Abstract
Wnt signals regulate differentiation of neural crest cells through the β-catenin associated with a nuclear mediator of the lymphoid-enhancing factor 1 (LEF-1)/T-cell factors (TCFs) family. Here we show the interaction between the basic helix–loop–helix and leucine-zipper region of microphthalmia-associated transcription factor (MITF) and LEF-1. MITF is essential for melanocyte differentiation and its heterozygous mutations cause auditory–pigmentary syndromes. Functional cooperation of MITF with LEF-1 results in synergistic transactivation of the dopachrome tautomerase (DCT) gene promoter, an early melanoblast marker. This activation depends on the separate cis-acting elements, which are also responsible for the induction of the DCT promoter by lithium chloride that mimics Wnt signaling. β-catenin is required for efficient transactivation, but dispensable for the interaction between MITF and LEF-1. The interaction with MITF is unique to LEF-1 and not detectable with TCF-1. LEF-1 also cooperates with the MITF-related proteins, such as TFE3, to transactivate the DCT promoter. This study therefore suggests that the MITF/TFE3 family is a new class of nuclear modulators for LEF-1, which may ensure efficient propagation of Wnt signals in many types of cells.
Keywords: dopachrome tautomerase/LEF-1/melanocyte/MITF/Wnt
Introduction
Transcription factors containing a basic helix–loop–helix and leucine-zipper (bHLH/LZ) structure play critical roles in regulatory networks of many developmental pathways, cell growth and differentiation (Murre et al., 1994). The basic region permits the bHLH/LZ proteins to bind to the E box motif (CANNTG) and the HLH/LZ region allows these proteins to form homodimers and/or heterodimers. Microphthalmia-associated transcription factor (Mitf), encoded by the mouse Mitf gene, belongs to an evolutionarily ancient family of the bHLH/LZ proteins (Atchley and Fitch, 1997). Mitf plays an important role in the differentiation of various cell types, including melanocytes of neural crest origin, optic cup-derived retinal pigment epithelium (RPE), and bone marrow-derived mast cells and osteoclasts (Hodgkinson et al., 1993). In addition, mutations in the MITF gene, the human counterpart of the Mitf gene, are associated with dominantly inherited auditory–pigmentary syndromes, which are characterized by sensorineural hearing loss and abnormal pigmentation of the hair and skin (Tassabehji et al., 1994; Nobukuni et al., 1996; Amiel et al., 1998).
Recent studies have revealed the isoform multiplicity of MITF/Mitf, which could account in part for various phenotypic consequences of MITF/Mitf mutations (reviewed in Yasumoto et al., 1998; Shibahara et al., 1999). In fact, MITF is composed of at least five isoforms with distinct N-termini, MITF-M, -H, -A, -B and -C (Amae et al., 1998; Mochii et al., 1998; Fuse et al., 1999; Udono et al., 2000). These isoforms share the entire downstream region, including the transcriptional activation domain and the bHLH/LZ domain. In addition, MITF shares significant amino acid sequence similarity with transcription factors such as TFE3, TFEB and TFEC, especially in the bHLH/LZ region (Beckmann et al., 1990; Carr and Sharp, 1990; Yasumoto and Shibahara, 1997). MITF-M is expressed specifically in melanocytes and melanoma cells, although other isoforms are expressed in various tissues and cultured cell lines (Amae et al., 1998; Yasumoto et al., 1998; Fuse et al., 1999). In cultured cells, MITF-M transactivates the melanogenesis enzyme genes, tyrosinase and tyrosinase-related protein-1 (TRP-1), through the cis-acting DNA elements containing a CATGTG motif, such as M box (Yasumoto et al., 1994, 1995, 1997; reviewed in Goding, 2000).
Wnt, a group of secretory signaling molecules, evokes a signal to regulate melanocyte differentiation (Patapoutian and Reichardt, 2000). The binding of Wnt to its receptor Frizzled leads to inactivation of glycogen synthase kinase-3β (GSK3β), followed by the accumulation of β-catenin and its translocation to the nucleus. lymphoid-enhancing factor 1 (LEF-1)/T-cell factor (TCF) transcription factors can bind to the β-catenin and the complexes formed transactivate the target genes (Cadigan and Nusse, 1997; Barker et al., 2000). A recent study has shown that injection of β-catenin mRNA into zebrafish embryos increases the population of pigment cells of the neural crest origin (Dorsky et al., 1998). Direct gene transfer of Wnt1 or β-catenin to mouse neural crest cells resulted in melanocyte expansion and differentiation (Dunn et al., 2000). We have shown that exogenously added Wnt-3a protein to cultured murine melanocytes increased the expression of endogenous MITF mRNA and transactivated the melanocyte-specific M promoter of the MITF gene through the LEF-1 site (Takeda et al., 2000). These results suggest that the Wnt signaling pathway regulates the differentiation of melanocytes from neural crest cells by activating the M promoter. In fact, selective requirement of Mitf-M for melanocyte development was verified by the molecular analysis of recessive black-eyed white Mitfmi–bw mice that are deficient in Mitf-M expression (Yajima et al., 1999). Moreover, in zebrafish embryos, Wnt signaling directly activates nacre, a zebrafish MITF homolog, which is required for the formation of neural crest-derived pigment cells (Dorsky et al., 2000). Expression of dopachrome tautomerase (DCT) is almost entirely absent from neural crest cells in nacre–/– embryos, and conversely, misexpression of nacre induced ectopic expression of DCT in wild-type and mutant embryos (Lister et al., 1999). In mice, the DCT gene has been established as an early melanoblast marker (Steel et al., 1992). Taken together, these results suggest that the DCT gene may be a downstream target of Wnt signaling.
Here we show the functional interaction of MITF-M and LEF-1 using the DCT gene promoter, which represents a novel mechanism by which Wnt signaling leads to transcriptional activation of target genes.
Results
Transactivation of the DCT promoter by MITF-M and LEF-1
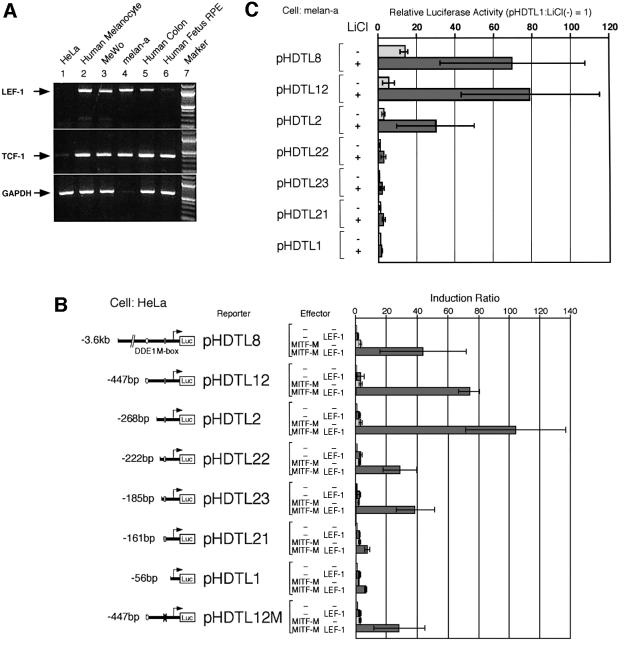
Expression of LEF-1 mRNA was analyzed in various cell types by RT–PCR (Figure 1A). LEF-1 mRNA is expressed in melanin-producing cells, such as melanocytes, melanoma cells and RPE, and in other cell types examined, but not in HeLa cells. The lack of LEF-1 expression in HeLa cells is consistent with the previous report by Giese et al. (1995). In contrast, TCF-1 mRNA was detected in all cell types, but its expression level seems to be lower in HeLa cells. We therefore performed transient cotransfection assays in HeLa cells to analyse the effect of LEF-1 on the human DCT promoter. Both LEF-1 and MITF-M showed no or only marginal effects on DCT promoter activity (Figure 1B), despite the fact that the promoter contains the M box, which is bound by MITF-M (Yasumoto et al., 1997), and a potential LEF-1-binding site in DDE1 (Amae et al., 2000). Unexpectedly, synergistic transactivation of the DCT promoter was observed when LEF-1 and MITF-M were coexpressed. Deletion studies suggest that the two separate regions, –268 to –222 and –185 to –161, are involved in the transactivation by MITF-M and LEF-1. Moreover, the mutation at the M box (–138 to –128) reduced the degree of transactivation by ∼2- to 3-fold.
