Abstract
Drosophila Mi-2 (dMi-2) is the ATPase subunit of a complex combining ATP-dependent nucleosome remodelling and histone deacetylase activities. dMi-2 contains an HMG box-like region, two PHD fingers, two chromodomains and a SNF2-type ATPase domain. It is not known which of these domains contribute to nucleosome remodelling. We have tested a panel of dMi-2 deletion mutants in ATPase, nucleosome mobilization and nucleosome binding assays. Deletion of the chromodomains impairs all three activities. A dMi-2 mutant lacking the chromodomains is incorporated into a functional histone deacetylase complex in vivo but has lost nucleosome-stimulated ATPase activity. In contrast to dHP1, dMi-2 does not bind methylated histone H3 tails and does not require histone tails for nucleosome binding. Instead, the dMi-2 chromodomains display DNA binding activity that is not shared by other chromodomains. Our results suggest that the chromodomains act at an early step of the remodelling process to bind the nucleosome substrate predominantly via protein–DNA interactions. Furthermore, we identify DNA binding as a novel chromodomain-associated activity.
Keywords: chromatin remodelling/chromodomain/dMi-2
Introduction
Fundamental nuclear processes requiring access of factors to DNA involve ATP-dependent chromatin remodelling and histone modifying complexes. These enzymes regulate the interaction of DNA binding factors with their cognate sites by modulating chromatin structure. Whereas ATP-driven complexes can directly remodel nucleosomes (Becker and Hörz, 2002), histone modifying complexes create specific patterns of histone tail modifications. This ‘histone code’ is subsequently ‘read’ through the binding of factors which in turn alter chromatin structure (Turner, 2000; Imhof and Becker, 2001; Jenuwein and Allis, 2001).
Several chromatin remodelling and histone modifying complexes share a number of domains, including PHD fingers (Aasland et al., 1995), bromodomains (Tamkun et al., 1992; Horn and Peterson, 2001) and chromodomains (Paro and Hogness, 1991; Eissenberg, 2001). Our understanding of how these domains function is incomplete. Bromodomains and chromodomains have been suggested to read the histone code by binding to acetylated and methylated histone tails, respectively (Horn and Peterson, 2001). The HP1 chromodomain binds to H3 tails methylated at lysine 9 (K9), an interaction that is important for proper localization of HP1 to heterochromatic regions of the genome (Rea et al., 2000; Bannister et al., 2001; Jacobs et al., 2001; Lachner et al., 2001; Nakayama et al., 2001; Peters et al., 2001; Jacobs and Khorasanizadeh, 2002). It is unlikely that methyl–lysine binding is the only function of chromodomains: the chromodomain of Polycomb (Pc) is important for targeting of the Pc group (PcG) complex (Messmer et al., 1992; Platero et al., 1995). There is no evidence that targeting involves histone tail binding. Instead, the Pc chromodomain is mainly involved in protein interactions required for the assembly of the PcG complex (Strutt and Paro, 1997; Breiling et al., 1999). A region with high similarity to the chromodomain in MOF, a histone acetylase involved in dosage compensation in Drosophila, specifically binds roX RNA, a non-coding RNA that coats the X chromosome (Akhtar et al., 2000). Chromodomains appear to target proteins to their sites of action in chromatin by interacting with different chromatin components, including histones, non-histone proteins and RNA (Eissenberg, 2001).
The chromatin remodeller Drosophila Mi-2 (dMi-2), a member of the CHD subgroup of the SNF2 ATPase superfamily (Eisen et al., 1995; Woodage et al., 1997), harbours several chromatin-related domains (Kehle et al., 1998). It contains a region with similarity to the HMG-box, a pair of PHD fingers and a pair of chromodomains preceding a central ATPase domain (Figure 1A). Whereas HMG-boxes, PHD fingers and ATPase domains are also found in ISWI and SWI2/SNF2 remodelling complexes, chromodomains are unique to the CHD proteins. We have previously shown that dMi-2 is a nucleosome-stimulated ATPase that binds to and mobilizes nucleosomes along a linear DNA fragment (Brehm et al., 2000). In this study we investigate which dMi-2 domains are critical for nucleosome mobilization. We find that deletion of the chromodomains abrogates nucleosome binding, mobilization and ATPase functions. The chromodomains display nucleosome and DNA binding activities. We propose that the chromodomains function during an early step of the remodelling process to bind the nucleosome substrate, mainly via contacts with nucleosomal DNA.
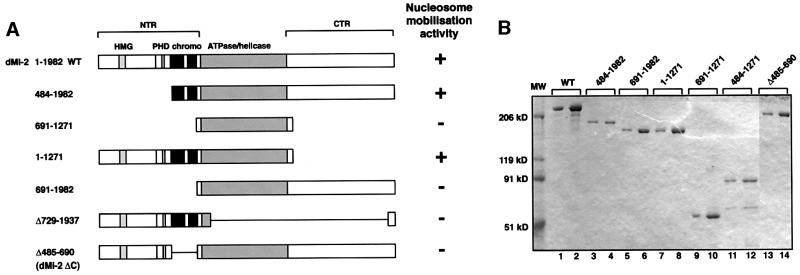
Fig. 1. The dMi-2 chromodomain region is important for nucleosome mobilization. (A) Schematic representation of dMi-2 mutants used. Domains are indicated on top. HMG, HMG box-like region; PHD, PHD fingers; chromo, chromodomains; NTR, N-terminal region; CTR, C-terminal region; WT, wild type. (B) Coomassie Blue-stained SDS polyacrylamide gel of recombinant dMi-2 mutants. Three hundred (odd-numbered lanes) and 600 ng (even-numbered lanes) of each protein were loaded as indicated. MW, molecular weight marker. (C) Nucleosome mobilization assay. Increasing amounts of recombinant dMi-2 proteins were incubated with a radioactively labelled end-positioned 248 bp nucleosome in the absence (–) or presence (+) of ATP as indicated. Nucleosomes were resolved by native PAGE. Positions of endpositioned (open circle) and centrally positioned (filled circle) nucleosomes are indicated.
Results
We have created a panel of deletion mutants to determine which regions of dMi-2 function in ATP-dependent chromatin remodelling (Figure 1A). Mutants were expressed using the baculovirus system and affinity purified via a C-terminal flag tag (Figure 1B). Equivalent amounts of purified proteins were used in the nucleosome sliding assay (Längst et al., 1999). We have used this assay previously to demonstrate that recombinant dMi-2 mobilizes mononucleosomes in an ATP-dependent manner (Brehm et al., 2000).
