Abstract
We have previously described the isolation of a replication competent (RC) complex from calf thymus, containing DNA polymerase α, DNA polymerase δ and replication factor C. Here, we describe the isolation of the RC complex from nuclear extracts of synchronized HeLa cells, which contains DNA replication proteins associated with cell-cycle regulation factors like cyclin A, cyclin B1, Cdk2 and Cdk1. In addition, it contains a kinase activity and DNA polymerase activities able to switch from a distributive to a processive mode of DNA synthesis, which is dependent on proliferating cell nuclear antigen. In vivo cross-linking of proteins to DNA in synchronized HeLa cells demonstrates the association of this complex to chromatin. We show a dynamic association of cyclins/Cdks with the RC complex during the cell cycle. Indeed, cyclin A and Cdk2 associated with the complex in S phase, and cyclin B1 and Cdk1 were present exclusively in G2/M phase, suggesting that the activity, as well the localization, of the RC complex might be regulated by specific cyclin/Cdk complexes.
Keywords: Cdk/cell cycle/cyclin/DNA polymerase/DNA replication regulation
Introduction
DNA replication in eukaryotic cells takes place during a restricted period of the cell cycle: the S phase. The transition from G1 into S phase in mammalian cells is regulated by at least two cyclin-dependent kinases (Cdks), cyclin E/Cdk2 and cyclin A/Cdk2. Cyclin E/Cdk2 activity peaks in late G1, while cyclin A/Cdk2 activity appears later, at the onset of DNA synthesis. Cyclin A is also required later for the S/G2 transition (Pagano et al., 1992). The initiation of chromosomal DNA replication in eukaryotes can be divided into two major events (Diffley, 1996; Dutta and Bell, 1997). The first event takes place during G1 phase, when a pre-initiation complex is formed at the origin of replication by the ORC proteins. The second event occurs during G1/S transition, when Cdks promote the conversion of the pre-initiation complex into an active replication form through the sequential recruitment of several proteins such as Cdc6, minichromosome maintenance proteins (MCMs), Cdc45, replication protein A (RP-A) and DNA polymerase (pol) α. The Cdks appear also to exert a negative effect on DNA replication by catalysing a series of phosphorylation events that render these replication factors unable to re-enter the pre-initiation state, thus preventing re-replication of the genome (Jallepalli and Kelly, 1997). Thus, a physical link between Cdk/cyclin complexes and essential replication factors at the origin of replication has been proposed (Fotedar and Roberts, 1991; Jaumot et al., 1994; Jallepalli and Kelly, 1997). DNA replication occurs in specific nuclear compartments, termed replication factories (Hozak et al., 1993, 1994; Montecucco et al., 1995), containing not only replication factors (Montecucco et al., 1998), but also other proteins that are not directly involved in DNA replication, such as cell cycle regulator factors (Cardoso et al., 1993; Sobczak-Thepot et al., 1993) and DNA methyl transferase (Leonhardt et al., 1992). Each replication factory undergoes an assembly/disassembly cycle reflecting the recruitment of replicative factors to the replicons and the subsequent release of the same factors upon the completion of DNA synthesis (Dimitrova and Gilbert, 2000; Leonhardt et al., 2000; Montecucco et al., 2001).
The essential function of replicating DNA in eukaryotic cells is performed by a network of enzymes and proteins, which co-operate together to rapidly and accurately duplicate the genetic information (Hübscher et al., 1996, 2002; Wang, 1996). Several reports have described the isolation of large macromolecular complexes of replication proteins from the extracts of eukaryotic cells (Jackson and Cook, 1986; Vishwanatha et al., 1986; Hickey et al., 1988). A fully functional 18–21S multiprotein complex called DNA synthesome was isolated from HeLa cells (Malkas et al., 1990; Applegren et al., 1995), HL60 cells (Lin et al., 1997b) and MDA MB-468 human breast cancer cells (Coll et al., 1996, 1997). Using a combination of immunoblot analysis and functional assays, pol δ, DNA ligase I, DNA topoisomerase I (topo I) and II, replication factor C (RF-C), RP-A, proliferating cell nuclear antigen (PCNA), DNA helicase, pol α/primase and poly(ADP–ribose) polymerase (PARP) were found to be part of these complexes. The DNA synthesome was shown to be fully competent to support origin-specific simian virus 40 (SV40) DNA replication in vitro. Studies on the organizational status of the DNA synthesome either in terminal differentiated or temporarily G1-arrested HL60 cells (Lin et al., 1997a) showed that it remained inactive but intact in temporarily growth-arrested cells, but disassembled in differentiated cells. Characterization of a replication-competent (RC) multiprotein complex from calf thymus containing pol α, pol δ and RF-C has been also described, and its biochemical and enzymological analysis suggested the existence of an asymmetric DNA polymerase complex in mammalian cells (Maga and Hübscher, 1996).
In an attempt to study the composition and regulation of the RC complex in human cells, we isolated from HeLa cells synchronized in G1/S, S and G2/M phases of the cell cycle a multiprotein complex that contains DNA replication proteins such as pol α, pol δ, DNA ligase I, RP-A, RF-C, topo I and PARP, together with cyclins and Cdks. In particular, we showed that cyclin A and Cdk2 associated with the complex in S phase, whereas cyclin B1 and Cdk1 were present only in G2 phase. This complex was capable of RF-C dependent and independent DNA synthesis activity and also displayed histone H1 kinase activity. Cross-linking of proteins to DNA in vivo in synchronized HeLa cells demonstrated the association of this complex to the chromatin and allowed the study of its composition along the cell cycle.
Results
Isolation of an RC complex associated with cell cycle regulatory proteins from HeLa cells
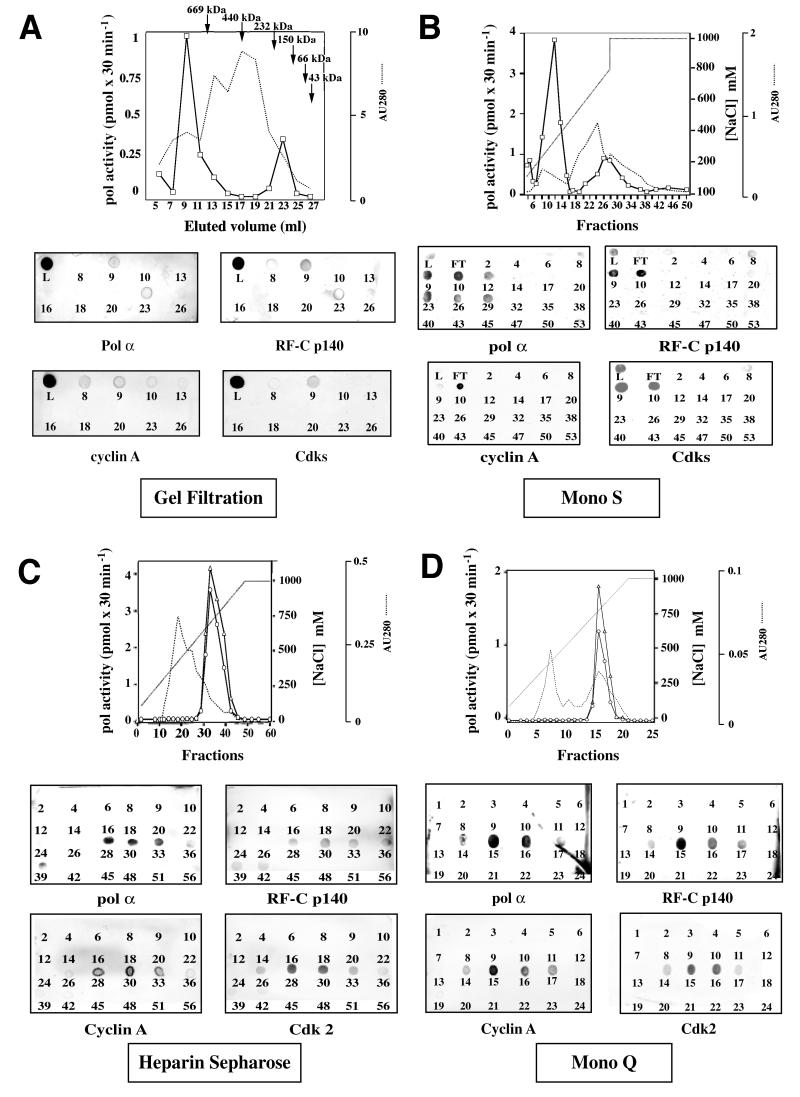
NaCl nuclear extracts (0.2 M) from asynchronous HeLa cells were first loaded on a gel filtration column and the presence of the different proteins involved in DNA synthesis and cell cycle regulation (Cdk/cyclins) was systematically followed throughout the purification by immunoblot analysis. As shown in Figure 1A, DNA replication proteins (RF-C, pol α) co-eluted with cell cycle regulation factors such as cyclin A and Cdks with a retention volume of 9 ml, corresponding to a protein complex with an apparent mol. wt >669 kDa. The same proteins also eluted according to their respective weight: for example, RF-C (p140) and pol α (p180) large sub units were found also in fraction 23, corresponding to a molecular weight range of 150–200 kDa (Figure 1A). When pol activity was assayed (see Materials and methods), two peaks of activity were detected in fractions 9 and 23, respectively, matching the elution profile of pol α (Figure 1A).