Fig. 1. Effects of LEF-1 on the DCT promoter activity. (A) Expression profiles of LEF-1 and TCF-1 mRNAs determined by RT–PCR. Note the faint band of GAPDH in mouse melan-a cells. (B) Promoter-context dependent transactivation of the DCT promoter by MITF-M and LEF-1. Each of the DCT reporter plasmids was coexpressed in HeLa cells with MITF-M, LEF-1 or a combination of MITF-M and LEF-1. An enhancer DDE1 (positions –447 to –416) and the M box (positions –138 to –128) are indicated. The magnitude of activation is presented as the ratio of normalized luciferase activity obtained with each plasmid and that with vector DNA (Induction Ratio). The results of at least three independent experiments are shown with standard deviations. (C) Activation of DCT promoter by LiCl. Melan-a cells, maintained in a 6-well plate, were transfected with the indicated constructs (1 µg each of reporter and effector and 0.05 µg of pCDNA3-His-LacZ), incubated for 20 h, and then treated with 30 mM LiCl for 24 h in fresh medium. The data are shown as a ratio to the basal luciferase activity obtained with pHDTL1. Other conditions were the same as in (B).
To explore the role of Wnt signaling in the observed activation of the DCT promoter, we analyzed the effects of LiCl, an inhibitor of GSK3β, which mimics Wnt signaling (Klein and Melton, 1996). For this series of experiments, a mouse melanocyte cell line, melan-a, was used (Bennett et al., 1987), because melan-a cells are able to respond to Wnt signaling (Takeda et al., 2000) and express TCF-1 and LEF-1 mRNA endogenously (Figure 1A). Treatment with LiCl resulted in activation of the DCT promoter through a cis-acting region (–268 to –222) that is also required for the transactivation by MITF-M and LEF-1 (Figure 1B and C). Notably, no activation by LiCl was detected with a construct pHDTL22, carrying the 222-bp promoter region, unlike the effect of coexpression of MITF-M and LEF-1.
We also attempted to identify the core sequence of another region (–185 to –161) (Figure 1B) by using four pHDTL12-derived constructs carrying various base changes that cover the entire 25-bp region. However, we were unable to detect any significant effects of the base changes on the promoter activation by MITF-M and LEF-1. The 25-bp element lacks the E box and the potential binding sites for LEF-1/TCFs, and is not required for activation by LiCl (Figure 1C). This 25-bp element may be less physiologically important, and its function was detected only when the identified cis-regulatory region (–268 to –222) is deleted, as in the case of pHDTL22 (Figure 1B and C).
A cis-acting element that is required for activation by MITF-M and LEF-1
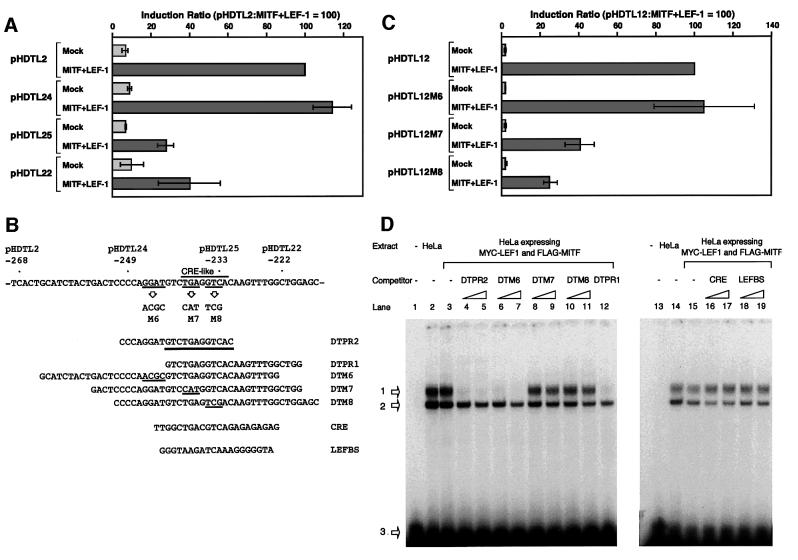
We then identified the cis-acting region (–249 to –233) that is required for the transactivation by cooperation of MITF-M and LEF-1 (Figure 2A and B). This 17-bp region contains a motif similar to a cAMP-responsive element (CRE), termed the CRE-like motif (Bertolotto et al., 1998), but does not contain the potential E box or the binding sites for LEF-1/TCFs. This region is also required for activation by LiCl in melan-a cells (data not shown). Base changes were then introduced into the 17-bp segment of construct pHDTL12 (Figure 2B and C). Synergistic effects of MITF-M and LEF-1 were detected with a mutant construct carrying the intact CRE-like motif but reduced by ∼2.5- to 4-fold with the constructs carrying the base changes at the CRE-like motif. Thus, the 5′-TGA-3′ and 5′-GTC-3′ sequences of the CRE-like motif are required for the activation by MITF-M and LEF-1.
Fig. 2. Identification of the cis-acting element that is required for the activation of DCT promoter by LEF-1 and MITF-M. (A) Deletion studies of the DCT promoter. HeLa cells were cotransfected with the indicated reporter plasmids. (B) The cis-acting region in the DCT promoter. The 12-bp cis-acting element is underlined in DTPR2. Also shown is the strategy for the functional analysis in HeLa cells (C) and EMSAs (D). Base changes shown were introduced into construct pHDTL12. Nuclear extracts of HeLa cells were incubated with a 32P-end labeled DTPR2 in the absence (lanes 2, 3, 14 and 15) or presence of an indicated competitor (200- and 500-fold excesses, shown as triangles). LEFBS represents a LEF-1-binding site. Lanes 1 and 13 represent a control lacking nuclear extracts. Arrows 1 and 2 indicate the specific and unspecific protein–DNA complexes, respectively. Unbound probes are indicated by arrow 3.
We then analyzed whether nuclear proteins bind the 17-bp segment (–249 to –233) by electrophoretic mobility shift assays (EMSAs) (Figure 2D). A synthetic primer DTPR2 of 20 bp was bound by nuclear extracts prepared from the HeLa cells expressing LEF-1 and MITF-M or the mock-transfected cells, and no difference in the DNA-binding activity was detected between these two nuclear extracts (lanes 2 and 3, 14 and 15). Thus, the detected protein–DNA complex did not contain LEF-1 and MITF-M, which are deficient in HeLa cells. The DNA-binding activity was competed for by a competitor DTPR2, DTPR1 or DTM6 (lanes 4–7 and 12), but not by DTM7 (lanes 8 and 9), DTM8 (lanes 10 and 11), a synthetic CRE (lanes 16 and 17), a LEF-1-binding site (lanes 18 and 19) or the M box (data not shown). Thus, a third factor specifically binds the 12-bp element that is shared by DTPR2 and DTPR1, and is different from MITF-M and LEF-1. Similar DNA-binding activity was also detected in melanoma nuclear extracts (data not shown). It is noteworthy that the DNA-binding activity was not competed for by a synthetic CRE, suggesting that this CRE-like motif-binding protein may be different from CRE-binding proteins.
