Abstract
Nucleolar localization of vertebrate box C/D snoRNA involves transit through Cajal bodies, but the significance of this event is unkown. To define better the function of this compartment, we analyzed here the maturation pathway of mammalian U3. We show that 3′-extended U3 precursors possess a mono-methylated cap, and are not associated with fibrillarin and hNop58. Importantly, these precursors are detected at both their transcription sites and in Cajal bodies. In addition, mature U3, the core box C/D proteins and the human homolog of the methyltransferase responsible for U3 cap tri-methylation, hTgs1, are all present in Cajal bodies. In yeast, U3 follows a similar maturation pathway, and equivalent 3′-extended precursors are enriched in the nucleolus and in the nucleolar body, a nucleolar domain that concentrates Tgs1p under certain growth conditions. Thus, spatial organization of U3 maturation appears to be conserved across evolution, and involves specialized and related nuclear compartments, the nucleolus/nucleolar body in yeast and Cajal bodies in higher eukaryotes. These are likely places for snoRNP assembly, 3′ end maturation and cap modification.
Keywords: Cajal bodies/nucleolus/RNA processing/RNA trafficking/snoRNA
Introduction
Maturation of most types of RNA requires many complex biochemical reactions, which often occur in an ordered manner that can be described as a processing pathway. RNA maturation can also be associated with a series of specific localizations taken by the RNA, and there are now many examples of RNA targeting pathways occurring in multiple steps, in particular for stable ribonucleoproteins (RNPs). Yeast tRNA, signal recognition particle (SRP), telomerase RNA, vertebrate RNase P and U6 snRNA all transiently localize to the nucleolus (for reviews see Pederson, 1998; Scheer and Hock, 1999; Olson et al., 2000). These transient localizations are likely to reflect the place where specific biochemical reactions take place, but direct evidence for this is often lacking. In the case of vertebrate snRNA, however, it has been shown that they travel through the cytoplasm and the Cajal bodies, and that final 3′ end trimming, Sm protein assembly and cap trimethylation occur in the cytoplasm (for a review see Will and Lührmann, 2001). Other types of RNA such as mRNA and rRNA are believed to have a simpler localization pathway, consisting of a single step leading from the transcription site to the nuclear pores and to the cytoplasm, with most of the RNA maturation steps occuring at or near the RNA transcription sites (Bataille et al., 1990; Woolford, 1991; Zhang et al., 1994). Never theless, in the case of rRNA, it is established that maturation follows a complex pathway which is tightly linked to the structural organization of the nucleolus, and that maturation proceeds while the RNA moves from one nucleolar subdomain to another (Thiry et al., 2000 and references therein).
The link between RNA localization and maturation is thought to be particularly important in the nucleus. However, the function of a number of nuclear compartments is still unknown, and this is particularly true for Cajal bodies, which have attracted much attention during the past decade (for reviews see Matera, 1999; Gall, 2000). This structure is present in many organisms, including vertebrates, plants and possibly Drosophila and Caeno rhabditis elegans. Cajal bodies are biochemically and spatially linked to nucleoli, sharing many components and often being localized in proximity to or even within nucleoli (Ramón y Cajal, 1903; Ochs et al., 1994; Bohmann et al., 1995). Cajal bodies also contain some specific markers, such as RNA polymerase II holoenzyme, coilin, survival of motor neuron (SMN) protein, snRNA and their associated proteins (for reviews see Matera, 1999; Gall, 2000). In addition, snRNA genes often localize next to Cajal bodies. They have thus been proposed to play various roles in small RNA metabolism and function, but none has been demonstrated yet. Cajal bodies could be involved in snRNA storage and negative transcriptional feedback, snRNA base modification or recycling of snRNP following splicing.
Cajal bodies are also likely to be involved in some aspects of snoRNA metabolism. Box C/D snoRNAs pass through Cajal bodies while en route to the nucleolus (Narayanan et al., 1999), and snoRNA genes are also associated with these structures (Gao et al., 1997). Unlike snRNA, snoRNA can be synthesized via several pathways, depending on the structural organization of the gene: intronic, multi- or mono-cistronic (Filipowicz et al., 1999; Weinstein and Steitz, 1999; Fatica et al., 2000; Villa et al., 2000; Kiss, 2001). Localization in Cajal bodies is, however, likely to reflect a step common to all pathways, since the box C/D motif is sufficient to confer this property (Samarsky et al., 1998).