Fig. 1. Isolation of a RC complex from S-phase HeLa cells. (A) Gel filtration elution profile. Pol activity was assayed in the gel filtration fractions of 0.2 M NaCl nuclear extract from HeLa cells and dot-blot analysis of this fractions was performed. L, loading. (B) Mono S elution profile. Fractions from the gel filtration containing DNA polymerase activity and proteins of interest were pooled and loaded onto a Mono S column, which was eluted with a non-linear gradient. The Mono S fractions were tested for pol activity and immunoblot analysis with antibodies against pol α, RF-C and cyclin A, and Cdks (anti-PSTAIRE). L, loading; FT, flow-through. (C) Heparin–Sepharose elution profile. Active fractions from the Mono S column were pooled and were loaded onto a heparin column. Immunoblot analysis and pol activity profile of heparin fractions are shown. Pol activity was assayed in the absence (triangles) or presence (circles) of PCNA. (D) Mono Q elution profile. Active fractions from the heparin–Sepharose column were pooled and were loaded onto a Mono Q column. Immunoblot analysis and pol activity profile of Mono Q fractions are shown. Pol activity was assayed in the absence (circles) or presence (triangles) of PCNA. All chromatographic steps were performed as described in Materials and methods.
Fractions containing to the peak of pol activity, were pooled and loaded onto a Mono S column. Proteins were eluted with a non-linear gradient from 0.1 to 1 M NaCl and the fractions were analysed as described above. Immuno blot analysis showed that pol α, cyclin A, RF-C and Cdks co-eluted between 0.25 and 0.3 M NaCl (Figure 1B). Pol activity, assayed on poly(dA)/oligo(dT), peaked in fraction 10 (Figure 1B). Histone H1 kinase activity was found to co-elute in the same fractions (data not shown). Fractions 8–12 were pooled and loaded on a heparin– Sepharose column. Proteins were eluted using a linear gradient of NaCl from 0.1 to 1 M. Pol α, RF-C, cyclin A and Cdk2, revealed by immunoblot analysis, co-eluted in fractions 28–33 (Figure 1C) and pol activity peaked in fraction 30 (Figure 1C). Histone H1 kinase activity was also found to co-elute in the same fractions (data not shown).
Finally, the active fractions from heparin–Sepharose were pooled and loaded onto a Mono Q column. As shown in Figure 1D, pol α, RF-C, cyclin A and Cdk2 co-eluted in fractions 14–17. Pol activity peaked in fraction 15. Histone H1 kinase activity was also found to co-elute in the same fractions (data not shown; see Figure 4B). The purification procedure is summarized in Table I. Thus, a multiprotein complex containing pol α, RF-C, cyclin A and Cdk2 could be isolated from a HeLa cell nuclear extract. This complex was a relatively stable entity and survived successive purification steps.
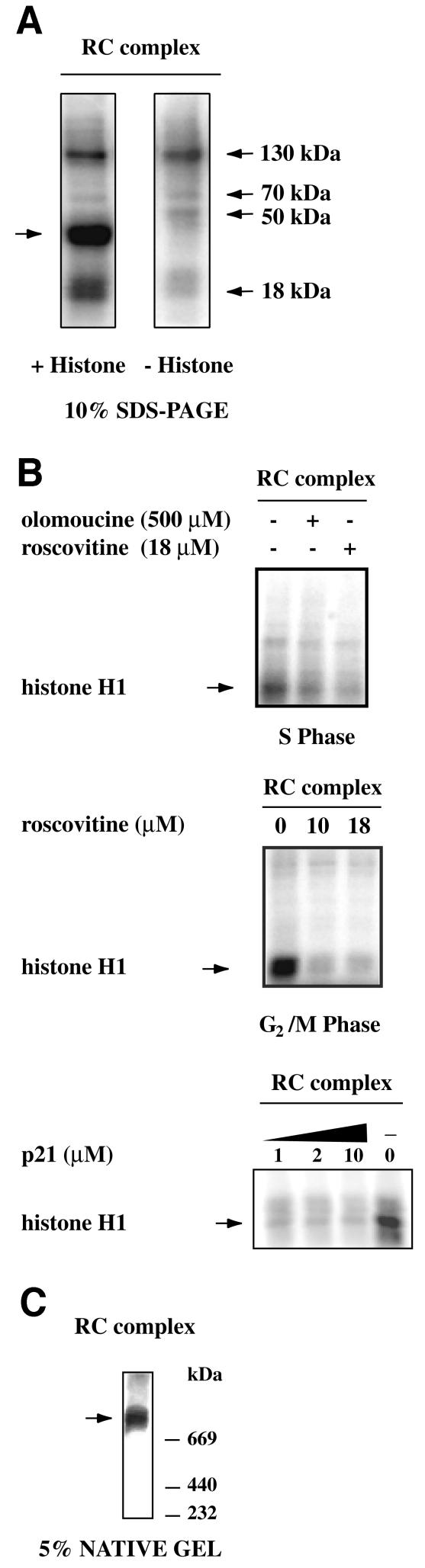
Fig. 4. The cyclin/Cdks present in the RC complex are active. (A) Kinase activity in fraction 9 of gel filtration of HeLa cells synchronized in S phase and G2 phase was assayed in presence of olomoucin and roscovitine. The kinase activity of the Mono Q fraction was assayed in the presence of p21. Proteins were then separated by 10% SDS–PAGE and radioactive bands were visualized with a Phosphor Imager. (B) Kinase activity was assayed in the Mono Q fraction, in presence or in the absence of histone H1 (0.4 mg/ml) as substrate. Proteins were then separated by 10% SDS–PAGE and radioactive bands were visualized with PhosphorImager. The arrows on the left indicate histone H1. (C) Kinase activity was assayed in fraction 9 of gel filtration and proteins were then separated by 5% native polyacrylamide gel and were visualized with a PhosphorImager.
Table I. Purification table of RC complex from HeLa cells.
| Fraction | Protein (mg) | Pol activity (U) | Pol specific activity (U/mg) | Pol purification (fold) | Kinase activity (U) | Kinase specific activity (U/mg) | Kinase purification (fold) |
|---|---|---|---|---|---|---|---|
| Crude extract | 9.1 | 6000a | 660 | 1 | 20a | 2.2 | 1 |
| HLS200 | 1.7 | 7500a | 4411 | 6.7 | 45a | 26.4 | 12 |
| Mono S | 0.25 | 7000 | 28 000 | 42.4 | 30 | 120 | 54.5 |
| HTHeparin | 0.07 | 6500 | 92 857 | 140.6 | 27.5 | 392.8 | 178.5 |
| Mono Q | 0.025 | 6000 | 240 000 | 363.6 | 22.5 | 900 | 409 |
aThe activity after the first purification step (gel filtration) was higher than the one measured in the crude extract, due to the removal of non-specific inhibitors.
DNA replication proteins and cell cycle regulatory factors co-sedimented in a sucrose gradient as a high molecular weight complex
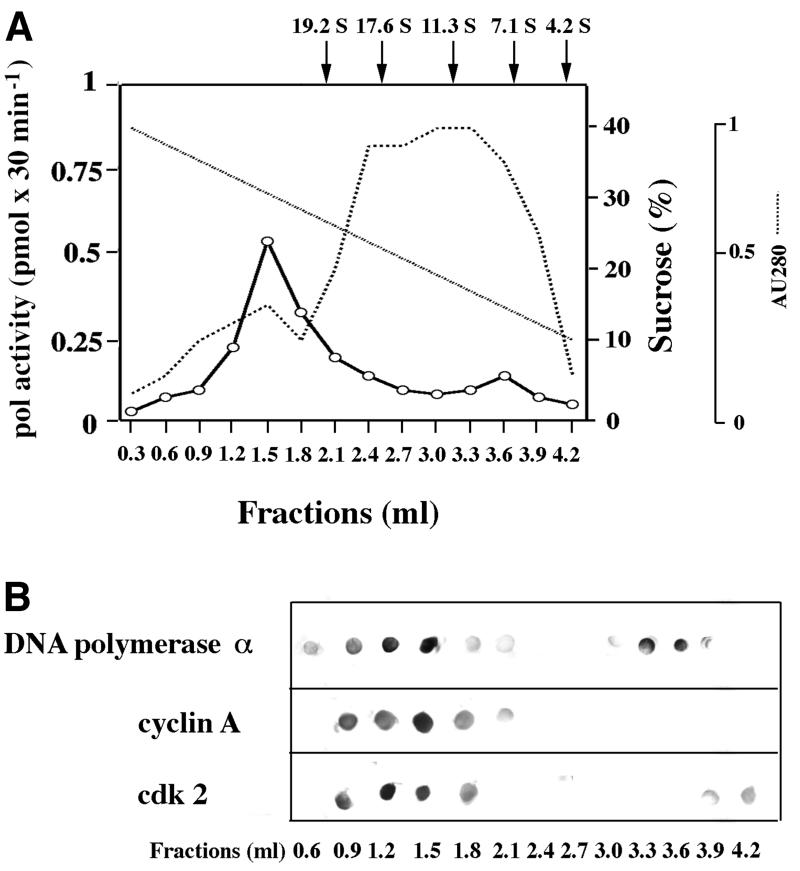
In order to confirm the presence of a multiprotein complex containing DNA replication and cell cycle regulatory factors, a 0.2 M NaCl nuclear extract from asynchronous HeLa cells was layered on a pre-formed 10–40% sucrose gradient (see Materials and methods). After ultracentrifugation, the gradient was fractionated from the bottom of the tube and the protein distribution profile was monitored spectrophotometrically. As shown in Figure 2A, two peaks of polymerase activity were detected, one sedimenting faster than the thyroglobulin marker (19.2S) and the second one between 11.3S and 7.1S. The dot-blot immunoassays shown in Figure 2B confirmed the presence of cyclin A, Cdk2 and pol α in the high molecular weight activity peak. In addition, pol α and Cdk2 were also sedimenting according to their native molecular weight.