Functional cooperation between MITF-M and LEF-1
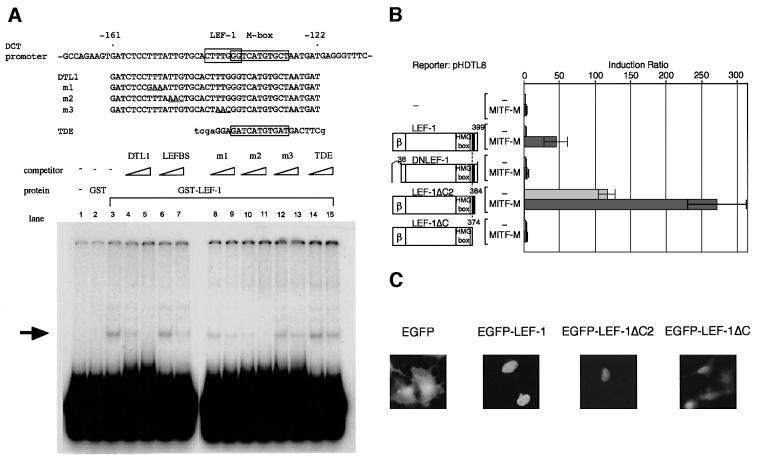
We next searched for the LEF-1-binding site in the DCT promoter region. The deletion study suggested that the downstream region (–161 to –56) is also required for the transactivation by LEF-1 and MITF-M (data not shown). Consequently, using several synthetic fragments covering the relevant region as EMSA probes, we have identified the CTTTGGG sequence (–143 to –137) as a LEF-1-binding site (Figure 3A); namely, this element was specifically bound by the full-length LEF-1 fused to glutathione S-transferase (GST). The LEF-1-binding activity of this element was also confirmed with nuclear extracts of HeLa cells expressing LEF-1 (data not shown).
Fig. 3. Functional cooperation of MITF-M with LEF-1. (A) EMSAs showing the LEF-1-binding site of the DCT promoter. GST–LEF-1 fusion protein was incubated with a 32P-end labeled DTL1 in the absence (lane 3) or presence of indicated competitors (200- and 500-fold excesses, shown as triangles). Lanes 1 and 2 represent a buffer control and GST control. TDE represents the distal enhancer of the tyrosinase gene that is bound by MITF-M (Yasumoto et al., 1994). An arrow indicates the specific protein–DNA complex. (B) Domains of LEF-1 required for the DCT promoter activation. HeLa cells were cotransfected with a DCT reporter plasmid, pHDTL8, and indicated LEF-1 plasmids. Shown are the N-terminal β-catenin-binding domain (β) and the NLS near the C-terminus (closed box). Reporter luciferase activity obtained was normalized with each β-galactosidase activity that represents an internal control. The magnitude of activation is presented as the ratio of normalized luciferase activity obtained with each effector plasmid and that with vector DNA. (C) Subcellular localization of EGFP–LEF-1 fusion proteins in COS-7 cells, as assessed by fluorescence.
To search for a domain of LEF-1 that is required for transactivation of the DCT promoter, we first analyzed the involvement of the N-terminus of LEF-1, which contains the β-catenin-binding domain (amino acid residues 2–37) (Behrens et al., 1996; Molenaar and van de Wetering, 1996). The degree of transactivation was reduced when MITF-M was coexpressed with dominant-negative LEF-1 (DNLEF-1), which lacks the β-catenin-binding domain (Figure 3B). It is noteworthy that DNLEF-1 shows a weak but significant activation of the DCT promoter with MITF-M (∼5-fold in Figure 3B). Moreover, β-catenin alone did not noticeably activate the DCT promoter (data not shown), but co-expression of β-catenin with LEF-1 and MITF-M significantly enhanced the synergistic activation of the DCT promoter by LEF-1 and MITF-M (4.13 ± 0.40-fold). Thus, β-catenin is involved in the efficient cooperation of LEF-1 with MITF-M on the DCT promoter.
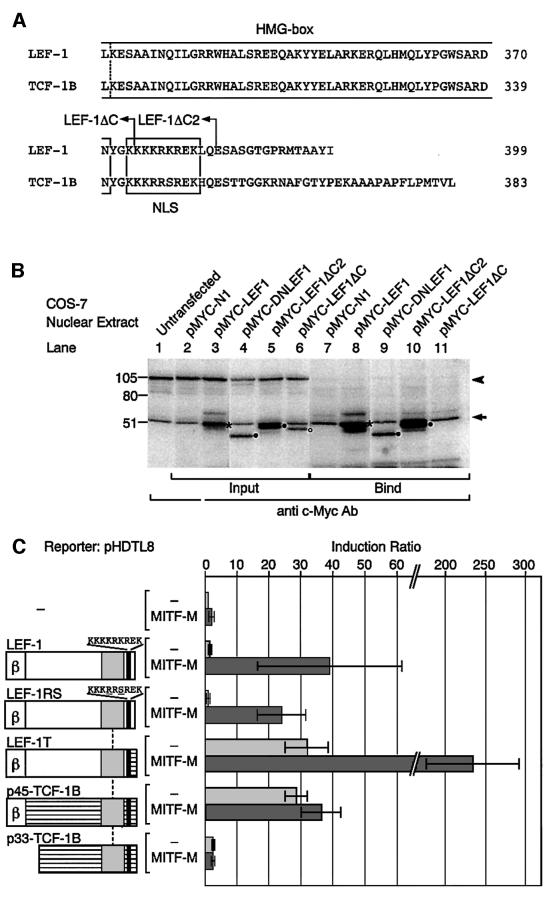
The C-terminal portion of LEF-1 was also analyzed (Figure 3B). LEF-1ΔC lacks the C-terminal 25 amino acid residues (positions 375–399), and LEF-1ΔC2 lacks the C-terminal 15 residues (385–399) but retains the nuclear localization signal (NLS) (Figure 4A). Surprisingly, LEF-1ΔC2 by itself transactivated the DCT promoter and the degree of transactivation by LEF-1ΔC2 was further enhanced by MITF-M (Figure 3B). LEF-1ΔC2, fused to enhanced green fluorescent protein (EGFP), was specifically targeted to the nucleus (Figure 3C). In contrast, no activation was detected with LEF-1ΔC, which lacks the NLS, despite its noticeable expression in the nucleus. In fact, ∼34% of expressed tagged-LEF-1ΔC were detected in the nuclear extracts (Figure 4B). Thus, the C-terminal domain of 15 residues, deleted in LEF-1ΔC2, appears to repress the function of LEF-1. In addition, the 10 residues (positions 375–384), which are deleted in LEF-1ΔC, are required for the functional cooperation with MITF-M or with certain endogenous coactivators. The lack of transactivation by LEF-1ΔC is consistent with the crucial role for the NLS, which is known to be responsible for DNA binding (Carlsson et al., 1993).
Fig. 4. Interaction of MITF-M with the C-terminal region of LEF-1. (A) Alignment of the C-termini. Broken line represents the position where LEF-1 cDNA was replaced by TCF-1B cDNA. (B) Western blot analysis of the c-Myc-tagged LEF-1 proteins bound to MITF-M. Tagged LEF-1 (asterisks) comigrated with endogenous c-Myc, present in COS-7 nuclear extracts (lanes 1–11), indicated by an arrow. The unspecific signal of 105 kDa is indicated by an arrowhead (lanes 1–6). Mutant LEF-1 proteins are shown by closed circles and LEF-1ΔC, an open circle. (C) Functions of LEF-1–TCF-1 chimeric proteins in HeLa cells. Other conditions were the same as in Figure 2A.
