Abstract
Cajal (coiled) bodies are conserved subnuclear organelles that are present in the nucleoplasm of both animal and plant cells. Although Cajal bodies were first described nearly 100 years ago, their function has remained largely speculative. Here, we describe a novel class of human small nuclear RNAs that localize specifically to Cajal bodies. The small Cajal body- specific RNAs (scaRNAs) are predicted or have already been demonstrated to function as guide RNAs in site-specific synthesis of 2′-O-ribose-methylated nucleotides and pseudouridines in the RNA polymerase II-transcribed U1, U2, U4 and U5 spliceosomal small nuclear RNAs (snRNAs). Our results provide strong support for the idea that the Cajal body, this mysterious nuclear organelle, provides the cellular locale for post-transcriptional modification of spliceosomal snRNAs.
Keywords: Cajal body/RNA modification/scaRNA/spliceosomal snRNA/snoRNA
Introduction
In eukaryotic cells, biogenesis of cytoplasmic mRNAs, tRNAs, rRNAs, small nuclear RNAs (snRNAs) and nucleolar RNAs (snoRNAs) takes place in the nucleus. The interphase nucleus contains at least a dozen morphologically distinct subdomains, called nuclear bodies, which are believed to function in the synthesis, processing, modification or transport of various cellular RNAs. However, with the exception of the nucleolus, which has long been known as the site of the synthesis and maturation of cytoplasmic rRNAs, the precise function of nuclear bodies remained largely elusive (Lamond and Earnshaw, 1998; Matera, 1999; Lewis and Tollervey, 2000). Besides the nucleolus, the Cajal (coiled) body is the most prominent and most extensively studied nucleoplasmic organelle (Matera, 1998; Gall, 2000). Cajal bodies are enriched with spliceosomal and nucleolar ribonucleoproteins (snRNPs and snoRNPs) and components of the basal transcription machinery. But, despite the rapidly growing list of its molecular components, the precise function of the Cajal body is still unknown.
The removal of intronic sequences of precursor mRNAs (pre-mRNAs) is catalysed by the spliceosome, a dynamic assembly of five snRNAs and numerous protein factors. The U1, U2, U4, U5 and U6 spliceosomal snRNAs play a fundamental role in pre-mRNA splicing: they select the correct splice sites and orchestrate formation of the active spliceosome (Burge et al., 1999; Will and Lührmann, 2001). The spliceosomal snRNAs contain many post-transcriptionally synthesized 2′-O-ribose-methylated nucleotides and pseudouridines (Reddy and Busch, 1988; Massenet et al., 1998). These modified nucleotides are confined to the functionally important regions of snRNAs, which are known to interact with pre-mRNAs, other snRNAs or spliceosomal proteins. Most likely, the modified nucleotides facilitate formation of the specific and dynamic molecular interactions required for the correct function of the spliceosome. Consistent with this view, modification of the U2 snRNA is fundamental for the assembly of the active spliceosome (Pan and Prives, 1989; Ségault et al., 1995; Yu et al., 1998).
The mechanism responsible for the site-specific synthesis of modified nucleotides in snRNAs has long remained an enigma. Recently, it has been established that the nucleolus contains an enormous number of snoRNAs that direct 2′-O-methylation and pseudouridylation of rRNAs (Smith and Steitz, 1997; Kiss, 2001). The 2′-O-methylation guide snoRNAs carry the conserved 5′-terminal C (RUGAUGA) and 3′-terminal D (CUGA) boxes as well as imperfect internal copies of these elements, called C′ and D′ boxes. To select the correct methylation site, sequences preceding the D or D′ box of the snoRNA form a 10–21 bp helix with rRNAs (Cavaillé et al., 1996; Kiss-László et al., 1996). The pseudouridylation guide snoRNAs fold into a consensus ‘hairpin– hinge–hairpin–tail’ structure and contain the conserved H (ANANNA) and ACA boxes (Balakin et al., 1996; Ganot et al., 1997b). They possess bipartite rRNA recognition motifs, which form two short helices with ribosomal sequences preceding and following the substrate uridine (Ganot et al., 1997a; Ni et al., 1997). The conserved box C/D and H/ACA motifs play an essential role in snoRNA-guided RNA modification reactions as well as in the processing and nucleolar accumulation of snoRNAs. The C/D and H/ACA boxes accomplish these jobs through binding of specific snoRNP proteins. Thus far, four box C/D (fibrillarin, Nop56p, Nop58p and Snu13p) and four box H/ACA (Gar1p, dyskerin, Nhp2p and Nop10p) snoRNA-associated proteins have been identified (Kiss, 2001). Most likely, fibrillarin catalyses the box C/D snoRNA-guided 2′-O-methylation reaction (Tollervey et al., 1993; Wang et al., 2000) and dyskerin is the pseudouridine synthase in box H/ACA snoRNPs (Lafontaine et al., 1998; Zebarjadian et al., 1999).
Recent evidence indicates that synthesis of the eight 2′-O-methylated nucleotides and the three pseudouridines in the RNA polymerase (pol) III-synthesized U6 spliceosomal snRNA takes place in the nucleolus and that it is directed by box C/D and H/ACA snoRNAs (Tycowski et al., 1998; Ganot et al., 1999). The U6 snRNA, which is transcribed in the nucleoplasm, is transported to the nucleolus to undergo snoRNA-mediated modification (Lange and Gerbi, 2000). Less is known about modification of the pol II-transcribed U1, U2, U4 and U5 snRNAs. Following nucleoplasmic synthesis, these RNAs are transported to the cytoplasm to associate with seven Sm proteins and undergo cap hypermethylation and 3′-end formation (Will and Lührmann, 2001). The newly assembled snRNPs are re-imported to the nucleoplasm, where they accumulate in interchromatin granule clusters (Lamond and Ernshow, 1998; Sleeman and Lamond, 1999b; Matera, 1999; Lewis and Tollervey, 2000). However, a fraction of snRNPs accumulates transiently in Cajal bodies (Carmo-Fonseca et al., 1992; Matera and Ward, 1993; Spector 1993; Sleeman and Lamond, 1999a) and, under certain conditions, in the nucleolus (Lyon et al., 1997; Sleeman and Lamond, 1999a; Lange and Gerbi, 2000; Yu et al., 2001), raising the possibility that these nuclear bodies might have a function in the biogenesis, trafficking or storage of snRNPs.
Microinjection experiments demonstrated that internal modification of the U2 snRNA occurs in the nucleus rather than in the cytoplasm of Xenopus oocytes (Yu et al., 2001). The same experiments favoured the nucleolus as the intranuclear locale for U2 modification. This conclusion was seemingly supported by identification of the U85 box C/D–H/ACA composite guide RNA that functions in both 2′-O-methylation and pseudouridylation of the U5 snRNA (Jády and Kiss, 2001). In this study, we have characterized six novel putative guide RNAs that are predicted to direct 2′-O-methylation and pseudouridylation of the RNA pol II-specific U1, U2, U4 and U5 snRNAs. Unexpectedly, the new guide RNAs as well as the previously characterized U85 RNA accumulate specifically in Cajal bodies and therefore they comprise a novel class of small nuclear RNAs, called the small Cajal body-specific RNAs (scaRNAs). These results indicate that the Cajal body functions in the post-transcriptional modification of pol II-specific spliceosomal snRNAs.