Nucleosome mobilization activity
In agreement with our previous results, addition of wild-type dMi-2 and ATP to a mononucleosome positioned at the end of the 248 bp DNA fragment resulted in the movement of nucleosomes to a more central position (Figure 1C). Nucleosome mobilization is visualized by the appearence of a nucleosome that migrates more slowly during native PAGE (lanes 2–5). The isolated dMi-2 ATPase domain was not active (dMi-2 691–1271, lanes 18–21), demonstrating that this domain alone is not sufficient for mobilization. A dMi-2 mutant consisting of the ATPase domain and the N-terminal region [amino acids (aa) 1–690, hereafter referred to as the NTR] efficiently mobilized the nucleosome (dMi-2 1–1271; lanes 14–17). In contrast, a mutant consisting of the ATPase domain and the C-terminal region (aa 1271–1982, hereafter referred to as the CTR) was inactive (dMi-2 691–1982; lanes 10–13). We conclude that whereas the NTR is required, the CTR is dispensable for nucleosome mobilization. The NTR is, however, not sufficient: as expected, a mutant retaining the NTR but lacking the ATPase domain was inactive (dMi-2 Δ729–1937; lanes 22–25), indicating that NTR and ATPase domain must act in concert.
The NTR harbours an HMG-box homology region, a pair of PHD fingers and a pair of chromodomains (Figure 1A). To dissect which of these was critical for nucleosome mobilization we tested a mutant lacking the HMG box-like region and the PHD fingers but retaining the chromodomains. This mutant was active in our assay (dMi-2 484–1982; Figure 1C, lanes 6–9), suggesting that residues preceding the chromodomains are not critical for mobilization. Further deletion of the chromodomains abrogated activity (dMi-2 691–1982; lanes 10–13). Moreover, a mutant carrying an internal deletion of the chromodomain region (dMi-2 Δ485–690, hereafter referred to as dMi-2 ΔC) was compromised for nucleosome mobilization (lanes 26–29). Taken together, these results suggest that the chromodomains of dMi-2 are important for ATP- dependent nucleosome mobilization.
Nucleosome-stimulated ATPase activity
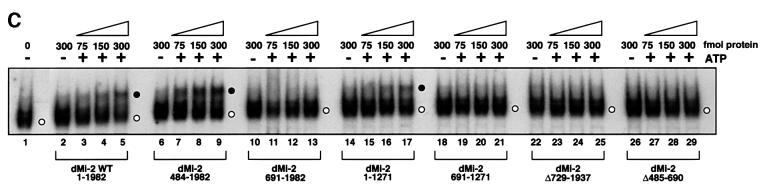
ATP-dependent nucleosome mobilization is likely to be a multi-step process that involves binding of the nucleosome and ATP substrates, ATP hydrolysis, movement of the histone octamer relative to DNA and dissociation of the enzyme from the nucleosome. To determine at which of these steps the chromodomains might function, we tested the mutants in ATPase and nucleosome binding assays. Recombinant dMi-2 has low ATPase activity that is strongly stimulated by nucleosomes (Brehm et al., 2000). Unlike most other chromatin remodellers, dMi-2 is poorly stimulated by free DNA, a property that has also been reported for the Xenopus Mi-2 complex (Guschin et al., 2000). We subjected dMi-2 mutants to ATPase assays in the presence of nucleosomes or DNA (Figure 2A). Two mutants retained strong ATPase activity: dMi-2 484–1982 (lacking the HMG-box/PHD finger region but retaining the chromodomains) displayed somewhat higher activity than the wild type in the absence of added effectors. Activity was stimulated by addition of nucleosomes but was not significantly affected by the addition of DNA. dMi-2 1–1271 (lacking the CTR) also showed higher activity in the absence of added effectors. Unexpectedly, this mutant was stimulated by free DNA as well as by nucleosomes, in striking contrast to the wild-type protein. No mutant lacking the chromodomain region (dMi-2 691–1271, dMi-2 691–1982 and dMi-2 ΔC) was stimulated by DNA or nucleosomes, although they retained low but detectable ATPase activity. Increasing the DNA and nucleosome concentrations also failed to stimulate the chromodomain mutants (data not shown). We thus observe a strict correlation between presence of the chromodomain region, nucleosome-stimulated ATPase and nucleosome mobilization activity (Figures 1 and 2).
Fig. 2. ATPase activity and nucleosome binding of recombinant dMi-2 proteins. (A) Recombinant dMi-2 (90 fmol) was incubated with γ-32P-labelled ATP in the presence of 100 ng DNA or 100 ng nucleosomes as indicated. ATP hydrolysis was detected by thin-layer chromatography and quantified by phosphoimager analysis. Activity is plotted relative to the nucleosome-stimulated ATPase activity of wild-type dMi-2, which is set to 100. The data presented are derived from three independent experiments. (B) Recombinant dMi-2 proteins (150, 300, 450 fmol) were incubated with a radioactively labelled 146 bp mononucleosome as indicated. Complexes were resolved by native PAGE. Empty circles denote positions of dMi-2–nucleosome complexes.
Nucleosome binding activity
dMi-2 forms a stable complex with a nucleosome containing 146 bp DNA that withstands electrophoresis through native polyacrylamide gels (Brehm et al., 2000; Figure 2B). dMi-2 mutants retaining the chromodomain region formed one or more complexes with the nucleosome probe (dMi-2 484–1982, lanes 8–10; dMi-2 1–1271, lanes 14–16; and dMi-2 Δ729–1937, lanes 17–19). Mutants lacking the chromodomain region failed to form specific complexes (dMi-2 ΔC, lanes 5–7; dMi-2 691– 1982, lanes 11–13; and dMi-2 691–1271, lanes 20–22); suggesting that the chromodomains are important for nucleosome binding. Failure of chromodomain mutants to hydrolyse ATP and to mobilize nucleosomes is likely to be a consequence of failure to bind the nucleosome substrate.
Loss of nucleosome binding activity could be caused by two mechanisms: the chromodomains could function as nucleosome binding domains mediating the interaction between enzyme and substrate. Alternatively, the chromodomain region might be critical for maintaining the correct three dimensional structure of dMi-2. In the latter scenario their removal would result in misfolding of dMi-2 into in inactive conformation.
Chromodomain function in the dMi-2 complex
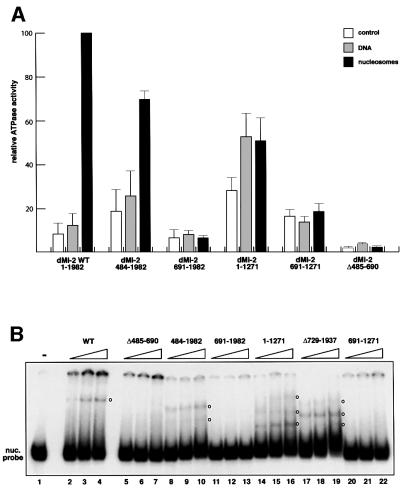
To probe directly the conformation of recombinant dMi-2 we used partial proteolysis. This technique allows dissection of multidomain proteins into independently folded fragments (i.e. domains). Domains are generally more resistant to digestion with specific proteases than unstructured or hinge regions. If deletion of the chromodomains resulted in misfolding, one would expect a higher sensitivity to proteolysis and a changed pattern of protease resistent fragments. However, we did not detect differences in protease sensitivity between dMi-2 wild type (WT) and dMi-2 ΔC (Figure 3A). Moreover, we obtained the same αdMi-2 antibody-reactive protease resistent fragments (compare lanes 1–3 with lanes 4–6). ATPase domains generally adopt a globular fold and often display high levels of resistance towards protease digestion (Hopfner et al., 2000; Kambampati et al., 2000). Two of the protease-resistent fragments co-migrated with the recombinant dMi-2 ATPase region, consistent with the notion that the ATPase domains of dMi-2 WT and dMi-2 ΔC are properly folded (compare lanes 3, 6 and 7). We conclude that removal of the chromodomains does not drastically change dMi-2 domain structure.