Physical interactions between MITF-M and LEF-1
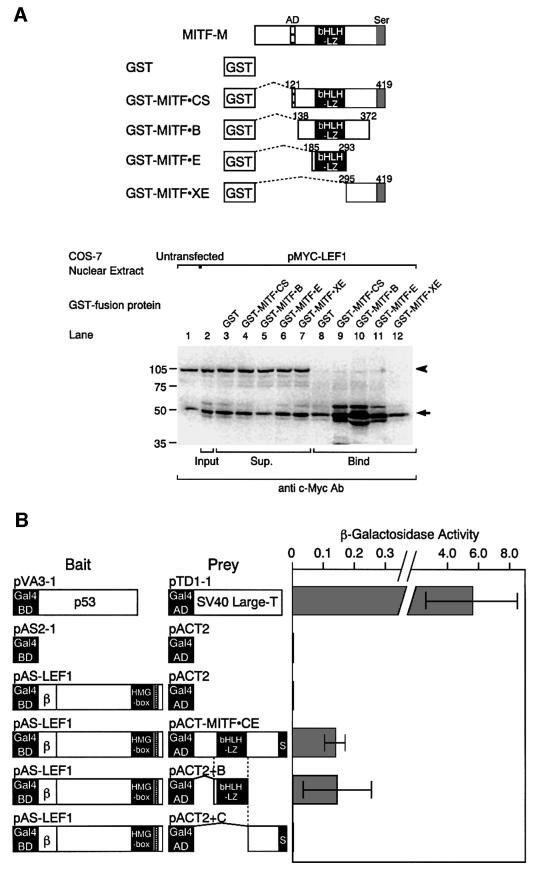
We next examined the physical interaction between MITF-M and the C-terminus of LEF-1 by pull-down assays. Both LEF-1 and TCF-1B possess an identical HMG box and share a similar NLS (van de Wetering et al., 1996) (Figure 4A). The mobility of c-Myc-tagged LEF-1 was similar to that of the endogenous c-Myc protein, and was detected as enhanced signals in the nuclear extracts (Figure 4B, lanes 1–3). Tagged LEF-1 mutants, including LEF-1ΔC lacking the NLS, were detected in the nuclear extracts at the expected sizes (lanes 4–6). LEF-1 and DNLEF-1 were also detected in the bound fraction (lanes 8 and 9). Thus, LEF-1 and DNLEF-1 can bind MITF-M, indicating that the β-catenin interaction domain is not required for the interaction with MITF-M. Moreover, MITF-M was able to bind LEF-1ΔC2 retaining the NLS (lane 10), but not LEF-1ΔC lacking the NLS (lane 11).
To confirm the functional significance of the C-terminus of LEF-1, we analyzed the effects of TCF-1B on the DCT promoter (Figure 4C). Interestingly, p45-TCF-1B, a full-length isoform of TCF-1, transactivated the DCT promoter, but no enhancing effects were observed when coexpressed with MITF-M. A dominant-negative isoform of TCF-1B (DNTCF-1B), p33-TCF-1B, lacking the β-catenin-binding domain (van de Wetering et al., 1996), showed no noticeable effects on the DCT promoter. These results suggest that the functional cooperation with MITF-M is unique to LEF-1 and is not a general feature of TCFs. In addition, TCF-1B transactivates the DCT promoter by cooperating with β-catenin but not with MITF-M. Accordingly, we analyzed the functions of LEF-1–TCF-1B chimeric proteins. LEF-1RS contains the NLS of TCF-1B instead of the LEF-1 NLS, and LEF-1T contains the C-terminus of TCF-1B. The function of LEF-1RS is indistinguishable from that of LEF-1, despite the two amino acid differences in their NLSs. Interestingly, like p45-TCF-1B, LEF-1T transactivated the DCT promoter, but unlike p45-TCF-1B, LEF-1T did cooperate with MITF-M. Thus, the C-terminus of LEF-1, deleted in LEF-1T, may function as a repression domain, which is consistent with the activation of the DCT promoter by LEF-1ΔC2 (see Figure 3). In addition, the NLS of TCF-1B is able to mimic the function of the LEF-1 NLS, indicating that the interaction with MITF-M depends on the middle portion of LEF-1, located between the β-catenin interaction domain and the HMG box.
Functional difference between LEF-1 and TCF-1B
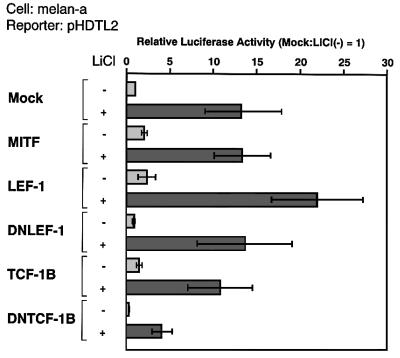
We analyzed the effect of a dominant-negative form of LEF-1 or TCF-1B on the basal promoter and the LiCl-mediated activation of the DCT promoter in melan-a cells (Figure 5). In this melanocyte cell line, MITF-M, LEF-1 or TCF-1B only marginally transactivated the DCT promoter activity (∼2-fold). These results suggest that MITF-M and LEF-1 could interact with endogenous LEF-1 and MITF-M, respectively. Interestingly, DNTCF-1B, but not DNLEF-1, significantly reduced the DCT promoter activity (∼3-fold). Thus, TCF-1B may compete for the binding site with endogenous LEF-1. Moreover, remarkably, only DNTCF-1B reduced the LiCl-mediated activation of the DCT promoter. These results suggest that both LEF-1 and TCF-1B share the binding sites but the mechanism by which TCF-1B transactivates the DCT promoter is different from that by LEF-1, which is consistent with our proposal that MITF-M is able to interact with LEF-1 and even with DNLEF-1, but not with TCF-1B.
Fig. 5. Functional difference between LEF-1 and TCF-1B. Melan-a cells were transfected with pHDTL2 and the indicated constructs, and then treated for 24 h with 30 mM LiCl. Other conditions were the same as in Figure 1C.
The bHLH/LZ region of MITF-M as an interacting domain with LEF-1
We then localized the domain of MITF-M, which is required for the interaction with LEF-1, by pull-down assays (Figure 6A). Tagged LEF-1 was detectable in the fractions bound to truncated MITF-M proteins containing the bHLH/LZ domain (lanes 9–11), whereas no tagged LEF-1 was detected in the fraction bound to GST (lane 8) or truncated MITF-M carrying only its C-terminal region (lane 12). These results indicate that the bHLH/LZ domain of MITF-M is required for the interaction with LEF-1.
Fig. 6. Interaction of the bHLH/LZ region of MITF-M with LEF-1. (A) In vitro interaction between LEF-1 and the bHLH/LZ domain of MITF. COS-7 nuclear extracts contained endogenous c-Myc (lanes 1–12), as indicated by an arrow. Tagged LEF-1 was detected as enhanced signals in the fractions, bound to truncated MITF-M proteins containing the bHLH/LZ domain (lanes 9–11). An arrowhead indicates the unspecific protein binding (lanes 1–7). (B) Interaction between LEF-1 and the bHLH/LZ region of MITF-M in yeast cells.
To confirm the involvement of the bHLH/LZ region in the interaction with LEF-1 in vivo, we next performed yeast two-hybrid experiments (Figure 6B). LEF-1 was chosen as bait, because a strong transactivation domain is located near the N-terminal region of MITF-M (Sato et al., 1997) and LEF-1 itself does not act as a transcriptional activator in yeast cells (Prieve et al., 1998). When the full-length LEF-1 plasmid was introduced into yeast cells together with the plasmid, carrying a large portion of MITF-M, significant β-galactosidase activity was detected. Similarly, β-galactosidase activity was detected with a plasmid, carrying only the bHLH/LZ region. In contrast, no β-galactosidase activity was detected with the parent vector or the plasmid, containing the C-terminal region of MITF-M. These results support the notion that the bHLH/LZ region of MITF-M is responsible for the association with LEF-1 in vivo.
Effects of mutations in the bHLH/LZ region on the interaction with LEF-1
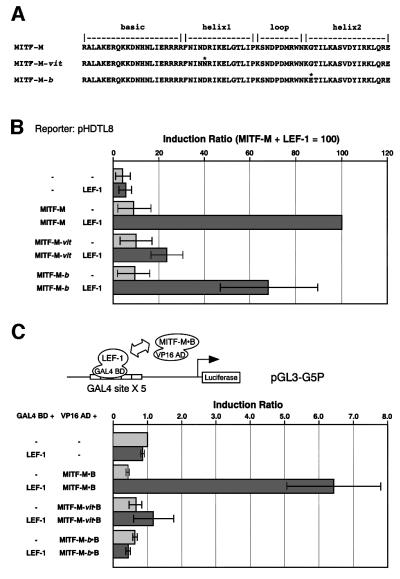
To confirm the crucial role for the bHLH/LZ region of MITF-M in the interaction with LEF-1, we introduced an Asp222Asn substitution in helix 1 and a Gly244Glu substitution in helix 2, respectively (Figure 7A). These amino acid substitutions represent molecular lesions of the two Mitf mutant mice, recessive Mitfvitiligo (Mitfvit) (Lerner et al., 1986; Steingrímsson et al., 1994) and semidominant Mitfbrownish (Mitfb) (Steingrímsson et al., 1996). Both mutant mice are associated with pigmentary defects and late-onset retinal degeneration. Asp222 and Gly244 are predicted to lie on the outside face of the bHLH/LZ domain dimer (Steingrímsson et al., 1994) and to lie near the protein–DNA interface (Steingrímsson et al., 1996), respectively. In fact, the Mitf-Mvit protein was shown to bind in vitro to DNA as either a homodimer or a heterodimer (Hemesath et al., 1994), whereas Mitf-Mb protein possesses greatly reduced DNA-binding activity but retains its dimerization potential (Steingrímsson et al., 1996). Consistent in part with these properties, the tyrosinase promoter, a well-known target for MITF-M, was transactivated in HeLa cells by MITF-M (16.3 ± 8.7-fold), MITF-Mvit protein (5.4 ± 2.3-fold), and MITF-Mb protein (1.9 ± 0.6-fold), respectively. Western blot analysis confirmed the expression of similar amounts of wild-type and mutant MITF-M proteins in transfected HeLa cells (data not shown).