Results
Identification of novel composite box C/D–H/ACA RNAs
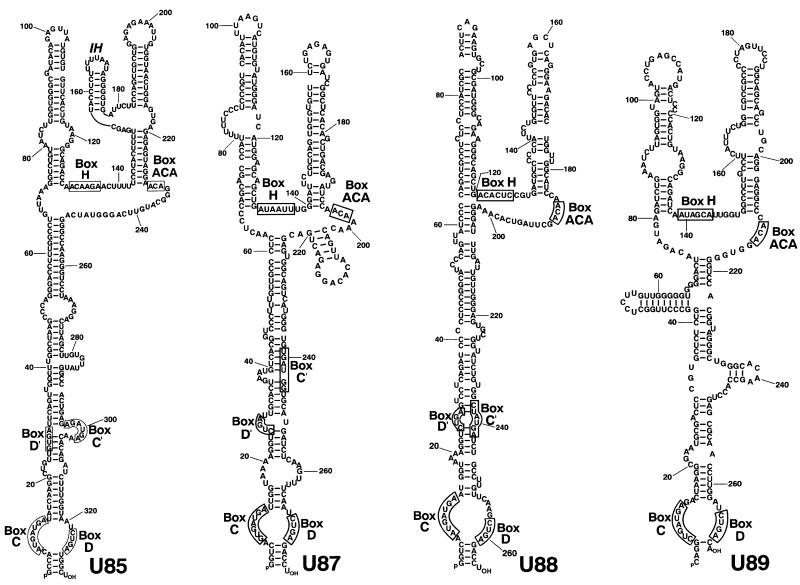
We have recently characterized an unusual modification guide RNA, the U85 RNA, which functions both in 2′-O-methylation and pseudouridylation of the U5 snRNA (Jády and Kiss, 2001). The U85 RNA is composed of a box C/D and a box H/ACA snoRNA-like domain (Figure 1), and is associated with both fibrillarin and Gar1p, which are specific protein components of box C/D and box H/ACA snoRNPs, respectively. To identify novel box C/D–H/ACA composite RNAs, human HeLa RNAs were immunoprecipitated with anti-fibrillarin antibody and size-fractionated on a sequencing gel. RNAs migrating between labelled DNA markers of 234 and 368 nucleotides were recovered and used as a template for cDNA synthesis (Kiss-László et al., 1996). In brief, the 5′ and 3′ termini of the purified RNAs were tagged with a phosphorylated oligonucleotide by using T4 RNA ligase. The double-tagged RNAs were amplified by RT–PCR and cloned into a plasmid vector. Sequence analyses of several individual clones identified three putative snoRNAs, called U87, U88 and U89, which possessed C and D box motifs, but otherwise lacked significant similarity to any known RNAs (Figure 1). Northern blot analyses confirmed that the new RNAs are expressed in HeLa cells (data not shown). The genomic copies of the new RNAs were found within introns of putative protein-coding genes, indicating that they are generated by intron processing (see legend to Figure 1).
Fig. 1. Computer-predicted two-dimensional structures of human box C/D–H/ACA snoRNAs. The evolutionarily conserved box C, D, H, ACA and the putative box C′ and D′ sequence motifs are boxed. The structure of the U85 snoRNA has been adopted from Jády and Kiss (2001). The genomic copies of the U85 and U89 snoRNAs are in the fourth and sixth introns of the structural maintenance of chromatin (D63880) and the B cell-associated protein (U75511) genes, respectively. The U87 and U88 snoRNAs are encoded within the ninth and twelfth introns of a putative protein-coding gene (BC000061).
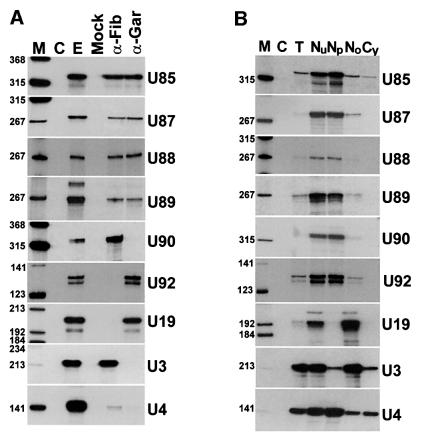
A computer-aided structural analysis revealed that the U87, U88 and U89 RNAs can be folded into a two-dimensional structure that is highly reminiscent of the architecture of the U85 box C/D–H/ACA RNA characterized previously (Figure 1). In each RNA, the 5′- and 3′-terminal regions carrying the C and D boxes can form a long stem structure, whereas the middle region folds into the consensus ‘hairpin–hinge–hairpin–tail’ structure of box H/ACA snoRNAs. Moreover, the single-stranded hinge and tail regions carry potential box H and ACA sequences, respectively. To assess whether the novel putative box C/D–H/ACA snoRNAs are associated with both box C/D and H/ACA snoRNP proteins, immunoprecipitation experiments were performed from a human HeLa cell extract using anti-fibrillarin and anti-GAR1 antibodies (Figure 2A). RNase A/T1 protection analyses demonstrated that the U87, U88 and U89 RNAs, like the U85 box C/D–H/ACA RNA, were precipitated by both antibodies. Please notice that immunological properties of the human U90 and U92 RNPs, although tested in this experiment, will be discussed later. As expected, the U19 box H/ACA snoRNP was precipitated only by the anti-GAR1 and the U3 box C/D snoRNP was recognized only by the anti-fibrillarin antibody and neither of the two antibodies reacted with the U4 spliceosomal snRNP. We concluded that the newly identified U87, U88 and U89 RNAs, together with U85, comprise a new group of snoRNAs that are composed of a box C/D and a H/ACA snoRNA domain, and are associated with both box C/D and box H/ACA snoRNP proteins.
Fig. 2. Characterization of the novel RNAs. (A) Immunoprecipitation of human snoRNPs. An extract prepared from HeLa cells was incubated with protein A–Sepharose saturated with anti-fibrillarin (α-Fib) or anti-GAR1 (α-Gar) antibodies. Upon collection of Sepharose beads by centrifugation, RNAs were recovered by proteinase K treatment and phenol extraction. Distribution of RNAs was determined by RNase A/T1 mapping using sequence-specific antisense RNA probes as indicated on the right. Control mappings performed with E.coli tRNA (C) or RNAs obtained from the HeLa cell extract (E) are also shown. Mock represents control reaction with protein A–Sepharose alone. Lane M, size markers (terminally labelled HaeIII- and TaqI-digested pBR322). (B) Intracellular localization. RNA isolated either from HeLa cells (T), or from nuclear (Nu), nucleoplasmic (Np), nucleolar (No) or cytoplasmic (Cy) fractions of HeLa cells were analysed by RNase A/T1 mapping using sequence-specific RNA probes. Lane C represents control mapping with E.coli tRNA.
