Abstract
The spindle checkpoint protein Mad1 recruits Mad2 to unattached kinetochores and is essential for Mad2–Cdc20 complex formation in vivo but not in vitro. The crystal structure of the Mad1–Mad2 complex reveals an asymmetric tetramer, with elongated Mad1 monomers parting from a coiled-coil to form two connected sub-complexes with Mad2. The Mad2 C-terminal tails are hinged mobile elements wrapping around the elongated ligands like molecular ‘safety belts’. We show that Mad1 is a competitive inhibitor of the Mad2–Cdc20 complex, and propose that the Mad1–Mad2 complex acts as a regulated gate to control Mad2 release for Cdc20 binding. Mad1–Mad2 is strongly stabilized in the tetramer, but a 1:1 Mad1–Mad2 complex slowly releases Mad2 for Cdc20 binding, driven by favourable binding energies. Thus, the rate of Mad2 binding to Cdc20 during checkpoint activation may be regulated by conformational changes that destabilize the tetrameric Mad1–Mad2 assembly to promote Mad2 release. We also show that unlocking the Mad2 C-terminal tail is required for ligand release from Mad2, and that the ‘safety belt’ mechanism may prolong the lifetime of Mad2–ligand complexes.
Keywords: anaphase/Cdc20/Mad1/Mad2/mitotic spindle checkpoint
Introduction
The spindle assembly checkpoint monitors sister chromatid bi-orientation in mitosis and prevents loss of sister chromatid cohesion in the presence of a damaged spindle (Hardwick, 1998; Shah and Cleveland, 2000). Mad1, Mad2, Bub1, BubR1/Mad3 and Bub3 associate with kinetochores in prophase and prometaphase to monitor attachment of sister chromatids to the spindle (Chen et al., 1996, 1998; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998). These proteins are conserved from yeast to metazoans, suggesting that the signal transduction pathways originating at unattached kinetochores are functionally and mechanistically similar in all eukaryotes. The target of the spindle checkpoint is the anaphase-promoting complex (APC), a multi-subunit ubiquitin ligase, whose activity ultimately determines loss of sister chromatid cohesion at the metaphase– anaphase transition (Zachariae and Nasmyth, 1999). APC inhibition requires a direct interaction of Mad2 with Cdc20, a positive regulator of the APC (Li et al., 1997; Fang et al., 1998; Hwang et al., 1998; Kallio et al., 1998; Kim et al., 1998; Wassmann and Benezra, 1998). Cdc20 mutants impaired in Mad2 binding do not arrest in metaphase upon Mad2 or Mps1 over-expression in fission and budding yeast, respectively (Hwang et al., 1998; Kim et al., 1998). The importance of Mad2 is underlined by the observation that its gene is essential in mammals, and that the disruption of one Mad2 allele causes enhanced rates of chromosome loss, impaired checkpoint function, and may promote tumorigenesis (Dobles et al., 2000; Michel et al., 2001).
Mad2 also interacts with Mad1, a 718-residue coiled-coil protein (Hardwick and Murray, 1995; Chen et al., 1998,1999; Jin et al., 1998). The Cdc20 and Mad1 complexes with Mad2 are exclusive, and occur in the absence of other proteins in vitro (Sironi et al., 2001; Zhang and Lees, 2001; Luo et al., 2002). Precisely how these complexes relate to each other is unclear, but a functional link is indicated by the inability of Mad2 and Cdc20 to interact in budding yeast strains lacking Mad1 (Hwang et al., 1998; Fraschini et al., 2001), and by the observation that Mad1 depletion severely affects the spindle checkpoint in mammalian cells (Luo et al., 2002). Thus, although Mad2 can interact effectively with Cdc20 in the absence of other proteins in vitro, binding of Mad2 to Mad1 makes this interaction possible in vivo. Dynamic studies indicated that Mad2 cycles at the kinetochore with a rapid turnover (Howell et al., 2000, 2001). Conceivably, Cdc20 may associate with Mad2 at unattached kinetochores and depart to prevent loss of cohesion on already bi-oriented sister chromatids. The Mad2 binding sites on Mad1 and Cdc20 have been mapped using genetic and biochemical approaches (Hwang et al., 1998; Jin et al., 1998; Kim et al., 1998; Luo et al., 2000; Zhang and Lees, 2001), and recently it was shown that these otherwise unrelated proteins share an ∼10-residue Mad2 binding motif (Luo et al., 2002). This suggests that Mad2 is transferred from one ligand to the other via competition. Binding of Mad2 to peptides encompassing the Mad1 and Cdc20 binding sites promotes a similar dramatic conformational change in the Mad2 C-terminal tail (Luo et al., 2000, 2002), raising the possibility that Mad1 may prepare the Mad2 binding site for Cdc20 entry, increasing the on-rate for this interaction (Luo et al., 2002).
We determined the crystal structure of the tetrameric Mad1–Mad2 core complex and analysed its structural and biochemical properties to find out how this complex may favour the Mad2–Cdc20 interaction. We find that Mad1 is a competitive inhibitor of Cdc20, and that the Mad1–Mad2 complex is engineered to slow down the rate of formation of Mad2–Cdc20. The Mad2 C-terminal tail locks in the Mad1 ligand, and release of Mad2 likely requires its unfolding. We show consistently that previous binding of Mad2 to Mad1 decreases the rate of the Mad2–Cdc20 interaction in vitro relative to unbound (apo) Mad2. The Mad1 binding sites are buried at the interface of the Mad1–Mad2 tetramer, and an unperturbed Mad1–Mad2 tetramer will not release Mad2. A 1:1 complex, however, will allow a slow exchange of Mad2 from Mad1 for Cdc20 binding. These results suggest that an appropriate conformational change in the Mad1–Mad2 complex may allow regulated release of Mad2 for Cdc20 binding, which our thermodynamic studies indicate as a favourable interaction.
Results
Structure determination
A soluble complex between the C-terminal region of human Mad1 (residues 485–718, Mad1485–718) and histidine-tagged (His) human Mad2wt was purified from bacteria (Sironi et al., 2001). The complex eluted in a single peak from a size-exclusion chromatography (SEC) column, but further analyses established that it contains a super-stoichiometric amount of Mad2wt, probably due to Mad2 aggregation (Sironi et al., 2001). Consistently, Mad1485–718–Mad2wt was polydisperse (not shown) and did not crystallize. Mad2R133A, a point mutant of Mad2 with a substitution of Arg133 by alanine, effectively binds Mad1 and Cdc20 (Sironi et al., 2001). Its interaction with Mad1 is tight and results in a monodisperse complex (not shown) devoid of the excess Mad2 observed in the complex containing Mad2wt (Sironi et al., 2001). We refer to the resulting complex as the Mad1–Mad2 core complex. Although we were unable to crystallize Mad1485–718–Mad2R133A (large Mad1–Mad2 core complex), a complex containing a shorter fragment of Mad1 (Mad1485–584–Mad2R133A: short Mad1–Mad2 core complex) yielded a crystallizable sample. The structure was determined at 2.05 Å resolution using SIRAS methods on a selenomethionyl derivative (Table I) as described in Materials and methods.