Fig. 7. Identification of a crucial amino acid in the bHLH/LZ region of MITF-M for protein interaction. (A) The amino acid substitutions in the bHLH/LZ region (asterisks). (B) Effects on functional cooperation. HeLa cells were cotransfected with a DCT-luciferase reporter plasmid (pHDTL8), LEF-1 and the indicated MITF-M proteins. The results of three independent experiments are shown with standard deviations. (C) Effects on physical interaction. Two-hybrid assays were performed in 293 human embryonic kidney cells using the fusion proteins containing a bHLH/LZ region of MITF-M or its mutant proteins.
Unexpectedly, MITF-Mvit protein and MITF-Mb protein showed differential effects on the cooperative activation of the DCT promoter by LEF-1 (Figure 7B). The degree of synergistic activation by LEF-1 and MITF-Mvit protein or MITF-Mb protein was ∼25 or 70% of the control value obtained by the combination of LEF-1 and MITF-M, respectively. MITF-Mb protein may act on the DCT promoter as a non-DNA-binding cofactor for LEF-1 because it is deficient in the DNA-binding activity. In this context, the vit and b mutations did not noticeably impair the in vitro interaction of mutant MITF proteins with LEF-1, as judged by the pull-down assays (data not shown). These results suggest that the mutations may differentially influence the interaction between MITF-M, LEF-1, a CRE-like motif-binding protein, and other factors, such as CBP/p300, which were reported to associate with MITF-M (Sato et al., 1997) (see Figure 9).
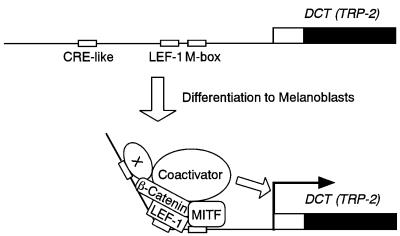
Fig. 9. Proposed model for transcriptional activation of the DCT gene. The relevant cis-acting elements are shown schematically. Note that a CRE-like motif-binding protein (X) is endogenously expressed in HeLa cells and melanoma cells (see Figure 2D and the relevant text).
The functional consequences of the vit and b mutations were also assessed by mammalian two-hybrid assays (Figure 7C). The bHLH/LZ region of MITF-M (residues 138–372; see Figure 6A) was fused to the transactivation domain of VP16 protein, and LEF-1 was fused to the GAL4 DNA-binding domain. A reporter luciferase gene was under the control of the promoter containing five copies of the GAL4 DNA-binding site. The expressed luciferase activities were near the background levels when the MITF–VP16 fusion protein carries the vit or b mutation. Thus, these mutations impair the interaction between the bHLH/LZ region and LEF-1 on the GAL4 promoter, probably due to the profound conformational changes of the fusion protein or the lack of the CRE-like motif in the GAL4 promoter.
The bHLH/LZ region as a common interacting domain with LEF-1
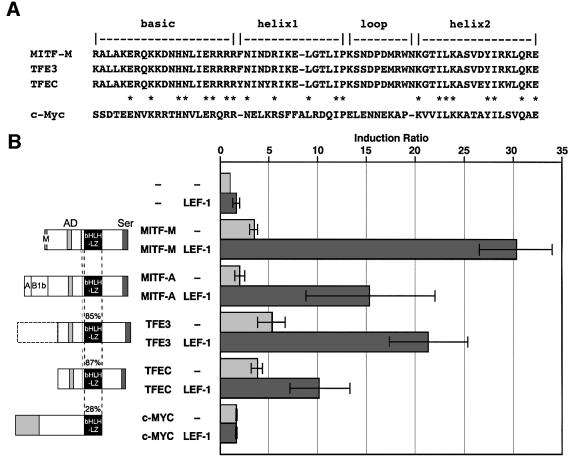
To assess whether interaction with LEF-1 is a general feature of bHLH/LZ proteins, we examined effects of other bHLH/LZ transcription factors, including TFE3, TFEC and c-Myc (Watt et al., 1983) (Figure 8A). MITF-A, an isoform of MITF-M, possesses a different N-terminus from MITF-M, but shares the same bHLH/LZ region. The bHLH/LZ regions of TFE3 and TFEC show >85% identity with that of MITF-M. Every combination of LEF-1 either with MITF-A, TFE3 or TFEC was able to activate the DCT gene promoter, whereas c-Myc exerted no noticeable effects (Figure 8B). These results suggest a novel role for MITF/TFE3 proteins as downstream modulators of Wnt signaling. Thus, multiple MITF isoforms and other family members, such as TFE3, may be involved in efficient propagation of Wnt signals in certain cell types, depending on the expression levels of a given MITF isoform or other family member.
Fig. 8. Functional cooperation of LEF-1 with MITF-related proteins. (A) Comparison of the bHLH/LZ regions. (B) Cooperation of LEF-1 with various bHLH/LZ proteins. The amino acid identity is shown as a percentage of the MITF bHLH/LZ region. HeLa cells were cotransfected with pHDTL8 and the indicated combination of LEF-1 and various bHLH/LZ proteins. An equal amount of MITF constructs was used (2 µg each), and the total amounts of plasmid DNA were maintained at 4 µg with the vector DNA (pRc/CMV). The data are presented as the ratio of normalized luciferase activity obtained with each combination and that obtained with vector DNA.
Discussion
Here we provide evidence for a novel mechanism by which MITF regulates gene transcription through LEF-1, thereby ensuring efficient propagation of Wnt signals in melanocytes. Thus, MITF-M could function in melanocytes as a target, as well as a nuclear effector of Wnt signaling. Moreover, we have shown that functional cooperation of MITF-M and LEF-1 requires a hitherto unidentified factor that binds the CRE-like motif (–242 to –231) of the DCT promoter (summarized in Figure 9). The functional importance of this 12-bp motif is also supported by the presence of the same sequence at the equivalent position of the mouse DCT gene promoter (Budd and Jackson, 1995). In this context, DCT mRNA expression is upregulated by forskolin, a cAMP-elevating agent, in human melanoma cells (Udono et al., 2001) and mouse melanoma cells (Bertolotto et al., 1998), but not in human retinoblastoma cells (Udono et al., 2001). Future studies will be aimed at exploring the role of cAMP for the observed functional cooperation of MITF-M, LEF-1 and the CRE-like motif-binding protein.
DCT is characterized by its early expression in migrating melanoblasts of mouse embryos (Steel et al., 1992) as well as in certain types of human tumors, such as retinoblastoma (Udono et al., 2001) and glioblastoma (Suzuki et al., 1998). In mouse embryos, the onset of LEF-1 mRNA expression is detected at 7.5 embryonic days (Oosterwegel et al., 1993), which precedes the onset of Mitf expression (∼9.5–10.5 days) (Nakayama et al., 1998) and DCT mRNA expression (∼10 days) (Steel et al., 1992). Thus, the expression profiles of DCT, LEF-1 and Mitf mRNAs are consistent in part with our proposal that DCT expression is directed by the functional cooperation of MITF-M and LEF-1.