Box C/D–H/ACA RNAs can direct post-transcriptional modification of pol II-specific spliceosomal snRNAs
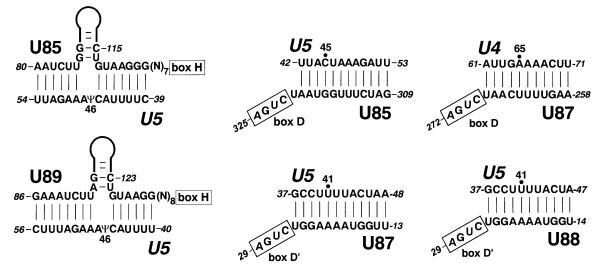
Examination of the sequences of the new box C/D–H/ACA RNAs showed that they lack significant sequence complementarity to rRNA sequences, making it unlikely that they function in rRNA modification. We have recently demonstrated that the human U85 box C/D–H/ACA RNA directs 2′-O-methylation of the C45 and pseudouridylation of the U46 residues in the U5 spliceosomal snRNA (Figure 3). We therefore investigated whether the novel box C/D–H/ACA RNAs could also function in snRNA modification. We found that sequences preceding the D and the putative D′ boxes of the U87 RNA could, in principle, direct 2′-O-methylation of the A65 and U41 residues in the U4 and U5 snRNAs, respectively (Figure 3). Indeed, both of these nucleotides are known to be 2′-O-methylated in vertebrate U4 and U5 snRNAs (Reddy and Busch, 1988; Massenet et al., 1998). Like U87, the U88 RNA was also predicted to function in 2′-O-methylation of the U41 residue in the U5 snRNA, although apart from the putative U5 recognition elements, the two RNAs show no significant sequence similarity. Likewise, the U89 RNA and the previously characterized U85 RNA also seem to share a common function, namely directing pseudouridylation of the U46 residue in the U5 snRNA. Again, the U85 and U89 RNAs, disregarding the short U5 recognition motifs, possess no overall sequence similarity. Our observations strongly suggest that the box C/D–H/ACA RNAs function in the modification of pol II-specific spliceosomal snRNAs and that synthesis of 2′-O-methylated nucleotides and pseudouridines in snRNAs is frequently achieved by functionally redundant snoRNPs.
Fig. 3. Potential base-pairing interactions between human box C/D–H/ACA RNAs and spliceosomal snRNAs. Nucleotides predicted to be selected for 2′-O-methylation (closed circle) or pseudouridylation (Ψ) in the human snRNAs are indicated. The 5′-terminal hairpins of the box H/ACA snoRNA-like domains of the U85 and U89 RNAs are schematically represented. Positions of the H, D and D′ boxes are indicated.
Human U85 RNA accumulates in Cajal bodies
So far, all small RNAs carrying the box C/D or H/ACA motifs have been found to accumulate in the nucleolus (Smith and Steitz, 1997; Tollervey and Kiss, 1997; Weinstein and Steitz, 1999). In fact, the conserved C/D and H/ACA boxes are the key determinants of the nucleolar targeting of snoRNAs (Lange et al., 1998; Samarsky et al., 1998; Narayanan et al., 1999a, b). Therefore, the U85, U87, U88 and U89 box C/D–H/ACA RNAs were assumed to accumulate in the nucleolus. However, during exploration of cDNA libraries of human nucleoplasmic or nucleolar RNAs, we noticed that partial or full-length cDNAs of the U85 RNA frequently appeared in the nucleoplasmic library, but were not found in the nucleolar one (our unpublished results). To test the significance of this unexpected observation, human HeLa cells were fractionated into nuclear, nucleoplasmic, nucleolar and cytoplasmic fractions, and distribution of the U85 RNA was determined by RNase A/T1 mapping (Figure 2B). To our surprise, the U85 RNA was found mostly in the nucleoplasmic fraction. As expected, the U19 box H/ACA and the U3 box C/D snoRNAs showed a predominantly nucleolar accumulation, while the U4 spliceosomal snRNA appeared mostly in the nucleoplasmic fraction.
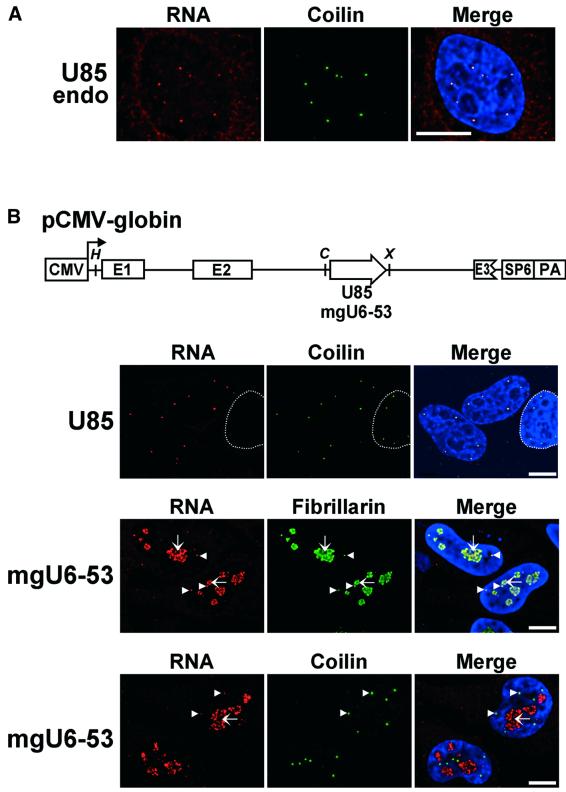
The fascinating discovery that the U85 box C/D–H/ACA RNA co-purifies with the nucleoplasmic fraction of HeLa cells prompted us to examine further the localization of this RNA using fluorescent in situ hybridization microscopy (Figure 4A). Probing of HeLa cells with a U85-specific fluorescent antisense RNA revealed that the U85 RNA, instead of showing a uniform distribution, localizes to a few sharp, dot-like structures in the nucleoplasm. Since this distribution pattern of U85 was highly reminiscent of that of Cajal bodies (Matera, 1998; Gall, 2000), the cells were also stained with an antibody directed against p80 coilin, a protein marker of the Cajal body (Andrade et al., 1991). The U85 RNA and p80 coilin co-localized perfectly, demonstrating that the nucleoplasmic dots revealed by the U85 probe correspond to Cajal bodies.
Fig. 4. Human U85 box C/D–H/ACA RNA localizes to Cajal bodies. (A) In situ localization of the endogenous U85 RNA. Human HeLa cells were probed with a fluorescent antisense RNA complementary to the human U85 RNA from position 1 to 76 and with an antibody directed against p80 coilin as indicated above the panels. The merged image shows that the U85 RNA co-localizes with p80 coilin. Scale bar, 10 µm. (B) In situ localization of transiently overexpressed U85 and mgU6-53 RNAs in HeLa cells. The schematic structure of the pCMV-globin expression construct is shown. The exons (E1, E2 and E3) and the polyadenylation site (PA) of the human β-globin gene are indicated. The transcription initiation site of the cytomegalovirus promoter (CMV) is indicated. The coding regions of the human U85 and mgU6-53 RNAs in the second intron of the globin gene are represented by an open arrow. Relevant restriction sites are shown (H, HindIII; C, ClaI; X, XhoI). The pCMV-globin-mgU6-53 plasmid was co-transfected with an expression construct producing a GFP-tagged version of the human fibrillarin protein. The overexpressed U85 and mgU6-53 RNAs were visualized by sequence-specific fluorescent RNA probes and Cajal bodies were specifically stained by anti-p80 coilin. Arrows indicate nucleolar stained structures enriched in mgU6-53 and fibrillarin. Arrowheads point to Cajal bodies, which clearly contain mgU6-53, fibrillarin and p80 coilin. The contour of the nucleus of a non-transfected HeLa cell is highlighted.
