 721–731. [DOI] [PubMed] [Google Scholar]
- Peracchi A., Beigelman,L., Scott,E.C., Uhlenbeck,O.C. and Herschlag,D. (1997) Involvement of a specific metal ion in the transition of the hammerhead ribozyme to its catalytic conformation. J. Biol. Chem., 272, 26822–26826. [DOI] [PubMed] [Google Scholar]
- Scott W.G., Murray,J.B., Arnold,J.R., Stoddard,B.L. and Klug,A. (1996) Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science, 274, 2065–2069. [DOI] [PubMed] [Google Scholar]
- Shan S.O. and Herschlag,D. (1999) Probing the role of metal ions in RNA catalysis: kinetic and thermodynamic characterization of a metal ion interaction with the 2′-moiety of the guanosine nucleophile in the Tetrahymena group I ribozyme. Biochemistry, 38, 10958–10975. [DOI] [PubMed] [Google Scholar]
- Shan S.O. and Herschlag,D. (2000) An unconventional origin of metal-ion rescue and inhibition in the Tetrahymena group I ribozyme reaction. RNA, 6, 795–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan S., Yoshida,A., Sun,S., Piccirilli,J.A. and Herschlag,D. (1999) Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc. Natl Acad. Sci. USA, 96, 12299–12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan S., Kravchuk,A.V., Piccirilli,J.A. and Herschlag,D. (2001) Defining the catalytic metal ion interactions in the Tetrahymena ribozyme reaction. Biochemistry, 40, 5161–5171. [DOI] [PubMed] [Google Scholar]
- Shelton V.M., Sosnick,T.R. and Pan,T. (2001) Altering the intermediate in the equilibrium folding of unmodified yeast tRNAPhe with monovalent and divalent cations. Biochemistry, 40, 3629–3638. [DOI] [PubMed] [Google Scholar]
- Smith D. and Pace,N.R. (1993) Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry, 32, 5273–5281. [DOI] [PubMed] [Google Scholar]
- Thomas B.C., Gao,L., Stomp,D., Li,X. and Gegenheimer,P.A. (1995) Spinach chloroplast RNase P: a putative protein enzyme. Nucleic Acids Symp. Ser., 33, 95–98. [PubMed] [Google Scholar]
- Wadkins T.S., Shih,I., Perrotta,A.T. and Been,M.D. (2001) A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J. Mol. Biol., 305, 1045–1055. [DOI] [PubMed] [Google Scholar]
- Wang S., Karbstein,K., Peracchi,A., Beigelman,L. and Herschlag,D. (1999) Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of a cleavage site phosphorothioate. Biochemistry, 38, 14363–14378. [DOI] [PubMed] [Google Scholar]
- Warnecke J.M., Furste,J.P., Hardt,W.D., Erdmann,V.A. and Hartmann,R.K. (1996) Ribonuclease P (RNase P) RNA is converted to a Cd2+-ribozyme by a single Rp-phosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc. Natl Acad. Sci. USA, 93, 8924–8928. [DOI] [PMC free article] [PubMed] [Google Scholar]