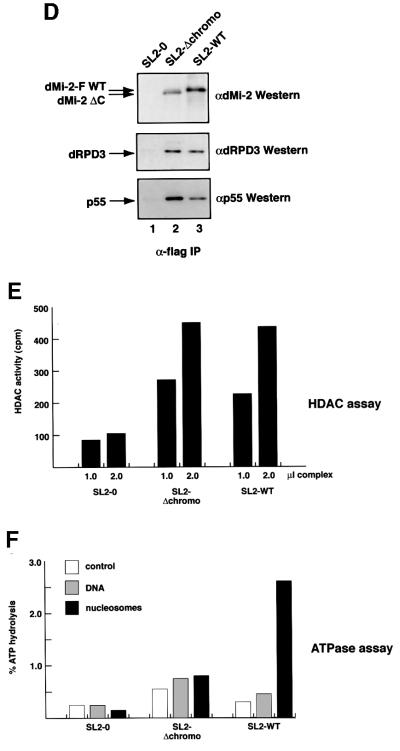
Fig. 3. Chromodomains are required for nucleosome-stimulated ATPase activity of the dMi-2 complex. (A) Limited proteolysis. One microgram of recombinant wild-type dMi-2 (lanes 1–3) or dMi-2 ΔC (lanes 4–6) were digested with increasing amounts of trypsin as indicated. Tryptic fragments were visualized by SDS–PAGE and western blotting using αdMi-2-specific antiserum (αdMi2-C). As a control, recombinant dMi-2 691–1271 (ATPase domain) was loaded in lane 7. Molecular weights are shown on the left. (B) GST pull-down. GST (lanes 2, 5 and 8) or a GST–dRPD3 fusion (lanes 3, 6 and 9) immobilized on beads were incubated with 35S-labelled in vitro translated (IVT) luciferase, dMi-2 WT and dMi-2 ΔC as indicated on top of the figure. Beads were washed extensively and bound material was visualized by SDS–PAGE and autoradiography. In vitro translated proteins were loaded as an input control (10%; lanes 1, 4 and 7). Positions of dMi-2 WT, dMi-2 ΔC and luciferase are indicated by arrows. (C) Co-infection/co-immunoprecipitation. Sf9 cells were co-infected with different combinations of recombinant baculoviruses as indicated on the top of the panels. Extracts from infected cells were immunoprecipitated with immobilized α-flag antibody, beads were extensively washed, subjected to SDS–PAGE and western analysis using specific antibodies as indicated on the right of the panels (lanes IP). To control for expression from the recombinant baculoviruses, an aliquot of cell extracts was loaded for comparison (lanes IN). (D) The dMi-2 chromodomain region is not required for formation of dMi-2 histone deacetylase complexes in vivo. Whole-cell extracts from SL2 cell lines stably expressing flag-tagged dMi-2 (SL2-WT), flag-tagged dMi-2 Δ484–691 (SL2-Δchromo) and from a control SL2 line (SL2-0) were fractionated by cation exchange chromatography. The dMi-2 containing fraction (500 mM KCl eluate) was subjected to affinity purification with immobilized α-flag antibody. Flag-tagged complexes were eluted with an excess of flag peptide. Eluted complexes were subjected to western analysis using antisera directed against dMi-2 (upper panel), dRPD3 (middle panel) and p55 (lower panel). (E) Eluted complexes were assayed for histone deacetylase activity. One and 2 µl of each complex preparation were used, as indicated. (F) Eluted complexes were subjected to ATPase assays in the presence or absence of 100 ng DNA or nucleosomes, as indicated. ATP hydrolysis was detected and quantified as above.
We also tested binding of dMi-2 ΔC to known subunits of the dMi-2 histone deacetylase complex. Zhang and colleagues have shown that recombinant human Mi2β binds HDAC1 in a cell-free system, suggesting that both proteins interact directly (Zhang et al., 1998). We have reproduced this interaction using in vitro translated dMi-2 and an immobilized GST–dRPD3 fusion (Figure 3B). In this assay, GST–dRPD3 specifically bound dMi-2 WT and dMi-2 ΔC with the same apparent efficiencies (compare lanes 6 and 9). We conclude that the domains required for dRPD3 binding are not affected by removal of the chromodomain region. We further verified this result using baculovirus-expressed proteins. We co-infected Sf9 cells with viruses driving expression of flag-tagged dMi-2 and untagged dRPD3, followed by immunoprecipitation with α-flag antibody (Figure 3C). Recombinant dRPD3 failed to co-immunoprecipitate with α-flag antibody unless flag-tagged dMi-2 was co-expressed (compare lanes 4 and 6). Co-expression of dRPD3 and flag-tagged dMi-2 ΔC likewise resulted in efficient co-immunoprecipitation of dRPD3, confirming that the dMi-2 ΔC mutant retains dRPD3 binding activity (compare lanes 10 and 12). We performed an analogous co-infection/co-immunoprecipitation analysis using flag-tagged dMi-2 and untagged p55, another subunit of the dMi-2 complex (Brehm et al., 2000). p55 co-immunoprecipitated equally well with dMi-2 WT and dMi-2 ΔC (Figure 3C, compare lanes 4, 6, 10 and 12). We conclude that removal of the chromodomain region does not impede binding to dRPD3 and p55.
What is chromodomain function in the multi-subunit dMi-2 complex? We established two Drosophila SL2 cell lines stably expressing flag-tagged dMi-2 proteins to address this issue. The SL2-WT line stably expresses wild-type flag-tagged dMi-2, and the SL2-Δchromo line stably expresses flag-tagged dMi-2 ΔC. We affinity-purified flag-tagged dMi-2 complexes from these lines and tested for co-purification of dRPD3 and p55 (Figure 3D; Brehm et al., 2000). Both dRPD3 and p55 co-purified with flag-tagged dMi-2 from SL2-WT cells but not from SL2-0 control cells (Figure 3D, compare lanes 1 and 2). dRPD3 and p55 also co-purified with flag-tagged dMi-2 ΔC (lane 3). Moreover, dMi-2 complexes isolated from the SL2-WT and SL2-Δchromo lines displayed comparable histone deacetylase activities (Figure 3E). These results suggest that structural integrity and histone deacetylase function of the dMi-2 complex do not depend on the chromodomains in vivo. We also tested dMi-2 complexes in ATPase assays (Figure 3F). Wild-type dMi-2 complex showed robust nucleosome-stimulated ATPase activity. We failed to detect nucleosome-stimulated ATPase activity with the dMi-2 ΔC complex, confirming that the chromodomains are important for nucleosome-stimulated ATPase activity. Taken together, our results indicate that removal of the chromodomain region has no detectable effect on protein structure and interaction with dMi-2 complex subunits. Rather, it specifically abrogates nucleosome-stimulated ATPase, nucleosome binding and mobilization activities, suggesting that the chromodomains directly interact with the nucleosome substrate.