To exclude the possibility that a fraction of U85 RNA accumulates outside the Cajal bodies, we investigated the distribution of U85 RNA after overproduction in HeLa cells. To this end, the human U85 RNA gene was inserted into the second intron of the human β-globin gene, which had been placed under the control of the cytomegalovirus (CMV) promoter (Figure 4B). The human U85 RNA was efficiently and correctly expressed in human HeLa and simian COS-7 cells transfected with the pCMV-globin-U85 expression construct (Jády and Kiss, 2001; data not shown). The in situ labelling patterns of the U85 RNA probe and the p80 coilin antibody were compared in transfected HeLa cells (Figure 4B). Upon hybridization with the U85 fluorescent probe, a strong signal was observed in cells that overproduced the U85 RNA. With the same exposure time, endogenous U85 RNA was not detected in the nucleus of non-transfected cells (Figure 4B, circled). Double labelling with anti-p80 coilin clearly showed that even after its overproduction, the U85 RNA accumulated exclusively in Cajal bodies. Since RNase A/T1 mappings failed to detect 5′- and/or 3′-extended precursors of U85 (Jády and Kiss, 2001; data not shown), we concluded that the U85-specific sequences accumulating in the Cajal bodies represent fully processed U85 RNAs. As a control, the human mgU6-53 box C/D snoRNA, which directs 2′-O-methylation of the RNA pol III-transcribed U6 spliceosomal snRNA (Ganot et al., 1999), was also overproduced in HeLa cells. In situ hybridization revealed that the mgU6-53 snoRNA accumulated in large areas of the nucleus, as well as in small dots. It clearly co-localized with a green fluorescent protein (GFP)-tagged version of human fibrillarin, an abundant nucleolar protein that is also present in Cajal bodies (Gall, 2000). Therefore, the mgU6-53 snoRNA accumulated mainly in the nucleolus of transfected cells and, to a smaller extent, in the Cajal bodies, as confirmed by staining with the anti-coilin antibody. This observation was fully consistent with our previous cell fractionation experiments, in which the mgU6-53 snoRNA was localized mainly to the nucleolar fraction of human HeLa cells (Ganot et al., 1999). In summary, we concluded that in both transfected and non-transfected HeLa cells, mature U85 RNA localizes specifically to Cajal bodies.
Box C/D–H/ACA RNAs reside in Cajal bodies
We then investigated the subcellular localization of the newly discovered box C/D–H/ACA RNAs. Cell fractionation experiments revealed that the U87, U88 and U89 RNAs, like U85, accumulate mostly in the nucleoplasmic fraction of HeLa cells (Figure 2B). To facilitate in situ localization, the U87, U88 and U89 RNAs were overexpressed in HeLa and COS-7 cells using the pCMV-globin expression vector as described for the U85 RNA. RNase A/T1 mappings demonstrated that the U87, U88, and U89 RNAs were efficiently and faithfully expressed in transfected HeLa and COS-7 cells (data not shown).
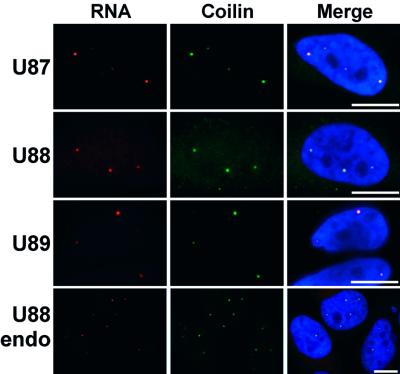
To determine the localization of the overproduced U87, U88 and U89 RNAs in HeLa cells, double labelling experiments were carried out with sequence-specific fluorescent RNA (U87, U89) or oligonucleotide (U88) probes and the anti-p80 coilin antibody (Figure 5). For each RNA, a perfect co-localization was observed with coilin, demonstrating that the overexpressed RNAs accumulate in the Cajal bodies of HeLa cells. The same results were obtained by in situ localization of the transiently expressed human U87, U88 and U89 RNAs in COS-7 cells (data not shown). Moreover, probing of non-transfected HeLa cells with the U88 probe specifically stained Cajal bodies, indicating that endogenous U88 RNA also resides in this nuclear organelle. Our results demonstrate that the human U85, U87, U88 and U89 box C/D–H/ACA RNAs comprise a novel class of cellular RNAs that localize specifically to the Cajal body. The new group of snRNAs was collectively named small Cajal body-specific RNAs (scaRNAs).
Fig. 5. In situ localization of human U87, U88 and U89 box C/D–H/ACA RNAs. Human U87, U88 and U89 RNAs were transiently over expressed in HeLa cells using the pCMV-globin expression construct. The intracellular distribution of the overproduced U87, U88 and U89 RNAs as well as the endogenous HeLa U88 RNA (U88endo) was investigated by fluorescent in situ hybridization. Cajal bodies were visualized by staining with anti-p80 coilin antibody. For other details, see the legend to Figure 4.
Guide RNAs implicated in modification of pol II-specific spliceosomal snRNAs reside in Cajal bodies
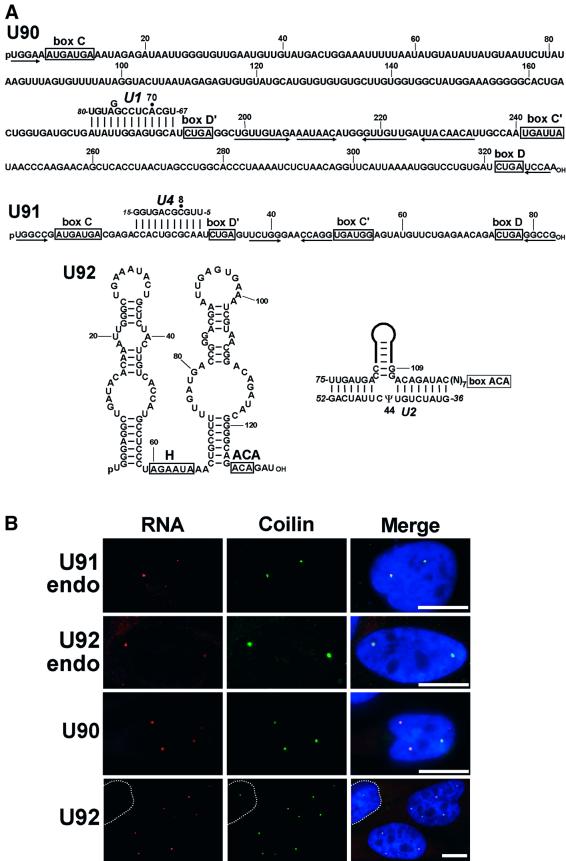
The results presented thus far indicate that scaRNAs directing modification of snRNAs are composed of a box C/D and a H/ACA snoRNA domain. In cDNA libraries of human fibrillarin- and Gar1p-associated snoRNAs, we have identified three novel RNAs, called U90, U91 and U92 (Figure 6A). The U90 and U91 RNAs possess the characteristic core motif of box C/D snoRNAs, which includes the conserved C and D boxes and a short 5′, 3′-terminal helix. Immunoprecipitation experiments demonstrated that the U90 (Figure 2A) and U91 (data not shown) RNAs represent bona fide fibrillarin-associated box C/D snoRNAs and are not associated with the Gar1 box H/ACA snoRNP protein. The U90 and U91 RNAs also carry putative C′ and D′ boxes, which are juxtaposed by short internal helices. This indicates that they probably function in RNA 2′-O-methylation, since positioning the C′ and D′ boxes in close proximity to each other is essential for the box D′-dependent 2′-O-methylation reaction (Kiss-László et al., 1998). Indeed, we found that sequences preceding the D′ boxes of U90 and U91 can position the A70 and C8 residues in the human U1 and U4 snRNAs, respectively, for 2′-O-methylation (Figure 6A). Both of these nucleotides are in fact 2′-O-methylated in mammalian U1 and U4 snRNAs (Reddy and Busch, 1988). In contrast to U90 and U91, the human U92 RNA lacks C and D boxes, but it folds into the consensus ‘hairpin–hinge–hairpin–tail’ structure of box H/ACA snoRNAs (Figure 6A). The U92 RNA binds the Gar1, but not the fibrillarin snoRNP protein (Figure 3A), demonstrating that it is an authentic box H/ACA snoRNA. We noticed that the U92 box H/ACA RNA, in theory, is capable of directing pseurouridylation of the U44 residue in the U2 snRNA (Figure 6A). During the course of this study, a large-scale characterization of mouse non-coding nuclear RNAs revealed partial sequences of the apparent mouse orthologues of the human U91 (MBII-119) and U92 (MBI-57) RNAs, showing that these putative snRNA modification guide RNAs are conserved in mammalian cells (Hüttenhofer et al., 2001).
