Abstract
Membrane-bound receptors such as tyrosine kinases and ionotropic receptors are associated with large protein networks structured by protein–protein interactions involving multidomain proteins. Although these networks have emerged as a general mechanism of cellular signalling, much less is known about the protein complexes associated with G-protein-coupled receptors (GPCRs). Using a proteomic approach based on peptide affinity chromatography followed by mass spectrometry and immunoblotting, we have identified 15 proteins that interact with the C- terminal tail of the 5-hydroxytryptamine 2C (5-HT2C) receptor, a GPCR. These proteins include several synaptic multidomain proteins containing one or several PDZ domains (PSD95 and the proteins of the tripartite complex Veli3–CASK–Mint1), proteins of the actin/spectrin cytoskeleton and signalling proteins. Coimmunoprecipitation experiments showed that 5-HT2C receptors interact with PSD95 and the Veli3–CASK–Mint1 complex in vivo. Electron microscopy also indicated a synaptic enrichment of Veli3 and 5-HT2C receptors and their colocalization in microvilli of choroidal cells. These results indicate that the 5-HT2C receptor is associated with protein networks that are important for its synaptic localization and its coupling to the signalling machinery.
Keywords: 5-HT2C receptor/PDZ domain/proteomic analysis/scaffold protein
Introduction
In the classical model of G-protein-coupled receptor (GPCR)-mediated signalling, the receptor is stabilized under an active conformation able to activate heterotrimeric G-proteins following ligand binding. This activation consists of a catalytic GDP/GTP exchange on the α subunit, triggering its dissociation from βγ subunits. Both α-GTP and βγ stimulate and/or inhibit a large variety of intracellular effectors (Hepler and Gilman, 1992). Over the last decade, several studies have shown that other intracellular proteins are physically associated with GPCRs (Hall et al., 1999). These proteins interact directly, or indirectly, via scaffold proteins, with intracellular domains of GPCRs including their C-terminal tail. These interactions are important for some GPCR functions such as clustering, compartmentalization, optimization of transduction and signalling in the absence of G-protein coupling (Scott and Zuker, 1998; Dev et al., 2001; Milligan and White, 2001). A new concept of ‘signaling at zero G’ mediated by functional multiprotein complexes associated with GPCRs has emerged from these findings (Brzostowski and Kimmel, 2001).
There is accumulating evidence that ionotropic receptors and ionic channels are also clustered in supramolecular complexes, maintained via networks of protein–protein interactions that delimitate specialized sub-membrane microdomains. Some of these complexes have been extensively characterized by proteomic analyses. Protein components of the N-methyl-d-aspartic acid (NMDA) receptor-associated complex, including scaffold proteins, channel subunits, signalling, adhesion and cytoskeletal proteins, have been identified by a combination of mass spectrometry and large-scale immunoblotting (Husi et al., 2000; Husi and Grant, 2001). The large number of protein interactions revealed by this study indicated a more complicated degree of organization of this complex than that predicted by other approaches. Similarly, a complex associated with the P2X7 ATP receptor has been isolated recently by immunoprecipitation, allowing the identification by MALDI-TOF mass spectrometry of a set of proteins interacting with the receptor (Kim et al., 2001). While several proteins are associated with both NMDA receptor and P2X7 receptor complexes, many of them are specific to one of these complexes. This suggests a specificity in the protein composition and organization of protein networks associated with channel-linked receptors. In contrast, identification of proteins associated with a particular GPCR has, so far, only been achieved in a piecemeal fashion, often based on yeast two-hybrid screens (Hall et al., 1998; Ullmer et al., 1998; Zitzer et al., 1999a,b).
5-hydroxytryptamine type 2C (5-HT2C) receptors are broadly expressed in the central nervous system (CNS) (Abramowski et al., 1995; Clemett et al., 2000) and modulate a large variety of behavioural and physiological processes, such as nociception, motor behaviour, thermoregulation and modulation of appetite (Lucki et al., 1989; Fone et al., 1998). Activation of 5-HT2C receptors exerts a phasic and tonic inhibition of the mesocorticolimbic dopamine function. This suggests that 5-HT2C receptor antagonists may be useful for the treatment of negative schizophrenia symptoms (Di Matteo et al., 2001). Although initial studies of 5-HT2C receptor signalling showed that they activate phospholipase Cβ (Chen et al., 1994), the molecular events initiated by these receptors remain largely unknown. 5-HT2C receptors contain a C-terminal sequence (SSV) corresponding to one of the class 1 PDZ (PSD95/disc large/ZO-1) recognition motifs (X-S/T-X-I/L/V). MUPP1, a multivalent PDZ domain adaptor protein, has recently been identified as a binding partner of 5-HT2C receptors by a two-hybrid screen (Ullmer et al., 1998; Bécamel et al., 2001). This suggests that 5-HT2C receptors participate in a protein network organized around a PDZ domain-based scaffold. The present study was performed to identify additional proteins that interact with the C-terminal tail of 5-HT2C receptors. For that purpose, we used a proteomic approach based on peptide affinity chromatography followed by mass spectrometry and/or immunoblotting. We report that 5-HT2C receptors are associated with at least 15 proteins, including synapse-enriched multidomain proteins containing one or several PDZ domains, such as the Veli3–CASK–Mint1 complex and PSD95. We also provide evidence that these interactions take place in vivo.
Results
Isolation of proteins interacting with 5-HT2C receptors via a PDZ domain-based scaffold
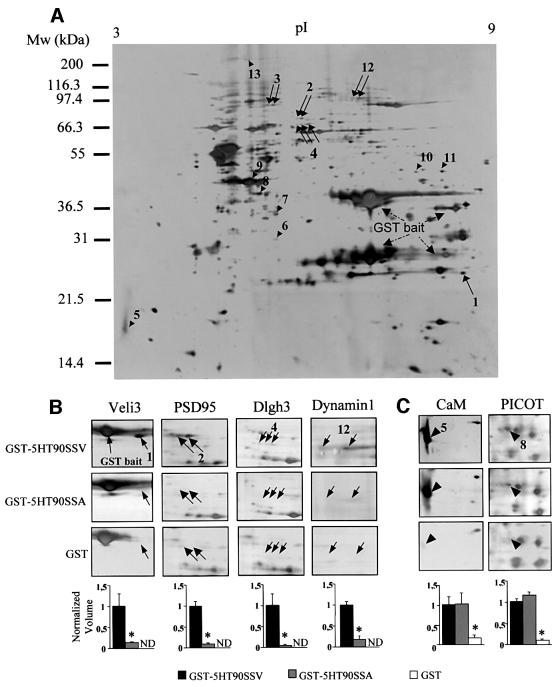
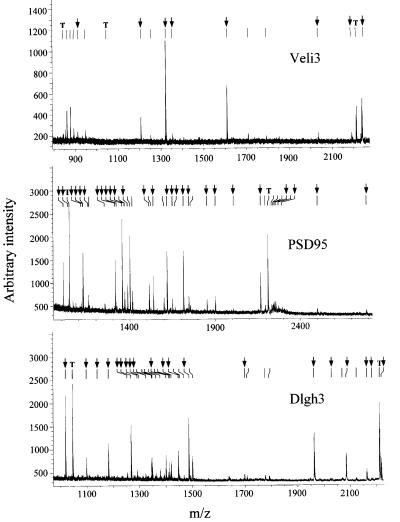
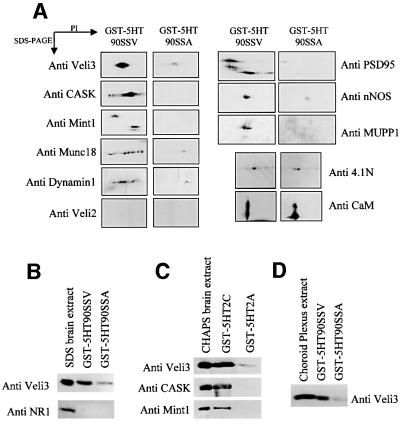
Proteins physically associated with the 5-HT2C receptor were purified by affinity chromatography using the entire C-terminal tail (90 amino acids) of the receptor fused to glutathione S-transferase (GST–5HT90SSV) as a bait. As 5-HT2C receptors display a wide but discrete distribution in the CNS (Abramowski et al., 1995), whole-brain extracts were prepared and loaded on to glutathione–Sepharose 4B beads coated with the GST– 5HT90SSV fusion protein. The bound proteins were eluted and separated by two-dimensional (2D) electrophoresis. A typical 2D gel is illustrated in Figure 1A. Numerous protein spots, including the GST fusion proteins and some bacterial proteins, were apparent on silver-stained 2D gels, due to the high sensitivity of this staining method. To detect proteins that interact specifically with the PDZ ligand of 5-HT2C receptors, we performed a control experiment using GST fused to the C-terminal tail of the receptor, which was mutated in this motif (GST– 5HT90SSA). The mutated residue is critical for the interaction with target PDZ domains (Bécamel et al., 2001). The analysis of protein patterns in the 2D gels obtained with the GST–5HT90SSV and GST–5HT90SSA baits indicated a marked difference in normalized volume of five spots or groups of spots (spots 1–4 and 12, indicated by arrows, Figure 1A). The differential binding of these proteins to the GST–5HT90SSV and GST–5HT90SSA baits is illustrated in detailed gels (Figure 1B). These proteins were unambiguously identified by MALDI-TOF mass spectrometry (see Table I). Figure 2 shows the peptide mass maps of three of them. Eight of the measured peptide masses obtained for spot 1 matched the theoretical tryptic peptide mass calculated for Veli3 (one of the vertebrate homologues of LIN-7), a 22 kDa protein containing one PDZ domain and enriched in the brain (Table II; Jo et al., 1999). The peptide masses exclude a match with the two other members of the Veli protein family, Veli1 and Veli2. As Veli2 and Veli3 display similar molecular weights and isoelectric points, we also performed 2D immunoblotting experiments using Veli2-and Veli3-specific antibodies. These experiments confirmed that the C-terminal tail of 5-HT2C receptors recruited Veli3 but not Veli2 (Figure 3A). Taken together, these results illustrate the power of MALDI-TOF technology to identify a protein isoform unambiguously. Spots 2 and 3 were identified as a single protein, the post-synaptic density-enriched protein PSD95, and spot 4 as a Dlgh3 protein, a brain-enriched scaffold protein of the p55 family (Figure 2, Tables I and II; Lin et al., 1998). Both proteins belong to a superfamily of modular proteins dubbed membrane-associated guanylate kinases (MAGUKs). These proteins contain an SH3 domain and a C-terminal guanylate kinase domain, in addition to one or several PDZ domains. The specific association of PSD95 with the wild-type bait, but not with the mutated one, was confirmed by immunoblotting (Figure 3A). Two major forms of the protein with different molecular weights and isoelectric points were detected by 2D immunoblotting. These immunoreactive signals matched spots 2 and 3, identified as PSD95 from their peptide mass fingerprint, on silver-stained gels (Figures 1A and 3A). The identification of dynamin 1 (spot 12) as a protein interacting with the 5-HT2C receptor through a PDZ domain-mediated scaffold was more unexpected, as this protein is devoid of PDZ domain. Nevertheless, the PDZ domain-dependent interaction of dynamin 1 with the 5-HT2C receptor was confirmed by immunoblotting (Figure 3A).