dMi-2–histone interactions
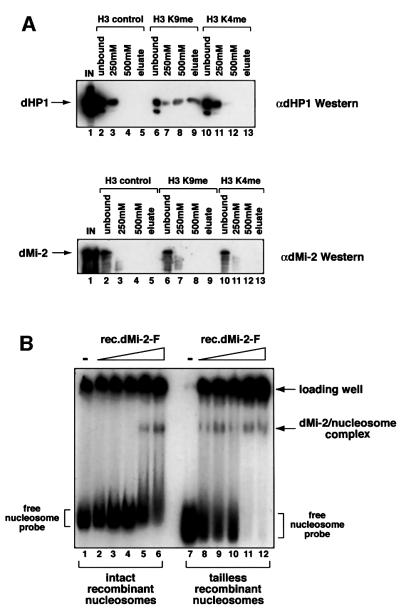
Recently, several groups have shown that the chromodomain of HP1 binds histones in a variety of assay systems (Bannister et al., 2001; Lachner et al., 2001; Nielsen et al., 2001). The HP1 chromodomain specifically recognizes K9-methylated histone H3 tails both in vitro and in vivo (Bannister et al., 2001; Lachner et al., 2001). These findings have led to the suggestion that chromodomains read the histone code by binding to histone methylation marks (Jenuwein and Allis, 2001). Given that the dMi-2 chromodomains play an important role in nucleosome binding we tested the possible involvement of methylated histone tails in this interaction. We compared binding of Drosophila HP1 and dMi-2 to methylated H3 tails. We incubated immobilized H3 peptides with Drosophila embryo nuclear extract. Figure 4A shows that a significant proportion of HP1 was retained on the K9-methylated peptide, even after successive washes with buffers containing 250 and 500 mM NaCl, respectively. In contrast, HP1 was removed from unmethylated and K4-methylated peptides by the 250 mM salt wash, confirming the specificity and high affinity of HP1 for the H3 K9 modification (Bannister et al., 2001; Lachner et al., 2001). Unlike HP1, dMi-2 failed to bind to any of the H3 peptides under these conditions, suggesting that the dMi-2 chromodomains do not share the high affinity of the HP1 chromodomain for K9-methylated H3 tails. However, we cannot rule out the possibility that dMi-2 binds other histone tails or other combinations of methylation marks. To more rigorously assess the contribution of histone tails to the dMi-2–nucleosome interaction, we employed 146 bp mononucleosomes reconstituted from recombinant histones lacking their N-termini (Brehm et al., 2000). ‘Intact’ and ‘tailless’ mononucleosomes were incubated with recombinant dMi-2 and subjected to the bandshift assay. As Figure 4B shows, dMi-2 bound to both nucleosomes. Interestingly, more dMi-2 protein was required to visualize a complex with intact recombinant nucleosomes than to shift tailless recombinant nucleosomes. However, differences in the quality of the two mononucleosome preparations (note material retained in the loading well) precludes us from accurately assessing the relative affinities of dMi-2. Nevertheless, these results suggest that dMi-2–histone tail interactions, if they occur, do not make a critical contribution to nucleosome binding. We also tested binding of dMi-2 mutants to the recombinant nucleosomes. Again, binding to both types of nucleosomes was sensitive to the removal of the chromodomain region (data not shown).
Fig. 4. dMi-2 does not require histone tails for nucleosome binding. (A) Drosophila embryo nuclear extract was incubated with immobilized methylated and unmethylated histone H3 peptides as indicated. Peptide beads were successively washed with buffer containing increasing concentration of NaCl and eluted with glycine buffer pH 2.5, as indicated on top of the panels. Proteins in unbound, wash and eluate fractions were detected by western analysis using antisera directed against dHP1 (upper panel) and dMi-2 (lower panel). IN, input; control, unmodified peptide; K9me, K9-methylated peptide; K4me, K4-methylated peptide. (B) dMi-2 nucleosome bandshift assay using a 146 bp nucleosome probe reconstituted from intact recombinant histones (lanes 1–6) or reconstituted from tailless recombinant histones (lanes 7–12). Increasing amounts of recombinant dMi-2 WT (lanes 2 and 8, 50 fmol; lanes 3 and 9, 100 fmol; lanes 4 and 10, 200 fmol; lanes 5 and 11, 300 fmol; lanes 6 and 12, 400 fmol) were incubated with radioactively labelled nucleosome probe. Complexes were resolved by native PAGE. The positions of free nucleosome probe, dMi-2–nucleosome complexes and nucleosome probe precipitated in the loading well are indicated.
DNA binding activity
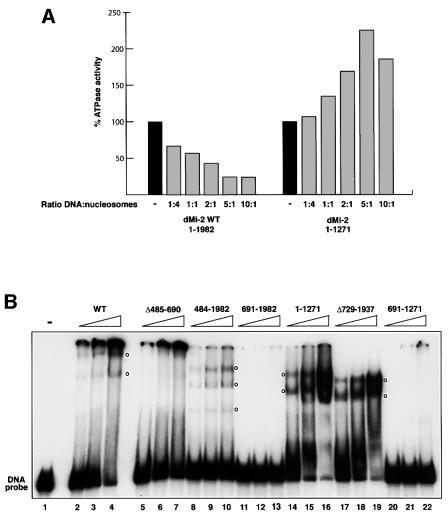
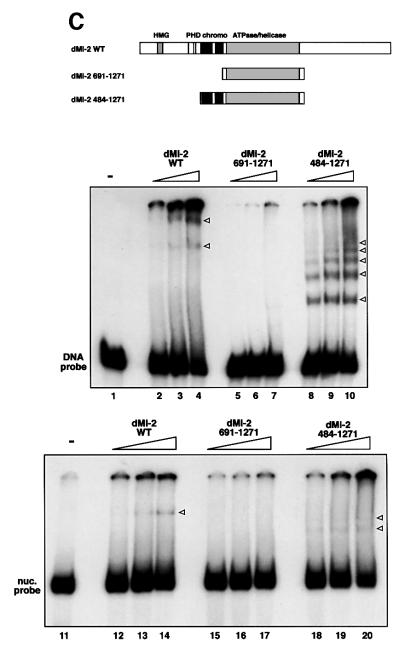
We sought to test whether dMi-2 binding to the nucleosome could involve nucleosomal DNA. We have shown previously that dMi-2 is retained on immobilized polynucleosomes but not on DNA when beads are subjected to a series of high salt washes (Brehm et al., 2000). However, we had noted that residual dMi-2 binding to immobilized DNA is detected when the salt concentration is reduced to 100 mM NaCl. To determine whether dMi-2 binds DNA under low salt conditions we measured nucleosome-stimulated ATPase activity in the presence of 50 mM KCl and increasing amounts of free DNA. Using this experimental strategy, the Kingston laboratory has recently demonstrated DNA binding by the nucleosome-stimulated SNF2H ATPase (Aalfs et al., 2001). Titration of DNA effectively inhibited dMi-2 nucleosome- stimulated ATPase activity, suggesting that dMi-2, like SNF2H, does indeed bind DNA in this assay (Figure 5A). The dMi-2 1–1271 mutant, which is stimulated by free DNA (Figure 2A), was not adversely affected by addition of free DNA. To look more directly at DNA binding, we performed bandshift assays using a 146 bp DNA probe (Figure 5B). dMi-2 WT formed two complexes entering the gel (lanes 2–4). Most of the probe was retained in the loading well, suggesting the formation of large protein– DNA complexes. dMi-2 does not appear to bind DNA in a sequence-specific manner and binds with equal apparent affinity to different DNA fragments unrelated in sequence (data not shown). dMi-2 484–1982 (lanes 8–10), dMi-2 1–1271 (lanes 14–16) and dMi-2 Δ729–1937 all formed multiple protein–DNA complexes. In contrast, no interaction between dMi-2 691–1982 (which lacks the NTR) and DNA was detected. Thus, the DNA binding activity of dMi-2 resides in the NTR. Internal deletion of the chromodomain region compromised formation of complexes migrating through the gel (dMi-2 ΔC, lanes 5–7). However, a significant fraction of the DNA probe was retained in the loading well. It is possible that other domains in the NTR, such as the HMG box-like region or the PHD fingers, cause the formation dMi-2–DNA complexes too large to enter the gel. These results suggest that the chromodomain region is important for the formation of dMi-2–DNA complexes that can be resolved in the bandshift assay.