Fig. 6. Localization of box C/D and H/ACA RNAs predicted to function in modification of pol II-specific spliceosomal snRNAs. (A) Structure and function of human U90, U91 box C/D and U92 box H/ACA RNAs. The conserved box C/D and the potential C′ and D′ motifs of the U90 and U91 RNAs are indicated. Inverted arrows under the sequences of U90 and U91 indicate nucleotides predicted to form terminal and internal helices. The predicted two-dimensional structure of the U92 RNA was obtained by computer folding. The H and ACA sequence motifs are boxed. Predicted base-pairing interactions of the U90, U91 and U92 RNAs with the U1, U4 and U2 snRNAs, respectively, are indicated. The 2′-O-methylated A70 and C8 nucleotides in the selected sequences of the U1 and U4 snRNAs are indicated by dots. The pseudouridine residue (Ψ) at position 44 in the human U2 snRNA is marked. (B) Localization of human U90, U91 and U92 RNAs in HeLa cells. Overexpressed (U90 and U92) and endogenous (U91endo and U92endo) modification guide RNAs were visualized with sequence-specific fluorescent probes. For other details, see legends to Figures 4 and 5.
To assess whether all guide RNAs implicated in modification of pol II-specific snRNAs accumulate in Cajal bodies, we investigated the intracellular localization of the newly identified U90, U91 and U92 RNAs. Encouragingly, cell fractionation experiments revealed a nucleoplasmic localization for the U90 box C/D and the U92 box H/ACA RNAs (Figure 2B). Next, probing of HeLa cells with fluorescent antisense probes specific for the U91 and U92 RNAs resulted in strongly staining dots in the nucleoplasm (Figure 6B, U91endo and U92endo). Double labelling with anti-p80 coilin antibody showed that the dots highlighted by the U91- and U92-specific probes corresponded to Cajal bodies. The U92 RNA, probably due to its low abundance, was not directly detectable in HeLa cells. Therefore, localization of this RNA and also the U90 RNA was investigated in HeLa cells transfected with pCMV-globin constructs expressing the U92 and U90 RNAs. U90 and U92 were correctly and efficiently expressed in transfected HeLa cells (data not shown) and, as in situ localization experiments demonstrated, localized specifically to Cajal bodies (Figure 6B, U90 and U92). We concluded that 2′-O-methylation and pseudouridylation guide RNAs that function in post-transcriptional modification of the RNA pol II-transcribed spliceosomal snRNAs, irrespective of their box elements, associated proteins and molecular sizes, specifically and exclusively accumulate in Cajal bodies.
Discussion
The interphase nucleus has an intricate structural and functional organization. It contains many distinct subdomains, also known as nuclear bodies, which represent specific and dynamic assemblies of protein and RNA factors involved in various aspects of nuclear gene expression. The Cajal body is a prominent and extensively studied nuclear organelle of unknown function (Bohmann et al., 1995; Lamond and Earnshaw, 1998; Matera, 1999; Gall, 2000; Lewis and Tollervey, 2000). Understanding of the function of the Cajal body has been hampered largely by the fact that all its known components are also distributed in the nucleoplasm or, frequently, they concentrate in other nuclear bodies. Thus far, the p80 coilin autoantigene is the only unambiguous molecular marker for the Cajal body, although most of this protein is dispersed in the nucleoplasm. The Cajal body is enriched in spliceosomal snRNPs and nucleolar snoRNPs, but lacks mRNAs and rRNAs, indicating that it does not function in mRNA splicing or rRNA maturation (Bohmann et al., 1995). Several lines of evidence indicate that there is a flux of newly synthesized snRNPs and snoRNPs through the Cajal body, leading to the idea that the Cajal body may function in the biogenesis and/or transport of snRNPs and snoRNPs (Sleeman and Lamond, 1999a; Gall, 2000).
In this report, we have described a novel class of human small nuclear RNAs, scaRNAs, which accumulate specifically in Cajal bodies. Apart from the fact that scaRNAs frequently contain both box C/D and H/ACA snoRNA domains, they are structurally indistinguishable from the canonical box C/D and box H/ACA snoRNAs (Figures 1 and 6A). Depending on their box elements, scaRNAs are associated with characteristic protein components of the nucleolar box C/D and H/ACA snoRNPs (Figure 2A). In situ hybridization experiments demonstrated that the U85 and U87–U92 scaRNAs localize specifically and exclusively to Cajal bodies. The scaRNAs are not detectable outside the Cajal bodies, even after massive overproduction in HeLa and COS-7 cells (Figures 4–6). Therefore, the U85 and U87–U92 scaRNAs represent the first molecular markers of the Cajal body, which partition exclusively to this nuclear body.
The exclusive localization of scaRNAs to the Cajal body suggests that they function in this nuclear organelle. We have demonstrated previously that the U85 scaRNA functions as a guide RNA in 2′-O-methylation and pseudouridylation of the U5 spliceosomal snRNA (Jády and Kiss, 2001). Intriguingly, the newly discovered scaRNAs are also predicted to direct 2′-O-methylation and/or pseudouridylation of the U1, U2, U4 and U5 snRNAs (Figures 3 and 6A). We propose that post-transcriptional modification of pol II-specific spliceosomal snRNAs is directed by a novel class of guide RNAs that reside in the Cajal body. Mammalian U1, U2, U4 and U5 snRNAs together carry 13 2′-O-methyl groups and 21 pseudouridine residues (Reddy and Busch, 1988). In this study, seven scaRNAs have been connected with the synthesis of five 2′-O-methylated nucleotides (U85, U87, U88, U90 and U91) and two pseudouridine residues (U85, U89 and U92) in pol II-specific snRNAs. Currently, we have identified another box H/ACA scaRNA, which is predicted to direct pseudouridylation of the U2 snRNA at the U54 position (our unpublished data). In a previous study, two box C/D (MBII-19 and MBII-382) and two box H/ACA (MBI-57 and MBI-125) RNAs have been implicated in the synthesis of three 2′-O-methylated nucleotides and two pseudouridines in the U2 snRNA (Hüttenhofer et al., 2001). Unfortunately, the cellular localization of these RNAs has not been explored. Nevertheless, thus far, 12 putative guide scaRNAs have been linked with the synthesis of 12 2′-O-methylated nucleotides and two pseudouridines in the U1, U2, U4 and U5 snRNAs. We can envisage that the synthesis of most, if not all, 2′-O-methylated nucleotides and pseudouridines in pol II-specific spliceosomal snRNAs is directed by scaRNAs.