Fig. 2. DNA replication proteins and cell cycle regulatory factors co- sedimented in a sucrose gradient as a high molecular weight complex. HeLa cell nuclear extract (900 µg) was layered on top of a 10–40% sucrose gradient and centrifuged as described in Materials and methods. (A) UV trace of the protein distribution profile along the gradient and pol activity in the gradient fractions. The positions of protein markers of known sedimentation coefficients along the gradient are indicated by arrows on top of the panel. (B) Dot-blot immunoassay of the gradient fractions with antibodies against pol α, cyclin A and Cdk2.
The RC complex isolated from synchronized HeLa cells contains both DNA replication proteins and cell cycle regulatory factors
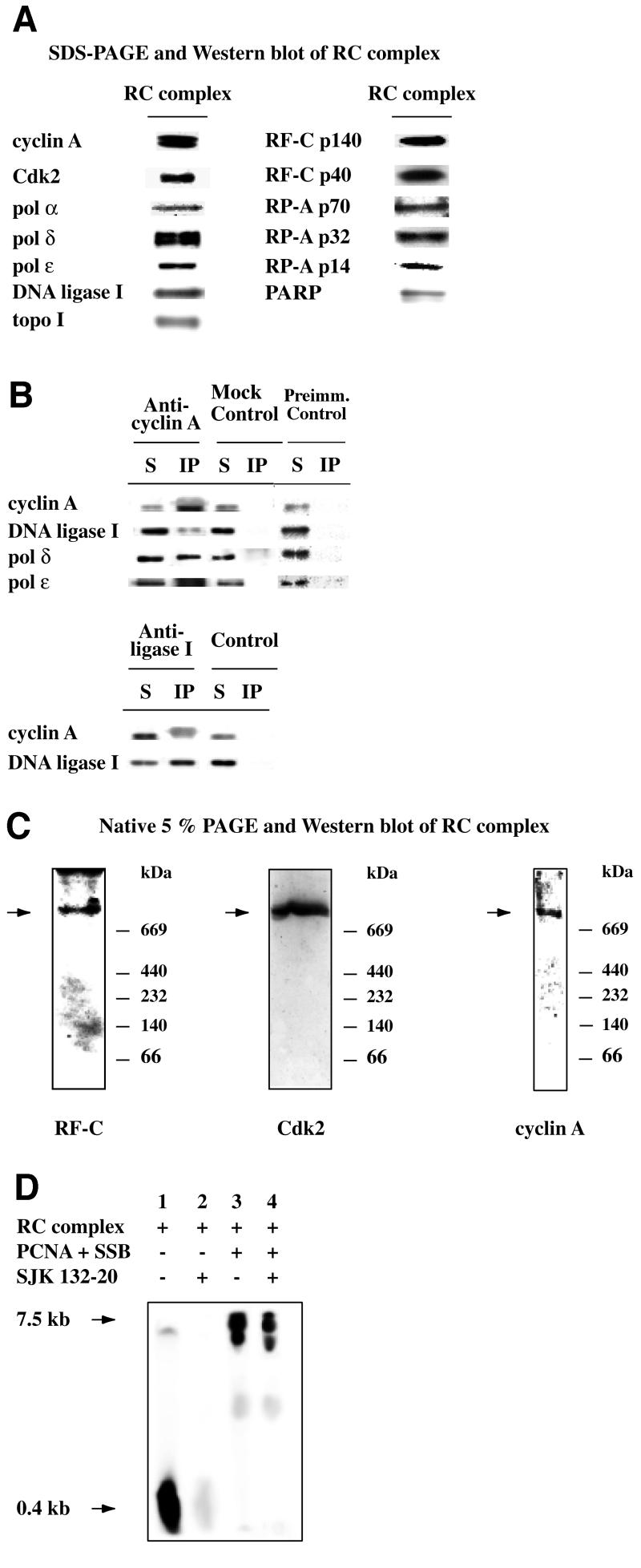
In order to determine the protein composition and to study the functional activities of the RC complex, we isolated the complex as described above from HeLa cells synchronized in S phase. The fractions of interest were analysed by SDS–PAGE and western blotting with different antibodies. As shown in Figure 3A, in the final purification step (Mono Q fraction) we detected the pol α p180, pol δ p125 and pol ε p210 large subunits, RF-C (subunits p140 and p40), RP-A (subunits p70, p32, p14), DNA ligase I, topo I, PARP together with cyclin A and Cdk2. No PCNA and cyclin E could be detected (data not shown). We confirmed the physical association of these proteins by two approaches. First, the Mono Q fraction was immunoprecipitated with antibodies against cyclin A and DNA ligase I. As shown in Figure 3B, anti-cyclin A antibodies co-immunoprecipitated the cognate antigen as well as other RC components, such as DNA ligase I, pol ε and pol δ. Control experiments shown in Figure 3B, bottom panel, demonstrated that antibodies against DNA ligase I also co-immunoprecipitated cyclin A. Secondly, aliquots of fraction 9 of gel filtration were electrophoresed through a 5% native polyacrylamide gel followed by immunoblotting with antibodies directed against RF-C p140, cyclin A and Cdk2 (Figure 3C). All the antibodies stained a band >669 kDa. Together, these results suggest that the proteins, purified through four different chromatographic steps, were part of high molecular weight multiprotein complexes.
Fig. 3. The RC complex contains DNA replication and cell cycle regulatory proteins and is active in DNA synthesis. (A) Western blot analysis of the Mono Q peak fractions that contained the RC complex with antibodies against PARP, pol α, pol δ, pol ε, RF-C, RP-A, topo I and DNA ligase I, and cell cycle regulatory factors like cyclin A and Cdk2. (B) Co-immunoprecipitation experiments with anti-cyclin A and with anti-DNA ligase I antibodies with Mono Q fraction. Reactions were performed as described in Materials and methods. The immunoprecipitated samples were analysed by western blot with antibodies against cyclin A, DNA ligase I, pol δ and pol ε. S, supernatant; IP, immunoprecipitated pellet; Mock Control, beads only; Preimm. Control, beads with pre-immune rabbit serum. (C) Thirty micrograms of fraction 9 of gel filtration were resolved through a 5% native polyacrylamide gel and electrophoretically transferred to nitrocellulose. The native western blots were probed with monoclonal antibodies against p140 of RF-C, cyclin A and Cdk2. (D) RF-C-dependent polymerase activity was assayed in the Mono Q fraction as described in Materials and methods. Synthesized products were visualized with a Phosphor Imager. Lane 1, 0.12 µg of Mono Q fraction; lane 2, 0.12 µg of Mono Q fraction, 100 ng of PCNA and 500 ng of SSB; lane 3, 0.12 µg of Mono Q fraction, 2 µg of SJK 132-20; lane 4, 0.12 µg of Mono Q fraction, 100 ng of PCNA, 500 ng of SSB and 5 µg of SJK 132-20.
The RC complex isolated from HeLa cells is active in DNA replication
Next, the pol activity within the RC complex was assayed on a singly primed (sp) M13 DNA template. In the presence of PCNA and Escherichia coli single-stranded DNA binding protein (SSB), this template can be replicated either in an RF-C-independent (pol α) distributive way or in a RF-C-dependent (pol δ or pol ε) processive manner. Reaction products were separated on a 4% polyacrylamide gel containing 5 M urea. As shown in Figure 3D, lane 1, in the absence of PCNA and SSB, only short products could be detected. The synthesis of these products was inhibited by SJK 132-20, a specific inhibitory antibody of pol α (compare lane 2 with lane 1), suggesting that they were due to the distributive activity of pol α. The addition of SSB and PCNA (lane 3) induced the synthesis of longer products and the concomitant decrease in the synthesis of short products. PCNA and SSB are known to inhibit pol α activity, whereas in the presence of ATP and RF-C they allow processive synthesis by pol δ and/or pol ε (Maga and Hübscher, 1996). Addition of the pol α inhibitory antibody under these conditions did not abolish the synthesis of the larger products, showing that they were not due to pol α synthesis (compare lane 4 with lane 3). These results indicated that the RC complex was able to perform a switch from a distributive to a processive mode of DNA synthesis, which was dependent on RF-C and PCNA, as has already shown with purified components (Maga et al., 2000).
Analysis of the cyclin/Cdk activity associated with the RC complex
Histone H1 phosphorylation activity was found in the final fraction of the RC complex (Figure 4A). To confirm that the phosphorylation was due to Cdk activity and not to other kinases present in the fractions, we treated the samples with olomoucin (500 µM; Vesely et al., 1994) or roscovitine (10 or 18 µM; De Azevedo et al., 1997), two potent inhibitors of Cdks that act as competitive inhibitors for ATP. The histone H1 phosphorylation activity in fractions obtained by gel filtration of S phase or G2/M phase HeLa cell extracts were inhibited by both drugs (Figure 4B). The most purified RC fraction (Mono Q) was also tested in a histone H1 phosphorylation assay in the absence or in the presence of the specific Cdk-inhibiting protein p21cip. As shown in Figure 4B, bottom panel, already at the lowest concentration tested p21 completely abolished kinase activity. As shown in Figure 4A, a few other proteins contained in these fractions were also phosphorylated. In several experiments, we always observed the presence of phosphorylated bands with apparent molecular weights of 130, 70 and 18 kDa, respectively. The phosphorylation patterns detected by SDS–PAGE with the RC complex purified at different stages of the cell cycle were also analysed. The phosphoprotein with molecular weight of 18 kDa was present from G1/S borders to G2/M, whereas the phosphoproteins with molecular weights of 70 and 130 kDa were present only during S phase. In addition, a phosphoprotein with a higher molecular weight (170 kDa) was found at the beginning of S phase only (data not shown). Fraction 9 of the gel filtration was also tested in a kinase assay and when analysed on a native gel showed one phosphorylated band with a molecular weight >669 kDa (Figure 4C).