Fig. 5. The chromodomain region of dMi-2 is required for DNA binding. (A) dMi-2 was pre-incubated with DNA prior to addition of 100 ng nucleosomes and ATP at different DNA:nucleosome ratios, as indicated. ATPase activity was determined as above and expressed as percentage nucleosome-stimulated ATPase activity. (B) dMi-2 proteins (40, 80 and 160 fmol) were incubated with a radioactively labelled 146 bp DNA fragment, as indicated. Complexes were resolved by native PAGE. Empty circles denote the position of complexes. (C) dMi-2 proteins (40, 80 and 160 fmol) were subjected to the bandshift assay using a 146 bp DNA fragment (upper panel) or a 146 bp nucleosome (lower panel) as a probe as indicated. Complexes are denoted by arrowheads.
Taken together, our histone tail, nucleosome and DNA binding results are consistent with the notion that the dMi-2 chromodomains mediate nucleosome binding not via histone tail interactions, but via interactions involving nucleosomal DNA. To further test this hypothesis we tested whether fusion of the chromodomain region would suffice to confer DNA binding activity to the ATPase domain (Figure 5B, lanes 20–22). Addition of the chromodomain region to the ATPase domain (dMi-2 484–1271) did indeed confer strong DNA binding activity (Figure 5C, upper panel, compare lanes 5–7 with lanes 8–10). dMi-2 484–1271 formed multiple protein–DNA complexes. Fusion of the chromodomain region to the ATPase domain did also confer nucleosome binding activity (Figure 5C, lower panel, compare lanes 15–17 with lanes 18–20). Despite the ability of dMi-2 484–1271 to bind DNA and nucleosomes we did not detect DNA- or nucleosome-stimulated ATPase or nucleosome mobilization activity with this mutant (data not shown). This argues that while the chromodomain region is sufficient to tether the ATPase to the nucleosome substrate, additional regions are required to translate nucleosome binding into increased ATP hydrolysis and nucleosome mobilization.
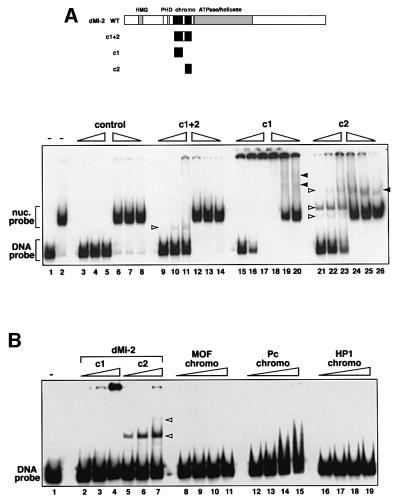
We next tested whether the isolated dMi-2 chromodomains bound DNA and nucleosomes. We expressed the chromodomain region (aa 488–673; c1+2) as well as the isolated chromodomains (aa 488–566, c1; and aa 608–673, c2) in Escherichia coli. Unlike the c2 peptide, the c1+2 and the c1 peptides were expressed in largely insoluble form (data not shown). We purified the histidine-tagged peptides and subjected them to bandshift assays using 146 bp DNA and 146 bp mononucleosome probes (Figure 6A). As a control, we also performed a mock purification from E.coli carrying the empty expression vector. The three peptide preparations displayed different DNA and nucleosome binding properties. The c1+2 peptide displayed weak DNA binding activity but formed a specific complex with the DNA probe (compare lanes 3–5 to lanes 9–11). However, we failed to detect significant binding to the mononucleosome (compare lanes 6–8 with lanes 12–14). The c1 peptide efficiently retained DNA in the loading well, but did not produce complexes migrating through the gel (lanes 15–17). The c1 peptide also retained much of the nucleosome probe in the loading well, but some c1–nucleosome complexes entering the gel were detectable (lanes 18–20). The c2 peptide displayed strong DNA and nucleosome binding activity and formed several distinct complexes (lanes 21–26). It is difficult to assess to what extent the different binding characteristics are a reflection of the way these domains interact with DNA and nucleosomes. The tendency of c1+2 and c1 to form inclusion bodies upon expression in E.coli indicates that these peptides might not adopt a fully functional conformation. Nevertheless, it is clear that at least the isolated c2 chromodomain has robust DNA and nucleosome binding activity.
Fig. 6. Chromodomains and DNA binding. (A) DNA and nucleosome bandshift assay using bacterially expressed dMi-2 chromodomain polypeptides. As a control, material obtained from a mock purification from E.coli transformed with empty expression vector was used (‘control’, lanes 3–8). c1+2, dMi-2 488–673; c1, dMi-2 488–566; c2, dMi-2 608–673. The amounts of chromodomain peptides used were as follows: lanes 9, 14, 15, 20, 21 and 26: 6 pmol; lanes 10, 13, 16, 19, 22 and 25: 12 pmol; lanes 11, 12, 17, 18, 23 and 24: 25 pmol. Complexes are denoted with open (chromodomain–DNA complexes) and closed (chromodomain–nucleosome complexes) arrowheads. (B) DNA bandshift assay using bacterially expressed chromodomain polypeptides. Position of chromodomain–DNA complexes are shown by open arrowheads. The amounts of chromodomain peptides used were as follows: lanes 2, 5, 8, 12 and 16: 6 pmol; lanes 3, 6, 9, 13 and 17: 12 pmol; lanes 4, 7, 10, 14 and 18: 25 pmol; lanes 11, 15 and 19: 50 pmol. Chromodomain–DNA complexes are denoted with open arrowheads.