Fig. 1. Two-dimensional analysis of the 5-HT2C receptor protein complex. (A) Proteins that bind to the C-terminal tail of the 5-HT2C receptor were purified by affinity chromatography using the 90-amino-acid C-terminal sequence of the receptor fused to GST (GST–5HT90SSV), separated by 2D electrophoresis and silver stained. A typical 2D gel is illustrated. Proteins that interact specifically with the PDZ ligand of the receptor (arrows) were detected by comparing protein patterns obtained with GST–5HT90SSV and a mutant bait in which the last residue was replaced by alanine (GST–5HT90SSA). Arrowheads indicate proteins that interact equally with both wild-type and mutated baits but that were less represented in gels from experiments using GST alone. (B and C) Areas of interest of gels obtained in experiments performed with GST–5HT90SSV, GST–5HT90SSA and GST alone. The quantification of proteins (spot volume relative to the volume of all spots) was performed with Image Master. Data (means ± SEM of values from four gels) were normalized for each spot to the value measured in experiments using the wild-type bait (ND, not detectable). *P < 0.05 versus GST–5HT90SSV (ANOVA followed by Student–Newman–Keul’s test).
Table I. Proteomic analysis of proteins interacting with 5-HT2C receptors.
| Spota | Protein identified | Accession numberb | Protein paramaters |
Observed |
MALDI-TOF MS |
PDZ domain | |||
|---|---|---|---|---|---|---|---|---|---|
| Mol. wt (kDa) | pI | Mol. wt (kDa) | pI | Matching peptides | Protein coverage (%) | ||||
| 1 | Veli3 | O88952 | 21.8 | 8.5 | 24 | 8.5 | 8 | 50.0 | + |
| 2 | Post-synaptic density protein 95 (PSD95) | P31016 | 80.5 | 5.6 | 75 | 5.8 | 27 | 45.4 | + |
| 3 | Post-synaptic density protein 95 (PSD95) | P31016 | 80.5 | 5.6 | 86 | 5.4 | 24 | 38.1 | + |
| 4 | Dlgh3 protein (MPP3) | O88910 | 64.5 | 5.8 | 66 | 5.8 | 20 | 43.0 | + |
| 5 | Calmodulin | P02593 | 16.7 | 4.1 | 17 | 3.5 | 7 | 59.5 | – |
| 6 | F-actin capping protein β subunit (CAPZ β) | P47757 | 31.3 | 5.5 | 30 | 5.4 | 13 | 47.0 | – |
| 7 | F-actin capping protein α-2 subunit (CAPZ α-2) | P47754 | 33.0 | 5.6 | 35 | 5.4 | 7 | 41.3 | – |
| 8 | PKCθ-interacting protein PICOT | Q9JLZ2 | 37.8 | 5.4 | 40 | 5.3 | 9 | 38.6 | – |
| 9 | Actin, cytoplasmic 1 (β-actin) | P02570 | 41.6 | 5.3 | 45 | 5.2 | 15 | 56.0 | – |
| 10 | 2810409H07Rik protein | Q9CWE2 | 44.8 | 7.6 | 50 | 7.7 | 7 | 21.5 | – |
| 11 | 2810409H07Rik protein | Q9CWE2 | 44.8 | 7.6 | 50 | 8.1 | 8 | 27.0 | – |
| 12 | Dynamin 1 | P39053 | 97.4 | 8.2 | 97 | 7.0 | 31 | 36.6 | – |
| 13 | Spectrin α II chain (α-fodrin) | O88663 | 285 | 5.2 | 250 | 5.2 | 36 | 22.2 | – |
aThe numbers correspond to those illustrated in Figure 1.
bSWISS-PROT and TrEMBL accession numbers are listed.
Fig. 2. MALDI-TOF peptide mass maps obtained from spots 1, 2 and 4. Ion signals with measured masses (Table II) that matched calculated masses of protonated tryptic peptides of mouse Veli3, PSD95 and Dlgh3 are indicated by arrows. T indicates the ion signals corresponding to the autolysis products of trypsin that were used for internal calibration of spectra (mol. wts 842.51, 1045.56 and 2211.10, respectively).
Table II. Veli3, PSD95 and Dlgh3 peptides identified from the MALDI-TOF peptide mass maps shown in Figure 2.