Analysis of the RC complex composition at different stages of the cell cycle
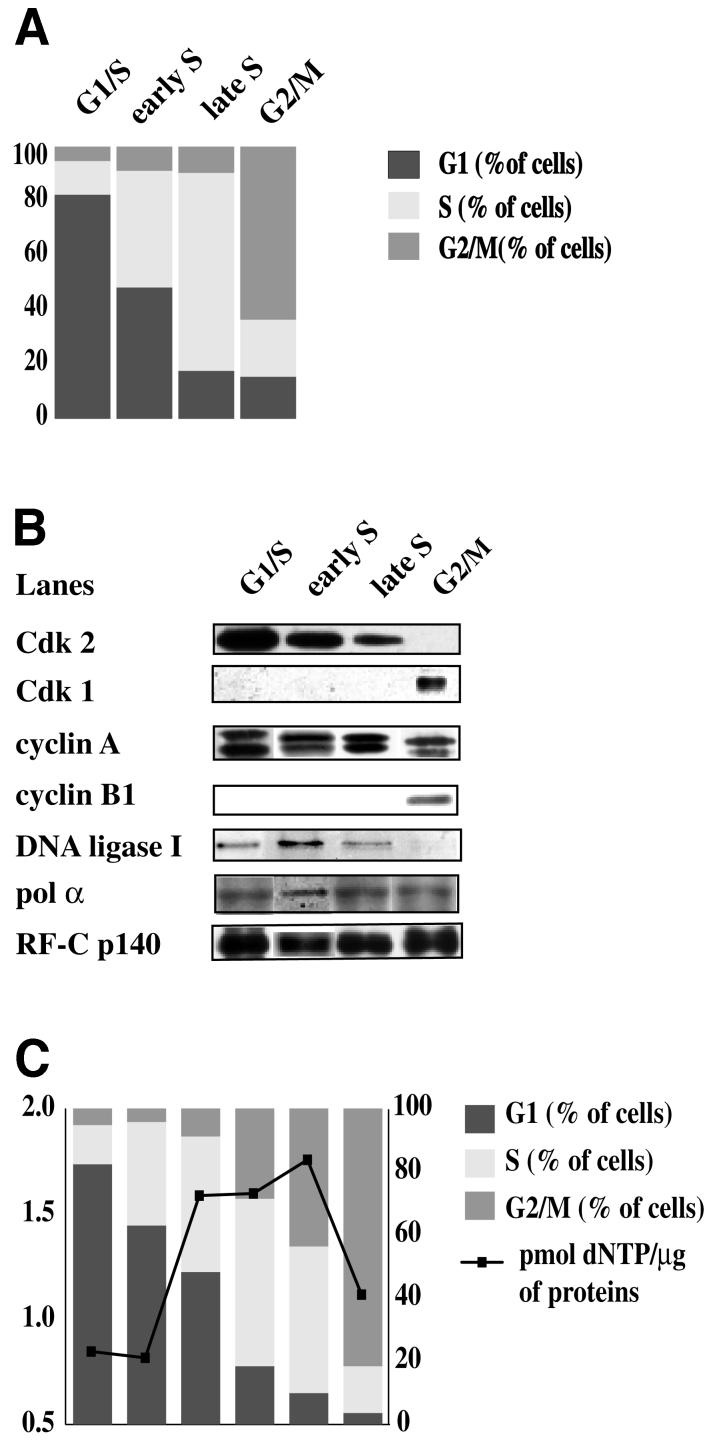
Next we studied the dynamics and structure of the multiprotein complex during the cell cycle. HeLa cells were synchronized using a double thymidine block. Aliquots were withdrawn at different times after block release and cell cycle distribution was determined by flow cytometry (Figure 5A). The RC complexes from 0.2 M NaCl nuclear extracts of HeLa cells synchronized in G1/S, early S, late S and G2/M phases were purified and analysed as described before.
Fig. 5. Analysis of the RC complex during the cell cycle. HeLa cells were synchronized using a double block of thymidine. RC complex from HeLa cells synchronized in G1/S, early S, late S and G2/M phase was then purified as described in Materials and methods. (A) Cell cycle distribution was determined by flow cytometry. (B) Fraction 9, which contains the multiprotein complex, was analysed by western blot using antibodies against pol α, pol δ, RP-A, RF-C and DNA ligase I, and cyclin A, cyclin B1, Cdk1 and Cdk2. (C) The pol activity of the multiprotein complex along the cell cycle was assayed as described in Materials and methods.
The cell cycle regulatory proteins showed a dynamic association with the complex (Figure 5B): cyclin A was detected within the complex from G1/S to G2 phase, while Cdk2 was associated with the complex along the S phase but disappeared in G2. Interestingly, Cdk1 and cyclin B1 associated with the complex only at G2/M. Pol α, DNA ligase I and RF-C were detected in the complex throughout the S phase. Interestingly, DNA ligase I was not found associated with the RC complex in G2/M. Total pol activity was measured in the samples obtained from synchronized HeLa cells and it steadily increased along the S phase and decreased in G2/M (Figure 5C).
Thus, the protein composition of the RC complex varied during the cell cycle, indicating that this higher order complex is a dynamic entity.
Cross-linking of the RC complex to chromatin in vivo
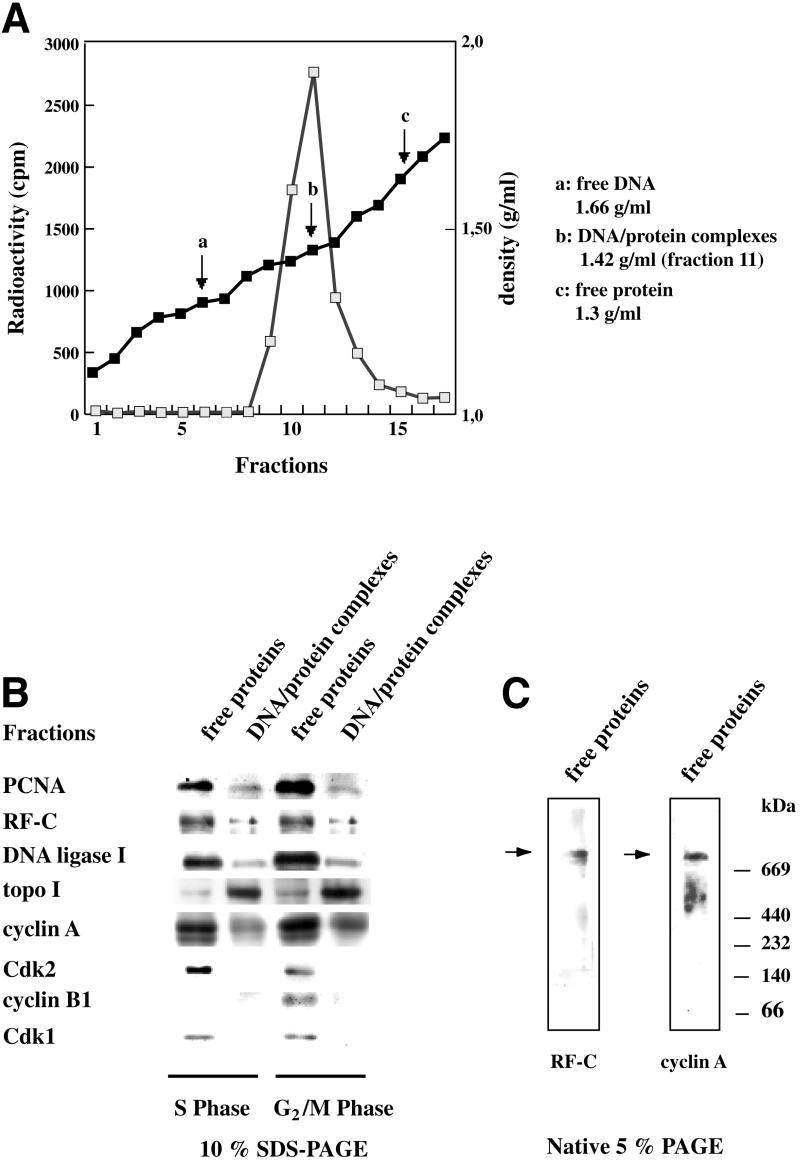
To investigate whether the multiprotein complex was associated with chromatin, we in vivo cross-linked proteins to DNA with formaldehyde in synchronized HeLa cells. We used a modified cross-linking procedure (Göhring and Fackelmayer, 1997) that minimizes the formation of non-specific cross-linking and largely excludes contamination with non-cross-linked material. DNA was labelled in vivo with tritiated thymidine and HeLa cells were synchronized in S phase or G2/M phase as described above, before treatment with formaldehyde, which prevents artefactual redistribution of proteins along the DNA by freezing the native conformation of chromatin. Then, DNA–protein complexes were isolated by centrifugation in CsCl gradients. Density of individual fractions was determined by refractometry and relative DNA concentrations as well as the position of DNA– protein complexes were measured by liquid scintillation counting. As shown in Figure 6A, DNA–protein complexes were found at a density of 1.4 g/ml corresponding to a mass ratio DNA–protein of 1:1, characteristic of native chromatin (Göhring and Fackelmayer, 1997). Purified complexes were de-cross-linked by boiling in SDS–PAGE sample buffer and analysed for their protein components by gel electrophoresis. Based on the intensities of the Coomassie Blue staining, we were able to verify that the amount of proteins loaded on the gel and subsequently analysed by western blot was comparable for all the cell cycle points. Immunoblot analysis was performed to determine whether the proteins of the RC complex were cross-linked to DNA in HeLa cells synchronized in S and G2 phase. RF-C, RP-A, PCNA, DNA ligase I and topo I were found in DNA–protein complexes as the cell cycle regulatory protein cyclin A. Interestingly Cdk1, Cdk2 and cyclin B1 were not found in the DNA–protein complexes and were detected in the fraction of free proteins (Figure 6B). The presence of the RC complex in the free protein fraction was confirmed by native gel electrophoresis (Figure 6C). The complex isolated from 0.2 M NaCl nuclear extracts (Figure 1), probably represents a fraction that is not stably associated with the chromatin. Thus, by comparing the proteins present in the chromatin-bound and in the free protein fractions of the CsCl gradient, with the proteins purified as a complex by chromatography, it appeared that the composition of the multiprotein complex seem to change when it was either unbound or associated with the chromatin. PCNA, on the other hand, was found within the complex bound to the chromatin (Figure 6B), whereas it cannot be detected in the unbound complex (Figure 5B). Cyclin A was always present (Figures 4B and 5B), whereas Cdk2 and Cdk1/cyclin B1 were found associated with this complex in the unbound form both in S phase and G2 phase (Figures 4B and 5B).