The processing of U3 snoRNA has recently been analyzed in detail in yeast (Kufel et al., 2000). In this work, we have examined the spatial distribution of snoRNA biosynthesis in both yeast and mammalian cells. Our data indicate that in both systems, several steps such as assembly with core snoRNP proteins, 3′ end maturation and cap hypermethylation probably occur in specific nuclear compartments: Cajal bodies in vertebrates, and a specialized nucleolar compartment in yeast.
Results
Tgs1p concentrates in the nucleolar body, a specific subregion of the yeast nucleolus
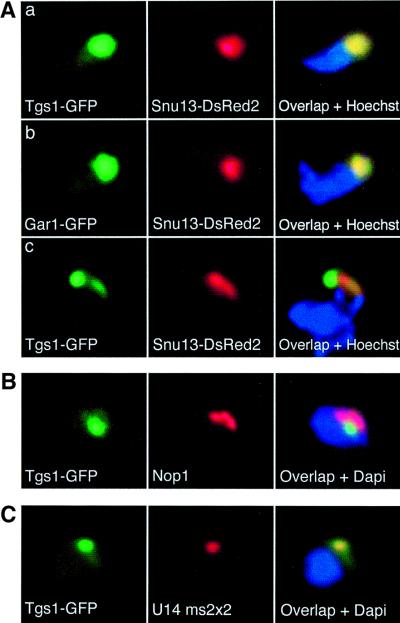
Recently, we have shown that Tgs1p is a conserved RNA methyltransferase that is required to convert the mono-methyl cap (m7G) of snRNA and snoRNA into the tri-methyl cap (m3G) of the mature forms (Mouaikel et al., 2002). When yeast cells were grown in liquid media, a Tgs1–green fluorescent protein (GFP) fusion expressed from the basal activity of the Met25 promoter was localized in the nucleolus (Mouaikel et al., 2002). Interestingly, in a few cells, Tgs1–GFP appeared highly concentrated in a small region of the nucleolus. This prompted us to analyze its localization under a variety of conditions. Tgs1–GFP was first cloned under its natural promoter, and its localization was observed in living cells. While the protein was always nucleolar, we observed that when cells were grown on solid medium, many of them had the Tgs1–GFP fusion protein concentrated in a dot within the nucleolus (∼25% of the cells; see Figure 1). This was not simply a result of confluent growth, because saturating liquid cultures did not show this phenotype and had a signal distributed throughout the nucleolus (data not shown). To confirm these observations, we performed several double label experiments: the nucleolus was visualized either by Nop1p staining following fixation, or directly in living cells by co-expressing a fusion between the DsRed2 fluorescent protein and Snu13p. The Snu13–DsRed2 fusion protein localized correctly to the nucleolus, as shown by co-localization with a Gar1p–GFP marker (Trumtel et al., 2000; Figure 1A, panel b). Again, when cells were grown on solid media, two patterns of localization were observed. In some cells, Tgs1–GFP was distributed throughout the nucleolus, and co-localized with either Nop1p or Snu13–DsRed2 (Figure 1A, panel a). In others, Tgs1–GFP was concentrated in a dot-like structure (Figure 1A, panel c, and B). This dot was within the nucleolar territory but, surprisingly, contained little if any Nop1p or Snu13–DsRed2. This suggested that Tgs1–GFP was accumulating in a nucleolar compartment not involved in ribosome biogenesis.