| Measuredmass | Matchingmass | Δmass(p.p.m.) | Missedcleavage | Modification | Position | Peptide | |
|---|---|---|---|---|---|---|---|
| Veli3 | 911.54 | 911.53 | –10.02 | 0 | 122–130 | IIPGGIADR | |
| 1206.56 | 1206.61 | 42.53 | 0 | 112–121 | EQNSPIYISR | ||
| 1322.60 | 1322.65 | 39.40 | 0 | Cys-CAM | 41–51 | VLQSEFCNAVR | |
| 1352.59 | 1352.65 | 45.48 | 0 | 99–111 | TEEGLGFNIMGGK | ||
| 1608.75 | 1608.79 | 23.43 | 0 | 77–92 | ATVAAFAASEGHSHPR | ||
| 2034.01 | 2033.99 | –10.53 | 0 | 137–155 | GDQLLSVNGVSVEGEHHEK | ||
| 2190.10 | 2190.09 | –4.66 | 1 | 136–155 | RGDQLLSVNGVSVEGEHHEK | ||
| |
2238.04 |
2238.06 |
7.18 |
0 |
|
52–70 |
EVYEHVYETVDISSSPEVR |
| PSD95 | 1010.49 | 1010.49 | –17.55 | 0 | 579–586 | DYHFVSSR | |
| 1037.52 | 1037.50 | –19.51 | 1 | 571–578 | REYEIDGR | ||
| 1114.53 | 1114.58 | 40.56 | 0 | 234–242 | NTYDVVYLK | ||
| 1123.47 | 1123.48 | 6.74 | 0 | 506–516 | DWGSSSGSQGR | ||
| 1125.58 | 1125.60 | 18.38 | 0 | 99–110 | IIPGGAAAQDGR | ||
| 1156.57 | 1156.58 | 16.68 | 0 | 300–309 | DLLGEEDIPR | ||
| 1252.63 | 1252.66 | 27.30 | 0 | 369–380 | NASHEQAAIALK | ||
| 1314.72 | 1314.74 | 13.75 | 1 | 654–664 | SLENVLEINKR | ||
| 1322.59 | 1322.60 | 15.26 | 1 | 504–516 | AKDWGSSSGSQGR | ||
| 1354.63 | 1354.69 | 44.30 | 0 | Cys-CAM | 625–636 | HCILDVSANAVR | |
| 1368.68 | 1368.67 | –5.86 | 1 | 2×CysCAM | 1–11 | MDCLCIVTTKK | |
| 1386.69 | 1386.69 | 6.11 | 1 | Cys-CAM | 559–570 | FGSCVPHTTRPK | |
| 1513.83 | 1513.83 | 1.70 | 1 | 355–368 | KGDQILSVNGVDLR | ||
| 1538.79 | 1538.78 | –7.16 | 1 | 300–312 | DLLGEEDIPREPR | ||
| 1618.86 | 1618.84 | –10.58 | 0 | 113–126 | VNDSILFVNEVDVR | ||
| 1650.84 | 1650.81 | –16.49 | 0 | 409–424 | EQLMNSSLGSGTASLR | ||
| 1666.86 | 1666.81 | –29.96 | 0 | MSO | 409–424 | EQLMNSSLGSGTASLR | |
| 1714.95 | 1714.92 | –17.1 | 0 | 707–721 | VIEDLSGPYIWVPAR | ||
| 1746.92 | 1746.82 | –62.15 | 0 | 476–491 | VHSDSETDDIGFIPSK | ||
| 1853.13 | 1853.10 | –17.72 | 1 | 638–653 | LQAAHLHPIAIFIRPR | ||
| 1902.96 | 1902.92 | –20.09 | 1 | 475–491 | RVHSDSETDDIGFIPSK | ||
| 2009.30 | 2009.20 | –50.55 | 2 | 637–653 | RLQAAHLHPIAIFIRPR | ||
| 2168.10 | 2168.10 | –1.62 | 1 | 381–399 | NAGQTVTIIAQYKPEEYSR | ||
| 2256.15 | 2256.10 | –20.31 | 0 | 598–617 | FIEAGQYNSHLYGTSVQSVR | ||
| 2294.12 | 2294.21 | 36.18 | 0 | 212–233 | ILAVNSVGLEDVMHEDAVAALK | ||
| 2501.25 | 2501.28 | 9.43 | 0 | 169–193 | GLGFSIAGGVGNQHIPGDNSIYVTK | ||
| |
2787.18 |
2787.36 |
64.09 |
0 |
|
71–98 |
GNSGLGFSIAGGTDNPHIGDDPSIFITK |
| Dlgh3 | 1018.50 | 1018.51 | 8.91 | 0 | 233–240 | ALFHYDPR | |
| 1098.60 | 1098.63 | 25.85 | 1 | 143–152 | NKEPLGATIR | ||
| 1139.55 | 1139.58 | 24.65 | 0 | 154–164 | DEHSGAVVVAR | ||
| 1181.58 | 1181.63 | 39.7 | 0 | 174–184 | SGLVHVGDELR | ||
| 1252.54 | 1252.45 | –76.16 | 0 | 323–332 | ETCDCDEYFK | ||
| 1267.62 | 1267.63 | 5.5 | 1 | 377–386 | YQHQPGERPR | ||
| 1290.65 | 1290.67 | 10.13 | 0 | 301–312 | TTGTLPSPQNFK | ||
| 1295.67 | 1295.68 | 6.93 | 1 | 153–154 | RDEHSGAVVVAR | ||
| 1323.65 | 1323.62 | –27.08 | 0 | 440–450 | QAFEADVHHNR | ||
| 1399.75 | 1399.72 | –17.6 | 0 | Cys-CAM | 478–489 | VCLVDVEPEALR | |
| 1419.84 | 1419.83 | –8.33 | 0 | 90–101 | ELLQLLSTPHLR | ||
| 1446.79 | 1446.77 | –15.78 | 1 | 300–312 | RTTGTLPSPQNFK | ||
| 1486.79 | 1486.75 | –26.19 | 0 | Cys-CAM | 244–256 | AIPCQEAGLPFQR | |
| 1699.07 | 1699.03 | –27.16 | 0 | 387–402 | LVVLIGSLGAHLHELK | ||
| 1962.08 | 1962.02 | –31.82 | 0 | 359–376 | VPTGAESQVLLTYEEVAR | ||
| 2029.07 | 2029.00 | –32.2 | 0 | 258–274 | QVLEVVSQDDPTWWQAK | ||
| 2084.16 | 2084.13 | –14.31 | 1 | 195–213 | RPDEISQILAQSQGSITLK | ||
| 2162.24 | 2162.20 | –17.26 | 2 | 193–510 | TPEFKPYVIFVKPAIQER | ||
| 2185.10 | 2185.10 | –0.19 | 1 | 257–274 | RQVLEVVSQDDPTWWQAK | ||
| 2218.14 | 2218.17 | 14.15 | 1 | 405–424 | VVAEDPQQFAVAVPHTTRPR |
MSO, oxidized methionine; Cys-CAM, carbamidomethyl cysteine.
Fig. 3. Detection of proteins interacting with the 5-HT2C receptor by western blotting. (A) CHAPS-solubilized proteins from whole brain, retained by the GST–5HT90SSV and GST–5HT90SSA baits, were resolved on 2D gels and transferred electrophoretically on to nitrocellulose membranes. (B) Proteins were solubilized with 1% SDS instead of CHAPS and incubated with the GST–5HT90SSV and GST–5HT90SSA baits. (C) CHAPS-solubilized proteins from whole brain were passed over affinity columns containing the C-terminal tails of 5-HT2C and 5-HT2A receptors fused to GST. (D) CHAPS-solubilized proteins from choroid plexus were incubated with the GST–5HT90SSV and GST–5HT90SSA baits. Immunoblotting was performed with antibodies raised against the indicated proteins. For each protein, the immunoreactive signals were found at the expected isoelectric points (A) and molecular weights. The data illustrated are representative of three experiments.
An immunoblotting screen was then performed to identify additional partners of 5-HT2C receptors. Veli proteins form a stable tripartite complex with two other neuron-enriched modular proteins: CASK, a MAGUK that contains an N-terminal calmodulin kinase II domain in addition to one PDZ domain; and Mint1, which contains two PDZ domains (Borg et al., 1998; Butz et al., 1998). Veli proteins, CASK and Mint1 are the mammalian orthologues of the Caenorhabditis elegans proteins LIN-7, LIN-2 and LIN-10. In C.elegans, this complex is required for the normal basolateral localization of the tyrosine kinase receptor LET23 in vulval cells. The whole complex is necessary for the proliferation and differentiation of vulval cells (Simske et al., 1996; Kaech et al., 1998). CASK binds directly to both Veli proteins and Mint1 through PDZ-independent interactions, leaving their PDZ domains free to interact with other proteins, such as cell adhesion molecules, receptors and signalling proteins (Butz et al., 1998). CASK and Mint1 were not detectable in silver-stained 2D gels obtained from the pull-down experiment using GST–5HT90SSV and GST–5HT90SSA fusion proteins. However, 2D immunoblotting indicated that both CASK and Mint1 were recruited by the wild-type bait but not by the mutated one (Figure 3A). This suggests that 5-HT2C receptors interact with the entire tripartite complex through a PDZ-based mechanism. Pull-down experiments performed with the C-terminal tail of the 5-HT2A receptor, a group 2 5-HT receptor expressed in the CNS and containing a PDZ ligand domain, indicated that the Veli3–CASK–Mint1 complex does not associate with this 5-HT2 receptor subclass (Figure 3C). This result further supports the idea that the proteins recruited by the C-terminal tail of 5-HT2C receptors are specific binding partners of these receptors rather than proteins interacting with any PDZ domain recognition motif. Mint1 can bind to Munc18, a presynaptic protein that is essential for exocytosis of synaptic vesicles (Okamoto and Südhof, 1997; Borg et al., 1999; Biederer and Südhof, 2000; Verhage et al., 2000). As shown by immunoblotting, Munc18 was also recruited by 5-HT2C receptors via a PDZ domain-dependent scaffold (Figure 3A).