Fig. 6. In vivo cross-linking of replicative complexes in HeLa cells synchronized either in S or in G2/M phase. Proteins were cross-linked to DNA with formaldehyde in vivo in HeLa cells synchronized in S and G2/M phases after in vivo radioactive labelling of newly replicated DNA. DNA–protein complexes were extracted as described in Materials and methods. (A) Density of individual fractions was determined by refractometry and relative DNA concentrations were measured by liquid scintillation counting. (B) Western blot analysis was performed on DNA–protein complexes and in fractions that contained free proteins using antibodies against pol α, pol δ, RP-A, RF-C, DNA ligase I and topo I, and cyclin A, cyclin B1, Cdk1 and Cdk2. (C) Thirty micrograms of free protein fraction of CsCl gradient were resolved through a 5% native polyacrylamide gel and electrophoretically transferred to nitrocellulose. The native western blots were probed with monoclonal antibodies against p140 of RF-C and cyclin A.
Discussion
It is known that upon hypotonic lysis of eukaryotic cells, the bulk of pol activity is in the cytosolic fraction, and this approach has been used, for example, to purify all the replicative pols from calf thymus (Weiser et al., 1991). Other DNA replication enzymes, for example RF-C, are retained in the nucleus and standard purification protocols include a high salt (0.35 M NaCl) nuclear extraction (Tsurimoto and Stillman, 1989). When we analysed HeLa cell cytosolic extracts by gel filtration, very little pol activity eluted with an apparent high molecular weight, and the three major replicative pols (α, δ and ε) eluted at a position corresponding to their native molecular weights, as confirmed by western blot analysis (data not shown). Gel filtration analysis of 0.35 M NaCl nuclear extracts showed a similar elution profile, with very little pol activity associated with high molecular weight material. On the other hand, we found that mild nuclear extraction conditions (0.2 M NaCl), as well as buffer optimization (for example with the inclusion of ATP), allowed the preservation of a high molecular weight complex with associated pol activity. These observations might explain why classical purification protocols for DNA replication enzymes failed to detect such large complexes. Thus, in this paper we present the purification of a RC complex containing DNA replication proteins like pol α, pol δ, RF-C, RP-A, DNA ligase I and topo I, and cell cycle regulatory factors like cyclin A, cyclin B1, Cdk2 and Cdk1 through different chromatographic columns (gel filtration, Mono S, heparin–Sepharose, Mono Q) from 0.2 M NaCl nuclear extracts of HeLa cells synchronized at different points of the cell cycle. Gel filtration and native PAGE showed that these proteins were associated in multiprotein complex with a molecular weight >600 kDa, which exists from the G1/S border throughout the entire S phase. This multiprotein complex displayed both an RF-C-independent and an RF-C-dependent polymerase activity, with a switch from a distributive to a processive mode of DNA synthesis that was dependent on PCNA. Immunoblot analysis showed that, while cyclin A was associated with the complex from G1/S to mitosis, Cdk2 was present in the complex only during S phase and disappeared in G2. It is very probable that in G2/M, Cdk2 is replaced by Cdk1, which was found to be one component of the complex, together with the cognate cyclin B1 only in G2/M phase. The complex contains a kinase activity able to phosphorylate exogenous histone H1 and different proteins within the complex itself. Altogether, our data suggest the existence of a multiprotein complex competent for DNA synthesis, the composition and activity of which could be regulated by specific cyclin/Cdk complexes during the cell cycle.
As already described for the RC complex isolated from calf thymus (Maga and Hübscher, 1996), we did not find PCNA associated with the complex. However, PCNA is known to interact with multiple protein partners (Jónsson and Hübscher, 1997; Loor et al., 1997; Warbrick, 2000), including the replication proteins pol δ (Zhang et al., 1999) and pol ε (Maga et al., 1999), RF-C (Mossi et al., 1997), Fen1 (Li et al., 1995), DNA ligase I (Levin et al., 1997; Montecucco et al., 1998), topo I and the cell-cycle regulation factors cyclin/Cdks (Xiong et al., 1992; Koundrioukoff et al., 2000), which are part of this complex. The complex isolated after 0.2 M NaCl nuclear extraction probably represents a soluble or loosely DNA-associated form. One possibility to account for the lack of PCNA is to hypothesize that PCNA remains bound to the chromatin, which is consistent with its proposed role to tether the RC complex to DNA. Previous experiments, have shown that the association of DNA ligase I with PCNA was cell cycle dependent (Rossi et al., 1999) and DNA ligase I could not be co-immunoprecipitated with PCNA in G2/M. Accordingly, DNA ligase I was not found in the RC complex in G2/M, suggesting a possible common regulatory mechanism for the association of DNA ligase I with replication proteins. Cross-linking of proteins to DNA in vivo in synchronized HeLa cells supported this hypothesis. These experiments revealed the presence of PCNA, along with the DNA replication proteins RF-C, DNA ligase I and topo I in the cross-linked chromatin. Interestingly, a cell cycle regulatory factor, cyclin A, was also found within the chromatin-associated complex. Cyclin A has been already found associated with replicating DNA (Fotedar and Roberts, 1991) and co-localized with replication foci in S-phase nuclei (Cardoso et al., 1993; Sobczak-Thepot et al., 1993). In addition, Kim and Kaelin (2001) have hypothesized that cyclin A and cyclin E might be concentrated at certain regions of genome in a temporally controlled manner by virtue of their ability to form stable complexes with specific proteins that directly or indirectly bind to DNA.
We did not find Cdk2 associated with the chromatin-bound complex in S phase, but only in the free complex. Similarly, Cdk1/cyclin B1 were associated exclusively with unbound complexes in G2/M. In a previous study carried out in asynchronous HeLa cells (Jaumot et al., 1994), it was shown that cyclin A and Cdk2 co-migrated with the replicase complex on sucrose gradient, whereas only small amounts of Cdk1/cyclin B1 were found in the same fractions. By enriching cells in, respectively, S and G2/M phases, we were able to distinguish which complex (either free or chromatin bound) contained Cdk1/cyclin B1 or Cdk2/cyclin A. Cdk2/cyclin A has been shown to co-localize at the nuclear foci (Cardoso et al., 1993) and to interact directly with PCNA. This PCNA–Cdk2/cyclin A complex was active in phosphorylating the PCNA binding region of RF-C p145 as well as DNA ligase I (Koundrioukoff et al., 2000). Thus, our results suggest the presence of two complexes: (i) one bound to the chromatin that contains replication proteins, cyclin A and no Cdks; and (ii) a soluble complex in the nucleus containing the same replicative proteins as the chromatin bound complexes, except PCNA, which is absent. This DNA-unbound complex is associated with Cdk2/cyclin A in S phase and Cdk1/cyclin A and B1 in G2 phase. This complex displays a kinase activity that is due to Cdks as confirmed by the inhibition by roscovitine, olomoucine and the Cdk inhibitor p21. Cdk/cyclins are known to phosphorylate several DNA replication proteins, such as SV40 T antigen (McVey et al., 1993), RP-A (Dutta and Stillman, 1992), pol α (Nasheuer et al., 1991), pol δ (Zeng et al., 1994) and PCNA (Prosperi et al., 1994). Accordingly, we have detected Cdk-dependent phosphorylation of different proteins within our complex. Cdk-dependent phosphorylation of DNA replication proteins appears to have a regulatory role. For example, cyclin A/Cdk2 has been shown to inhibit the replication activity of human pol α primase in an SV40 initiation assay, whereas the activities of pol α and the tightly associated primase were not impaired in simpler in vitro assays (Voitenleitner et al., 1997, 1999). In addition to the role in modulating the activity of DNA replication enzymes, our results seem to suggest a role for cyclin/Cdk complexes in regulating the association of replication complexes to chromatin during the cell cycle. It could be that a stable association of cyclin A to replication complexes during S phase has the role of recruiting Cdk2, which in turn can regulate the dynamic association of the replication proteins to the chromatin. This might represent an example of intra-phase regulation, perhaps correlated to a different timing of origin firing. Cyclin A could have an ‘informational’ role, independent of its association to a Cdk, as suggested by Kim and Kaelin (2001) for its role in the repression of transcription during the S phase. The appearance of Cdk1/cyclin B1 associated with replication complexes in G2/M, concomitantly with the disappearance of Cdk2, could reflect an inter-phase regulatory mechanism, which prevents re-binding of replication complexes to chromatin during G2/M phase. This hypothesis fits well with the so-called Cdk-driven ‘replication switch’ model (Kelly and Brown, 2000), which predicts that Cdk activity serves both to activate initiation complexes and to inhibit further initiation complex assembly.