Fig. 1. Tgs1p accumulates in the nucleolar body in yeast cells grown on solid medium. TGS1/tgs1::KAN diploid cells expressing a Tgs1–GFP fusion protein under its natural promoter were grown on solid medium for 24–48 h and imaged live (A) or following fixation (B and C). DNA of living and fixed cells was stained with Hoechst and DAPI, respectively. (A) Living cells expressing a DsRed2–Snu13 fusion protein. Tgs1–GFP co-localizes with DsRed2–Snu13 (red) in the majority of the cells (a), but not in ∼25% of cases (c). DsRed2–Snu13p always co-localizes with Gar1p (b). (B) Fixed cells stained for Nop1p (red). (C) Fixed cells overexpressing U14/MS2x2 and hybridized in situ with a probe specific for the artificial snoRNA (red). GFP signal (green) and DNA (blue). Each field is 16 µm2.
Recently, we have shown that overexpression of a box C/D snoRNA prevented its normal nucleolar localization, and resulted in its accumulation in a subnucleolar domain that we termed the nucleolar body (Verheggen et al., 2001). The nucleolar body was a potential intermediate in the localization pathway of snoRNAs, and, because some of them are substrates for Tgs1p (Mouaikel et al., 2002), we tested whether the nucleolar body and the Tgs1p-enriched nucleolar domain were the same. Yeast cells expressing the Tgs1–GFP protein and overexpressing U14/MS2x2, an artificial box C/D snoRNA (Verheggen et al., 2001), were grown in solid medium. In situ hybridization experiments indicated that the Tgs1–GFP fusion co-localized with the U14/MS2x2 artificial snoRNA, demonstrating that the two domains were identical (Figure 1C). We will thus hereafter refer to the Tgs1p-enriched domain of the nucleolus as the nucleolar body. Importantly, this result confirms that the nucleolar body is present in wild-type yeast cells (Verheggen et al., 2001), and establishes Tgs1p as the first nucleolar body-specific marker.
Unassembled yeast U3 precursors accumulate in the nucleolus and the nucleolar body
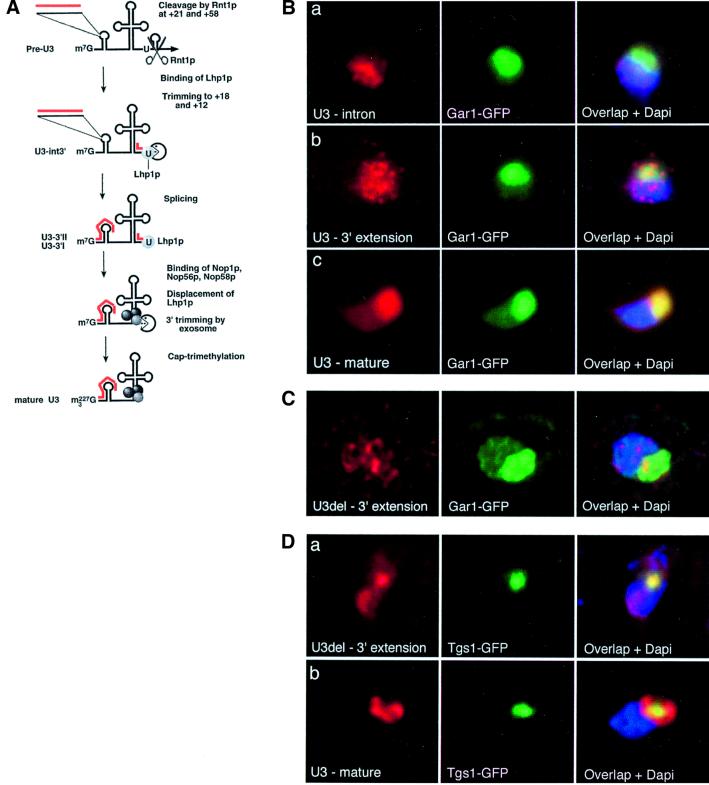
To determine whether Tgs1p catalyzes cap hypermethylation in the nucleolar body, or whether this is merely a storage place for the enzyme, we analyzed in detail the localization of one of its substrates, U3. In yeast, U3 maturation has recently been shown to follow a highly ordered pathway (Kufel et al., 2000; Figure 2A). The primary transcript is first cleaved ∼600 nucleotides downstream of the mature 3′ end (+600) before being digested by the yeast homolog of Escherichia coli RNase III (Rnt1p) at positions +21 and +58. These 3′-extended precursors are trimmed further to nucleotides +12 and +18, and stabilized against degradation by the binding of the yeast homolog of La (Lhp1p). U3 precursors are then spliced, to give rise to the most abundant precursor species, denoted U3-I and U3-II. These species have a m7G cap, and are not bound by the core box C/D snoRNP proteins, Nop1p, Nop56p and Nop58p. The final 3′ end processing and hypermethylation of the cap by Tgs1p are thought to be dependent on snoRNP assembly (Kufel et al., 2000; Mouaikel et al., 2002).