PSD95 can couple functionally specific signalling proteins to post-synaptic receptors. For example, PSD95 binds to neuronal NO-synthase (nNOS) via a PDZ– β-finger interaction and the C-terminal PDZ domain-binding motif of NMDA receptor subunits (NR2A and NR2B), allowing a physical coupling of nNOS to the NMDA receptor (Brenman et al., 1996; Christopherson et al., 1999; Tochio et al., 2000). Moreover, activation of 5-HT2 receptors, including the 5-HT2C subtype, induces cGMP production through activation of nNOS in various cell populations (Kaufman et al., 1995). We found that nNOS was specifically retained by the GST–5HT90SSV bait, suggesting that 5-HT2C receptors are physically associated with nNOS through a PSD95-dependent scaffold (Figure 3A). PSD95 is a major scaffolding protein involved in the assembly of NMDA receptor-associated protein complex (Husi et al., 2000). Thus, the 5-HT2C receptor, by interacting with PSD95, may also belong to this complex. To determine whether the 5-HT2C receptor associates with NMDA receptors, brain proteins were solubilized with 1% SDS instead of CHAPS (Ehlers et al., 1998) and incubated with the GST–5HT90SSV and GST–5HT90SSA baits. In this experimental condition, Veli3 was recruited by the PDZ ligand of 5-HT2C receptors (Figure 3B). However, the C-terminal tail of 5-HT2C receptors did not retain any detectable amount of the NMDA receptor subunit NR1, which is essential for the formation of functional NMDA receptors (Figure 3B). Finally, in agreement with our previous findings (Bécamel et al., 2001), the 5-HT2C receptor associated with MUPP1 via its C-terminal PDZ domain recognition motif (Figure 3A).
Isolation of proteins interacting with the 5-HT2C receptor independently of PDZ domain-based scaffolds
To identify proteins that bind to residues not located in the PDZ ligand of the 5-HT2C receptor, the 2D gel protein patterns resulting from pull-down experiments using GST–5HT90SSV and GST–5HT90SSA fusion proteins were compared with that obtained with GST alone (control). Using this approach, we detected eight proteins (indicated by arrowheads, Figure 1A) that were recruited to a similar extent by GST–5HT90SSV and GST– 5HT90SSA baits but were less represented in gels from experiments performed with GST alone (see, for example, the quantification of spots 5 and 8, Figure 1C). These comprised calmodulin, PICOT, a protein that was recently identified as a binding partner of protein kinase Cθ (PKCθ) (Witte et al., 2000) and cytoskeletal proteins [β-actin, spectrin α II chain (α-fodrin) and both α and β chains of CAPZ (Table I)]. CAPZ is a capping protein that binds as a dimer to the barbed end of actin filaments and inhibits the growth of actin microfilaments (Xu et al., 1999). These results suggest that 5-HT2C receptors are physically associated with actin/spectrin microfilaments. Association of actin filaments with tetrameric spectrin is promoted by the 4.1 proteins, a family of peripheral membrane proteins identified initially in erythrocytes (Conboy et al., 1986). 4.1 proteins are critical for the attachment of the actin cytoskeleton to the plasma membrane through interaction with integral membrane proteins such as glycophorin C (Takakuwa, 2000). 4.1 proteins can also interact with other proteins through specific domains, including calmodulin and MAGUKs of the p55 subfamily such as CASK (Hoover and Bryant, 2000). We thus examined by western blotting whether 5-HT2C receptors interact with the neuronal form of 4.1 protein (4.1N). Identical amounts of 4.1N were recruited by the wild type and the mutated C-terminal tail of 5-HT2C receptors (Figure 3A). This result is consistent with the aforementioned findings that the 5-HT2C receptor binds to cytoskeletal proteins through a PDZ domain-independent scaffold and suggests that 4.1N protein may be a critical component that anchors 5-HT2C receptors to the actin cytoskeleton. The two last spots recruited by both fusion proteins but not by GST were identified as a single protein with an unknown function (2810409H07Rik protein; Kawai et al., 2001).
Coimmunoprecipitation of PSD95 and the Veli3–CASK–Mint1 complex with 5-HT2C receptors
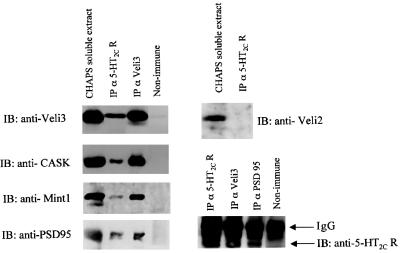
Taken together, our results suggest that 5-HT2C receptors are part of multiprotein complexes including synapse-enriched scaffold proteins, containing one or several PDZ domains, such as the Veli3–CASK–Mint1 complex and PSD95. In order to examine the association of these proteins with 5-HT2C receptors in brain extracts, we performed coimmunoprecipitation studies. CHAPS- soluble lysates of mice brains were immunoprecipitated with the anti-5-HT2C receptor 522 antibody. We found that PSD95, Veli3, CASK and, to a lesser extent, Mint1 coimmunoprecipitated with the 5-HT2C receptor (Figure 4). In contrast, Veli2 was not coimmunoprecipitated by the 5-HT2C receptor antibody, indicating that 5-HT2C receptors interact preferentially with Veli3 in vivo. This result is consistent with the specific recruitment of this Veli isoform in pull-down experiments and, thus, supports the specificity of this in vitro binding assay. In a similar manner, we immunoprecipitated Veli3 and PSD95 from brain extract with Veli3 and PSD95 antibodies. We found that 5-HT2C receptors coimmunoprecipitated with both proteins (Figure 4, bottom). CASK and Mint1 also coimmunoprecipitated with Veli3 (Figure 4). This is consistent with previous findings that demonstrate that CASK, Mint1 and Veli proteins form a stable tripartite complex (Borg et al., 1998; Butz et al., 1998). Taken together, these results indicate that 5-HT2C receptors are associated with both the Veli3–CASK–Mint1 complex and PSD95 in vivo. In agreement with a previous report indicating that Veli proteins are clustered with PSD95 and NMDA-type glutamate receptors (Jo et al., 1999), we also found that PSD95 coimmunoprecipitated with Veli3 (Figure 4).
Fig. 4. Association of PSD95 and the Veli3–CASK–Mint1 complex with 5-HT2C receptors of mice brain. Solubilized membranes of mice brain were immunoprecipitated with either the anti-5-HT2C, the anti-Veli3 or the anti-PSD95 antibody. Immunoprecipitated proteins were analysed by western blotting using antibodies to Veli3, CASK, Mint1, PSD95, Veli2 and the 5-HT2C receptor. Input (CHAPS-soluble extract) represents 10% of the total protein used for the immunoprecipitation.
PDZ domain-based interactions of the 5-HT2C receptor with Veli3 and PSD95 within intact cells
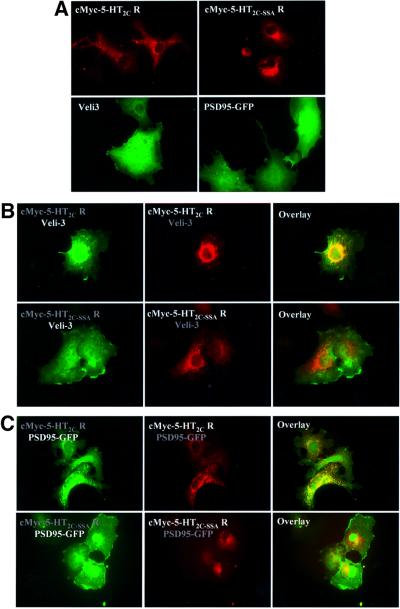
Next, we examined whether the entire 5-HT2C receptor interacts with PSD95 and Veli3 within intact cells. For that purpose, we performed immunostaining of COS-7 cells transiently expressing the c-Myc epitope-tagged version of the human 5-HT2C receptor or its mutated form 5-HT2C-SSA, in the presence and absence of either Veli3 protein or GFP-tagged PSD95. Staining COS-7 cells transfected with either the wild type or mutated 5-HT2C receptor with an anti-c-Myc antibody revealed a random distribution of 5-HT2C receptors and 5-HT2C-SSA receptors on membrane-type structures, including intracellular membranes, consistent with our previous findings (Figure 5A; Bécamel et al., 2001). Staining of COS-7 cells expressing Veli3 with a Veli3 antibody indicated a homogeneous distribution of the protein. Similarly, GFP-tagged PSD95 was homogeneously distributed throughout transiently transfected cells (Figure 5A). No staining was detectable with an anti-c-Myc, an anti-Veli3 or an anti-PSD95 antibody in non-transfected cells (data not shown). Coexpression of the c-Myc-tagged 5-HT2C receptor together with Veli3 revealed distinct distributions from those observed when they were transfected separately (Figure 5B). Moreover, both proteins were colocalized. This contrasts with the observations made on cells coexpressing Veli3 and the mutated c-Myc epitope-tagged 5-HT2C-SSA receptor (Figure 5B), indicating that Veli3 interacts with the C-terminal sequence of recognition of PDZ domains of the 5-HT2C receptor. Similarly, a redistribution of PSD95 was observed in COS-7 cells coexpressing the c-Myc epitope-tagged 5-HT2C receptor (Figure 5C). Both proteins were localized in the same regions in cells and formed many clusters. Expression of GFP-tagged PSD95 with the c-Myc epitope-tagged 5-HT2C-SSA receptor failed to form clusters, indicating that the interaction between these proteins also requires the PDZ ligand of the 5-HT2C receptor (Figure 5C).