So far, no other chromodomain has been shown to directly bind to DNA. To assess whether the DNA binding activity of the dMi-2 chromodomains is shared by other chromodomains, we subjected the chromodomains of Drosophila Polycomb, MOF and dHP1 to the DNA bandshift assay. As shown in Figure 6B, the chromodomains of HP1 and MOF did not bind DNA. Both proteins were expressed in soluble form in E.coli and were functional in H3 peptide (dHP1) and RNA (MOF) binding assays (data not shown). The Polycomb chromodomain retarded the DNA probe at high protein concentrations but failed to resolve into distinct protein–DNA complexes indicating that complexes are unstable and dissociate during the gel run. These results demonstrate that the DNA binding activity displayed by the dMi-2 chromodomains is not a common property of all chromodomains and provide further evidence for the diverse functional specialization within the chromodomain family.
Discussion
The presence of chromodomains defines the CHD family of ATP-dependent chromatin remodellers (Woodage et al., 1997). It is not known what their role is and whether and how they are involved in chromatin remodelling. We show here that the chromodomain region of dMi-2 plays an important role in ATP-dependent nucleosome mobilization, and that it binds the nucleosome via interactions with nucleosomal DNA. This notion is supported by six observations: (i) dMi-2 mutants lacking the chromodomain region fail to respond to DNA or nucleosomes in the ATPase assay; (ii) these mutants fail to bind efficiently to DNA and nucleosomes; (iii) the chromodomain region confers DNA and nucleosome binding activity when fused to the ATPase domain; (iv) the isolated chromodomains bind DNA and nucleosomes; (v) the chromodomains have higher affinity for DNA than for a 146 bp mononucleosome; and (vi) removal of the histone tails does not impede nucleosome binding. Chromodomain mutants that no longer interact with the nucleosome are compromised for ATPase activity and accordingly fail to mobilize mononucleosomes. Thus, the dMi-2 chromodomains seem to function at an early step of the chromatin remodelling process by mediating the interaction between the enzyme and its nucleosome substrate.
The best studied chromodomain proteins are Pc, HP1 and MOF. Their chromodomains interact with other protein subunits within a large complex (Pc), bind histone cores and methylated histone tails (HP1) and bind a specific RNA molecule (MOF) (Breiling et al., 1999; Akhtar et al., 2000; Bannister et al., 2001; Lachner et al., 2001; Nielsen et al., 2001). Our identification of the DNA binding activity of the dMi-2 chromodomains adds another activity to the growing list of chromodomain functions.
The dMi-2 chromodomains and complex assembly
The chromodomain of Pc is required for interaction with Polyhomeotic and for the proper assembly of the PcG complex (Strutt and Paro, 1997). In contrast, deletion of the dMi-2 chromodomains does not affect binding of the dRPD3 and p55 subunits of the dMi-2 complex in vitro and in vivo. This suggests that the chromodomains do not engage in protein interactions critical for the integrity of the dMi-2–dRPD3–p55 complex. This finding is consistent with previous work showing that HDAC1 binds to the human Mi2β PHD fingers (Zhang et al., 1998). It is not known which domain of dMi-2 is responsible for binding p55. We cannot rule out that association of other dMi-2 complex subunits, such as the recently identified dMBD-like and dMTA-like, is affected by deletion of the chromodomains (Ballestar et al., 2001).
dMi-2 and histone tail interactions
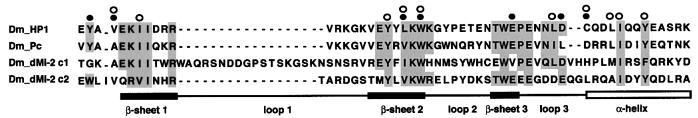
Based on the finding that the HP1 chromodomain specifically recognizes K9-methylated histone H3 tails (Bannister et al., 2001; Lachner et al., 2001), Jenuwein and Allis (2001) have suggested that this function might be shared by other chromodomains. Structural studies show that the K9-methylated H3 peptide interacts with a concave binding surface on one face of the chromodomain β-sheet structure (Ball et al., 1997; Jacobs et al., 2001). The seven residues forming the binding surface are highly conserved among HP1 proteins from different species [Y24, V26, L43, W45, E56, D62 and C63; numbering according to Jacobs et al. (2001)] (Figure 7). However, only some of these are conserved in non-HP1 chromodomains. Furthermore, those residues that are conserved are not only involved in peptide recognition by dHP1, but in addition form part of a hydrophobic core important for the structure of the chromodomain fold (V26, L43, W45 and C63; Ball et al., 1997). More recently, solution of the co-crystal structure of HP1 bound to K9-methylated histone H3 peptide revealed that three aromatic residues (Y24, W45 and Y48) form a three-walled cage recognizing the methylammonium group of K9 (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002). Of these three residues, only one is conserved in the dMi-2 chromo domains. It appears unlikely that the dMi-2 chromodomains recognize methylated histone H3 tails. Accordingly, we find that dMi-2 does not share the affinity of the dHP1 chromodomain for K9-methylated H3 tails in vitro. Moreover, staining of polytene chromosomes with antibodies directed against dHP1 and dMi-2, repectively, results in mutually exclusive staining patterns: dHP1 co-localizes with K9-methylated histone H3 and concentrates at the heterochromatic chromocentre (Jacobs et al., 2001). Instead, dMi-2 localizes to many sites within euchromatin and is notably absent from the chromocentre (Murawsky et al., 2001), in agreement with a role of dMi-2 in transcriptional regulation (Kehle et al., 1998). Recently, the human NuRD complex has been shown to bind unmodified H3 tails but not H3 tails methylated at K4 (Nishioka et al., 2002; Zegerman et al., 2002). It is not clear which subunit of NuRD mediates tail binding. Our peptide pull-down assays have failed to reveal any difference in affinity of dMi-2 for unmodified and K4 methylated H3 peptides. This may reflect differences in subunit composition and function between human and Drosophila Mi-2 complexes.
Fig. 7. Alignment of chromodomains. The chromodomain sequences of Drosophila HP1 (Dm_HP1; aa residues 23–74), Drosophila Polycomb (Dm_Pc) and the two dMi-2 chromodomains (Dm_dMi-2 c1 and Dm_dMi-2 c2) were aligned. Conserved residues are shaded. Positions of α-helix, β-sheets and loops are indicated below the alignment according to the HP1 structure as determined by Ball et al. (1997). HP1 residues forming the hydrophobic core of the structure are denoted with open circles above of the alignment. HP1 residues involved in methylated histone tail recognition are indictated by filled circles (Jacobs et al., 2001).
It has been speculated that dMi-2 chromodomains recognize methylated histone tails simultaneously carrying two modifications (Jenuwein and Allis, 2001). How ever, the finding that dMi-2–nucleosome binding does not require the histone tails at all does not support this notion. Moreover, histone tails are not required for nucleosome stimulated ATPase activity of dMi-2 and the Xenopus Mi-2 complex (Boyer et al., 2000; Brehm et al., 2000; Guschin et al., 2000).