Fig. 2. Yeast U3 precursors are present in the nucleolus and enriched in the nucleolar body. (A) Schematic of the maturation of yeast U3 with the position of the probes indicated in red (adapted from Kufel et al., 2000). (B) Localization of the indicated U3 species (red) by in situ hybridization in cells expressing U3 from a 2 µm plasmid, and grown in liquid medium. The U3-intron probe was used in a strain also lacking the exosomal component RRP6. The nucleolus was labeled with Gar1–GFP fusion protein (green) and the DNA was stained with DAPI (blue). (C) The same as in (B), but with cells expressing the U3del construct. (D) Localization of the indicated U3 species in cells expressing Tgs1–GFP and U3del, and grown on solid medium. Each field is 25 µm2.
We made three fluorescent oligonucleotide probes against specific U3 species (Figure 2A): (i) the U3-intron probe binds within the intron, and is thus specific for the first part of the U3 maturation pathway; (ii) the U3-3′ extension probe is complementary to the 12–18 nucleotide 3′ extensions, and thus also labels U3-I and U3-II; and (iii) the U3-mature probe base-pairs at the exon–exon junction, and should bind the mature U3, in addition to U3-I and U3-II. We first attempted to localize U3 precursors in cells grown in liquid cultures. In wild-type cells, we failed to detect any signal, most probably because of the low abundance of these species. We thus used a yeast strain that was transformed with a plasmid present in multiple copies per cell (2 µm) and expressing a wild-type U3 gene. In this case, we detected U3-I and U3-II, but not the unspliced U3 precursors. However, those became detectable in an RRP6 deletion mutant, which slows down exosome activity and RNA degradation. There was a striking progression in localization, while going through the U3 maturation pathway (Figure 2B). The earliest precursors, detected with the intronic probe, were mostly visible in the nucleoplasm. In contrast, the probe complementary to the 3′ extensions, which also labeled U3-I and U3-II, was detected in both the nucleoplasm and the nucleolus (Figure 2B, panel b). Finally, the exon–exon probe gave a signal primarily nucleolar, as expected from its ability to bind the mature U3. Altogether, these results showed that U3-I and U3-II, which have an m7G cap structure and which are not assembled with the core snoRNP proteins, were present in both the nucleoplasm and the nucleolus, while earlier precursor species were restricted to the nucleoplasm.
Because overexpression with multicopy plasmids may result in artifactual localization, we attempted to confirm these results with altered U3 genes expressed at physiological levels. We focused on a minimal version of U3 that was developed previously, U3del (Samarsky and Fournier, 1998). While fully functional, this U3 variant gives rise to high levels of U3del-I and U3del-II precursors, about a tenth of the levels of the mature molecules. U3del was expressed from a low copy plasmid (ARS/CEN), and the localization of the U3del RNAs was determined by in situ hybridization (Figure 2C). As expected, the intronic probe did not reveal any signal, while the U3-mature probe labeled the nucleolus (data not shown). We could also obtain a distinct signal with the U3-3′ extension probe, which can detect U3del-I and U3del-II. Importantly, the signal was present both in the nucleoplasm and in the nucleolus, a result previously obtained with the overexpressed U3 gene. In addition, in some cells, the signal was also slightly enriched in the nucleolus (Figure 2C).