Fig. 5. Interaction of 5-HT2C receptors with Veli3 and PSD95 via PDZ-based scaffolds in transiently transfected COS-7 cells. (A) Cells expressing the c-Myc-tagged 5-HT2C receptor or the c-Myc-tagged 5-HT2C-SSA receptor were immunostained using the anti-c-Myc antibody. Cells expressing Veli3 were immunostained using the anti-Veli3 antibody. PSD95 was revealed by its GFP tag. (B) COS-7 cells were cotransfected with Veli3 and either the c-Myc-tagged 5-HT2C receptor (top panels) or the c-Myc-tagged 5-HT2C-SSA receptor (bottom panels). The left panels illustrate Veli3 staining, medium c-Myc staining. Both proteins were colocalized in cells coexpressing the c-Myc-tagged 5-HT2C receptor and Veli3, in contrast with cells coexpressing the 5-HT2C-SSA receptor mutant and Veli3. (C) COS-7 cells were cotransfected with PSD95–GFP and either the c-Myc-tagged 5-HT2C receptor (top panels) or the c-Myc-tagged 5-HT2C-SSA receptor (bottom panels). The left panels illustrate GFP fluorescence, medium c-Myc staining. Both proteins were colocalized in clusters in cells coexpressing the c-Myc-tagged 5-HT2C receptor and PSD95, whereas PSD95 failed to form clusters with the 5-HT2C-SSA receptor.
Immunolocalization of the 5-HT2C receptor and Veli3 within the brain
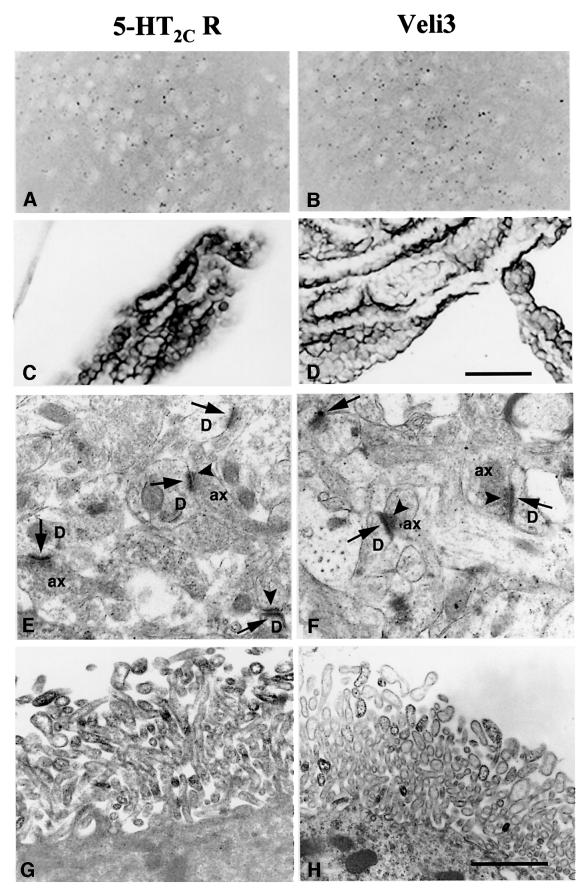
In all the sections immunostained with the 5-HT2C receptor or Veli3 antibody, only faint labelling could be detected on dot-structures dispersed within some brain regions, including the olfactory nuclei (Figure 6A and B), the olfactory bulb and the substantia nigra (data not shown). On the other hand, strong immunostaining with both antibodies was associated with the choroid plexus located within the brain ventricles (Figure 6C and D) and, to a lesser extent, the ependymocytes bordering the ventricles (data not shown). Consistent with this observation, the C-terminal tail of 5-HT2C receptors recruited significant amounts of Veli3 protein solubilized from the choroid plexus (Figure 3D). In order to characterize the cellular localization of 5-HT2C receptor and Veli3 proteins, electron microscope immunocytochemistry was performed on sections including the anterior olfactory nucleus and the choroid plexus. Again, very similar immunostaining patterns were observed with both antibodies (Figure 6E–H). In the anterior olfactory nucleus, both immunostainings were essentially associated with axo-dendritic synapses, electron-dense precipitates being mostly localized at post-synaptic and, to a lesser extent, presynaptic thickenings (Figure 6E and F). In the choroid plexus, both 5-HT2C receptor and Veli3 immunostainings were associated with the limiting membrane of distal portions of microvilli bordering the apical surface of choroidal cells (Figure 6G and H).
Fig. 6. Immunolocalization of the 5-HT2C receptor and Veli3 in the anterior olfactory nucleus and the choroid plexus. Light microscope observations (A–D) indicated similar distribution patterns for both 5-HT2C receptor and Veli3 immunostainings in the anterior olfactory nucleus (A and B) and the choroid plexus (C and D). Electron microscope observations (E–H) demonstrated that electron-dense precipitates corresponding to immunostainings for both antibodies were associated with the same subcellular structures, i.e. (i) post-synaptic (arrows) and, to a lesser extent, presynaptic (arrowheads) thickenings of axo-dendritic synapses in the olfactory nucleus (E and F); and (ii) the membrane of distal portions of microvilli in choroidal cells (G and H). D, dendritic profile; ax, axonal profile. Scale bars (A–D) = 50 µm, (E–H) = 1 µm.
Discussion
In this study, we have identified 15 proteins that bind to the C-terminal tail of 5-HT2C receptors. These comprise scaffold proteins, such as the Veli3–CASK–Mint1 complex and two MAGUKs, Dlgh3 and PSD95, proteins of the actin/spectrin cytoskeleton and signalling proteins. To our knowledge, this study provides the first global characterization of a multiprotein complex associated with the C-terminal tail of a GPCR and suggests that, like ionic channels, GPCRs are also part of protein networks maintained by protein–protein interactions involving multidomain adaptor proteins. Moreover, one cannot exclude that additional proteins interact with the cytosolic loops of the receptor.
Several pieces of evidence indicate that the proteins fished out in our in vitro binding assay are specific binding partners of 5-HT2C receptors rather than proteins interacting with any PDZ ligand domain. First, 2D analysis of proteins retained by the GST–5HT90SSV bait suggests that 5-HT2C receptors associate with a single isoform of Veli proteins. Indeed, we could only detect Veli3 on silver-stained 2D gels, whereas this isoform shows the most discrete and restricted expression in the brain (Misawa et al., 2001). It is noteworthy that 5-HT2C receptors and Veli3 show an overlapping distribution in several brain regions, including the olfactory bulb, the olfactory nuclei, the substantia nigra and the choroid plexus, in agreement with previous findings (Abramowski et al., 1995; Misawa et al., 2001). Secondly, coimmunoprecipitation experiments performed with some of the binding partners fished out with the GST–5HT90SSV bait (PSD95 and the Veli3–CASK–Mint1 complex) confirmed that 5-HT2C receptors associate with these proteins in vivo. Moreover, Veli2, which was not retained by the GST–5HT90SSV bait, did not coimmunoprecipitate with the 5-HT2C receptor. Finally, analysis of proteins interacting with another 5-HT receptor of group 2, the 5-HT2A receptor, which expresses a PDZ domain-binding motif similar to that of the 5-HT2C receptor (SCV instead of SSV), revealed a different profile of protein interaction. Indeed, the 5-HT2A receptor did not associate with the Veli3–CASK–Mint1 complex.
Coexpression of the 5-HT2C receptor with Veli3 or PSD95 in COS-7 cells indicated a possible interaction of its PDZ domain-binding motif (SSV) with either Veli3 or PSD95. This is consistent with biochemical analyses showing that the PDZ domains of PSD95 and Veli proteins belong to class I and interact with proteins terminating with a consensus X-S/T-X-I/L/V (Sheng and Sala, 2001). However, the preferential recruitment of Veli3 by 5-HT2C receptors suggests that additional molecular determinants contribute to this specificity and further support the idea that amino acids upstream of the –2 position determine optimal binding of PDZ domains to their targets. In contrast, the PDZ domain of CASK is class II (Daniels et al., 1998) and, thus, probably does not interact directly with the 5-HT2C receptor. CASK was retained in a much smaller quantity by immobilized GST–5HT90SSV than was Veli3 (CASK cannot be detected by silver staining), consistent with an indirect recruitment of CASK via Veli3. These results, together with the relatively small amount of Mint1 that coimmunoprecipitated with 5-HT2C receptors, strongly suggest that the 5-HT2C receptor is anchored to the Veli3–CASK–Mint1 complex via Veli3. Similarly, Dlgh3 protein can be recruited indirectly by 5-HT2C receptors via Veli3. Indeed, Dlgh3 belongs to a subfamily of MAGUKs closely related to p55, including CASK, Dlgh2 and proteins associated with Lin-7 types 1 and 2 (Pals 1 and 2), which share a highly conserved domain located N-terminally from the PDZ domain and involved in the binding to Veli proteins (Lin et al., 1998; Kamberov et al., 2000). The smaller amount of Dlgh3, compared with Veli3, recovered from immobilized GST–5HT90SSV is also consistent with an indirect recruitment of Dlgh3. However, one cannot exclude that Dlgh3 also interacts directly with the PDZ recognition motif of the 5-HT2C receptor.