DNLEF-1 could interact with MITF-M, leading to small activation of the DCT promoter (Figures 3B and 4B, lane 9). Thus, β-catenin is required for efficient activation of the DCT promoter, but dispensable for the interaction with MITF-M. This notion is of physiological significance, because even in the absence of Wnt signals, LEF-1 by itself could cooperate with MITF-M to maintain DCT gene transcription. DCT has been considered to play an important role in detoxification of melanin precursors (Steel et al., 1992). Taken together, we assume that Wnt signaling initially induces MITF-M expression, and then the expressed MITF-M cooperates with LEF-1 to initiate and maintain transcription of the DCT gene.
At least two separate regions of LEF-1 are required for the interaction with MITF-M: the middle portion located between the β-catenin-binding domain and the HMG box, and the NLS. The middle portion of LEF-1 is known as the context-dependent activation domain and is not conserved in TCF-1B. Interestingly, nuclear transport proteins bind to the NLS of LEF-1 but not TCF-1B (Prieve et al., 1998). These results suggest that the middle portion of LEF-1 may profoundly influence the protein-binding potential of LEF-1 NLS.
The observed functional difference between LEF-1 and TCF-1 may account for the complex phenotypes of LEF-1-deficient mice (van Genderen et al., 1994), compared with TCF-1-deficient mice (Verbeek et al., 1995), despite their overlapping expression profiles during embryonic development (Oosterwegel et al., 1993). The LEF-1–/– mice lack teeth, hair follicles, whiskers (vibrissae) and trigeminal nerves of neural crest origin, and die shortly after birth, whereas TCF-1–/– mice showed T-cell abnormality but appear healthy and are fertile. It is noteworthy that the LEF-1-deficient mice contain unpigmented melanocytes in the skin. Thus, LEF-1 is essential for hair follicle development and melanin production in melanocytes, but dispensable for melanocyte development during embryogenesis. These results suggest that a certain member of the TCF family, such as TCF-1, may be responsible for Mitf-M expression during fetal development of LEF-1-deficient mice. In fact, TCF-1 mRNA is expressed in human epidermal melanocytes (Figure 1A).
The homozygous Mitfvit mice appear normal when young with uniformly lighter color and congenital white spots, but show ageing-dependent melanocyte loss (Lerner et al., 1986). In addition, plucking hairs promotes the regrowth of amelanotic hairs due to melanocyte loss in the plucked areas. These phenotypes indicate a crucial role of Mitf-M in postnatal maintenance of follicular melanocytes. In fact, expression of Mitf mRNA becomes undetectable in most tissues, except for follicular melanocytes, in which Mitf mRNA expression is maintained even in adult mice (Nakayama et al., 1998). Here we suggest that the vit mutation may impair the formation of a stable complex on the DCT promoter involving MITF-M, LEF-1 and other factors (Figure 9), which leads to the defect in Wnt signal transduction in follicular melanocytes. In contrast, the b mutation does not severely affect such a protein–protein interaction on the DCT promoter, probably due to the property of MITF-Mb protein as a non-DNA-binding cofactor for LEF-1. The latter notion is consistent in part with the results that the base changes at the M box, the binding site for MITF-M, resulted in only a 2-fold reduction in the synergistic activation (see Figure 1B).
Materials and methods
RT–PCR analysis
Total RNA was prepared from culture cells and tissues as described (Amae et al., 1998), and the first strand cDNA was synthesized using oligo(dT)12–18 primer. A portion of the reverse transcription mixture was subjected to PCR (35 cycles of 30 s at 95°C, 30 s at 59°C and 2 min at 72°C) using AmpliTaq Gold DNA polymerase (PE Applied Biosystems). The PCR primers used were: 5′-gggatgTTCGCCGAGATCAGTCATCC-3′ and 5′-cggtacctgGATGTAGGCAGCTGTCATTC-3′ for LEF-1 (Waterman et al., 1991); 5′-GTTCACCCACCCATCCTTGATGC-3′ and 5′-CAGCCTGGGTATAGCTGCATGTG-3′ for TCF-1 (van de Wetering et al., 1996); and 5′-CCACCCATGGCAAATTCCATGGCA-3′ and 5′-TCTAGACGGCAGGTCAGGTCCACC-3′ for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The lower cases in these primers indicate additional nucleotides.
Plasmid construction
MITF expression plasmids, pRc/CMV-MITF-M and pRc/CMV-MITF-A, were described previously (Yasumoto et al., 1994; Amae et al., 1998). FL9B, a mammalian expression plasmid, contains the full-length human LEF-1 cDNA (Waterman et al., 1991). TCF-1B cDNAs were gifts from H.Clevers (van de Wetering et al., 1996). Reporter plasmids contain the firefly luciferase gene, linked to the 5′-flanking region of the human DCT gene (Yokoyama et al., 1994). pENL, a β-galactosidase expression vector, was used as an internal control for the transfection efficiency. LEF-1 cDNA and its deletion mutants were inserted into pEGFP-N1, encoding enhanced EGFP (Clontech) for the analysis of subcellular localization. These LEF-1 proteins are fused to GFP at their C-termini. LEF-1–TCF-1 chimeric cDNAs were constructed by replacing the C-terminal portion of LEF-1 by that of TCF-1B. Detailed procedures for construction of various DCT reporter plasmids, truncated LEF-1 mutants and chimeric proteins are available on request. Human c-Myc cDNA was a gift from M.Obinata and was cloned in a mammalian expression vector pRc/CMV (Invitrogen). All constructions used were confirmed by sequencing.
Cell cultures and transfection
HeLa human uterine cervical cancer cells and COS-7 monkey kidney cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS). Melan-a murine immortalized melanocytes were grown in Minimum Essential Medium supplemented with 10% fetal calf serum and 200 nM phorbol 12-myristate 13-acetate (Bennett et al., 1987). HeLa cells were transfected with each fusion plasmid and a β-galactosidase expression plasmid by the calcium phosphate precipitation method (Yasumoto et al., 1997). The amount of reporter DNA was kept at 4 µg and the total amount of DNA was kept constant (usually 9.4 µg/60-mm dish). At 29 h post-transfection, cells were harvested, and luciferase activity was measured with a PicaGene luciferase assay system (Toyo Ink) and a Lumat LB9507 (Berthold). The luciferase activity was normalized with each β-galactosidase activity that represents an internal control. The magnitude of activation is presented as the ratio of normalized luciferase activity and that with a vector DNA. The results of at least three independent experiments are shown with standard deviations. Melan-a cells and COS-7 cells were transfected using FuGENE 6 transfection reagent (Roche).
HeLa cells were transfected with an equal amount of LEF-1 and each bHLH/LZ protein construct (2 µg each), and the total amounts of plasmid DNA were maintained at 4 µg with the vector DNA (pRc/CMV). Expression vectors for TFE3 (Beckmann et al., 1990) and TFEC were constructed as described previously (Yasumoto and Shibahara, 1997). The data are presented as the ratio of normalized luciferase activity obtained with each combination and with vector DNA.
EMSA
Nuclear extracts were prepared from HeLa cells untransfected or transfected with MITF-M and LEF-1 by the method of Schreiber et al. (1989). EMSA was performed as described previously (Yasumoto et al., 1995), except that the binding reaction consisted of 12 mM HEPES–NaOH pH 7.9, 80 mM NaCl, 0.6 mM EDTA, 0.6 mM EGTA, 12% glycerol, 2 mM MgCl2 and 0.1 mg/ml poly(dIdC). GST–LEF-1 fusion protein was prepared as described previously for the GST–MITF-M fusion protein (Yasumoto et al., 1995) and used for EMSA.
In vitro protein–protein interactions
LEF-1 and its truncated proteins were fused to a c-Myc epitope tag at their C-termini. In vitro binding studies were performed using GST–MITF immobilized on the GST–Sepharose resin and COS-7 nuclear extracts, containing c-Myc-tagged LEF-1. COS-7 cells (5 × 106) were transfected with 8 µg of a LEF-1 expression vector and harvested 42 h post-transfection. GST–MITF fusion proteins were purified on GST– Sepharose 4B resin (Amersham-Pharmacia), according to the manufacturer’s instructions. The resin was preincubated with non-transfected COS-7 nuclear extracts at 4°C for 1 h to reduce the non-specific binding. Nuclear extracts of transfected COS-7 cells (300 µg of protein) were added to 30 µl of GST–MITF resin suspension and diluted with buffer C (20 mM HEPES pH 7.9, 133 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 20% glycerol and 0.1% NP-40) to adjust the protein concentration to 1 µg/µl. The sample was then incubated at 4°C for 90 min. The resin was washed with 700 µl of buffer C four times, and a final suspension of 10 µl was applied to SDS–PAGE. c-Myc-tagged LEF-1 was detected by western blot analysis with anti c-Myc antibody (Santa Cruz Biotechnology).