Table I. Structure determination.
| Data statistics | High resolution native | Low resolution native | Peak 1 | Peak 2 |
|---|---|---|---|---|
| Crystal form | I | II | II | II |
| Facility and beamline | ESRF ID14 | Elettra XRD1 | ESRF ID29 | Elettra XRD1 |
| Unit cell dimensions (Å) | a = 111.0b = 63.0c = 139.5 | a = 111.6b = 63.0c = 261.5 | a = 111.7b = 63.2c = 262.6 | a = 111.4b = 62.9c = 262.2 |
| Angles (°) | β = 111.6 | β = 90.3 | β = 90.6 | β = 90.4 |
| Wavelength (Å) | 0.9340 | 1.0000 | 0.9793 | 0.9809 |
| Resolution (Å) | 25–2.05 | 25–3.2 | 25–3.3 | 25–3.5 |
| Observations/unique reflections | 368 557/113 010 | 228 813/58 325 | 169 263/54 062 | 154 312/48 031 |
| Completeness (last shell) (%) | 99.2 (90.0) | 95.3 (79.8) | 82.6 (62.2) | 95.2 (84.1) |
| Rsyma (last shell) (%) | 3.8 (34.1) | 6.3 (37.5) | 5.5 (8.3) | 4.5 (7.6) |
| I/σI (last shell) | 18.2 (2.9) | 16.9 (2.9) | 13.4 (8.0) | 14.7 (8.3) |
| Number of Mad1–Mad2 in asymmetric unit |
4–4 |
8–8 |
8–8 |
8–8 |
| Refinement statistics | ||||
| Protein atoms | 9237 | |||
| Water molecules | 711 | |||
| Rcrystb (% of data) | 24.0 (95) | |||
| Rfreec (%) (% of data) | 26.8 (5) | |||
| R.m.s. bonds (Å) | 0.015 | |||
| R.m.s. angles (°) | 1.88 | |||
Overall view of the Mad1–Mad2core complex
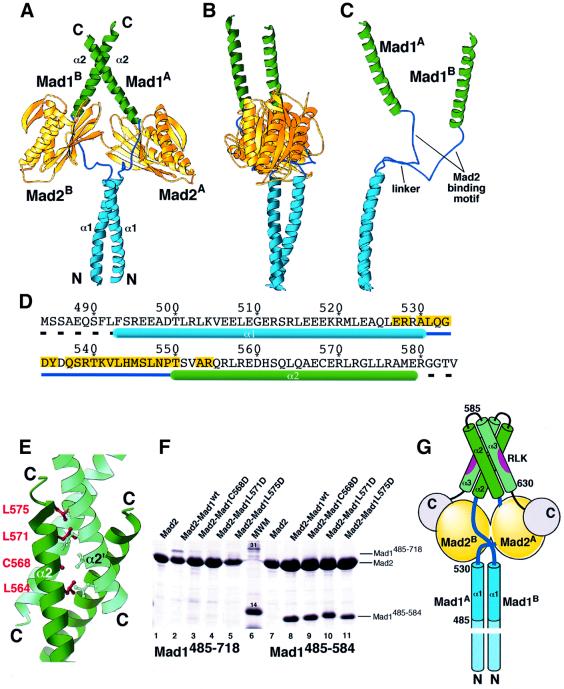
Mad1–Mad2core is a tetramer containing two copies of each subunit (Mad2A and Mad2B, yellow in Figure 1; and Mad1A and Mad1B). The Mad1 molecules are elongated, and are tightly held together by a parallel coiled-coil involving the N-terminal α1 helices (cyan). The Mad1 chains part in opposite directions at the end of the coiled coil, and point towards their Mad2 ligands, from which they emerge ∼20 residues later with another helical segment (α2, in green). The segment between the α1 and α2 helices of Mad1 (dark blue) contains the critical residues for Mad2 binding. The first part of this segment (the ‘linker’; residues 531–539) is flexible and adopts different conformations in Mad1A and Mad1B, as revealed by superposition of the α1 helices (Figure 1C). As a result, the Mad1–Mad2 core complex is significantly asymmetric. The second part (residues 540–549) contains the Mad2 binding motif, which supports essentially identical interactions defining Mad1A–Mad2A and Mad1B–Mad2B sub-complexes. The C-terminal tails (residues 161–205) in both Mad2 monomers undergo a conformational change similar to that observed in the solution structure of Mad2 with the Mad2 binding peptide 1 (MBP1) (Luo et al., 2002).
Fig. 1. Structure of the Mad1–Mad2 complex. (A and B) Ribbon diagrams of the Mad1–Mad2 tetramer viewed from orthogonal directions. Mad2A and Mad2B (yellow) are two Mad2 monomers in the complex. The Mad1 chains (Mad1A and Mad1B) interact via the N-terminal coiled-coil (cyan). The dark blue segments contain the Mad2 binding sites. A helical conformation is resumed in the C-terminal region (green). The 2-fold symmetry of the coiled coil breaks at the linker, and the complex is asymmetric. (C) Superposition of the N-terminal regions of Mad1A and Mad1B shows their structural differences. The diagrams were generated with Ribbons (Carson, 1991). (D) Sequence of Mad1. Numbering refers to full-length human (Hs) Mad1. The N-terminal methionine was added for translation. Assignment of secondary structure refers to molecule Mad1A in the first tetramer. Yellow boxes label Mad1A residues within 4.5 Å of any Mad2A atom. (E) The anti-parallel coiled-coil interaction between the α2 helices of Mad1 (α2′ is the equivalent helix in a different tetramer) in the crystals with ball-and-stick model (dark red) of residues at the helical interface. (F) Immobilized Metal Affinity Chromatography (IMAC) pull-down assays on bacterial lysates expressing His-Mad2 (lanes 1 and 7), His-Mad2–Mad1485–718 and indicated mutants (lanes 2–5), or His-Mad2–Mad1485–584 and indicated mutants (lanes 8–11). (G) Scheme showing the predicted overall structure of the Mad1–Mad2 complex. Hs Mad1 positions cited in the text are shown. RLK is the sequence motif contributing to Bub1–Bub3 binding. The putative α3 helix and the C-terminal domain are shown in light green and grey, respectively.
The Paircoil server (Berger et al., 1995) suggests an uninterrupted coiled coil for the N-terminal segment of Hs Mad1 not included in our structure (residues 1–484). There are essential segments in this region, which may be required to localize Mad1 at kinetochores (Chen et al., 1999). The prediction also suggests that after a short interruption around residue 584 (which was chosen to end our construct), a segment with a high propensity for coiled-coil formation will resume until residue 630. The geometry of the α2 helices of Mad1A and Mad1B as they emerge from their cognate Mad2 ligands is incompatible with the formation of a parallel coiled coil. In the crystals, these helices pack with an anti-parallel orientation with equivalent segments (α2′) in a second tetramer (Figure 1E). Although this interaction is weak (because Mad1485–584–Mad2 remains tetrameric in solution), it suggests that the α2 helices may form an intra-molecular anti-parallel coiled coil with a speculative α3 helix lying between residues 590 and 630, with a connecting loop tentatively fitted to residues 579–589.