To determine whether U3del-I and U3del-II accumulated in the nucleolar body, we repeated the experiment above but with cells grown on solid medium and expressing the Tgs1–GFP fusion protein (Figure 2D). As before, the U3-3′ extension probe labeled both the nucleoplasm and the nucleolus but, in some cells that had Tgs1–GFP concentrated in the nucleolar body, the U3del precursors were also detected primarily in this structure. This was in sharp contrast to the results obtained with the U3-mature probe, which resulted in a nucleolar signal mostly excluded from the nucleolar body. Thus, U3del-I and U3del-II were enriched specifically in the nucleolar body.
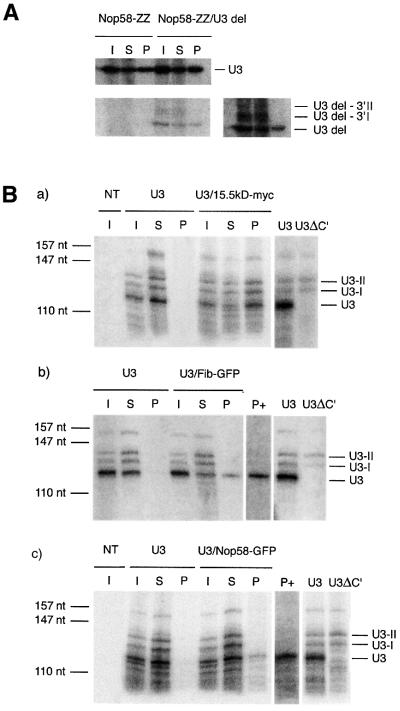
It was shown previously that U3-I and U3-II are not bound by the core snoRNP proteins Nop1p, Nop56p and Nop58p (Kufel et al., 2000). To determine if U3del followed a similar maturation pathway, we determined whether Nop58p was bound or not to U3del-I and U3del-II. The centromeric U3del plasmid was transformed in strains expressing ZZ-tagged versions of Nop58p. Following immunoprecipitation with IgG–Sepharose beads, the bound RNA was purified and analyzed by northern blot with a U3 probe (Figure 3A). As expected, both mature U3 and U3del were co-precipitated with ZZ-Nop58p. In sharp contrast, the precursors U3del-I and U3del-II were not pelleted. Thus, U3del is likely to follow a maturation pathway similar to that of wild-type U3.
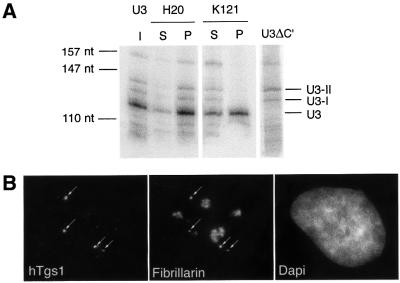
Fig. 3. Yeast and mammalian U3 precursors are not associated with all the core box C/D proteins. (A) RNA co-immunoprecipitation with yeast cells expressing a chromosomal ZZ-Nop58 protein alone or together with U3del. Upper panel: wild-type endogeneous U3. Lower panels: U3del. Left and right: two exposures of the same northern blot. The same amount of material was loaded in each lane. (B) RNA co- immunoprecipitation in mammalian cells. HeLa (a and b) or 293T (c) cells were transfected with the rU3B.7 gene alone or together with the indicated construct, and the RNA was co-immunoprecipitated with the relevant antibody and analyzed by RNase protection assays. I, input; S, supernatant; P, pellet (five times the input). Non-transfected cells (NT) showed no signal, while a U3 gene lacking box C′ (U3ΔC′) allowed identification of both U3 precursors, denoted U3-I and U3-II, and mature U3. For (b) and (c), P+ corresponds to the pellet lane after longer exposure of the gel.