Yeast two-hybrid assay
Yeast two-hybrid assay was performed using MATCHMAKER Two-Hybrid System 2 (Clontech), according to the manufacturer’s instructions. LEF-1 was chosen as a bait, because LEF-1 itself does not act as a transcriptional activator in yeast cells (Prieve et al., 1998) and a strong transactivation domain is located near the N-terminal region of MITF-M (Sato et al., 1997). LEF-1 cDNA and portions of MITF-M cDNA fragments were inserted into pAS2-1, encoding the GAL4 DNA-binding domain, and pACT2, encoding the GAL4 transactivation domain, respectively.
Mammalian two-hybrid assay
To assess the effect of mutation in the bHLH/LZ region of MITF-M on the interaction with LEF-1, we performed mammalian two-hybrid assay using Mammalian MATCHMAKER Two-Hybrid Assay Kit (Clontech). The DNA segment, containing five consensus GAL4-binding sites and an adenovirus E1b minimal promoter region, was isolated from pG5CAT vector and inserted into the multiple cloning site of pGL3-Basic (Promega), generating a reporter plasmid pGL3-G5P. Full-length human LEF-1 cDNA was inserted in the pM cloning vector, generating pM-LEF-1 that codes for the LEF-1 fused to GAL4 DNA-binding domain. The bHLH/LZ region (amino acids 138–372) of MITF-M or its mutant (MITF-Mvit or MITF-Mb) was fused to an activation domain derived from the VP16 protein of herpes simplex virus. The blunt-ended BamHI fragment, containing each bHLH/LZ region, was inserted at the blunt-ended SalI site of pVP16 vector. The resulting plasmids pVP-MI·wt·B, pVP-MI·vit·B and pVP-MI·b·B encode the fusion proteins MITF-M·B MITF-M-vit·B and MITF-M-b·B, respectively (see Figure 7C). 293 human embryonic kidney cells were grown in Eagle’s medium α-modification supplemented with 10% FBS. 293 cells were transfected using FuGENE 6 transfection reagent. The amount of total DNA was kept at 4 µg per 60 mm dish (1 µg of reporter and 1.5 µg each of effector DNA). Transfected cells were then incubated for 43 h at 37°C, and were harvested for luciferase assay.
Acknowledgments
Acknowledgements
We thank K.A.Jones, H.Clevers, T.Kadesch and M.Obinata for cDNA materials, and M.Yoshizawa for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research (B), for Exploratory Research, and for Encouragement of Young Scientist (to K.Y.) from the Ministry of Education, Science, Sports and Culture of Japan. This work was also supported in part by the grants provided by Uehara Memorial Foundation, Ichiro Kanehara Foundation, the Kao Foundation for Arts and Sciences, and the Cosmetology Research Foundation.
References
- Amae S. et al. (1998) Identification of a novel isoform of microphthalmia-associated transcription factor that is enriched in retinal pigment epithelium. Biochem. Biophys. Res. Commun., 247, 710–715. [DOI] [PubMed] [Google Scholar]
- Amae S., Yasumoto,K., Takeda,K., Udono,T., Takahashi,K. and Shibahara,S. (2000) Identification of a composite enhancer of the human tyrosinase-related protein 2/DOPAchrome tautomerase gene. Biochim. Biophys. Acta, 1492, 505–508. [DOI] [PubMed] [Google Scholar]
- Amiel J., Watkin,P.M., Tassabehji,M., Read,A.P. and Winter,R.M. (1998) Mutation of the MITF gene in albinism–deafness syndrome (Tietz syndrome). Clin. Dysmorphol., 7, 17–20. [PubMed] [Google Scholar]
- Atchley W.R. and Fitch,W.M. (1997) A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl Acad. Sci. USA, 94, 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Morin,P.J. and Clevers,H. (2000) The yin-yang of TCF/ β-catenin signaling. Adv. Cancer Res., 77, 1–24. [DOI] [PubMed] [Google Scholar]
- Beckmann H., Su,L.-K. and Kadesch,T. (1990) TFE3: a helix– loop–helix protein that activates transcription through the immunoglobulin enhancer µE3 motif. Genes Dev., 4, 167–179. [DOI] [PubMed] [Google Scholar]
- Behrens J., von Kries,J.P., Kühl,M., Bruhn,L., Wedlich,D., Grosschedl, R. and Birchmeier,W. (1996) Functional interaction of β-catenin with the transcription factor LEF-1. Nature, 382, 638–642. [DOI] [PubMed] [Google Scholar]
- Bennett D.C., Cooper,P.J. and Hart,I.R. (1987) A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int. J. Cancer, 39, 414–418. [DOI] [PubMed] [Google Scholar]
- Bertolotto C., Buscà,R., Abbe,P., Bille,K., Aberdam,E., Ortonne,J.-P. and Ballotti,R. (1998) Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: pivotal role of M boxes (GTCATGTGCT) and of microphthalmia. Mol. Cell. Biol., 18, 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd P.S. and Jackson,I.J. (1995) Structure of the mouse tyrosinase-related protein-2/dopachrome tautomerase (Tyrp2/Dct) gene and sequence of two novel slaty alleles. Genomics, 29, 35–43. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M. and Nusse,R. (1997) Wnt signaling: a common theme in animal development. Genes Dev., 11, 3286–3305. [DOI] [PubMed] [Google Scholar]
- Carlsson P., Waterman,M.L. and Jones,K.A. (1993) The hLEF/TCF-1α HMG protein contains a context-dependent transcriptional activation domain that induces the TCRα enhancer in T cells. Genes Dev., 7, 2418–2430. [DOI] [PubMed] [Google Scholar]
- Carr C.S. and Sharp,P.A. (1990) A helix–loop–helix protein related to the immunoglobulin E box-binding proteins. Mol. Cell. Biol., 10, 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky R.I., Moon,R.T. and Raible,D.W. (1998) Control of neural crest cell fate by the Wnt signalling pathway. Nature, 396, 370–373. [DOI] [PubMed] [Google Scholar]
- Dorsky R.I., Raible,D.W. and Moon,R.T. (2000) Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev., 14, 158–162. [PMC free article] [PubMed] [Google Scholar]
- Dunn K.J., Williams,B.O., Li,Y. and Pavan,W.J. (2000) Neural crest-directed gene transfer demonstrates Wnt1 role in melanocyte expansion and differentiation during mouse development. Proc. Natl Acad. Sci. USA, 97, 10050–10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse N. et al. (1999) Molecular cloning of cDNA encoding a novel microphthalmia-associated transcription factor isoform with a distinct amino-terminus. J. Biochem., 126, 1043–1051. [DOI] [PubMed] [Google Scholar]
- Giese K., Kingsley,C., Kirshner,J.R. and Grosschedl,R. (1995) Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein–protein interactions. Genes Dev., 9, 995–1008. [DOI] [PubMed] [Google Scholar]
- Goding C.R. (2000) Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev., 14, 1712–1728. [PubMed] [Google Scholar]
- Hemesath T.J. et al. (1994) microphthalmia, a critical factor in melanocyte development, defines a discrete transcription family. Genes Dev., 8, 2770–2780. [DOI] [PubMed] [Google Scholar]
- Hodgkinson C.A., Moore,K.J., Nakayama,A., Steingrímsson,E., Copeland, N.G., Jenkins,N.A. and Arnheiter,H. (1993) Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix–loop–helix-zipper protein. Cell, 74, 395–404. [DOI] [PubMed] [Google Scholar]
- Klein P.S. and Melton,D.A. (1996) A molecular mechanism for the effect of lithium on development. Proc. Natl Acad. Sci. USA, 93, 8455–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner A.B., Shiohara,T., Boissy,R.E., Jacobson,K.A., Lamoreux,M.L. and Moellmann,G.E. (1986) A mouse model for vitiligo. J. Invest. Dermatol., 87, 299–304. [DOI] [PubMed] [Google Scholar]