So far, the Cajal body is the only common nuclear locale where both spliceosomal snRNAs and their modification guide RNAs have been demonstrated to accumulate. Therefore, it seems unlikely that the Cajal body is only a storage place for scaRNAs, and scaRNA-directed snRNA modification occurs in another nuclear compartment. Previously, the nucleolus has been implicated in post-transcriptional modification of both the RNA pol III-transcribed U6 (Tycowski et al., 1998; Ganot et al., 1999; Lange and Gerbi, 2000) and the pol II-specific U2 spliceosomal snRNAs (Yu et al., 2001). The idea that modification of the U6 snRNA occurs within the nucleolus is supported by the fact that all trans-acting factors, most likely snoRNPs, that accomplish the synthesis of the eight 2′-O-methylated nucleotides and the three pseudouridines of the U6 snRNA are present and are functionally active in the nucleolus (Tycowski et al., 1998; Ganot et al., 1999; Figure 4B). Upon microinjection into the nucleoplasm of Xenopus oocyte, the U6 snRNA appears transiently in the nucleolus, suggesting that it transits through the nucleolus to undergo snoRNA-directed modification (Lange and Gerbi, 2000). Although maturation of the RNA pol II-synthesized U1, U2, U4 and U5 snRNAs includes a cytoplasmic phase (Will and Lührmann, 2001), internal modification of the pol II-transcribed U2 snRNA occurs in the nucleus (Yu et al., 2001). In Xenopus oocytes, modification of the U2 snRNA was found to require an intact Sm motif (Yu et al., 2001). Lack of a functional Sm site could be complemented by addition of a 5′,3′-terminal box C/D snoRNA core structure. As the resulting chimeric U2–box C/D RNA was efficiently modified and it accumulated in the nucleolus, it was suggested that the nucleolus provides the cellular locale for U2 modification. However, knowing that box C/D snoRNAs appear transiently in Cajal bodies before accumulating in the nucleolus (Samarsky et al., 1998; Narayanan et al., 1999b; see also Figure 4B), these experiments could not rule out the possibility that the modification of the chimeric U2–box C/D RNA happened in Cajal bodies (Yu et al., 2001). Previously, we found that U2 snRNA sequences synthesized within the nucleolus by RNA pol I were not modified, but U6-specific sequences expressed under the same conditions were correctly and efficiently modified (Ganot et al., 1999). This indicated that modification of the U2 and probably other pol II-specific snRNAs, in contrast to U6, occurs outside the nucleolus. Consistently, in situ hybridization experiments failed to provide any evidence supporting accumulation of the U85 and U87–U92 guide RNAs in the nucleolus, while the mgU6-53 box C/D snoRNA directing 2′-O-methylation of the U6 snRNA accumulated mostly in the nucleolus (Figures 4–6).
A notion that the Cajal body provides the cellular locale for modification of pol II-specific snRNAs raises the question of whether the scaRNA-guided modification of snRNAs occurs before or after the cytoplasmic assembly of Sm snRNPs. Upon microinjection into the cytoplasm of Xenopus oocytes, in vitro synthesized U2 RNA accumulates in the nucleoplasm and undergoes pseudouridylation and 2′-O-methylation (Yu et al., 2001). This demonstrates that modification of the U2 snRNA can occur in the nascent Sm snRNP after its re-entry into the nucleus. Indeed, the newly made snRNPs accumulate transiently in Cajal bodies after re-entering the nucleus (Carvalho et al., 1999; Sleeman and Lamond, 1999a). However, we cannot exclude that at least some modifications are introduced into the U2 snRNA before its export to the cytoplasm, since nascent precursor U2 snRNAs have also been reported to appear in Cajal bodies (Smith and Lawrence, 2000).
Since scaRNAs possess all those elements necessary and sufficient to direct the nucleolar accumulation of box C/D and H/ACA snoRNAs (reviewed in Tollervey and Kiss, 1997; Weinstein and Steitz, 1999), the molecular mechanism directing scaRNAs to the Cajal body, rather than to the nucleolus, is conjectural. The U85, U87, U88 and U89 scaRNAs are composed of a box C/D and a box H/ACA snoRNA domain. Therefore, these RNAs, in theory, possess two independent nucleolar localization signals, any of which should be able to direct these RNAs into the nucleolus. Apparently this is not the case. Since box C/D snoRNAs are known to transit through Cajal bodies before accumulating in the nucleolus (Samarsky et al., 1998; Narayanan et al., 1999b), we can hypothesize that the C/D motif targets the box C/D (U90, U91) and the composite box C/D–H/ACA (U85, U87, U88 and U89) scaRNAs to the Cajal body, where a putative retention factor inhibits the nucleolar export of these RNAs. However, identification of the U92 box H/ACA scaRNA clearly demonstrates that RNAs lacking C and D boxes can also accumulate in the Cajal body (Figure 6B). Whether the Cajal body-specific accumulation of the box C/D and box H/ACA scaRNAs is supported by two different mechanisms remains uncertain.
In summary, demonstration that the molecular machinery mediating the 2′-O-methylation and pseudouridylation of the RNA pol II-transcribed U1, U2, U4 and U5 spliceosomal snRNAs is sequestered into the Cajal body sheds new light on the cellular function of this mysterious nucleoplasmic organelle and reveals new details of the spatial organization of the biogenesis of spliceosomal snRNPs. In the future, dissection of the cis-acting elements and identification of the trans-acting factors responsible for the Cajal body-specific accumulation of scaRNAs will provide us with new insights into the molecular mechanism underlying the nuclear compartmentalization of eukaryotic RNA biogenesis.