The dMi-2 chromodomains are DNA binding modules
The binding of dMi-2 and a dMi-2 chromodomain to DNA was initially surprising to us, as we had previously shown that dMi-2 is not efficiently retained on DNA immobilized on beads in a pull-down experiment (Brehm et al., 2000). The pull-down experiment involves a series of high salt wash steps. In contrast, the bandshift reactions were performed in low salt, and the resulting complexes immediately subjected to gel electrophoresis, conditions that are likely to favour dMi-2–DNA complex formation and stability. The c2 chromodomain efficiently binds DNA, unlike the chromodomains of Pc, dHP1 and MOF. It is interesting to note that based on structural studies, Ball et al. (1997) have suggested an evolutionary relationship between the HP1 chromodomain and two archaebacterial DNA binding proteins. They noted, however, that the charge distribution on the exterior of the two domains is not conserved, indicating that the HP1 chromodomain functions as a protein interaction rather than a DNA binding motif. Indeed, neither the HP1 chromodomain nor the chromo-shadow domain are sufficient to bind a 146 bp DNA probe in the bandshift assay (Figure 6B; Zhao et al., 2000). A weak DNA binding activity has been indirectly inferred for the Pc chromodomain (Breiling et al., 1999). Accordingly, the Pc chromodomain displays weak affinity for DNA in our bandshift assay. The MOF chromodomain is a dedicated RNA binding module, and MOF chromo domain–RNA complexes are resistent to competition with DNA (Akhtar et al., 2000). Consistent with these results, the MOF chromodomain does not bind DNA in our assay. It is an intriguing possibility that some chromodomains may have retained an ancient DNA binding function (dMi-2), whereas others have evolved to recognize RNA (MOF) or to function as protein interaction domains (Pc, HP1). It is important to note that DNA binding does not elicit increased dMi-2 ATPase activity. Presumably, other regions of dMi-2 recognize the nucleosome structure following initial binding and then activate the ATPase domain.
Role of other regions of dMi-2
Fusion of the chromodomain region and the ATPase domain is sufficient for DNA and nucleosome binding, but not for nucleosome-stimulated ATPase and nucleosome mobilization activities. It follows that additional regions outside these domains make critical contributions to nucleosome remodelling. Conversion of the chromo domain–ATPase domain fusion (dMi-2 484–1271) to an active nucleosome remodeller can be achieved by addition of the remainder of the NTR or by addition of the CTR. This implies that both NTR and CTR provide activities that are redundant in our assay. The CTR binds the repression domains of hunchback and tram-track 69 (Ttk69) (Kehle et al., 1998; Murawsky et al., 2001). The C-terminus of mammalian Mi2β interacts with the KAP-1 co-repressor (Schultz et al., 2001). Our results suggest that CTR function is not restricted to transcription factor binding. Instead, it plays an active role in ATPase regulation: although deletion of the CTR does not affect nucleosome mobilization it makes the ATPase responsive to DNA. In this respect, the dMi-2 1–1271 mutant resembles ATPases of the SWI/SNF subgroup (Becker and Hörz, 2002). This observation suggests that the CTR is directly involved in regulation of the ATPase domain: it is required to suppress activity in presence of the ‘wrong’ effector (DNA), when no remodelling substrate (nucleosome) is available. It is conceivable that the CTR might undergo a change in conformation following nucleosome recognition, which then allows the ATPase domain to function.
We have presented the first biochemical analysis of chromodomain function in an ATP-dependent chromatin remodelling factor. Our results suggest that the chromodomain region of dMi-2 serves as a DNA and nucleosome binding module. It will now be important to determine whether the two dMi-2 chromodomains are redundant or whether they make different contributions to chromatin remodelling.
Materials and methods
Expression and purification of recombinant proteins
dMi-2 mutants were generated by PCR using appropriate sets of primers and transferred into the pVL1392 transfer vector. Baculovirus and protein purification procedures have been described previously (Brehm et al., 2000). dMi-2 chromodomains were amplified by PCR using appropriate primers and subcloned into pET15b. His-tagged dMi-2, MOF (aa 366–450) and Polycomb (aa 19–84) chromodomain polypeptides were expressed in E.coli BL21(DE3) and purified with Talon™ (Clontech) resin according to the manufacturer’s instructions, followed by dialysis against EX50 buffer (Längst et al., 1999). The HP1 chromodomain containing peptide (aa 1–134) was expressed and purified using the intein system according to the manufacturer’s instructions. All constructs were verified by DNA sequencing.
Enzymatic assays
ATPase, deacetylase and nucleosome assembly assays have been described previously (Brehm et al., 2000). Assembly was verified by micrococcal nuclease digestion. For DNA inhibition experiments, dMi-2 and DNA were pre-incubated for 5 min at 26°C. Recombinant histones were expressed in E.coli, purified and reconstituted into octamers as described previously (Luger et al., 1997). For HDAC assays, recombinant histones were acetylated with recombinant Hat1p and yGcn5 using [3H]acetylCoA and purified by ion exchange chromatography. Acetylation was monitored by acid–urea gel electrophoresis.
Electrophoretic mobility shift and nucleosome mobility assay
These assays were carried out as described previously (Brehm et al., 2000). Briefly, proteins and 32P-labelled probes were incubated on ice for 10 min in EX50 buffer prior to native PAGE. Complexes and probe were visualized by autoradiography.
Limited proteolysis
Recombinant proteins were obtained from baculovirus-infected Sf9 cells by fractionation over HiTrap SP Sepharose (Amersham Pharmacia). Extracts were applied in buffer A250 (25 mM HEPES pH 7.6, 250 mM KCl, 10% glycerol). Recombinant dMi-2 was eluted with buffer A500 (500 mM KCl). Trypsin digestions (20 µl) were performed in buffer T (25 mM HEPES pH 7.6, 350 mM KCl, 1 mM CaCl2, 10% glycerol) for 15 min at 26°C. Reactions were stopped by addition of SDS–PAGE loading buffer and heating to 95°C and analysed by SDS–PAGE and western blot using αdMi2-C antiserum.
Peptide pull-down experiments
H3 peptides were synthesized (Peptide Speciality Laboratories) and coupled to Sulfolink™ Sepharose (Bio-Rad) beads according to the manufacturers’ instructions. Beads were incubated with Drosophila embryo nuclear extract in HEMG100 buffer (Brehm et al., 2000) for 90 min and subjected to sequential washes in HEMG250 and HEMG500, followed by elution with 0.1 M glycine pH 2.5. Washes and eluates were analysed by SDS–PAGE and western blot using αdMi-2 antiserum (Brehm et al., 2000) and αHP1 antiserum (kind gift of S.Elgin; James and Elgin, 1986).
Establishment of stable SL2 lines and purification of dMi-2 from SL2 cells
cDNAs encoding flag-tagged dMi-2 and flag-tagged dMi-2 ΔC were subcloned into pPacFlag (Chen et al., 1999), and co-transfected with pUChsneo (Steller and Pirrotta, 1986) into SL2 cells using the Effectene (Qiagen) reagent according to the manufacturer’s instructions. Two days post-transfection cells were selected for 3–4 weeks using 1 mg/ml G418. Approximately 109 cells were harvested, lysed in buffer A250 containing protease inhibitors (0.5 mM PMSF, 1 µM leupeptin, 1 µM pepstatin, 0.3 µM aprotinin), sonicated and cleared by centrifugation. Cleared extracts were applied to HiTrap SP Sepharose (Amersham Pharmacia). dMi-2 complexes were eluted with buffer A500 (500 mM KCl), subjected to flag-affinity purification and complexes were eluted using an excess of flag peptide as described previously (Brehm et al., 2000).