Materials and methods
Chemicals
[3H]dTTP (25 Ci/mmol), [3H]dTTP (30 Ci/mmol), [3H]dATP (73 Ci/mmol), [γ-32P]ATP (3000 Ci/mmol) and [α-32P]dTTP (3000 Ci/mmol) were from Amersham Pharmacia Biotech. GF/C fibre glass filters were purchased from Schleicher & Schuell. Nitrocellulose membrane (Hybond ECL) was from Amersham Pharmacia Biotech. Olomoucin and protease inhibitor cocktail were purchased from Sigma and roscovitine was a gift from E.Prosperi (CSI-CNR, Pavia, Italy). All other reagents were of analytical grade and purchased from Sigma, Fluka and BDH.
Buffers
Buffer A: 1 mM KH2PO4, 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA pH 8.0, 10% (v/v) glycerol, 0.5 mM phenylmethylsulfonyl fluoride (PMSF). Buffer B: buffer A containing 0.35% (v/v) Triton X-100. Buffer C: phosphate-buffered saline (PBS) containing 5 mM MgCl2, 1 mM EDTA pH 8.0, 10% (v/v) glycerol, 0.5 mM PMSF. Buffer D: 50 mM Tris–HCl pH 7.5, 0.35 M NaCl, 1 mM EDTA pH 8.0, 1 mM dithiothreitol (DTT), 10% (v/v) glycerol, 1 mM PMSF, 1/1000 protease inhibitor cocktail. Buffer E: buffer D containing 0.01% (v/v) Nonidet P-40 (NP-40). Buffer F: 25 mM bis-Tris pH 6.6, 50 mM NaCl, 0.5 mM ATP, 1 mM DTT, 5% (v/v) glycerol, 1 mM PMSF. Buffer TDB: 50 mM Tris–HCl pH 7.5, 1 mM DTT, 10 mM MgCl2, 0.2 mg/ml bovine serum albumin (BSA).
Cell culture
HeLa cells S3 were grown as monolayers in Dulbecco’s modified Eagle’s medium (DMEM; Sigma–Aldrich) supplemented with 10% fetal bovine serum (Sigma), 50 µg/ml gentamicin and 2 mM l-glutamine. Cells were grown at 37°C in a humidified atmosphere containing 5% CO2. Synchronization of Hela S3 cells at the G1/S border was obtained by growing cells in 2 mM thymidine as described by Stein (1989). After thymidine removal, cells were washed twice with DMEM, harvested immediately (G1/S border) or further cultured in complete medium for 1.5 h (early S phase), 3 h (late S phase) or 6 h (G2/M phase). Cells were then harvested and washed three times with PBS. The cells were pelleted by low-speed centrifugation (1500 r.p.m., 5 min, 4°C), and the cell pellets stored at –80°C until use.
Isolation of the RC complex
Isolation of HeLa cell nuclei. Cells (1 × 108) were resuspended in 1 ml of buffer A and lysed for 1 h on ice in 9 ml of buffer B. The suspension was centrifuged at 900 g for 10 min at 4°C in a Sorvall RC-5B centrifuge, and the pellet containing the nuclei washed with 1.5 ml of buffer C and centrifuged at 900 g for 10 min at 4°C.
Preparation of nuclear extracts. Nuclei were resuspended in 1 ml of buffer D containing 0.2 M NaCl, stirred for 30 min at 4°C and centrifuged at 12 000 g for 30 min at 4°C. The supernatant was kept as the 0.2 M NaCl nuclear extract. The pellet was resuspended in 1 ml of buffer E containing 0.35 M NaCl, stirred for 1 h at 4°C and centrifuged at 10 000 g for 20 min at 4°C. The supernatant was kept as the 0.35 M NaCl nuclear extract.
Size exclusion chromatography. Half a millilitre of 0.2 M NaCl nuclear extract from HeLa cells was loaded onto a Superdex 200 HR 10/30 gel filtration column (Pharmacia) equilibrated with buffer F. The column was previously calibrated with blue dextran (2000 kDa), thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), alcohol dehydrogenase (150 kDa), BSA (66 kDa), ovalbumin (43 kDa) and myoglobulin (17.5 kDa) as molecular weight standards. Each calibration was performed twice.
Mono S column chromatography. Gel filtration fractions that contained the proteins of interest were loaded on a Mono S column (Pharmacia) equilibrated with buffer F. Proteins were eluted with a non-linear gradient from 0.1 to 1 M NaCl.
Heparin–Sepharose chromatography. Active fractions from Mono S column were pooled and loaded onto a HiTrap heparin–Sepharose column (Pharmacia) equilibrated with buffer F. The column was eluted with a linear gradient from 0.1 to 1 M NaCl.
Mono Q chromatography. Active fractions from heparin–Sepharose column were pooled and loaded onto a Mono Q column (Pharmacia) equilibrated with buffer F. The column was eluted with a linear gradient from 0.1 to 1 M NaCl.
Sucrose gradient analysis. Nine hundred micrograms of 0.2 M NaCl HeLa nuclear extract were layered on top of a 4.5 ml 10–40% sucrose gradient formed in buffer F (without glycerol) in a 13 × 51 mm Beckman polyallomer ultracentrifuge tube. The gradient was centrifuged in a Beckman SW-50 rotor using a Beckman L8-50 M/E ultracentrifuge at 40 000 r.p.m. for 16 h at 4°C. In a parallel run, molecular weight markers were loaded onto an identical gradient. Both gradients were fractionated from the bottom of the tube and the protein distribution monitored spectrophotometrically.
Immunoblot analysis of column fractions (spot test)
For spot test analysis, 4 µl of each fraction were spotted on a nitrocellulose membrane without electrophoretic separation and the membrane was treated as in standard immunoblot protocols.
For SDS–PAGE and western blot analysis, aliquots of the fractions that contained proteins of interest were electrophoresed in a 7.5 or 10% SDS–polyacrylamide gel and transferred by standard procedures to a nitrocellulose filter. The membrane was blocked for 1 h with PBS containing 10% newborn calf serum and 0.2% Tween (PTN) and then incubated for 3 h with the different primary antibodies. After washing with PBS containing 0.2% Tween-20, the membrane was incubated with either anti-mouse (Amersham) or anti-rabbit (Pierce) IgG conjugated to horseradish peroxidase. Immunodetection was carried out using a light-enhanced chemiluminescence (ECL) detection system according to manufacturer’s instructions (Amersham, Arlington Heights, IL).
Native PAGE
Gel filtration fractions (30 µg) were resolved using a 5% native polyacrylamide gel with the exclusion of SDS in the polyacrylamide gel, running, sample buffer as described in Tom et al. (1996). 2-mercaptoethanol was excluded from the sample buffer. All samples were electrophoresed at 4°C. Proteins were electrophoretically transferred to nitrocellulose at 12 V for 12–16 h at 4°C. Immunoblotting was carried out as described previously. Thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa) and BSA (66 kDa) were used as molecular weight standards (Amersham Pharmacia).
Immunoprecipitation
Mono Q fraction (5 µg) was ajusted to 10 mM Tris–HCl pH 7.4, 50 mM NaCl, 2.5 mM MgCl2 and protease inhibitor cocktail (IP buffer) and then were incubated for 1 h at 4°C with a suspension of protein A–Sepharose beads (Bio-Rad) coupled with polyclonal L-β antibodies against DNA ligase I (Rossi et al., 1999) or cyclin A. After centrifugation, the pellet was washed three times with IP buffer. The supernatant and the immunoprecipitated material were analysed by western blot analysis.
Antibodies
Antibodies against Cdc2 p34 sc-54, Cdk2 sc-748, Cdk2 sc-6248, cyclin B1 sc-245 and PCNA (clone PC10) were purchased from Santa Cruz Biotechnology. The antibody against cyclin A (clone CY-A1) was purchased from Sigma. The polyclonal antibody against the 40 kDa subunit of human RF-C was a gift from J.Hurwitz (Memorial Sloan-Kettering Cancer Center, NY). Antibody against calf thymus pol α catalytic subunit was a gift from A.M.Holmes (Holmes et al., 1986). Antibody against the 140 kDa subunit of human RF-C was a gift from B.Stillman (Cold Spring Harbor Laboratory, NY). Monoclonal antibody against the large subunit pol δ was a gift from H.P.Nasheuer (Abteilung Biochemie, Jena, Germany). Monoclonal antibody against human DNA ligase I (2B1) was produced by Rossi et al. (1999). Antiboby against RP-A was as described by Georgaki and Hübscher (1993). Monoclonal antibody against PARP was a gift from G.Poirier (Université Laval, Québec, Canada). mAb 6B5 against DNA topo I was a gift from A.I.Scovassi (IGBE-CNR, Pavia, Italy). The polyclonal antibody against the cyclin A was a gift from J.Sobczak-Thépot (Université Paris 6, Paris, France).
Enzymatic assays
Pol α/pol ε. A final volume of 25 µl contained the following components: buffer TDB, 8 µM [3H]dNTP (1.5 Ci/mmol), 10 mM MgCl2 and 0.5 µg of poly(dA)/poly(dT).