3′-extended precursors of mammalian U3 are bound by the 15.5 kDa protein, but not by fibrillarin or hNop58
Whereas the vertebrate U3 maturation pathway is not known in detail, it is likely to occur in at least two steps, with the formation of 3′-extended species similar to yeast U3-I and U3-II (Stroke and Weiner, 1985). In order to analyze the mammalian U3 processing pathway, we cloned the rat U3B.7 gene variant, which gives rise to precursors bearing 8 and 15 nucleotide extensions (Stroke and Weiner, 1985). HeLa cells were transiently transfected with rU3B.7, and the resulting RNAs were analyzed by RNase protection assay with a probe that covers the 3′ end of U3 (transcribed nucleotides +98 to +236). Control cells showed no signal, while several forms of U3 were observed with transfected cells (Figure 3B). The shorter RNA species was the most abundant, and corresponded to the size of mature U3. Two 10–15 nucleotide longer transcripts were also detected, which could correspond to the 3′-extended precursors described previously. In addition, a very long form covered most of the probe, and was likely to be the primary transcript. To identify unambiguously the bands corresponding to the mature and precursor forms of U3, we used a variant that had an inactivated box C, and that does not support snoRNA maturation (Caffarelli et al., 1996; Samarsky and Fournier, 1998). With this mutant, only the shorter, abundant form of U3 disappeared, confirming that this band corresponded to the mature RNA (Figure 3B).
To determine which proteins are bound to the precursor forms of U3, we co-transfected the rU3B.7 gene with several tagged protein genes, the 15.5 kDa protein (the homolog of Snu13p), fibrillarin, hNop56 and hNop58. The fusion proteins were then immunoprecipitated with antibodies specific for their tag, and the bound RNA analyzed by RNase protection. Transfection with rU3B.7 alone did not allow co-immunoprecipitation of any U3 RNA. In contrast, co-transfection with the myc-tagged 15.5 kDa protein allowed immunoprecipitation of both the mature and the precursor forms of U3, with a similar efficiency (Figure 3B, panel a). Fibrillarin and hNop58 were tagged with GFP, and the fusion proteins allowed efficient precipitation of the mature form of U3, but the 3′-extended precursors were not detectable in the pellet, even after long exposure (Figure 3B, panels b and c). We tested in a similar way GFP- and protein A-tagged versions of hNop56, but the efficiency of immunoprecipitation was low, and we could not determine unambiguously whether the U3 precursors were bound or not by hNop56. In any case, these results show that 3′-extended U3 precursors are not associated with fibrillarin and hNop58, two proteins of the mature snoRNP.
3′-extended precursors of mammalian U3 are present at their transcription sites and in Cajal bodies
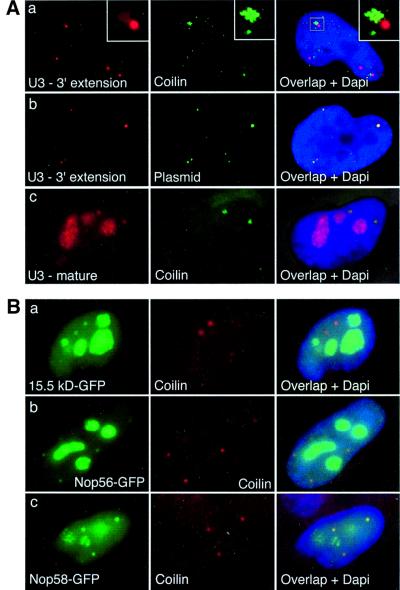
The fact that box C/D snoRNAs travel through Cajal bodies on their way to nucleoli (Narayanan et al., 1999) prompted us to determine the localization of the mature and 3′-extended forms of mammalian U3 (Figure 4A). In HeLa cells transiently transfected with the rU3B.7 gene, the probe specific for the 3′ extension did not label the nucleolus, but revealed some bright focus in the nucleoplasm, as well as a faint and diffuse signal that was often connected to a bright focus. Double labeling with antibodies against coilin showed that the latter bright foci were often localized next to a Cajal body, and that the diffuse signal was within Cajal bodies. U3 genes have been reported to associate with Cajal bodies (Gao et al., 1997) and, indeed, double labeling of the U3 precursors with a probe binding to the non-transcribed portion of the transfected plasmid confirmed that the bright foci were the snoRNA transcription sites (Figure 4A, panel b). While the plasmids were often detected next to a Cajal body, no plasmid was, however, detected within them (data not shown).