To test this hypothesis, we introduced hydrophilic residues in place of the hydrophobic residues involved in the stabilization of the α2–α2′ interface observed in the crystals (Figure 1F). While the wild-type (wt) sequence of Mad1485–718 was pulled down by His-Mad2 from the bacterial lysates after co-expression in bacteria (lane 2), the resulting Mad1485–718 mutants were unable to interact with Mad2 (lanes 3–5). The same mutations, however, were completely silent when introduced into Mad1485–584 (lanes 9–11). This striking difference could be explained if the mutated residues were exposed to solvent in Mad1485–584 and buried in Mad1485–718, because in this case hydrophobic to hydrophilic substitutions would be expected to cause structural destabilization in the latter but not in the former. These results, together with the structural prediction indicating the possible existence of an α3 segment, suggest that the α2 helices may indeed participate in an intra-molecular α2–α3 anti-parallel interaction similar to that observed in the crystals. A conserved RLK (Arg-Leu-Lys) motif in the putative α3 helix mediates binding of Bub1–Bub3 to Mad1–Mad2 in budding yeast (the corresponding motif in Hs Mad1 is located at residues 617–619), and mutational abrogation of this interaction prevents checkpoint activation without impairing Mad2 binding (Brady and Hardwick, 2000). The cartoon in Figure 1G shows that Mad1 may consist of a long parallel coiled coil leading into two head domains, each containing a Mad2 binding site followed by an anti-parallel coiled coil, and by a C-terminal ∼90-residue region (grey) previously identified as a target of mutations abolishing the checkpoint (Chen et al., 1999). Resistance to limited proteolysis (not shown) suggests that the C-terminal domain of Mad1 folds into a stable domain. We believe that the 2–2 tetrameric assembly revealed by our crystal structure is the functional Mad1–Mad2 species in vivo.
A ‘safety belt’ binding mechanism
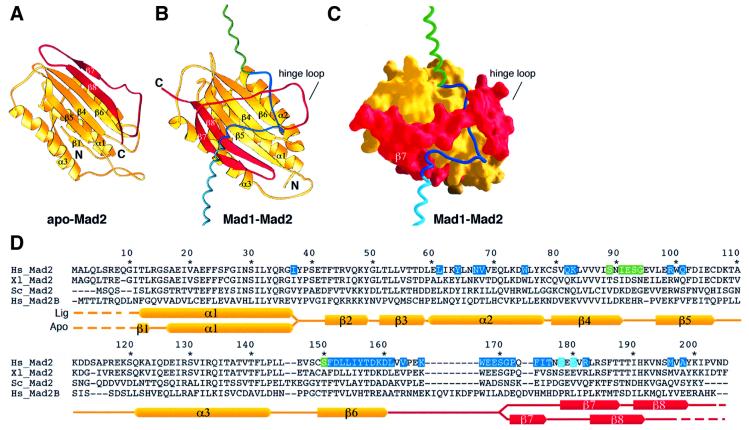
The structure of the Mad1–Mad2 complex confirms the recent finding that Mad1 and Cdc20 share a common Mad2 binding motif (Luo et al., 2002). The details of the interaction of Mad2 with the two Mad1 motifs in the core complex are essentially identical, and binding promotes a remarkable reorganization of the Mad2 C-terminal tail (residues 161–205; the red segment in Figure 2A) compared with the structure of apo-Mad2 reported by Wagner and collaborators (Luo et al., 2000). The first 160 residues of Mad2 are structurally invariant in apo- or Mad1-bound Mad2, but the C-terminal region (starting from the β6–β7 loop, the ‘hinge loop’ in Figure 2B and C) occupies the opposite edges of the large exposed β-sheet in these structures. In the complex, the Mad2 C-terminal tail crosses the entire surface of the β-sheet and locks Mad1 in such a way that it resembles a fastened safety belt. This is clarified by the surface representation in Figure 2C, showing how the Mad1 chain is ‘threaded’ through Mad2.
Fig. 2. Conformational rearrangement of the Mad2 C-terminal tail. (A) Ribbon diagram of apo-Mad2 as reported by Luo et al. (2000). The C-terminal region of Mad2 is displayed in red. (B) The conformation of the C-terminal tail is changed in the Mad1–Mad2 complex. Mad1A–Mad2A sub-complex is shown. Mad1B and Mad2B have been omitted to improve clarity but undergo the same rearrangement. This starts from the ‘hinge loop’. A short N-terminal segment is also rearranged in the two structures. (C) Molecular surface of Mad2 using the same colour coding used in (A and B), and a worm representation for Mad1 (Nicholls et al., 1991). Complex disruption must involve the opening of the Mad2 C-terminal tail. (D) Most contacts of Mad2 with Mad1 (defined and colour-coded as in Figure 1) map to the C-terminal segment of Mad2 (contacts of Mad2A are shown). Hs: Homo sapiens; Xl: Xenopus laevis; Sc: Saccharomyces cerevisiae. Hs Mad2B/Rev7 is also shown. The secondary structure of apo-Mad2 (apo) and Mad1-bound Mad2 (lig) are depicted. Numbering refers to Hs Mad2. Dashed lines indicate regions that are invisible in the electron density map. The N-terminal polyhistidine tag was omitted, as it is invisible in the electron density map.
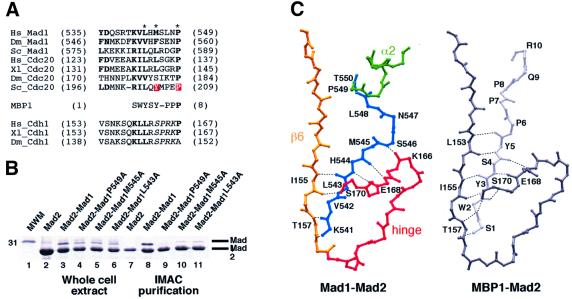
A structural alignment of the Mad2-binding motif with the MBP1 peptide, whose solution structure in complex with Mad2 was recently reported (Luo et al., 2002), is shown in Figure 3A. We note a less conserved version of the motif in the APC activator CDH1, a target of Mad2B/Rev7 (Chen and Fang, 2001; Pfleger et al., 2001). Point mutations in the Mad2 binding motif abolish the Mad2–Cdc20 interaction and the spindle checkpoint in budding yeast (Hwang et al., 1998; Kim et al., 1998). To test whether analogous mutations in Mad1 affected Mad2 binding, residues Lys541, Leu543, Met545 or Pro549 were mutated into alanine, and the amount of wild-type and mutant Mad1485–718 co-purifying with immobilized His-Mad2 was evaluated. Mutation of Lys541 (not shown) and Pro549 totally abolished binding, while the other two mutants displayed a partial but significant reduction in binding (Figure 3B).
Fig. 3. Interaction of Mad2 with Mad2 binding motifs. (A) Mad2 binding motifs in Mad1, Cdc20 and MBP1. Dm: Drosophila melanogaster, other abbreviations as in Figure 2. The motif is also present in CDH1, and contains a potential CDK or MAPK target site (italics). Red boxes indicate mutation sites affecting Cdc20–Mad2 and the asterisks Mad1 mutations analysed in our study. (B) His-Mad2–Mad1485–718 and indicated mutants were co-expressed in bacteria, IMAC-purified, and analysed by SDS–PAGE. Lane 1, molecular weight marker; lanes 2–6, total bacterial lysates of indicated expression tests, showing overall protein production; lanes 7–11, IMAC-purified samples. (C) Comparison of Mad1 and MBP1 binding to Mad2. Mad1–Mad2 is shown with the colour scheme of Figure 2B. Most structural elements were removed to improve readability, and only main chain atoms are shown. Equivalent region of MBP1 (light grey) and Mad2 (dark grey) are also shown. Hydrogen bonds are shown as dashed lines. Ligand and Mad2 residues involved in main chain hydrogen bonding are labelled.