Our results demonstrate that 5-HT2C receptors are physically associated with the actin/spectrin cytoskeletal network. The C-terminal tail of 5-HT2C receptors also recruited 4.1N, which is localized at dendritic spines and associated with post-synaptic densities (Walensky et al., 1999). A recent study has suggested that 4.1N serves to cross-link AMPA receptors to the actin cytoskeleton at excitatory synapses and to stabilize surface expression of AMPA receptors (Shen et al., 2000). Similarly, 4.1N may anchor 5-HT2C receptors to the submembrane actin cytoskeletal network. 4.1 proteins interact with a consensus polybasic sequence located in the HOOK domain of MAGUKs of the p55 family, including CASK and p55 (Hoover and Bryant, 2000). CASK and p55 have also been identified as important organizers of the actin/spectrin cytoskeleton in neuronal and non-neuronal cells, respectively (Cohen et al., 1998; Biederer and Südhof, 2001). We found that the interaction of 5-HT2C receptors with cytoskeletal proteins was not mediated by PDZ domain-based scaffolds, ruling out the implication of CASK as an intermediate in anchoring cytoskeletal proteins to 5-HT2C receptors. Alternatively, CASK may cross-link the Veli3–CASK–Mint1 complex to 4.1N and actin/spectrin cytoskeleton and, thus, cooperate with 4.1N to stabilize interactions between the receptor and the cytoskeleton. In contrast, the Dlgh3 protein cannot bind 4.1N because it lacks the consensus polybasic binding sequence within its HOOK domain (Lin et al., 1998).
Electron microscopy experiments strongly support the interaction of the 5-HT2C receptor with protein complexes that include Veli3 within the brain. Indeed, these experiments indicate that the 5-HT2C receptor and Veli3 are localized in the same subcellular structures in specific brain regions. The observation that the 5-HT2C receptor and Veli3 are associated with post-synaptic structures is consistent with a previous study demonstrating that Veli proteins are enriched in the post-synaptic density, where they are clustered together with PSD95 (Jo et al., 1999). On the other hand, the 5-HT2C receptor and Veli3 were also detected in presynaptic terminals. This result fits recent findings: (i) a direct stimulatory effect of striatal 5-HT2C receptors on local dopamine release and, thus, a presynaptic function of 5-HT2C receptors (Lucas and Spampinato, 2000); and (ii) the presynaptic localization of the Veli–CASK–Mint1 complex, based on the known interactions of CASK and Mint1 with presynaptic protein components (Okamoto and Südhof, 1997; Borg et al., 1999; Biederer and Südhof, 2000; Verhage et al., 2000). Finally, we provide evidence that in the choroid plexus, which expresses high levels of the 5-HT2C receptor and Veli3, both proteins are colocalized in the microvilli bordering the apical surface of choroidal cells. Taken together, these results demonstrate the biological relevance of the interaction between both protein species and, therefore, the assembly of 5-HT2C receptors into multiprotein complexes.
Materials and methods
Antibodies and plasmids
Production, characterization and purification of the rabbit polyclonal antibody 522 raised against the 5-HT2C receptor have been described previously (Abramowski et al., 1995). The anti-c-Myc antibody was a gift from Dr B.Mouillac (Montpellier, France). The rabbit polyclonal anti-Veli3 antibody was purchased from Zymed Laboratories (San Francisco, CA). Mouse monoclonal anti-CASK, anti-Mint1, anti-Munc18, anti-4.1N protein, anti-dynamin 1 and anti-PSD95 antibodies were purchased from Transduction Laboratories (Lexington, KY). The Veli2 antibody was obtained by immunization of rabbits with a synthetic peptide derived from the Veli2 sequence (QHHSYSSLESRG) as described previously (Misawa et al., 2001). This antibody recognized a single polypeptide around 24 kDa in western blots of mouse brain extracts. Secondary antibodies used in immunofluorescence experiments were goat Alexa green- or red-conjugated anti-rabbit or anti-mouse antibodies (Jackson Immuno Research, PA).
Mammalian expression vectors c-Myc-h5-HT2C/pRK5 and c-Myc-h5-HT2C-SSA/pRK5 have been described previously (Bécamel et al., 2001). Veli3 cDNA, purchased from IMAGE, was subcloned in PRK7 into the BamHI and HindIII sites. PSD95–GFP construct was kindly provided by Dr E.D.Gundelfinger (Magdeburg, Germany).
Membrane preparations and GST pull-down
Mice brains were homogenized with a polytron in 20 ml of phosphate buffered saline (PBS) and centrifuged at 200 g for 3 min. Pellets were resuspended in ice-cold lysis buffer containing 50 mM Tris–HCl pH 7.4, 1 mM EDTA and a protease inhibitor cocktail (Roche), homogenized 20 times on ice with a glass-Teflon homogenizer and centrifuged at 10 000 g for 30 min. Choroid plexus samples were directly homogenized in lysis buffer as above. The membrane pellets were resuspended in CHAPS extraction buffer (50 mM Tris–HCl pH 7.4, 0.05 mM EDTA, 10 mM CHAPS and protease inhibitors) for 3 h in rotation at 4°C. In experiments performed to determine whether the 5-HT2C receptor associates with NMDA receptor subunits, brain proteins were solubilized with 1% SDS instead of CHAPS (Ehlers et al., 1998). Samples were then centrifuged for 1 h at 10 000 g.
GST and GST fusion proteins were expressed in Escherichia coli strain BL21 as described previously (Bécamel et al., 2001) and immobilized (50 µg each) on glutathione–Sepharose beads (Amersham Pharmacia Biotech, Uppsala, Sweden). Solubilized proteins of mice brains were incubated with immobilized GST fusion proteins overnight at 4°C. Samples were washed five times with 150 mM NaCl and eluted with 10 mM reduced glutathione. Samples were then precipitated with 10% ice-cold trichloroacetic acid (TCA) for 2 h and precipitates were washed three times with diethyl ether.
High-resolution 2D gel electrophoresis
TCA precipitates were resuspended in 350 µl of isoelectrofocusing medium containing 7 M urea, 2 M thiourea, 4% (w/v) CHAPS, ampholines (pre-blended, pI 3.5–9.5, 8 mg/ml; Amersham Pharmacia Biotech), 100 mM dithiothreitol (DTT,), 0.2% (w/v) Tergitol NP7 (Sigma) and traces of bromophenol blue. Proteins were first separated according to their isoelectric point along linear immobilized pH-gradient (IPG) strips (pH 3–10, 18 cm long) using the IPGphor apparatus (Amersham Pharmacia Biotech). Sample loading for the first dimension was performed by passive in-gel re-swelling. After the first dimension, the IPG strips were equilibrated for 10 min in a buffer containing 6 M urea, 50 mM Tris–HCl pH 6.8, 30% (v/v) glycerol, 2% (w/v) SDS, 10 mg/ml DTT and bromophenol blue, and then for 15 min in the same buffer containing 15 mg/ml iodoacetamide instead of DTT. For the second dimension, the strips were loaded on to vertical 12.5% SDS–polyacrylamide gels. The gels were silver stained according to the procedure of Shevchenko et al. (1996).
Image acquisition and 2D gel spot pattern analysis
Gels to be compared were always processed and stained in parallel. Gels were scanned using a computing densitometer (Amersham Pharmacia Biotech). Spot detection, gel alignment and spot quantification were performed using the Image Master 2D Elite software (Amersham Pharmacia Biotech). Quantitative variations of proteins were expressed as volumes of spots. To correct for variability resulting from silver staining, results were expressed as relative volumes of all spots in each gel. Data are the means of values from four gels originating from different pull-down experiments.