Materials and methods
General procedures
Unless indicated otherwise, all techniques used for manipulating RNA, DNA and oligonucleotides were according to Sambrook et al. (1989). The following oligonucleotides were used in this study: 1, AATAAAGCGGCCGCGGATCCAA; 2, TTGGATCCGCGGCCGCTTTAT; 3, ATAATCGATGGAAGGTGTTTGTTATC; 4, ATACTCGAGTTTCACTCACTTCTTTC; 5, ATAATCGATAGTCCCACTCCACTCCTGTG; 6, ATACTCGAGGGTGACCAAACCTTTTACCC; 7, ATAATCGATACATCAGTGAATACCTTCTG; 8, ATACTCGAGAACATCAGGACTCCTTATGT; 9, ATAATCGATCTCAGCCCAGCCCCTAGGGC; 10, ATACTCGAGCCTGGCCCTGTCCTTACCAC; 11, ATAATCGATTCTCCATAACAAGCATTAAT; 12, ATACTCGAGTAACTAATAAGTTTTACTCT; 13, ATAATCGATTGGGAGGCTGATACACAAATTGG; 14, ATACTCGAGATCTGTCTGCCCCGTATCTG; 15, ATAATCGATATCTCCCAATGGTACCTGAAC; 16, ATACTCGAGTCAGTCATGATGGAATGGGG; 17, GCCACATGATGATATCAAGGC; 18, GTAATACGACTCACTATAGGGGACCTTTAACAGGCCAAAGG; 19, TAAGTCATGTGTATGGGATC; 20, AATACGACTCACTATAGGGGGTTTGTTGGATACTCGTC; 21, AGATCTGAAATCTTAGTGGT; 22, AATACGACTCACTATAGGGGGCAGCACCAGAAATGAAGGC; 23, GCTGATATTGGAGTGCATCTG; 24, AATACGACTCACTATAGGGGGGTGCCAGGCTAGTTAGGTG; 25, TGGGAGGCTGATACACAAATTGG; 26, AATACGACTCACTATAGGGGGGATCTGTCTGCCCCGTATCTG; 27, TCCCAATGATGAGTTGCC; 28, AATACGACTCACTATAGGGGGACCCCTCAGATCTTCATGTG; 29, CT*GGGATGCCGGGAGGGGAT*CTGAGGACT*CAGACCTTTTACCATT*C; 30, CGGCCT*CAGTCAGTTGT*CAGAAGATACT*CCAT*CACCTGGTTC. Sequences corresponding to the T7 RNA polymerase promoter are underlined. Amino-allyl-modified T residues are marked by asterisks.
Plasmid construction
Synthesis and cloning of cDNAs of human scaRNAs were performed as described by Kiss-László et al. (1996). RNAs immunoprecipitated from a HeLa cell extract by anti-fibrillarin or anti-GAR1 antibodies were size-fractionated on a 6% sequencing gel. The appropriate RNA fractions were recovered and incubated with an excess of 5′-end-phosphorylated oligonucleotide 1 in the presence of T4 RNA ligase. The ligation product was used as a template for cDNA synthesis by using oligonucleotide 2 as a primer and AMV reverse transcriptase. The resulting first strand cDNA was used as a template for PCR amplification by Vent polymerase using oligonucleotides 1 and 2 as primers. The amplified DNA was digested by BamHI and inserted into the same site in pBluescribe (Stratagene).
To obtain pCMV-globin, the HindIII–EcoRI fragment of the human β-globin gene carrying three artificial restriction sites (ClaI, MluI and XhoI) in its second intron was excised from the pGCXM expression vector (Kiss and Filipowicz, 1995) and inserted into the same sites of the pcDNA3 vector (Invitrogen). Removal of the EcoRI–XbaI fragment of the resultant pcDNA3-globin construct yielded pCMV-globin. DNA fragments containing the coding genes of the human U85, U87, U88, U89, U90 and U92 scaRNAs were PCR amplified using human genomic DNA as a template, and oligonucleotides 3/4, 5/6, 7/8, 9/10, 11/12 and 13/14 as primers, respectively. The amplified DNAs were digested with ClaI and XhoI and were inserted into the same sites of pCMV-globin. Likewise, a fragment of the human genome encoding the mgU6-53 snoRNA was amplified (oligonucleotides 15 and 16), and cloned into the ClaI–XhoI sites of pCMV-globin. Transfection of human HeLa and COS-7 cells was performed as described before, except that the transfection reagent Fugene (Roche) was used (Kiss and Filipowicz, 1995).
RNA extraction and analysis
Guanidinium thiocyanate/phenol–chloroform extraction was used to isolate RNA from human HeLa and simian COS-7 cells, and from the nuclear, nucleolar and nucleoplasmic fractions of HeLa cells (Goodall et al., 1990). RNAs were extracted from the cytoplasmic fraction of HeLa cells and the Sepharose beads of immunoprecipitation reactions by proteinase K treatment followed by phenol–chloroform extraction. RNase A/T1 protection assays were performed as described (Goodall et al., 1990). Synthesis of antisense RNA probes for mapping of U4, U3 and U19 RNAs have been described (Ganot et al., 1997b). For synthesis of sequence-specific probes for the U85, U87, U88, U89, U90, U91 and U92 scaRNAs, the appropriate pCMV-globin expression construct was linearized by HindIII and used as template for transcription by the SP6 RNA polymerase. After synthesis, each RNA probe was purified on a 6% sequencing gel.
Immunoprecipitation and cell fractionation
Human HeLa S3 cells were grown in a suspension culture in Joklik’s modified Eagle’s medium (Gibco) containing 5% newborn calf serum. About 5 × 106 cells were washed in ice-cold TBS (150 mM NaCl, 40 mM Tris–HCl pH 7.4), sonicated in 200 mM NaCl, 40 mM Tris–HCl pH 7.4 containing 0.05% Nonidet P-40 as reported by Tyc and Steitz (1989) and centrifuged for 10 min at 10 000 g at 4°C. The supernatant was incubated with 2 mg of protein A–Sepharose beads swollen in the sonication buffer and saturated with anti-fibrillarin (72B9) or anti-hGAR1 antibodies, which were kindly provided by Drs J.A.Steitz and W.Filipowicz, respectively. After immunoprecipitation, the beads were washed four times with 10 vol of sonication buffer. Isolation of nuclei from HeLa cells, and fractionation of nuclei into nucleoplasmic and nucleolar fractions were performed as described (Tyc and Steitz, 1989).
Preparation of probes for in situ hybridization
Synthesis and labelling of antisense RNA probes were performed according to the protocol of Dr R.Singer (http://singerlab.aecom.yu.edu). To produce probes specific for U85, U87, U89, U90, U92 and mgU6-53 RNAs, fragments of these RNAs were amplified by PCR with oligonucleotides 17/18, 19/20, 21/22, 23/24, 25/26 and 27/28, respectively. The 3′-end-specific primers carried the sequence of the T7 RNA polymerase promoter. The resulting DNA fragments were used as templates for in vitro transcription by T7 RNA polymerase in the presence of 5-(3-aminoallyl) uridine 5′-triphosphate. About 2–5 µg of RNA purified on a 6% sequencing gel was resuspended in 70 µl of 0.1 M Na-carbonate buffer pH 8.8 and mixed with 30 µl of dimethylsulfoxide containing a vial of FluoroLink Cy3-monofunctional dye (Amersham). Labelling was performed for 48 h in the dark, at room temperature, with occasional vortexing. The unreacted dye was removed by ethanol precipitation of the RNA. Specific activity of the probes was determined by absorption spectroscopy. To detect U88 and U91, amino-modified antisense oligonucleotides 29 and 30 were synthesized, respectively, and reacted with Cy3 monofunctional dye.