A comparison of the Mad2 complexes with the MBP1 peptide (Luo et al., 2002) and with Mad1 identifies substantial similarities, but also important differences (Figure 3C). Besides forming a number of van der Waals and polar contacts with Mad2, both ligands bind by adding an (anti-parallel) β-strand at the end of the exposed β-sheet of Mad1, a binding mechanism defined as β-augmentation (Harrison, 1996). The MBP1 peptide, however, adopts a more extended and relaxed conformation relative to Mad1. The structural alignment shows that Ser1 (S1) and Pro8 (P8) of MBP1 occupy essentially identical positions to those of Lys541 (K541) and Pro549 (P549) of Mad1. Thus, nine Mad1 residues must be accommodated within the Mad2 binding site, rather than eight in MBP1. The Mad1 chain bulges out around residues 545–548, pushing against the Mad2 hinge loop (Figure 3C). As a result, the anti-parallel pairing of the Mad1 chain against the β6 strand of Mad2 and against a segment of the Mad2 hinge loop is imperfect, and restricted to a smaller number of inter-chain hydrogen bonds relative to the Mad2–MBP1 complex (Figure 3C). This might explain at least in part why Mad1 and Cdc20 synthetic peptides, which have a similar sequence and identical spacing between the lysine and proline residues (K541 and P549 in human Mad1), are significantly weaker Mad2 ligands than the MBP1 peptide (Luo et al., 2002). The fact that the phage display strategy used by Luo et al. (2002) identified sequences (such as MBP1’s) that are distinct from those in Mad1 and Cdc20 may suggest that the interaction of Mad2 with the Mad1 and Cdc20 Mad2 binding motifs is designed to be sub-optimal.
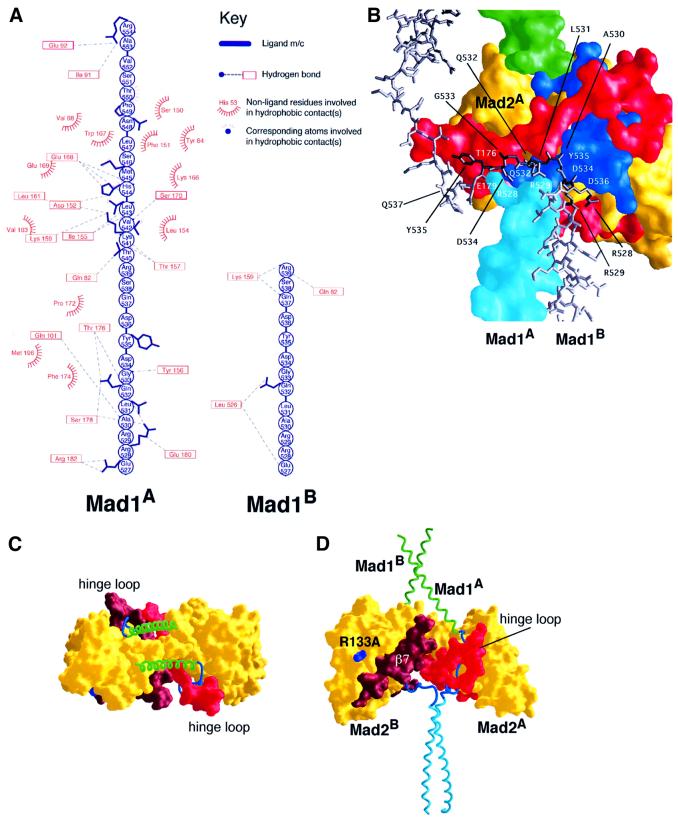
It should be noted, however, that the Mad1–Mad2 interaction is stabilized by a number of additional contacts in the tetramer, several of which involve the Mad1 linker region upstream of the conserved portion of the Mad2 binding motif. Because the linker regions are asymmetric, these contacts are different in the two sub-complexes (Figure 4A and B). Mad1A residues 527–539 are involved in a number of additional interactions with Mad2A (Figure 4A, left), which are not observed in the Mad1B–Mad2B sub-complex (Figure 4A, right). The Mad1B linker (residues 530–538), however, compensates for the fewer contacts by packing tightly against the Mad1A–Mad2A interface as shown in Figure 4B. Despite the asymmetric nature of the complex, whose origin remains unclear but which seems to rely on the flexibility of the linker, both ligand binding sites of Mad2A and Mad2B are deeply buried at the interface between the Mad1–Mad2 sub-complexes in the tetramer (Figure 4C and D). As shown below, this organization significantly increases the stability of the Mad1–Mad2 complex. We predict that the release of Mad2 from the tetrameric complex implies opening of the structure to expose the Mad2 C-terminal tails. This may occur along a preferential ‘fracture’ line determined by the asymmetry of the complex. Thus, although we do not demonstrate here that the asymmetry of the core complex is functionally important, we propend in favour of this hypothesis.
Fig. 4. Organization of the Mad1–Mad2 tetramer. (A) Contacts in Mad1A–Mad2A (left), and in Mad1B–Mad2B (right). Mad1B residues 540–554 form essentially identical contacts with Mad2B, and were omitted. The linker region of Mad1B forms very few contacts with Mad2B when compared with Mad1A–Mad2A. Figure created with LIGPLOT (Wallace et al., 1995). (B) The linker region (dark grey) of Mad1B (light grey) contacts a composite surface consisting of residues from Mad1A and Mad2A (colouring scheme as in Figures 1 and 2). To preserve clarity, only the Mad1A and Mad2A residues forming H-bonds with Mad1B are shown, but a number of van der Waals contacts are also present. The view is similar to that adopted in Figure 2, and Mad2B was omitted for clarity. (C and D) The molecular surface of Mad2 in the tetramer was coloured as in Figure 2 (the C-terminal region of Mad2B was coloured with a darker red to distinguish it from the equivalent region of Mad2A). The view in (D) is similar to the one in Figure 1A, while in (C) the molecule is observed from the top. The C-terminal regions of Mad2A and Mad2B (the hinge loop is indicated) are at the interface between Mad1–Mad2 sub-complexes.
Insights into the Mad2 exchange reaction
The finding that Mad1 and Cdc20 share a conserved Mad2 binding motif argues that the Mad1-dependent reaction that takes Mad2 on to Cdc20 occurs via a direct competitive mechanism. The structure suggests that Mad1 cannot be released without prior unlocking of the C-terminal tail. Mad2 residues Val181, Leu183, Phe186, Ile190, Val193, Val197 and Tyr199 pack against the Mad2 hydrophobic core stabilizing the closed conformation. Two of these residues (Val197 and Tyr199), and another two (Met196 and Ala198) that are directly involved in the interaction with Mad1, fall within a C-terminal 10-residue segment whose deletion (Mad2ΔC) abolishes Mad1 and Cdc20 binding to Mad2 (Luo et al., 2000; Sironi et al., 2001). In apo-Mad2, these residues are disordered in solution, and are not required for structural stabilization (Figure 2D; Luo et al., 2000). This likely explains why Mad2ΔC is structurally stable but impaired in ligand recognition (Luo et al., 2000; Sironi et al., 2001). As expected from the presence of a similar Mad2 binding site, the C-terminal tail of Mad2 may adopt a similar conformation in the Mad1 and Cdc20 complexes (Luo et al., 2002). Unless tail opening occurred on a fast time scale, the transition from a Mad1–Mad2 to a Cdc20–Mad2 complex would be limited by the dynamics of tail opening, independently of the relative affinity of Mad2 for Cdc20 and Mad1. Because the two Mad1–Mad2 binding sites are buried, they may be inaccessible to Cdc20 within the tetrameric complex (Figure 4). These considerations raise the question of whether the postulated exchange reaction taking Mad2 from Mad1 to Cdc20 occurs spontaneously. To answer this, we designed a set of experiments to dissect the contribution to the binding mechanism of (i) the relative preference of Mad2 for the Mad1 and Cdc20 binding motifs, (ii) Mad1 locking by the Mad2 C-terminal tail, and (iii) stabilization of the Mad1–Mad2 interaction as it might occur in the tetramer.