Protein identification by MALDI-TOF mass spectrometry
Proteins of interest were excised and digested in gel using trypsin (sequencing grade; Promega, Madison, WI), as described previously (Shevchenko et al., 1996). Digest products were completely dehydrated in a vacuum centrifuge and resuspended in 10 µl of formic acid (2% w/v), desalted using Zip Tips C18 (Millipore, Bedford, MA), eluted with 10 µl of acetonitrile:trifluoroacetic acid (TFA), (80:0.01%) and concentrated to 2 µl. Aliquots of analyte solutions (0.5 µl) were mixed with the same volume of α-cyano-4-hydroxy-trans-cinnamic acid (10 mg/ml in acetonitrile:TFA, 50:0.01%) and loaded on the target of a Biflex III MALDI-TOF mass spectrometer (Bruker-Franzen Analytik, Bremen, Germany). Analysis was performed in reflectron mode with an accelerating voltage of 20 kV and a delayed extraction of 400 ns. Spectra were analysed using the XTOF software (Bruker-Franzen Analytik), and auto-proteolysis products of trypsin (mol. wts 842.51, 1045.56, 2211.10) were used as internal calibrates. Identification of proteins was performed using both Mascot and PeptIdent softwares, available online at http://www.matrixscience.com and http://www. expasy.org/tools/peptident.html, respectively. A mass deviation of 100 p.p.m. was allowed for database interrogation, but the mass accuracy of our analyses was usually better than 50 p.p.m.. Coverage of the full-length protein exceeding 15% was considered to be sufficient, unless there were some obvious conflicts between the experimental molecular weight or isoelectric point and those of the identified protein (Garin et al., 2001). Matching peptides with one missed cleavage were considered as pertinent only when there were two consecutive basic residues or when arginine and lysine residues were followed by a proline or acidic residues inside the peptide amino acid sequence.
Western blotting
Proteins, resolved on 1D or 2D gels, were transferred electrophoretically to nitrocellulose membranes (Hybond-C; Amersham Pharmacia Biotech). Membranes were incubated overnight with primary antibodies. Immunoreactivity was detected with an enhanced chemiluminescence method (Renaissance Plus; NEN DuPont, Boston, MA).
Coimmunoprecipitation
CHAPS-soluble proteins from brain extracts (500 µg per experiment) were incubated overnight at 4°C with either the anti-5-HT2C receptor 522 antibody, the anti-PSD95 antibody or the anti-Veli3 antibody (1 µg each). Samples were incubated for 1 h at 4°C with 50 µl of protein A–Sepharose beads (Amersham Pharmacia Biotech). After five washes with homogenization buffer, immunoprecipitated proteins were eluted in Laemmli sample buffer, resolved by SDS–PAGE and detected by western blotting.
Cell culture and transfection
COS-7 cells (60% confluence) were transfected with Transfast transfection reagent (Promega) according to the manufacturer’s instructions, using 1 µg of each cDNA. Cells were grown in Dulbecco’s modified Eagle’s medium (Life Technologies, Cergy Pontoise, France) containing 10% dialysed fetal calf serum (Life Technologies).
Immunocytochemistry
COS-7 cells grown on 35 mm dishes were fixed 24 h after transfection in paraformaldehyde, 4% (w/v) in PBS, for 20 min at room temperature. They were washed three times in PBS supplemented with glycine (0.1 M) and permeabilized with 0.05% (w/v) Triton X-100 for 5 min. Cells were washed in PBS containing 0.2% gelatin and incubated overnight at 4°C with the primary antibody (1:500 dilution) in 0.2% gelatin/PBS. Cells were washed and incubated for 1 h at room temperature with the secondary antibody (1:1000 dilution) in 0.2% gelatin/PBS. After three washes, the cells were mounted on glass slides using gel mount (Biomeda, Foster City, CA) and viewed on a Zeiss Axioplan 2 microscope.
Immunohistochemistry/electron microscopy
Animals, anaesthetized with sodium pentobarbital (50 mg/kg), were perfused through the ascending aorta successively with PBS and 300 ml of fixative composed of 4% paraformaldehyde and 0.5% glutaraldehyde in PBS. The brain was then dissected and fixed by immersion in the fixative without glutaraldehyde for 12 h at 4°C. The rostral part of the brain was then cut frontally with a vibratome into 40–50 µm thick sections. After careful rinsing in PBS, sections were successively incubated: (i) for 48 h at 4°C with either the rabbit antibody against 5-HT2C (1:250 dilution) or the rabbit antibody against Veli3 (1:400 dilution); (ii) for 12 h at 4°C with a peroxidase-linked Fab fragment of goat anti-rabbit IgG (Biosys, Compiègne, France, 1:1000 dilution); and (iii) with 0.1% 3,3′-diaminobenzidine diluted in 0.05 M Tris buffer pH 7.3, in the presence of 0.2% H2O2. The primary and secondary antibodies were diluted in PBS containing 1% BSA, 1% normal goat serum and 0.1% saponin. Immunostained sections were either mounted in permount and observed using light microscopy or further treated for electron microscopy. They were rinsed in 0.1 M cacodylate buffer pH 7.3, post-fixed in 1% OsO4 in the same buffer, dehydrated in graded concentrations of ethanol and embedded in araldite. Punches of 1.5 mm diameter were cut through the anterior olfactory nucleus or the choroid plexus and mounted on araldite blocks. After being cut into ultrathin sections, they were observed in an electron microscope (Hitachi H 7110) without counterstaining.
Acknowledgments
Acknowledgements
We would like to thank Dr D.Abramowski for the 5-HT2C receptor antibody, Dr B.Mouillac for the c-Myc antibody and Dr E.D.Gundelfinger for the PSD95 construct. We are also grateful to Professor P.Mangeat and Dr S.Gavarini for critical reading of the manuscript. This work was supported by grants from the CNRS, the Génopôle de Montpellier and the Région Languedoc-Roussillon. Electron microscopy was realized using the facilities at CRIC (Montpellier).
References
- Abramowski D., Rigo,M., Duc,D., Hoyer,D. and Staufenbiel,M. (1995) Localization of the 5-hydroxytryptamine2C receptor protein in human and rat brain using specific antisera. Neuropharmacology, 34, 1635–1645. [DOI] [PubMed] [Google Scholar]
- Bécamel C., Figge,A., Poliak,S., Dumuis,A., Peles,E., Bockaert,J., Lubbert,H. and Ullmer,C. (2001) Interaction of serotonin 5-hydroxytryptamine type 2C receptors with PDZ10 of the multi-PDZ domain protein MUPP1. J. Biol. Chem., 276, 12974–12982. [DOI] [PubMed] [Google Scholar]
- Biederer T. and Südhof,T.C. (2000) Mints as adaptors. Direct binding to neurexins and recruitment of Munc18. J. Biol. Chem., 275, 39803–39806. [DOI] [PubMed] [Google Scholar]
- Biederer T. and Südhof,T.C. (2001) Cask and protein 4.1 support F-actin nucleation on neurexins. J. Biol. Chem., 276, 47869–47876. [DOI] [PubMed] [Google Scholar]
- Borg J.P., Straight,S.W., Kaech,S.M., de Taddeo-Borg,M., Kroon,D.E., Karnak,D., Turner,R.S., Kim,S.K. and Margolis,B. (1998) Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J. Biol. Chem., 273, 31633–31636. [DOI] [PubMed] [Google Scholar]
- Borg J.P., Lopez-Figueroa,M.O., de Taddeo-Borg,M., Kroon,D.E., Turner,R.S., Watson,S.J. and Margolis,B. (1999) Molecular analysis of the X11-mLin-2/CASK complex in brain. J. Neurosci., 19, 1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenman J.E. et al. (1996) Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell, 84, 757–767. [DOI] [PubMed] [Google Scholar]
- Brzostowski J.A. and Kimmel,A.R. (2001) Signaling at zero G: G-protein-independent functions for 7-TM receptors. Trends Biochem. Sci., 26, 291–297. [DOI] [PubMed] [Google Scholar]
- Butz S., Okamoto,M. and Südhof,T.C. (1998) A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell, 94, 773–782. [DOI] [PubMed] [Google Scholar]
- Chen Y., Baez,M. and Yu,L. (1994) Functional coupling of the 5-HT2C serotonin receptor to G proteins in Xenopus oocytes. Neurosci. Lett., 179, 100–102. [DOI] [PubMed] [Google Scholar]
- Christopherson K.S., Hillier,B.J., Lim,W.A. and Bredt,D.S. (1999) PSD-95 assembles a ternary complex with the N-methyl-d-aspartic acid receptor and a bivalent neuronal NO synthase PDZ domain. J. Biol. Chem., 274, 27467–27473. [DOI] [PubMed] [Google Scholar]