Pol δ. A final volume of 25 µl contained the following components: buffer TDB, 8 µM [3H]dNTP (1.5 Ci/mmol), 10 mM MgCl2, 0.5 µg of poly(dA)/poly(dT) (10:1 base ratio), in the presence of PCNA (100 ng). All reactions were incubated for 30 min at 37°C and precipitated with 10% trichloroacetic acid. Insoluble radioactivity was counted in a Beckman liquid scintillation counter and all values were corrected by subtracting the blank values.
RF-C-dependent polymerase assay. A final volume of 10 µl contained the following components: 50 mM Tris–HCl, pH 7.5, 1 mM DTT, 0.2 mg/ml BSA, 10 mM MgCl2, 130 ng of sp DNA, 1 mM ATP, 10 mM MgCl2, dATP, dCTP, dGTP each at 100 µM and 0.1 µM [α-32P]dTTP (3000 Ci/mmol). One hundred nanograms of PCNA and 500 ng of SSB, either alone or in combination, were added to the mixture. All reactions were incubated for 45 min at 37°C. The reaction was stopped by adding 1% SDS and 20 mM EDTA pH 8.0. Two microlitres of GBL 5X (10% w/v sucrose, 0.2% w/v Bromophenol Blue, 0.2% w/v xylene cyanol and 95% formamide) were added and samples were heated for 3 min at 95°C. Samples were loaded on a 4% polyacrylamide gel for 25 min at 250 V. The gel was fixed with 12% (v/v) methanol and 10% (v/v) acetic acid. DNA synthesis was visualized by PhosphorImager (Molecular Dynamics).
Kinase assay. This was carried out in a final volume of 10 µl containing histone-kinase buffer [50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.5 µM [γ-32P]ATP (3000 mCi/mmol), 0.4 mg/ml histone H1]. Reactions were carried out for 20 min at 37°C. Proteins were then separated by 10% SDS–PAGE and radioactive bands were visualized by PhosphorImager (Molecular Dynamics). For kinase inhibition assay, 0.5 µg of protein from gel filtration or 0.15 µg of Mono Q fraction were incubated with olomoucin (500 µM), roscovitine (10 and 18 µM) or p21cip (1, 2 and 10 µM) for 5 min at room temperature, and then kinase assay was performed as described above. One unit of kinase activity was defined as the amount of kinase incorporating 1 nmol of ATP into histone H1 in 60 min at 37°C under the conditions described above. For determination of the specific activity of the nuclear extract and of the active fractions throughout the purification, a recombinant Cdk2/cyclin A complex (0.4 mg/ml, 48 000 U/ml) was used as the standard reference.
Experiments were performed in duplicate. Three independent experiments were carried out.
In vivo cross-linking experiments
HeLa S3 cells were grown on 145 mm plastic dishes in DMEM with 5% fetal calf serum (Sigma). For DNA labelling, the medium of semi-confluent cultures was replaced by 20 ml of medium containing 15 µCi of [methyl-3H]thymidine (25 Ci/mmol), and cells were incubated for 24 h under normal culture conditions. Then cells were synchronized using a double thymidine block.
Formaldehyde cross-linking in vivo and DNA–protein complexes preparation were performed as described in Göhring and Fackelmayer (1997), with some modifications. Briefly, five dishes (145 mm) of HeLa cells, one of which contained labelled cells, were washed once with 37°C warm PBS and incubated for 4 min at room temperature in 20 ml of DMEM medium (without serum) containing 1% formaldehyde (freshly added from a 37% stock solution; Merck). The reaction was stopped with 2.2 ml/dish of a 7.5% solution of glycin in PBS. Cross-linked cells were rinsed twice with cold PBS, scraped off the Petri dishes in 10 ml PBS with a rubber policeman, and centrifuged for 5 min at 800 g in a swinging bucket rotor. After resuspension in 10 ml of RSB buffer (10 mM Tris–HCl pH 8.0, 10 mM NaCl and 3 mM MgCl2 pH 8.0), cells were homogenized on ice by 15 strokes in a chilled Dounce homogenizer and nuclei were collected by centrifugation (8 min, 800 g). After nuclei were washed twice with RSB, and proteins were extracted in 10 ml of buffer E (10 mM Tris–HCl pH 8.0, 10 mM Na2S2O5, 1 M NaCl, 0.1% NP-40, 1 mM EDTA, protease inhibitor cocktail). After extraction, nuclei were pelleted as above, resuspended in 2.7 ml of buffer E with 0.1 M NaCl, and lysed by addition of 0.3 ml of 20% sodium sarkosyl solution. The sample was then layered over a preformed CsCl step gradient consisting of 3 ml of 1.75 g/ml CsCl solution, 3.5 ml of 1.5 g/ml CsCl solution and 3 ml of 1.3 g/ml CsCl solution in a SW40 polyallomer ultracentrifugation tube; all CsCl solutions were prepared in 20 mM Tris–HCl pH 8.0, 1 mM EDTA and 0.5% sodium sarkosyl. The gradient was centrifuged for 24 h at 34 000 r.p.m. in an SW40 rotor at 20°C and then fractionated from the top in 18 aliquots of 700 µl each. DNA–protein complexes sedimented at a density of ∼1.4 g/ml and could usually be seen as a slightly turbid band by visual inspection of the gradient. Aliquots of the fraction were sheared by brief sonication, and the density of individual fractions was determined by refractometry. Relative DNA concentrations were measured by liquid scintillation counting. DNA-containing fractions were briefly sonicated and dialysed against a buffer containing 20 mM Tris–HCl pH 8.0, 50 mM NaCl and 2 mM EDTA overnight. SDS sample buffer was then added and cross-links were cleaved by incubation in a boiling water bath for 40 min. Samples were then analysed by SDS–PAGE and western blot as described above.
Acknowledgments
Acknowledgements
The authors thank E.Prosperi (Centro di Studio per l’Istochimica del CNR, Pavia, Italy) for the cell cycle distribution analysis by flow cytometry and Dr J.Breton (Pharmacia & Upjohn, Nerviano, Italy) for the recombinant Cdk 2/cyclin A complex. This work was supported by the EU-TMR Grant (ERBMRXCT 970125) to S.S. and U.H., by the CNR-Target Project on Biotechnology to S.S., by a grant from Associazione Italiana per la Ricerca sul Cancro to A.M., and from Ministero dell’Università e della Ricerca Scientifica e Tecnologica-Consiglio Nazionale delle Ricerche ‘Biomolecole per la salute umana’ L. 95/95 to G.B. I.F. was supported by a fellowship from the EU-TMR Network (grant ERBMRXCT 970125).
References
- Applegren N. et al. (1995) Further characterization of the human cell multiprotein DNA replication complex. J. Cell. Biochem., 59, 91–107. [DOI] [PubMed] [Google Scholar]
- Cardoso M.C., Leonhardt,H. and Nadal-Ginard,B. (1993) Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell, 74, 979–992. [DOI] [PubMed] [Google Scholar]
- Coll J.M., Sekowski,J.W., Hickey,R.J., Schnaper,L., Yue,W., Brodie,A.M., Uitto,L., Syvaoja,J.E. and Malkas,L.H. (1996) The human breast cell DNA synthesome: its purification from tumor tissue and cell culture. Oncol. Res., 8, 435–447. [PubMed] [Google Scholar]
- Coll J.M., Hickey,R.J., Cronkey,E.A., Jiang,H.Y., Schnaper,L., Lee,M.Y., Uitto,L., Syvaoja,J.E. and Malkas,L.H. (1997) Mapping specific protein–protein interactions within the core component of the breast cell DNA synthesome. Oncol. Res., 9, 629–639. [PubMed] [Google Scholar]
- De Azevedo W.F., Leclerc,S., Meijer,L., Havlicek,L., Strnad,M. and Kim,S.H. (1997) Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human Cdk2 complexed with roscovitine. Eur. J. Biochem., 243, 518–526. [DOI] [PubMed] [Google Scholar]
- Diffley J.F. (1996) Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev., 10, 2819–2830. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S. and Gilbert,D.M. (2000) Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nature Cell Biol., 2, 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A. and Stillman,B. (1992) Cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA and activate DNA replication. EMBO J., 11, 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A. and Bell,S.P. (1997) Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell. Dev. Biol., 13, 293–332. [DOI] [PubMed] [Google Scholar]
- Fotedar R. and Roberts,J.M. (1991) Association of p34Cdc2 with replicating DNA. Cold Spring Harb. Symp. Quant. Biol., 56, 325–333. [DOI] [PubMed] [Google Scholar]
- Georgaki A. and Hübscher,U. (1993) DNA unwinding by replication protein A is a property of the 70 kDa subunit and is facilitated by phosphorylation of the 32 kDa subunit. Nucleic Acids Res., 21, 3659–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhring F. and Fackelmayer,F.O. (1997) The scaffold/matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry, 36, 8276–8283. [DOI] [PubMed] [Google Scholar]
- Hickey R.J., Malkas,L.H., Pedersen,N., Li,C. and Baril,E.F. (1988) DNA Replication and Mutagenesis. American Society of Microbiology Publications, Washington, DC.
- Holmes A.M., Cheriathundam,E., Bollum,F.J. and Chang,L.M. (1986) Immunological analysis of the polypeptide structure of calf thymus DNA polymerase–primase complex. J. Biol. Chem., 261, 11924–11930. [PubMed] [Google Scholar]
- Hozak P., Hassan,A.B., Jackson,D.A. and Cook,P.R. (1993) Visualization of replication factories attached to nucleoskeleton. Cell, 73, 361–373. [DOI] [PubMed] [Google Scholar]
- Hozak P., Jackson,D.A. and Cook,P.R. (1994) Replication factories and nuclear bodies: the ultrastructural characterization of replication sites during the cell cycle. J. Cell Sci., 107, 2191–2202. [DOI] [PubMed] [Google Scholar]
- Hübscher U., Maga,G. and Podust V.N. (1996) DNA replication accessory proteins. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cell. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–543.