First, we determined thermodynamic parameters for the interaction of Mad2 with synthetic peptides corresponding to the isolated Mad2 binding motifs of Mad1 and Cdc20 using isothermal titration calorimetry (ITC; Table II). The Mad1 sequence used in these studies is unlikely to support self-oligomerization, as it lacks the coiled-coil region. It has also been shown that the interaction of Mad2 with Cdc20 and Mad1 peptides gives rise to simple 1:1 complexes (Sironi et al., 2001; Luo et al., 2002). Thus, our thermodynamic studies likely measure 1:1 interactions between Mad2 and its ligands. We refer to these interactions as ‘monovalent’ to distinguish them from those occurring in the Mad1–Mad2 tetramer, which are significantly tighter (see above and below). As shown in Table II, Mad2 binds the Cdc20 motif with 10-fold higher affinity (KD = 100 nM) than the Mad1 motif (KD = 1.0 µM). We also assessed whether the R133A substitution in Mad2 had any effect on ligand binding. Table II shows that Mad2wt and the monomeric Mad2R133A bind their Mad1 and Cdc20 ligands with essentially identical affinity, confirming the non-invasive nature of the R133A mutation. Although a structural explanation of why Mad2 binds the Cdc20 peptide more tightly than Mad1 is not available, the calorimetric determinations argue that Mad2 exchange to Cdc20 from a monovalent Mad1–Mad2 complex is a favoured reaction, provided no further stabilization of Mad1–Mad2 was present.
Table II. Thermodynamic parameters of Mad2 interactions with Mad1 and Cdc20.
| Interaction | KB (106 M–1) | KD (10–6 M) | ΔHobs (kcal/mol) | TΔS (kcal/mol) | ΔGobs (kcal/mol) | N |
|---|---|---|---|---|---|---|
| Mad1 + Mad2wt | 0.96 | 1.04 | –13.1 | –4.9 | –8.2 | 0.95 |
| Mad1 + Mad2R133A | 2.0 | 0.5 | –14.4 | –5.8 | –8.6 | 1.15 |
| Cdc20 + Mad2wt | 9.4 | 0.1 | –13.9 | –4.4 | –9.5 | 0.90 |
| Cdc20 + Mad2R133A | 9.2 | 0.1 | –16.0 | –6.6 | –9.4 | 0.96 |
Mad1 and Cdc20 are synthetic peptides corresponding to human Mad1523–550 and Cdc20111–138. KB, binding constant; KD, dissociation constant; ΔHobs, observed binding enthalpy; ΔS, entropy; ΔGobs, observed Gibbs’ free energy; N, binding stoichiometry.
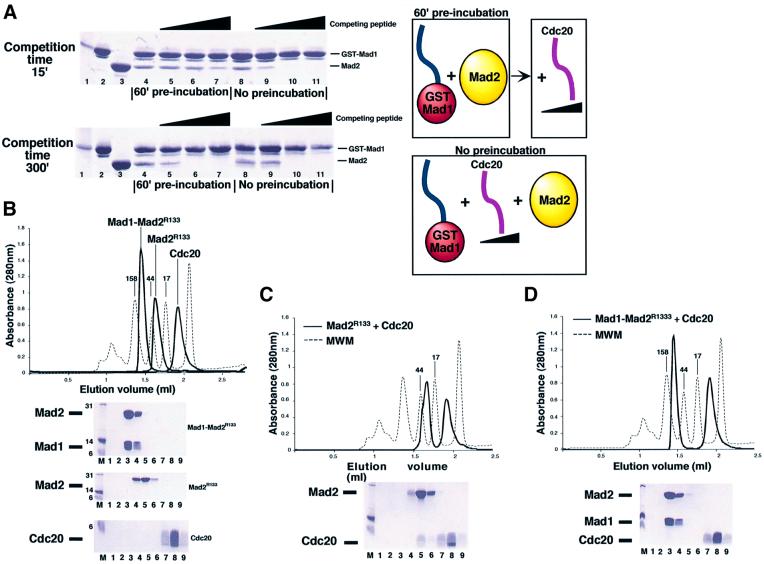
Preliminary unfolding of the Mad2 C-terminus may be required for ligand release, and the closed conformation is expected to delay the disruption of the Mad2–ligand complex. To test this, we investigated whether the ability of a Cdc20 synthetic peptide to compete with a monovalent Mad1–Mad2 complex was sensitive to the initial state, locked or unlocked, of the C-terminal tail. We considered apo-Mad2 (Figure 2A) as representative of the unlocked state, while the locked state was represented by a Mad2 sample already bound to Mad1. A graphical representation of the experiment is shown on the right-hand side of Figure 5A. GST–Mad1523–550 immobilized on a solid phase was mixed directly with recombinant apo-Mad2R133A (Mad2 unlocked) in the presence of different concentrations of Cdc20 peptide, or pre-incubated for 60 min with apo-Mad2R133A to allow complex formation (Mad2 locked), and only then incubated with Cdc20. The competition reaction was protracted for either 15 or 300 min for both samples, at the end of which the bound proteins were isolated by centrifugation, washed and examined by SDS–PAGE (Figure 5, left panels). In the absence of pre-incubation, the patterns were identical after 15 or 300 min (compare lanes 8–11 in the two gels), suggesting that the competition reactions reached equilibrium within the 15 min time frame. Pre-incubation of GST–Mad1523–550 with Mad2, however, significantly decreased the ability of the Cdc20 peptide to compete for Mad1-bound Mad2 within the 15 min time frame, and even after an incubation of 60 min we detected a significant difference in the pre- and non-pre-incubated samples (data not shown). These differences were completely eliminated after 300 min incubations (left bottom panel), indicating that this time is sufficient to equilibrate the competition reaction. Similar levels of Mad2 binding were observed in the pre-incubated and non-pre-incubated samples in the absence of competing peptide (compare lanes 4 and 8) even after 15 min, showing that this time is sufficient for Mad2 to bind Mad1. Release of pre-incubated Mad2 from Mad1 at 15 min was also dependent on the concentration of competing peptide, suggesting that besides tail opening, the release of the bound ligand is also limiting for the exchange reaction. Thus, the off-rate of the Mad1–Mad2 interaction must be significantly slower than the on-rate of the Cdc20–Mad2 interaction, even if this is a rather slow reaction per se (Sironi et al., 2001; Luo et al., 2002). The result also shows that Cdc20 and Mad1 are respective competitive inhibitors of their complexes with Mad2, and suggests that the conformational change of the C-terminal tail is designed to slow down the release of a bound ligand.