Fluorescent in situ hybridization and image acquisition and processing
HeLa cells were fixed in phosphate-buffered saline (PBS) buffer (100 mM Na2HPO4, 20 mM KH2PO4, 137 mM NaCl, 27 mM KCl pH 7.4) containing 4% formaldehyde for 30 min at room temperature. The cells were rinsed twice with PBS and permeabilized by incubation overnight in 70% ethanol. After rehydration in 2 × SSC (300 mM NaCl, 30 mM sodium citrate pH 7.0) containing 50% formamide, cells were hybridized overnight at 37°C in 40 µl of a mixture containing 10% dextran sulfate, 2 mM vanadyl–ribonucleoside complex, 0.02% RNase-free bovine serum albumin (BSA), 40 µg of Escherichia coli tRNA, 2 × SSC, 50% formamide, 20 ng of labelled probe. Cells probed with fluorescent antisense RNA were washed twice for 30 min in 0.1 × SSC, 50% formamide at 50°C, while oligonucleotide-treated cells were rinsed with 2 × SSC, 50% formamide, at 37°C. p80 coilin was detected with polyclonal rabbit anti-coilin antibody (1/100 dilution, kindly provided by Dr A.Lamond) followed by incubation with anti-rabbit antibodies conjugated to fluorescein (1/300 dilution, Sigma). Slides were incubated for 1 h at 37°C in PBS containing 1% BSA and washed twice for 15 min in PBS at room temperature. Slides were mounted in mounting media containing 90% glycerol, 1 × PBS, 0.1 µg/ml of 4′,6-diamidino-2-phenylindole and 1 mg/ml p-phenylendiamine. Images were acquired on a DMRA microscope equipped for epifluorescence (Leica), and with a CoolSnap camera (Photometrics) controlled by the software Metamorph (Universal Imaging). Images were then pseudo-coloured with Photoshop (Adobe Systems).
Accession numbers
The DDBJ/EMBL/GenBank accession numbers of human U87, U88, U89, U90, U91 and U93 scaRNAs are AY077737, AY77738, AY77739, AY77740, AY77741 and AY77742, respectively.
Acknowledgments
Acknowledgements
We thank M.Weber for critical reading of the manuscript. We are grateful to Y.de Preval for synthesis of oligodeoxynucleotides. X.D. and C.V. were supported by la Fondation pour la Recherche Médicale and Association pour la Recherche contre le Cancer, respectively. B.E.J. and A.M.K. were funded by the French Government and the Hungarian Academy of Sciences. This work was supported by grants from Association pour la Recherche contre le Cancer, la Ligue Nationale contre le Cancer, the French MNRT (ACI) and the Hungarian Research Foundation (OTKA, T29042 and T31738).
References
- Andrade L.E., Chan,E.K., Raska,I., Peebles,C.L., Roos,G. and Tan,E.M. (1991) Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med., 173, 1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- Bohmann K., Ferreira,J., Santama,N., Weis,K. and Lamond,A.I. (1995) Molecular analysis of the coiled body. J. Cell Sci., 19, Suppl., 107–113. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosome. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Carmo-Fonseca M., Pepperkok,R., Carvalho,M.T. and Lamond,A.I. (1992) Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol., 117, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Almeida,F., Calapez,A., Lafarga,M., Berciano,M.T. and Carmo-Fonseca,M. (1999) The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol., 147, 715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J., Nicoloso,M. and Bachellerie,J.P. (1996) Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature, 383, 732–735. [DOI] [PubMed] [Google Scholar]
- Gall J.G. (2000) Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol., 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Ganot P., Bortolin,M.-L. and Kiss,T. (1997a) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997b) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- Ganot P., Jády,B.E., Bortolin,M.-L., Darzacq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G.J., Wiebauer,K. and Filipowicz,W. (1990) Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol., 181, 148–161. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A., Kiefmann,M., Meier-Ewert,S., O’Brien,J., Lehrach,H., Bachellerie,J.P. and Brosius,J. (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády B.E. and Kiss,T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. and Filipowicz,W. (1995) Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev., 9, 1411–1424. [DOI] [PubMed] [Google Scholar]
- Kiss-László Z., Henry,Y., Bachellerie,J.-P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Kiss-László Z., Henry,Y. and Kiss,T. (1998) Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J., 17, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.L., Bousquet-Antonelli,C., Henry,Y., Caizergues-Ferrer,M. and Tollervey,D. (1998) The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev., 12, 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I. and Earnshaw,W.C. (1998) Structure and function in the nucleus. Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Lange T.S. and Gerbi,S.A. (2000) Transient nucleolar localization of U6 small nuclear RNA in Xenopus laevis oocytes. Mol. Biol. Cell, 11, 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T.S., Borovjagin,A., Maxwell,E.S. and Gerbi,S.A. (1998) Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J., 17, 3176–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D. and Tollervey,D. (2000) Like attracts like: getting RNA processing together in the nucleus. Science, 288, 1385–1389. [DOI] [PubMed] [Google Scholar]
- Lyon C.E., Bohmann,K., Sleeman,J. and Lamond,A.I. (1997) Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp. Cell Res., 230, 84–93. [DOI] [PubMed] [Google Scholar]
- Massenet S., Mougin,A. and Branlant,C. (1998) Posttranscriptional modifications in the U small nuclear RNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington DC, pp. 201–227.
- Matera A.G. (1998) Of coiled bodies, gems, and salmon. J. Cell. Biochem., 70, 181–192. [PubMed] [Google Scholar]
- Matera A.G. (1999) Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol., 9, 302–309. [DOI] [PubMed] [Google Scholar]
- Matera A.G. and Ward,D.C. (1993) Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol., 121, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A., Lukowiak,A., Jády,B.E., Dragon,F., Kiss,T., Terns,R.M. and Terns,M.P. (1999a) Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J., 18, 5120–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A., Speckmann,W., Terns,R. and Terns,M.P. (1999b) Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell, 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Tien,A.L. and Fournier,M.J. (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- Pan Z.Q. and Prives,C. (1989) U2 snRNA sequences that bind U2-specific proteins are dispensable for the function of U2 snRNP in splicing. Genes Dev., 3, 1887–1898. [DOI] [PubMed] [Google Scholar]
- Reddy R. and Busch,H. (1988) Small nuclear RNAs: RNA sequences, structure, and modifications. In Birnstiel,M.L. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag, Berlin, pp. 1–37.
- Samarsky D.A., Fournier,M.J., Singer,R.H. and Bertrand,E. (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J., 17, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ségault V., Will,C.L., Sproat,B.S. and Lührmann,R. (1995) In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J., 14, 4010–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E. and Lamond,A.I. (1999a) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol., 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Sleeman J.E. and Lamond,A.I. (1999b) Nuclear organization of pre-mRNA splicing factors. Curr. Opin. Cell Biol., 11, 372–377. [DOI] [PubMed] [Google Scholar]
- Smith K.P. and Lawrence,J.B. (2000) Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: Evidence for preU2 within Cajal bodies. Mol. Biol. Cell, 11, 2987–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- Spector D.L. (1993) Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol., 9, 265–315. [DOI] [PubMed] [Google Scholar]
- Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen,H., Jansen,R., Kern,H. and Hurt,E.C. (1993) Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell, 72, 443–457. [DOI] [PubMed] [Google Scholar]
- Tyc K. and Steitz,J.A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- Wang H., Boisvert,D., Kim,K.K., Kim,R. and Kim,S.H. (2000) Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J., 19, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein L.B. and Steitz,J.A. (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- Yu Y.T., Shu,M.D. and Steitz,J.A. (1998) Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J., 17, 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.T., Shu,M.D., Narayanan,A., Terns,R.M., Terns,M.P. and Steitz, J.A. (2001) Internal modification of U2 small nuclear (sn)RNA occurs in nucleoli of Xenopus oocytes. J. Cell Biol., 152, 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebarjadian Y., King,T., Fournier,M.J., Clarke,L. and Carbon,J. (1999) Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol., 19, 7461–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]