- Lister J.A., Robertson,C.P., Lepage,T., Johnson,S.L. and Raible,D.W. (1999) nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development, 126, 3757–3767. [DOI] [PubMed] [Google Scholar]
- Mochii M., Mazaki,Y., Mizuno,N., Hayashi,H. and Eguchi,G. (1998) Role of Mitf in differentiation and transdifferentiation of chicken pigmented epithelial cell. Dev. Biol., 193, 47–62. [DOI] [PubMed] [Google Scholar]
- Molenaar M. and van de Wetering,M. (1996) XTcf-3 transcription factor mediates β-catenin-induced axis formation in xenopus embryos. Cell, 86, 391–399. [DOI] [PubMed] [Google Scholar]
- Murre C. et al. (1994) Structure and function of helix–loop–helix proteins. Biochim. Biophys. Acta, 1218, 129–135. [DOI] [PubMed] [Google Scholar]
- Nakayama A., Nguyen,M.T., Chen,C.C., Opdecamp,K., Hodgkinson, C.A. and Arnheiter,H. (1998) Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech. Dev., 70, 155–166. [DOI] [PubMed] [Google Scholar]
- Nobukuni Y., Watanabe,A., Takeda,K., Skarka,H. and Tachibana,M. (1996) Analyses of loss-of-function mutations of the MITF gene suggest that haploinsufficiency is a cause of Waardenburg syndrome type 2A. Am. J. Hum. Genet., 59, 76–83. [PMC free article] [PubMed] [Google Scholar]
- Oosterwegel M., van de Wetering,M., Timmerman,J., Kruisbeek,A., Destree,O., Meijlink,F. and Clevers,H. (1993) Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development, 118, 439–448. [DOI] [PubMed] [Google Scholar]
- Patapoutian A. and Reichardt,L.F. (2000) Roles of Wnt proteins in neural development and maintenance. Curr. Opin. Neurobiol., 10, 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieve M.G., Guttridge,K.L., Munguia,J. and Waterman,M.L. (1998) Differential importin-α recognition and nuclear transport by nuclear localization signals within the high-mobility-group DNA binding domains of lymphoid enhancer factor 1 and T-cell factor 1. Mol. Cell. Biol., 18, 4819–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Roberts,K., Gambino,G., Cook,A., Kouzarides,T. and Goding, C.R. (1997) CBP/p300 as a co-factor for the microphthalmia transcription factor. Oncogene, 14, 3083–3092. [DOI] [PubMed] [Google Scholar]
- Schreiber E., Matthins,P., Müller,M.M. and Schaffner,W. (1989) Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res., 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara S., Yasumoto,K., Amae,S., Fuse,N., Udono,T. and Takahashi,K. (1999) Implications of isoform multiplicity of microphthalmia-associated transcription factor in the pathogenesis of auditory–pigmentary syndromes. J. Invest. Dermatol. Symp. Proc., 4, 101–104. [PubMed] [Google Scholar]
- Steel K.P., Davidson,D.R. and Jackson,I.J. (1992) TRP-2/DT, a new early melanoblast marker, shows that steel growth factor (c-kit ligand) is a survival factor. Development, 115, 1111–1119. [DOI] [PubMed] [Google Scholar]
- Steingrímsson E. et al. (1994) Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nature Genet., 8, 256–263. [DOI] [PubMed] [Google Scholar]
- Steingrímsson E. et al. (1996) The semidominant Mib mutation identifies a role for the HLH domain in DNA binding in addition to its role in protein dimerization. EMBO J., 15, 6280–6289. [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Takahashi,K., Yasumoto,K., Amae,S., Yoshizawa,M., Fuse, N. and Shibahara,S. (1998) Role of neurofibromin in modulation of the expression of the tyrosinase-related protein 2 gene. J. Biochem., 124, 992–998. [DOI] [PubMed] [Google Scholar]
- Takeda K., Yasumoto,K., Takada,R., Takada,S., Watanabe,K., Udono, T., Saito,H., Takahashi,K. and Shibahara,S. (2000) Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J. Biol. Chem., 275, 14013–14016. [DOI] [PubMed] [Google Scholar]
- Tassabehji M., Newton,V.E. and Read,A.P. (1994) Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nature Genet., 8, 251–255. [DOI] [PubMed] [Google Scholar]
- Udono T. et al. (2000) Structural organization of the human microphthalmia-associated transcription factor gene containing four alternative promoters. Biochim. Biophys. Acta, 1491, 205–219. [DOI] [PubMed] [Google Scholar]
- Udono T., Takahashi,K., Yasumoto,K., Takeda,K., Yoshizawa,M., Abe,T., Tamai,M. and Shibahara,S. (2001) Expression of tyrosinase-related protein 2/DOPAchrome tautomerase in the retinoblastoma. Exp. Eye Res., 72, 225–234. [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Castrop,J., Korinek,V. and Clevers,H. (1996) Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol. Cell. Biol., 16, 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Genderen C., Okamura,R.M., Fariñas,I., Quo,R.-G., Parslow,T.G., Bruhn,L. and Grosschedl,R. (1994) Development of several organs that require inductive epithelial–mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev., 8, 2691–2703. [DOI] [PubMed] [Google Scholar]
- Verbeek S. et al. (1995) An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature, 374, 70–74. [DOI] [PubMed] [Google Scholar]
- Waterman M.L., Fischer,W.H. and Jones,K.A. (1991) A thymus-specific member of the HMG protein family regulates the human T cell receptor Cα enhancer. Genes Dev., 5, 656–669. [DOI] [PubMed] [Google Scholar]
- Watt R., Stanton,L.W., Marcu,K.B., Gallo,R.C., Croce,C.M. and Rovera,G. (1983) Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature, 303, 725–728. [DOI] [PubMed] [Google Scholar]
- Yajima I., Sato,S., Kimura,T., Yasumoto,K., Shibahara,S., Goding,C.R. and Yamamoto,H. (1999) An L1 element intronic insertion in the black-eyed white (Mitfmi–bw) gene: the loss of a single Mitf isoform responsible for the pigmentary defect and inner ear deafness. Hum. Mol. Genet., 8, 1431–1441. [DOI] [PubMed] [Google Scholar]
- Yasumoto K. and Shibahara,S. (1997) Molecular cloning of a cDNA encoding a human TFEC isoform, a newly identified transcriptional regulator. Biochim. Biophys. Acta, 1353, 23–31. [DOI] [PubMed] [Google Scholar]
- Yasumoto K., Yokoyama,K., Shibata,K., Tomita,Y. and Shibahara,S. (1994) Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol. Cell. Biol., 14, 8058–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto K., Mahalingam,H., Suzuki,H., Yoshizawa,M., Yokoyama,K. and Shibahara,S. (1995) Transcriptional activation of the melanocyte-specific genes by the human homolog of the mouse Microphthalmia protein. J. Biochem., 118, 874–881. [DOI] [PubMed] [Google Scholar]
- Yasumoto K., Yokoyama,K., Takahashi,K., Tomita,Y. and Shibahara,S. (1997) Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J. Biol. Chem., 272, 503–509. [DOI] [PubMed] [Google Scholar]
- Yasumoto K., Amae,S., Udono,T., Fuse,N., Takeda,K. and Shibahara,S. (1998) A big gene linked to small eyes encodes multiple Mitf isoforms: many promoters make light work. Pigment Cell Res., 11, 329–336. [DOI] [PubMed] [Google Scholar]
- Yokoyama K., Yasumoto,K., Suzuki,H. and Shibahara,S. (1994) Cloning of the human DOPAchrome tautomerase/tyrosinase-related protein 2 gene and identification of two regulatory regions required for its pigment cell-specific expression. J. Biol. Chem., 269, 27080–27087. [PubMed] [Google Scholar]