- Hübscher U., Maga,G. and Spadari,S. (2002) Eukaryotic DNA polymerases. Annu. Rev. Biochem., 71, 133–163. [DOI] [PubMed] [Google Scholar]
- Jackson D.A. and Cook,P.R. (1986) Different populations of DNA polymerase α in HeLa cells. J. Mol. Biol., 192, 77–86. [DOI] [PubMed] [Google Scholar]
- Jallepalli P.V. and Kelly,T.J. (1997) Cyclin-dependent kinase and initiation at eukaryotic origins: a replication switch? Curr. Opin. Cell Biol., 9, 358–363. [DOI] [PubMed] [Google Scholar]
- Jaumot M., Grana,X., Giordano,A., Reddy,P.V., Agell,N. and Bachs,O. (1994) Cyclin/Cdk2 complexes in the nucleus of HeLa cells. Biochem. Biophys. Res. Commun., 203, 1527–1534. [DOI] [PubMed] [Google Scholar]
- Jónsson Z.O. and Hübscher,U. (1997) Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. BioEssays, 19, 967–975. [DOI] [PubMed] [Google Scholar]
- Kelly T.J. and Brown,G.W. (2000) Regulation of chromosome replication. Annu. Rev. Biochem., 69, 829–880. [DOI] [PubMed] [Google Scholar]
- Kim T.Y. and Kaelin,W.G.,Jr (2001) Differential control of transcription by DNA-bound cyclins. Mol. Biol. Cell., 12, 2207–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundrioukoff S., Jónsson,Z.O., Hasan,S., de Jong,R.N., van der Vliet,P.C., Hottiger,M.O. and Hübscher,U. (2000) A direct interaction between proliferating cell nuclear antigen (PCNA) and Cdk2 targets PCNA-interacting proteins for phosphorylation. J. Biol. Chem., 275, 22882–22887. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Page,A.W., Weier,H.U. and Bestor,T.H. (1992) A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell, 71, 865–873. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Rahn,H.P., Weinzierl,P., Sporbert,A., Cremer,T., Zink,D. and Cardoso,M.C. (2000) Dynamics of DNA replication factories in living cells. J. Cell Biol., 149, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D.S., Bai,W., Yao,N., O’Donnell,M. and Tomkinson,A.E. (1997) An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc. Natl Acad. Sci. USA, 94, 12863–12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Li,J., Harrington,J., Lieber,M.R. and Burgers,P.M. (1995) Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem., 270, 22109–22112. [DOI] [PubMed] [Google Scholar]
- Lin S., Hickey,R. and Malkas,L. (1997a) The biochemical status of the DNA synthesome can distinguish between permanent and temporary cell growth arrest. Cell Growth Differ., 8, 1359–1369. [PubMed] [Google Scholar]
- Lin S., Hickey,R.J. and Malkas,L.H. (1997b) The isolation of a DNA synthesome from human leukemia cells. Leuk. Res., 21, 501–512. [DOI] [PubMed] [Google Scholar]
- Loor G., Zhang,S.J., Zhang,P., Toomey,N.L. and Lee,M.Y. (1997) Identification of DNA replication and cell cycle proteins that interact with PCNA. Nucleic Acids Res., 25, 5041–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga G. and Hübscher,U. (1996) DNA replication machinery: functional characterization of a complex containing DNA polymerase α, DNA polymerase δ and replication factor C suggests an asymmetric DNA polymerase dimer. Biochemistry, 35, 5764–5777. [DOI] [PubMed] [Google Scholar]
- Maga G., Jónsson,Z.O., Stucki,M., Spadari,S. and Hübscher,U. (1999) Dual mode of interaction of DNA polymerase ε with proliferating cell nuclear antigen in primer binding and DNA synthesis. J. Mol. Biol., 285, 259–267. [DOI] [PubMed] [Google Scholar]
- Maga G., Stucki,M., Spadari,S. and Hübscher,U. (2000) DNA polymerase switching: I. Replication factor C displaces DNA polymerase α prior to PCNA loading. J. Mol. Biol., 295, 791–801. [DOI] [PubMed] [Google Scholar]
- Malkas L.H., Hickey,R.J., Li,C., Pedersen,N. and Baril,E.F. (1990) A 21S enzyme complex from HeLa cells that functions in simian virus 40 DNA replication in vitro. Biochemistry, 29, 6362–6374. [DOI] [PubMed] [Google Scholar]
- McVey D., Ray,S., Gluzman,Y., Berger,L., Wildeman,A.G., Marshak,D.R. and Tegtmeyer,P. (1993) Cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J. Virol., 67, 5206–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A., Savini,E., Weighardt,F., Rossi,R., Ciarrocchi,G., Villa,A. and Biamonti,G. (1995) The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. EMBO J., 14, 5379–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A. et al. (1998) DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J., 17, 3786–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A., Rossi,R., Ferrari,G., Scovassi,A.I., Prosperi,E. and Biamonti,G. (2001) Etoposide induces the dispersal of DNA ligase I from replication factories. Mol. Biol. Cell, 12, 2109–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossi R., Jónsson,Z.O., Allen,B.L., Hardin,S.H. and Hübscher,U. (1997) Replication factor C interacts with the C-terminal side of proliferating cell nuclear antigen. J. Biol. Chem., 272, 1769–1776. [DOI] [PubMed] [Google Scholar]
- Nasheuer H.P., Moore,A., Wahl,A.F. and Wang,T.S. (1991) Cell cycle-dependent phosphorylation of human DNA polymerase α. J. Biol. Chem., 266, 7893–7903. [PubMed] [Google Scholar]
- Pagano M., Pepperkok,R., Verde,F., Ansorge,W. and Draetta,G. (1992) Cyclin A is required at two points in the human cell cycle. EMBO J., 11, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosperi E., Scovassi,A.I., Stivala,L.A. and Bianchi,L. (1994) Proliferating cell nuclear antigen bound to DNA synthesis sites: phosphorylation and association with cyclin D1 and cyclin A. Exp. Cell Res., 215, 257–262. [DOI] [PubMed] [Google Scholar]
- Rossi R., Villa,A., Negri,C., Scovassi,I., Ciarrocchi,G., Biamonti,G. and Montecucco,A. (1999) The replication factory targeting sequence/PCNA-binding site is required in G1 to control the phosphorylation status of DNA ligase I. EMBO J., 18, 5745–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak-Thepot J., Harper,F., Florentin,Y., Zindy,F., Brechot,C. and Puvion,E. (1993) Localization of cyclin A at the sites of cellular DNA replication. Exp. Cell Res., 206, 43–48. [DOI] [PubMed] [Google Scholar]
- Stein G.S. and Stein,J.L. (1989) Cell synchronization. In Baserga,R. (ed.). Cell Growth and Division. IRL Press, Oxford, UK, pp. 133–137.
- Tom T.D., Malkas L.H. and Hickey R.J. (1996) Identification of multiprotein complexes containing DNA replication factors by native immunoblotting of HeLa cell protein preparation with T-antigen-dependent SV40 DNA replication activity. J. Cell. Biochem., 63, 259–267. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T. and Stillman,B. (1989) Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol. Cell. Biol., 9, 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesely J. et al. (1994) Inhibition of cyclin-dependent kinases by purine analogues. Eur. J. Biochem., 224, 771–786. [DOI] [PubMed] [Google Scholar]
- Vishwanatha J.K., Coughlin,S.A., Wesolowski-Owen,M. and Baril,E.F. (1986) A multiprotein form of DNA polymerase α from HeLa cells. Resolution of its associated catalytic activities. J. Biol. Chem., 261, 6619–6628. [PubMed] [Google Scholar]
- Voitenleitner C., Fanning,E. and Nasheuer,H.P. (1997) Phosphorylation of DNA polymerase α-primase by cyclin A-dependent kinases regulates initiation of DNA replication in vitro. Oncogene, 14, 1611–1615. [DOI] [PubMed] [Google Scholar]
- Voitenleitner C., Rehfuess,C., Hilmes,M., O’Rear,L., Liao,P.C., Gage,D.A., Ott,R., Nasheuer,H.P. and Fanning,E. (1999) Cell cycle-dependent regulation of human DNA polymerase α-primase activity by phosphorylation. Mol. Cell. Biol., 19, 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.S.-F. (1996) Cellular DNA polymerases. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 461–493.
- Warbrick E. (2000) The puzzle of PCNA’s many partners. BioEssays, 22, 997–1006. [DOI] [PubMed] [Google Scholar]
- Weiser T., Gassmann,M., Thommes,P., Ferrari,E., Hafkemeyer,P. and Hübscher,U. (1991) Biochemical and functional comparison of DNA polymerases α, δ and ε from calf thymus. J. Biol. Chem., 266, 10420–10428. [PubMed] [Google Scholar]
- Xiong Y., Zhang,H. and Beach,D. (1992) D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell, 71, 505–514. [DOI] [PubMed] [Google Scholar]
- Zeng X.R., Hao,H., Jiang,Y. and Lee,M.Y. (1994) Regulation of human DNA polymerase δ during the cell cycle. J. Biol. Chem., 269, 24027–24033. [PubMed] [Google Scholar]
- Zhang P., Mo,J.Y., Perez,A., Leon,A., Liu,L., Mazloum,N., Xu,H. and Lee,M.Y. (1999) Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase δ. J. Biol. Chem., 274, 26647–26653. [DOI] [PubMed] [Google Scholar]