- Clemett D.A., Punhani,T., Duxon,M.S., Blackburn,T.P. and Fone,K.C. (2000) Immunohistochemical localisation of the 5-HT2C receptor protein in the rat CNS. Neuropharmacology, 39, 123–132. [DOI] [PubMed] [Google Scholar]
- Cohen A.R., Woods,D.F., Marfatia,S.M., Walther,Z., Chishti,A.H.and Anderson,J.M.(1998) Human CASK/LIN-2 binds syndecan-2 and protein 4.1 and localizes to the basolateral membrane of epithelial cells. J. Cell Biol., 142, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy J., Kan,Y.W., Shohet,S.B. and Mohandas,N. (1986) Molecular cloning of protein 4.1, a major structural element of the human erythrocyte membrane skeleton. Proc. Natl Acad. Sci. USA, 83, 9512–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D.L., Cohen,A.R., Anderson,J.M. and Brunger,A.T. (1998) Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition. Nature Struct. Biol., 5, 317–325. [DOI] [PubMed] [Google Scholar]
- Dev K.K., Nakanishi,S. and Henley,J.M. (2001) Regulation of mglu(7) receptors by proteins that interact with the intracellular C-terminus. Trends Pharmacol. Sci., 22, 355–361. [DOI] [PubMed] [Google Scholar]
- Di Matteo V., De Blasi,A., Di Giulio,C. and Esposito,E. (2001) Role of 5-HT(2C) receptors in the control of central dopamine function. Trends Pharmacol. Sci., 22, 229–232. [DOI] [PubMed] [Google Scholar]
- Ehlers M.D., Fung,E.T., O’Brien,R.J. and Huganir,R.L. (1998) Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J. Neurosci., 18, 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fone K.C., Austin,R.H., Topham,I.A., Kennett,G.A. and Punhani,T. (1998) Effect of chronic m-CPP on locomotion, hypophagia, plasma corticosterone and 5-HT2C receptor levels in the rat. Br. J. Pharmacol., 123, 1707–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin J., Diez,R., Kieffer,S., Dermine,J.F., Duclos,S., Gagnon,E., Sadoul,R., Rondeau,C. and Desjardins,M. (2001) The phagosome proteome: insight into phagosome functions. J. Cell Biol., 152, 165–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A. et al. (1998) The β2-adrenergic receptor interacts with the Na+/H+-exchanger regulatory factor to control Na+/H+ exchange. Nature, 392, 626–630. [DOI] [PubMed] [Google Scholar]
- Hall R.A., Premont,R.T. and Lefkowitz,R.J. (1999) Heptahelical receptor signaling: beyond the G protein paradigm. J. Cell Biol., 145, 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler J.R. and Gilman,A.G. (1992) G proteins. Trends Biochem. Sci., 17, 383–387. [DOI] [PubMed] [Google Scholar]
- Hoover K.B. and Bryant,P.J. (2000) The genetics of the protein 4.1 family: organizers of the membrane and cytoskeleton. Curr. Opin. Cell Biol., 12, 229–234. [DOI] [PubMed] [Google Scholar]
- Husi H. and Grant,S.G. (2001) Proteomics of the nervous system. Trends Neurosci., 24, 259–266. [DOI] [PubMed] [Google Scholar]
- Husi H., Ward,M.A., Choudhary,J.S., Blackstock,W.P. and Grant,S.G. (2000) Proteomic analysis of NMDA receptor–adhesion protein signaling complexes. Nature Neurosci., 3, 661–669. [DOI] [PubMed] [Google Scholar]
- Jo K., Derin,R., Li,M. and Bredt,D.S. (1999) Characterization of MALS/Velis-1, -2, and -3: a family of mammalian LIN-7 homologs enriched at brain synapses in association with the postsynaptic density-95/NMDA receptor postsynaptic complex. J. Neurosci., 19, 4189–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Whitfield,C.W. and Kim,S.K. (1998) The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C.elegans EGF receptor LET-23 in vulval epithelial cells. Cell, 94, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamberov E., Makarova,O., Roh,M., Liu,A., Karnak,D., Straight,S. and Margolis,B. (2000) Molecular cloning and characterization of Pals, proteins associated with mLin-7. J. Biol. Chem., 275, 11425–11431. [DOI] [PubMed] [Google Scholar]
- Kaufman M.J., Hartig,P.R. and Hoffman,B.J. (1995) Serotonin 5-HT2C receptor stimulates cyclic GMP formation in choroid plexus. J. Neurochem., 64, 199–205. [DOI] [PubMed] [Google Scholar]
- Kawai J. et al. (2001) Functional annotation of a full-length mouse cDNA collection. Nature, 409, 685–690. [DOI] [PubMed] [Google Scholar]
- Kim M., Jiang,L.H., Wilson,H.L., North,R.A. and Surprenant,A. (2001) Proteomic and functional evidence for a P2X(7) receptor signalling complex. EMBO J., 20, 6347–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Peters,L.L., Ciciotte,S.L. and Chishti,A.H. (1998) cDNA sequence and chromosomal localization of mouse Dlgh3 gene adjacent to the BRCA1 tumor suppressor locus. Biochim. Biophys. Acta, 1443, 211–216. [DOI] [PubMed] [Google Scholar]
- Lucas G. and Spampinato,U. (2000) Role of striatal serotonin2A and serotonin2C receptor subtypes in the control of in vivo dopamine outflow in the rat striatum. J. Neurochem., 74, 693–701. [DOI] [PubMed] [Google Scholar]
- Lucki I., Ward,H.R. and Frazer,A. (1989) Effect of 1-(m-chlorophenyl)piperazine and 1-(m-trifluoromethylphenyl)piperazine on locomotor activity. J. Pharmacol. Exp. Ther., 249, 155–164. [PubMed] [Google Scholar]
- Milligan G. and White,J. (2001) Protein–protein interactions at G-protein-coupled receptors. Trends Pharmacol. Sci., 22, 513–518. [DOI] [PubMed] [Google Scholar]
- Misawa H., Kawasaki,Y., Mellor,J., Sweeney,N., Jo,K., Nicoll,R.A. and Bredt,D.S. (2001) Contrasting localizations of MALS/LIN-7 PDZ proteins in brain and molecular compensation in knockout mice. J. Biol. Chem., 276, 9264–9272. [DOI] [PubMed] [Google Scholar]
- Okamoto M. and Südhof,T.C. (1997) Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J. Biol. Chem., 272, 31459–31464. [DOI] [PubMed] [Google Scholar]
- Scott K. and Zuker,C.S. (1998) Assembly of the Drosophila phototransduction cascade into a signalling complex shapes elementary responses. Nature, 395, 805–808. [DOI] [PubMed] [Google Scholar]
- Shen L., Liang,F., Walensky,L.D. and Huganir,R.L. (2000) Regulation of AMPA receptor GluR1 subunit surface expression by a 4.1N-linked actin cytoskeletal association. J. Neurosci., 20, 7932–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M. and Sala,C. (2001) PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci., 24, 1–29. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Simske J.S., Kaech,S.M., Harp,S.A. and Kim,S.K. (1996) LET-23 receptor localization by the cell junction protein LIN-7 during C.elegans vulval induction. Cell, 85, 195–204. [DOI] [PubMed] [Google Scholar]
- Takakuwa Y. (2000) Protein 4.1, a multifunctional protein of the erythrocyte membrane skeleton: structure and functions in erythrocytes and nonerythroid cells. Int. J. Hematol., 72, 298–309. [PubMed] [Google Scholar]
- Tochio H., Mok,Y.K., Zhang,Q., Kan,H.M., Bredt,D.S. and Zhang,M. (2000) Formation of nNOS/PSD-95 PDZ dimer requires a preformed β-finger structure from the nNOS PDZ domain. J. Mol. Biol., 303, 359–370. [DOI] [PubMed] [Google Scholar]
- Ullmer C., Schmuck,K., Figge,A. and Lubbert,H. (1998) Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett., 424, 63–68. [DOI] [PubMed] [Google Scholar]
- Verhage M. et al. (2000) Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science, 287, 864–869. [DOI] [PubMed] [Google Scholar]
- Walensky L.D. et al. (1999) A novel neuron-enriched homolog of the erythrocyte membrane cytoskeletal protein 4.1. J. Neurosci., 19, 6457–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte S., Villalba,M., Bi,K., Liu,Y., Isakov,N. and Altman,A. (2000) Inhibition of the c-Jun N-terminal kinase/AP-1 and NF-κB pathways by PICOT, a novel protein kinase C-interacting protein with a thioredoxin homology domain. J. Biol. Chem., 275, 1902–1909. [DOI] [PubMed] [Google Scholar]
- Xu J., Casella,J.F. and Pollard,T.D. (1999) Effect of capping protein, CapZ, on the length of actin filaments and mechanical properties of actin filament networks. Cell. Motil. Cytoskel., 42, 73–81. [DOI] [PubMed] [Google Scholar]
- Zitzer H., Richter,D. and Kreienkamp,H.J. (1999a) Agonist-dependent interaction of the rat somatostatin receptor subtype 2 with cortactin-binding protein 1. J. Biol. Chem., 274, 18153–18156. [DOI] [PubMed] [Google Scholar]
- Zitzer H., Honck,H.H., Bachner,D., Richter,D. and Kreienkamp,H.J. (1999b) Somatostatin receptor interacting protein defines a novel family of multidomain proteins present in human and rodent brain. J. Biol. Chem., 274, 32997–33001. [DOI] [PubMed] [Google Scholar]