Acknowledgments
Acknowledgements
We thank A.Courey, B.Turner, T.Kouzarides and J.Müller for reagents, and R.Aasland and members of the Becker, Längst and Imhof laboratories for advice, discussion and critical reading of the manuscript. K.B. and A.M. were supported by a Deutsche Forschungsgemeinschaft grant (BR2102/1-1).
References
- Aalfs J.D., Narlikar,G.J. and Kingston,R.E. (2001) Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem., 276, 34270–34278. [DOI] [PubMed] [Google Scholar]
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Akhtar A., Zink,D. and Becker,P.B. (2000) Chromodomains are protein–RNA interaction modules. Nature, 407, 405–409. [DOI] [PubMed] [Google Scholar]
- Ball L.J. et al. (1997) Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J., 16, 2473–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestar E., Pile,L.A., Wassarman,D.A., Wolffe,A.P. and Wade,P.A. (2001) A Drosophila MBD family member is a transcriptional corepressor associated with specific genes. Eur. J. Biochem., 268, 5397–5406. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Becker P.B. and Hörz,W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., 71, 247–273. [DOI] [PubMed] [Google Scholar]
- Brehm A., Längst,G., Kehle,J., Clapier,C.R., Imhof,A., Eberharter,A., Muller,J. and Becker,P.B. (2000) dMi-2 and ISWI chromatin remodeling factors have distinct nucleosome binding and mobilization properties. EMBO J., 19, 4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Logie,C., Bonte,E., Becker,P.B., Wade,P.E., Wolffe,A.P., Wu,C., Imbalzano,A.N. and Peterson,C.L. (2000) Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem., 275, 18864–18870. [DOI] [PubMed] [Google Scholar]
- Breiling A., Bonte,E., Ferrari,S., Becker,P.B. and Paro,R. (1999) The Drosophila polycomb protein interacts with nucleosomal core particles in vitro via its repression domain. Mol. Cell. Biol., 19, 8451–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Sweder,K.S. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J.C. (2001) Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene, 275, 19–29. [DOI] [PubMed] [Google Scholar]
- Guschin D., Wade,P.A., Kikyo,N. and Wolffe,A.P. (2000) ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry, 39, 5238–5245. [DOI] [PubMed] [Google Scholar]
- Hopfner K.P., Karcher,A., Shin,D.S., Craig,L., Arthur,L.M., Carney,J.P. and Tainer,J.A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell, 101, 789–800. [DOI] [PubMed] [Google Scholar]
- Horn P.J. and Peterson,C.L. (2001) The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front. Biosci., 6, D1019–D1023. [DOI] [PubMed] [Google Scholar]
- Imhof A. and Becker,P.B. (2001) Modifications of the histone N-terminal domains. Evidence for an ‘epigenetic code’? Mol. Biotechnol., 17, 1–13. [DOI] [PubMed] [Google Scholar]
- Jacobs S.A. and Khorasanizadeh,S. (2002) Structure of HP1 chromo domain bound to a lysine 9-methylated histone H3 tail. Science, 295, 2080–2083. [DOI] [PubMed] [Google Scholar]
- Jacobs S.A., Taverna,S.D., Zhang,Y., Briggs,S.D., Li,J., Eissenberg,J.C., Allis,C.D. and Khorasanizadeh,S. (2001) Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J., 20, 5232–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T.C. and Elgin,S.C. (1986) Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol., 6, 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kambampati R., Pellegrino,C., Paiva,A., Huang,L., Mende-Mueller,L. and Chakraburtty,K. (2000) Limited proteolysis of yeast elongation factor 3. Sequence and location of the subdomains. J. Biol. Chem., 275, 16963–16968. [DOI] [PubMed] [Google Scholar]
- Kehle J., Beuchle,D., Treuheit,S., Christen,B., Kennison,J.A., Bienz,M. and Muller,J. (1998) dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science, 282, 1897–1900. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Längst G., Bonte,E.J., Corona,D.F. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- Luger K., Rechsteiner,T.J., Flaus,A.J., Waye,M.M. and Richmond,T.J. (1997) Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol., 272, 301–311. [DOI] [PubMed] [Google Scholar]
- Messmer S., Franke,A. and Paro,R. (1992) Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev., 6, 1241–1254. [DOI] [PubMed] [Google Scholar]
- Murawsky C.M., Brehm,A., Badenhorst,P., Lowe,N., Becker,P.B. and Travers,A.A. (2001) Tramtrack69 interacts with the dMi-2 subunit of the Drosophila NuRD chromatin remodeling complex. EMBO Rep., 2, 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Rice,J.C., Strahl,B.D., Allis,C.D. and Grewal,S.I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Nielsen A.L., Oulad-Abdelghani,M., Ortiz,J.A., Remboutsika,E., Chambon,P. and Losson,R. (2001) Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell., 7, 729–739. [DOI] [PubMed] [Google Scholar]
- Nielsen P.R., Nietlispach,D., Mott,H.R., Callaghan,J., Bannister,A., Kouzarides,T., Murzin,A.G., Murzina,N.V. and Laue,E.D. (2002) Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature, 416, 103–107. [DOI] [PubMed] [Google Scholar]
- Nishioka K., Chuikov,S., Sarma,K., Erdjument-Bromage,H., Allis,C.D., Tempst,P. and Reinberg,D. (2002) Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev., 16, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R. and Hogness,D.S. (1991) The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A.H. et al. (2001) Loss of the suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell, 107, 323–337. [DOI] [PubMed] [Google Scholar]
- Platero J.S., Hartnett,T. and Eissenberg,J.C. (1995) Functional analysis of the chromo domain of HP1. EMBO J., 14, 3977–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S. et al. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Schultz D.C., Friedman,J.R. and Rauscher,F.J.,III (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev., 15, 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H. and Pirrotta,V. (1986) P transposons controlled by the heat shock promoter. Mol. Cell. Biol., 6, 1640–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes D.G. and Perry,R.P. (1995) DNA-binding and chromatin localization properties of CHD1. Mol. Cell. Biol., 15, 2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H. and Paro,R. (1997) The polycomb group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol. Cell. Biol., 17, 6773–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J.W., Deuring,R., Scott,M.P., Kissinger,M., Pattatucci,A.M., Kaufman,T.C. and Kennison,J.A. (1992) brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell, 68, 561–572. [DOI] [PubMed] [Google Scholar]
- Turner B.M. (2000) Histone acetylation and an epigenetic code. BioEssays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- Woodage T., Basrai,M.A., Baxevanis,A.D., Hieter,P. and Collins,F.S. (1997) Characterization of the CHD family of proteins. Proc. Natl Acad. Sci. USA, 94, 11472–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Canas,B., Pappin,D. and Kouzarides,T. (2002) Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J. Biol. Chem., 277, 11621–11624. [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]
- Zhao T., Heyduk,T., Allis,C.D. and Eissenberg,J.C. (2000) Hetero chromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem., 275, 28332–28338. [DOI] [PubMed] [Google Scholar]