Fig. 5. Analysis of the postulated exchange mechanism. (A) Increasing concentrations of a Cdc20 synthetic peptide were incubated with Mad2, which had or had not been previously incubated with solid phase-bound GST–Mad1523–550. In the pre-incubation experiment the complex is allowed to form for 60 min, and the competing peptide is added afterwards. If no pre-incubation is allowed, the three components are mixed at the same time and incubated for 15 or 300 min. A diagrammatic representation is shown on the right-hand side. After incubation, beads and bound proteins are collected by centrifugation, washed and analysed by SDS–PAGE. Molecular weight marker, input GST–Mad1523–550 and Mad2 are shown on lanes 1–3. (B) Superimposed SEC profiles of isolated Mad1–Mad2, Mad2R133A and Cdc20111–150 synthetic peptide. The proteins/peptides run as single peaks by SEC. SDS–PAGE separation of collected fractions are also shown. Nine 100 µl fractions (1–9) were collected from 1.2 to 2.1 ml. (C) When Mad2R133A is incubated with a 5-fold excess of Cdc20111–150 and separated by SEC, a Mad2–Cdc20 complex forms [compare lanes 4–5 in (B) and (C)]. (D) When the Mad1–Mad2 complex is incubated with Cdc20111–150, no binding is observed. Furthermore, the peptide is unable to disrupt the complex, as no Mad2–Cdc20 complex or free Mad1 is detected.
The competition experiments were carried out with a monovalent Mad1 fragment. To establish whether the Mad1–Mad2 interaction was further stabilized in the tetramer, we asked whether the Cdc20 peptide was able to release Mad2 from Mad1–Mad2core. We incubated short Mad1–Mad2core (at a final concentration of ∼200 µM) with a 1 mM concentration of a synthetic peptide containing the Mad2 binding site of Cdc20, and after variable incubation times finally resolved the resulting species by size-exclusion chromatography (SEC). In control experiments, the short Mad1–Mad2core, Mad2R133A and the Cdc20 peptide eluted as single peaks from a SEC column, as shown by their superimposed elution profiles in Figure 5B. Recovery of Cdc20 peptide in the Mad2 peak (Figure 5C, lanes 4–6) indicates that the Mad2–Cdc20 complex can be easily detected using this approach. Even after 16 h incubations, the Cdc20 peptide did not associate with Mad1–Mad2, nor did it disrupt this complex, as it might have been revealed by the appearance of Mad2–Cdc20 or free Mad1 (Figure 5D). Similar experiments with the long Mad1–Mad2core gave identical results, with no association or disruption of the complex upon incubation with Cdc20 (Sironi et al., 2001). Lack of exchange is unlikely to result from the use of a truncated form of Cdc20, because Cdc20 fragments containing the Mad2 binding site are tighter Mad2 ligands than full-length Cdc20 (Tang et al., 2001; Zhang and Lees, 2001). This shows that the short Mad1–Mad2 core complex is very tight and indistinguishable from its larger counterpart, and confirms the structure-based prediction that tetrameric Mad1–Mad2 is significantly more stable than a monovalent complex.
Discussion
Mad1 interacts with Mad2 by β-augmentation, a frequent binding mechanism in protein–protein complexes. The very atypical feature in Mad1–Mad2 is the refolding of the Mad2 C-terminal tail over the Mad1 ligand in a way that is reminiscent of a fastened safety belt. We are unaware of a similar binding mechanism in other proteins. The Mad2–Cdc20 complex is expected to have an organization similar to that of Mad1–Mad2, with an elongated Cdc20 segment (residues 123–137) ‘threaded’ through the Mad2 C-terminal tail.
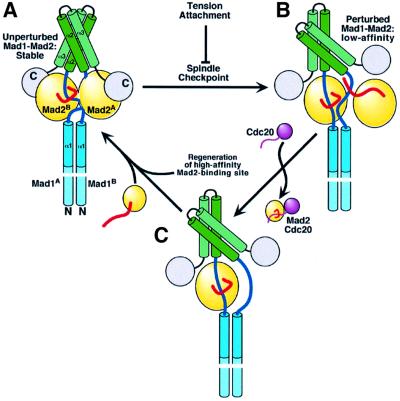
Important conclusions from this study are (i) that monovalent Mad1 is a weak competitive inhibitor of the Mad2–Cdc20 complex, (ii) that the conformation of the Mad2 C-terminal tail introduces a kinetic barrier towards the release of Mad2 from its ligand complexes, as a consequence of its required unfolding, and (iii) that the tetrameric assembly of the Mad1–Mad2 complex reinforces the interaction, preventing the release of Mad2. As expected for a competitive inhibitor, Mad1 opposes Mad2 binding to Cdc20 (Figure 5). Mad1 is essential to establish a Mad2–Cdc20 complex in vivo (Hwang et al., 1998; Fraschini et al., 2001), and the discovery that it acts as a competitive inhibitor of Mad2–Cdc20 is counterintuitive. Understanding why Cdc20 and Mad2 are unable to interact in vivo in the absence of Mad1, despite their proven record as direct ligands in vitro, is now a very urgent question. Mad2 binds poorly to full-length Cdc20, suggesting that the binding site is shielded in the full-length protein (Tang et al., 2001; Zhang and Lees, 2001). The activated Mad1–Mad2 complex may help Cdc20 to expose its Mad2 binding site. Furthermore, Cdc20 is associated with cellular chaperones, which may limit its ability to interact with Mad2 in vivo (Ho et al., 2002; Pines, 2002). Topological considerations may also be important to limit the Mad2–Cdc20 interaction. Mad1 recruits Mad2 to kinetochores, where the Mad2–Cdc20 interaction is expected to take place (Chen et al., 1998; Luo et al., 2002). It is unclear, however, why Cdc20, which is localized to kinetochores in a Mad1-independent manner (Shah and Cleveland, 2000; Sironi et al., 2001; Luo et al., 2002), is ineffective in directly recruiting Mad2 to these structures. The concentrations of Mad2 and Cdc20 may be insufficient to drive complex formation, while Mad1 may be able to recruit Mad2 to kinetochores through a high-affinity binding site. Subsequently, an external input of energy may destabilize the Mad1–Mad2 complex, allowing the release of Mad2 at a sufficiently high local concentration to support the interaction with Cdc20 (Figure 6). We suggest that the role of Mad1 as a matchmaker for the Mad2–Cdc20 complex in spite of its role as a competitive inhibitor of this interaction can be explained if funnelling through Mad1 is the only way in which Mad2 can effectively reach Cdc20 in vivo.
Fig. 6. Model for Mad1–Mad2 function in the spindle checkpoint. (A) Unperturbed Mad1–Mad2 will not exchange Mad2 with Cdc20. Our structure of Mad1–Mad2 may be related to this non-exchanging, high-affinity Mad1–Mad2 complex. (B) Perturbation by the spindle checkpoint decreases Mad2 affinity for Mad1. A monovalent Mad1–Mad2 interaction is low affinity, suggesting that decreased affinity may ensue if the Mad1–Mad2 heads become independent as an effect of reciprocal rotation. Cdc20 now competes with Mad1 for Mad2 binding. How Mad2 is re-loaded on to the complex is unclear, but it is reasonable to think that a new Mad2 binding site is generated to capture Mad2 with high affinity. (C) This high-affinity site would restore the unperturbed complex, and this cycle may continue until the attachment and/or tension dampen the checkpoint signal, preventing further release of Mad2 on to Cdc20.