Fig. 4. Mammalian 3′-extended U3 precursors are localized at their transcription sites and in Cajal bodies. (A) HeLa cells were transfected with the rU3B.7 gene and hybridized in situ with the indicated probe. (a) U3 3′-extended precursors (red) are present in Cajal bodies (green) and in bright foci often adjacent to them. Inset: enlargement of the boxed area. (b) U3 3′-extended precursors (red) are present at their transcription sites, which are visualized with a probe against the plasmid (green). (c) Mature U3B.7 RNA (red) is present in nucleoli and Cajal bodies (green). The image shown in (a) was deconvolved. (B) Localization of 15.5 kDa–GFP (a), hNop56–GFP (b) and hNop58–GFP (c) in nucleoli and Cajal bodies (red) of HeLa cells. DNA (blue) is stained with DAPI and GFP is green. Cajal bodies are visualized by indirect immunofluorescence with an antibody against coilin. Each field is 24 × 30 µm.
In contrast to the bright foci, the diffuse and faint signal revealed by the probe specific for the precursor was within Cajal bodies (Figure 4A). Within a cell, all the Cajal bodies were not labeled equally, possibly because of the variable proximity of actively transcribed U3 genes (Frey et al., 1999). To rule out the possibility that this diffuse signal resulted from out of focus light originating from a bright focus located above or below the Cajal body, three-dimensional image restoration was performed, and yielded similar results (Figure 4A, panel a). In addition, a diffuse signal in Cajal bodies not connected to bright foci was also observed (data not shown).
The probe that recognized the mature U3 revealed a strikingly different signal, and labeled essentially the nucleolus and the Cajal bodies, as expected (Jimenez-Garcia et al, 1994; Figure 4A, panel c). The signal in Cajal bodies was much brighter than the one obtained with the precursor probe, suggesting that it corresponded to mature molecules, and not to precursors. Altogether, this demonstrated that U3 precursors were localized at their transcription sites and in Cajal bodies, but not in nucleoli. In contrast, mature U3 is present in both Cajal bodies and nucleoli.
Cajal bodies contain fibrillarin (Raska et al., 1990; Gerbi and Borovjagin, 1997), but it was not determined previously whether they also contain the other box C/D snoRNA proteins. The GFP-tagged proteins described above were thus co-localized with coilin by immunofluorescence. The 15.5 kDa protein, hNop56 and hNop58 localized similarly to fibrillarin: they were highly enriched in the nucleolus but also present in Cajal bodies (Figure 4B).
Altogether, these results show that Cajal bodies contained mature U3 snoRNA, the complete set of snoRNP proteins and U3 precursors not assembled with the complete set of snoRNP proteins. This suggests that these organelles are most likely to be the assembly sites of U3 snoRNP.
The cap of mammalian U3 apparently is hypermethylated in Cajal bodies
In yeast, cap hypermethylation is one of the latest steps in the biogenesis of U3. It requires prior binding of Nop58p and/or Nop56p, ensuring that only the mature snoRNP is hypermethylated (Kufel et al., 2000; Mouaikel et al., 2002). Since we detected 3′-extended U3 precursors, mature U3, hNop56 and hNop58 in Cajal bodies of human cells, we decided to test whether the cap modification of U3 occurred in this organelle. We first analyzed the methylation status of U3 precursors. RNA purified from HeLa cells transiently transfected with rU3B.7 were immunoprecipitated with both monoclonal antibody (mAb) K121, which recognizes only m3G caps, and mAb H20, which recognizes both m7G and m3G cap structures (Mouaikel et al., 2002 and references therein). As shown in Figure 5A, mAb K121 precipitated only the mature form of U3, while mAb H20 precipitated both the mature and the 3′-extended forms. Thus, similarly to the yeast situation, 3′-extended forms of mammalian U3 possess a m7G cap structure, while the mature form contains an m3G cap. Since both forms are present in Cajal bodies, this is a likely place for such modification.