Previous studies have indicated that an unperturbed, stable Mad1–Mad2 complex exists during the cell cycle (Shah and Cleveland, 2000). As suggested above, this complex may be destabilized at kinetochores by the spindle checkpoint to promote the transfer of Mad2 to Cdc20, and the subsequent recruitment of new Mad2 from a cytoplasmic pool (Figure 6). Microtubule attachment may restore an inactive form of Mad1–Mad2, preventing further dispensing of Mad2 into APC inhibitory complexes, eventually dampening checkpoint signalling. Because the affinity of an isolated monovalent Mad1– Mad2 interaction is significantly lower than those occurring in the tetramer, the conversion of Mad1– Mad2core to a dissociating form may be promoted by a conformational change disrupting the interaction of the globular heads observed in the tetrameric assembly, and resulting in the exposure of the Mad2 ligand binding sites for ligand exchange. After Mad2 release, the regeneration of a high-affinity binding site may recruit new Mad2 to restore the high-affinity unperturbed Mad1–Mad2 core complex. The cycle may continue until the checkpoint is active, sustained by metabolic energy. The high turnover rate of Mad2 at kinetochores during checkpoint activation may reflect the kinetics of this reaction (Howell et al., 2000,2001). Positive regulators are expected to play an essential role in the postulated exchange reaction. The suggestion that the Bub1–Bub3 complex binds Mad1– Mad2 near the Mad2 binding site lends further credit to this kinase, which phosphorylates Mad1 in vitro (Seeley et al., 1999), as a possible regulator of Mad1–Mad2. Other actors in the exchange reaction may be the BubR1–Bub3 complex, which binds Cdc20 and may enter a quaternary complex also containing Mad2 (Fraschini et al., 2001; Sudakin et al., 2001; Tang et al., 2001), and the Mps1 kinase, which phosphorylates Mad1 in budding yeast (Hardwick et al., 1996). Our work shows that the C-terminal lock must open before a ligand can be released from Mad2. This observation rules out the proposal that Mad1 favours binding of Cdc20 by Mad2 by stabilizing a pre-formed template with a reorganized C-terminal tail (Luo et al., 2002). The ‘safety belt’ binding mechanism may be useful to delay the disruption of the Mad2–Cdc20 complex once this is formed at activated kinetochores. The mechanism may allow the generation at the kinetochore of a kinetically stable Mad2–Cdc20 complex, despite the low binding affinity required to prevent significant direct binding in the absence of an activated checkpoint.
An in vitro approach aiming at the mechanism by which Mad2 is routed from a soluble pool to Mad1 and then to Cdc20 will have to cope with the fact that simple mixing of detectable concentrations of Mad2 and Cdc20 in vitro will result in direct binding. The hypothesis that Mad2 is funnelled to Cdc20 via Mad1, however, presents several testable implications in vivo. The rate of Mad2 release from the Mad1–Mad2 complex must increase upon checkpoint activation, while a high binding on-rate would be required to replenish Mad2 at the kinetochore upon checkpoint activation. It should be possible to identify mutations in Mad1, Mad2 or other spindle checkpoint proteins that do not affect the interaction of Mad2 with Mad1, but rather the release of Mad2 from this complex to bind Cdc20. A likely candidate for such an ‘exchange mutant’ in Mad1 has been described above (Brady and Hardwick, 2000). Furthermore, because Cdc20 acts as the ultimate receptor for Mad2, the dynamic behaviour of Mad2 at kinetochores should be affected by Cdc20 depletion. Measurements of Mad1, Mad2 and Cdc20 dynamics at different stages of the cell cycle and with appropriate mutational strategies may be used to test this hypothesis.
Materials and methods
Structure determination
Mad1–Mad2 was generated and purified essentially as described (Sironi et al., 2001). The protein was concentrated to 7.5 mg/ml and crystallized by the hanging drop technique against a reservoir containing 100 mM ammonium sulfate, 100 mM sodium citrate pH 5.2 and 10 mM dithiothreitol at 20°C. A selenomethionine (SeMet) derivative of the complex crystallized under similar conditions. X-ray diffraction data (Table I) were collected at cryogenic temperatures in cryobuffer containing 35–40% glycerol, and processed with Denzo-Scalepack (Otwinowski, 1993). Although two related monoclinic crystal forms were identified under native conditions (Table I), we were unable to identify SeMet derivatives of the strongly diffracting crystal form I, and the initial solution to the phase problem was found using crystal form II. A subset of the 56 Se atoms in the asymmetric unit (AU) was identified with the SnB program (Weeks and Miller, 1999) from anomalous differences in two independently collected peak datasets. Phase refinement with MLPHARE (Collaborative Computational Project, 1994) was carried out against anomalous differences of the peak datasets, and their isomorphous differences with a low-resolution native dataset in crystal form II. After solvent flipping in CNS (Brünger et al., 1998), the maps displayed a clear solvent boundary and clear protein features. The model of apo-Mad2 (Luo et al., 2000) was tentatively fitted in the electron density, and this allowed the identification of subsets of heavy atom clusters belonging to the same Mad1–Mad2 complex. This eventually led to the identification of the non-crystallographic symmetry (NCS) operators relating the eight Mad2 monomers in the AU. The resulting density-modified 3.3 Å maps were of excellent quality and allowed the unequivocal fitting of a Mad2 monomer. The model was used to find the position of the Mad2 monomers in crystal form I by molecular replacement with AMoRe (Collaborative Computational Project, 1994). Only three of the four Mad2 monomers could be found. The Mad1 molecules, and the fourth Mad2 monomer (which has a significantly higher average B-factor than the other Mad2 models) were identified using program ARP/wARP in Molrep mode (Perrakis et al., 2001). Model building was carried out with program O (Jones et al., 1991). Several rounds of refinement with CNS and manual rebuilding were required to generate a final model containing residues 10–203 of the four Mad2 monomers, residues 493–579 of Mad1A and Mad1B in the first tetramer, and residues 487–579 and 485–584 of Mad1A and Mad1B in the second tetramer (Table I). 90.6% and 9.4% of residues reside within the most favoured and additional allowed regions of the Ramachandran plot, respectively, with no residues in disallowed regions.
Determination of Mad2 binding parameters
Calorimetric measurements were carried with protein samples purified as described (Sironi et al., 2001) and synthetic peptides corresponding to human Cdc20111–138 and Mad1523–550 at 25°C using a VP-ITC titration calorimeter (MicroCal Inc.). Each titration experiment consisted of a first injection of 2 µl followed by twenty-nine injections of 8 µl. Binding isotherms were corrected for dilution heats by subtraction of blank titrations and evaluated using the Origin software package provided with the instrument.
In vitro binding and competition assays
For the experiments shown in Figures 1 and 3, bacterial lysates containing His-Mad2, or His-Mad2 co-expressed with Mad1 and its mutants were incubated with Ni-NTA–agarose (Qiagen). Bound proteins were collected by centrifugation, washed and analysed by SDS–PAGE. The protein levels observed after IMAC purification reflect the fact that His-Mad2 is expressed at higher levels than Mad1 from the polycistronic vector (Sironi et al., 2001). For the assays in Figure 5A, a synthetic peptide encompassing residues 111–138 of human Cdc20 was added at concentrations of 0, 6, 30 and 120 µM (lanes 8–11, respectively), to solid-phase-bound GST–Mad1523–550 (2 µM; lane 1) that had been previously incubated with 6 µM Mad2 (lane 2) for 1 h at 4°C in the absence of the Cdc20 peptide. The experiments with no pre-incubation were carried out using identical concentrations of the reagents as in the pre-incubation experiments, but the competing Cdc20 peptide (lanes 3–6; same concentrations as in lanes 8–11) was added to solid-phase-bound GST–Mad1523–550 before addition of Mad2. After mixing the three reagents, both reactions were protracted for the indicated time at room temperature (RT). The beads were centrifuged, washed, and bound proteins analysed by SDS–PAGE. For the experiments of Figure 5B–D, the short Mad1–Mad2 core complex, Mad2R133A (Sironi et al., 2001), and a Cdc20111–150 synthetic peptide were analysed on a Superdex200 column on a SMART system (Amersham-Pharmacia). To evaluate peptide binding to the short Mad1–Mad2 core complex or Mad2R133A, these species were mixed and incubated at RT for different times, and finally separated on the same column.
Accession numbers
Coordinates of Mad1–Mad2 are available from the Protein Databank (ID code 1GO4).
Acknowledgments
Acknowledgements
Thanks to Kevin Hardwick, Kristian Helin, Jan-Michael Peters, Simonetta Piatti and members of the Musacchio group for critical reading of the manuscript and discussions, and to Hongtao Yu for sharing coordinates of Mad2–MBP1. Also thanks to Andy Thompson and Ed Mitchell (European Synchroton Radiation Facility) and Massimo Degano and Alberto Cassetta (Sincrotone Trieste) for support in data collection. A.M. is an EMBO Young Investigator and a Scholar of the Italian Foundation for Cancer Research. This work was supported by a grant from the Italian Association for Cancer Research to A.M. We also wish to acknowledge the generous support of the Giovanni Armenise–Harvard Foundation for Advanced Scientific Research.
References
- Berger B., Wilson,D.B., Wolf,E., Tonchev,T., Milla,M. and Kim,P.S. (1995) Predicting coiled coils by use of pairwise residue correlations. Proc. Natl Acad. Sci. USA, 92, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady D.M. and Hardwick,K.G. (2000) Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol., 10, 675–678. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carson M. (1991) Ribbons 2.0. J. Appl. Crystallogr., 24, 958–961. [Google Scholar]
- Chen J. and Fang,G. (2001) MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev., 15, 1765–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Waters,J.C., Salmon,E.D. and Murray,A.W. (1996) Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science, 274, 242–246. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko,A., Mann,M. and Murray,A.W. (1998) Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol., 143, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Brady,D.M., Smith,D., Murray,A.W. and Hardwick,K.G. (1999) The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell, 10, 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Dobles M., Liberal,V., Scott,M.L., Benezra,R. and Sorger,P.K. (2000) Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell, 101, 635–645. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu,H. and Kirschner,M.W. (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell, 2, 163–171. [DOI] [PubMed] [Google Scholar]
- Fraschini R., Beretta,A., Sironi,L., Musacchio,A., Lucchini,G. and Piatti,S. (2001) Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J., 20, 6648–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G. (1998) The spindle checkpoint. Trends Genet., 14, 1–4. [DOI] [PubMed] [Google Scholar]
- Hardwick K.G. and Murray,A.W. (1995) Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol., 131, 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Weiss,E., Luca,F.C., Winey,M. and Murray,A.W. (1996) Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science, 273, 953–956. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. (1996) Peptide–surface association: the case of PDZ and PTB domains. Cell, 86, 341–343. [DOI] [PubMed] [Google Scholar]
- Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Howell B.J., Hoffman,D.B., Fang,G., Murray,A.W. and Salmon,E.D. (2000) Visualization of Mad2 dynamics at kinetochores, along spindle fibers and at spindle poles in living cells. J. Cell Biol., 150, 1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., McEwen,B.F., Canman,J.C., Hoffman,D.B., Farrar,E.M., Rieder,C.L. and Salmon,E.D. (2001) Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol., 155, 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang L.H., Lau,L.F., Smith,D.L., Mistrot,C.A., Hardwick,K.G., Hwang,E.S., Amon,A. and Murray,A.W. (1998) Budding yeast Cdc20: a target of the spindle checkpoint. Science, 279, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Jin D.Y., Spencer,F. and Jeang,K.T. (1998) Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell, 93, 81–91. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y. and Cowan,S.W. (1991) Improved methods for building protein models in electron density map and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kallio M., Weinstein,J., Daum,J.R., Burke,D.J. and Gorbsky,G.J. (1998) Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol., 141, 1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin,D.P., Matsumoto,S., Kitazono,A. and Matsumoto,T. (1998) Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science, 279, 1045–1047. [DOI] [PubMed] [Google Scholar]
- Li Y. and Benezra,R. (1996) Identification of a human mitotic checkpoint gene: hsMAD2. Science, 274, 246–248. [DOI] [PubMed] [Google Scholar]
- Li Y., Gorbea,C., Mahaffey,D., Rechsteiner,M. and Benezra,R. (1997) MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc. Natl Acad. Sci. USA, 94, 12431–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Fang,G., Coldiron,M., Lin,Y., Yu,H., Kirschner,M.W. and Wagner,G. (2000) Structure of the mad2 spindle assembly checkpoint protein and its interaction with cdc20. Nature Struct. Biol., 7, 224–229. [DOI] [PubMed] [Google Scholar]
- Luo X., Tang,Z., Rizo,J. and Yu,H. (2002) The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell, 9, 59–71. [DOI] [PubMed] [Google Scholar]
- Michel L.S. et al. (2001) MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature, 409, 355–359. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1993) Oscillation data reduction program. Science and Engineering Research Council, Daresbury Laboratory, Warrington, UK.
- Perrakis A., Harkiolaki,M., Wilson,K.S. and Lamzin,V.S. (2001) ARP/wARP and molecular replacement. Acta Crystallogr. D, 57, 1445–1450. [DOI] [PubMed] [Google Scholar]
- Pfleger C.M., Salic,A., Lee,E. and Kirschner,M.W. (2001) Inhibition of Cdh1–APC by the MAD2-related protein MAD2L2: a novel mechanism for regulating Cdh1. Genes Dev., 15, 1759–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. (2002) Cell cycle trials in Salamanca: Workshop on G2/M progression and associated checkpoints. EMBO rep., 3, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley T.W., Wang,L. and Zhen,J.Y. (1999) Phosphorylation of human MAD1 by the BUB1 kinase in vitro. Biochem. Biophys. Res. Commun., 257, 589–595. [DOI] [PubMed] [Google Scholar]
- Shah J.V. and Cleveland,D.W. (2000) Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell, 103, 997–1000. [DOI] [PubMed] [Google Scholar]
- Sironi L., Melixetian,M., Faretta,M., Prosperini,E., Helin,K. and Musacchio,A. (2001) Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J., 20, 6371–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Chan,G.K. and Yen,T.J. (2001) Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20 and MAD2. J. Cell Biol., 154, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj,R., Li,B. and Yu,H. (2001) Mad2-independent inhibition of APC–Cdc20 by the mitotic checkpoint protein Bub1R. Dev. Cell, 1, 227–237. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha,E. and McKeon,F. (1998) The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol., 142, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S. and McKeon,F. (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell, 89, 727–735. [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Wassmann K. and Benezra,R. (1998) Mad2 transiently associates with an APC/p55Cdc complex during mitosis. Proc. Natl Acad. Sci. USA, 95, 11193–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks C.M. and Miller,R. (1999) The design and implementation of SnB v2.0. J. Appl. Crystallogr., 32, 120–124. [Google Scholar]
- Zachariae W. and Nasmyth,K. (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev., 13, 2039–2058. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Lees,E. (2001) Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol. Cell. Biol., 21, 5190–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]