Fig. 5. The cap of mammalian U3 is hypermethylated in Cajal bodies. (A) 3′-extended U3 precursors have an m7G cap. RNAs purified from HeLa cells transfected with the rU3B.7 gene were immunoprecipitated with K121, specific for the m3G cap, or H20, which recognize m7G and m3G cap structures. Details are as in the legend for Figure 3B. (B) hTgs1 is localized in Cajal bodies. HeLa cells were stained by immunofluorescence with anti-hTgs1 (left) and anti-fibrillarin antibodies (middle). DNA is stained with DAPI (right). Cajal bodies are indicated by arrows. In this image, hTgs1 is not detected in the cytoplasm. Indeed, optimal detection in the cytoplasm required permeabilization conditions different from those used to detect it in the nucleus. Each field is 30 × 36 µm.
If this was indeed the case, the enzyme responsible for cap hypermethylation should also be found in Cajal bodies. We thus investigated the localization of the human homolog of Tgs1, hTgs1, also known as PIMT (Zhu et al., 2001). By transient transfection of Cos cells with a replicative plasmid containing a GFP-tagged cDNA, it was shown previously that hTgs1–GFP is present in the nucleus in ill-defined structures (Zhu et al., 2001). To define these structures better, we raised two peptide antibodies against the human protein, and examined its intracellular localization by immunofluorescence in HeLa cells (Figure 5B). While pre-immune serum did not reveal any signal, both immune sera revealed a similar pattern. As expected from its predicted role in snRNA cap modification (Mattaj, 1986; Plessel et al., 1994), some signal was detected in the cytoplasm and this will be described in more detail elsewhere. In addition, the two antibodies labeled several bright spots in the nucleus, and double labeling with fibrillarin antibodies demonstrated that these were Cajal bodies (Figure 5B). To confirm this localization further, an hTgs1–GFP fusion was transiently expressed in HeLa cells, and we observed that it localized specifically in Cajal bodies (data not shown). Altogether, these results strongly suggested that Cajal bodies are the place where U3 cap tri-methylation takes place.
Discussion
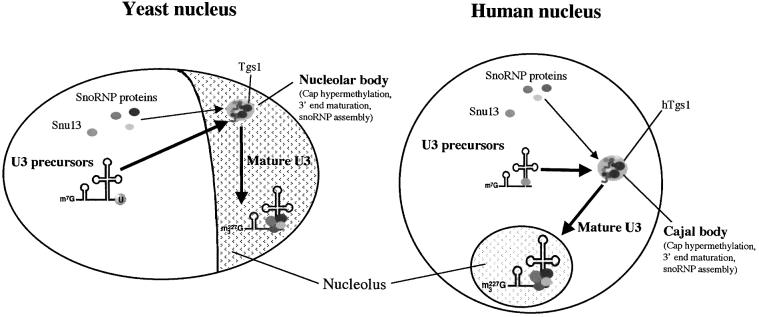
In Xenopus oocytes, it has been observed that U3 transiently localizes to Cajal bodies before being targeted to nucleoli (Narayanan et al., 1999). However, the significance of this localization was not known. In this work, we obtained evidence that this temporal localization pattern reflects the maturation pathway of U3. In vertebrates snoRNP assembly, 3′ end formation and cap tri-methylation are likely to occur in Cajal bodies. In yeast, these steps probably take place in the nucleolus or in a nucleolar domain showing similarities to Cajal bodies, the nucleolar body. Thus, the spatial organization of snoRNA maturation appears to have been conserved during evolution.
A relationship between the yeast nucleolar body and Cajal bodies
By overexpressing an artificial snoRNA in yeast, we previously uncovered a novel nucleolar domain that we called the nucleolar body. Based on the specific localization of human SMN protein in this domain, we hypothesized that the nucleolar body was related to Cajal bodies, and that it was involved in snoRNA maturation (Verheggen et al., 2001). Data presented herein strengthen this view. First, we show that a Tgs1–GFP fusion protein expressed at its normal levels localizes to the nucleolar body. As shown in Figure 1, this is also the case in wild-type yeast that do not overexpress any snoRNA, demonstrating that the nucleolar body indeed exists in physiological conditions. Secondly, the human homolog of Tgs1 is localized in Cajal bodies, and thus reinforces the link between the nucleolar body and mammalian Cajal bodies. Thirdly, both the nucleolar body and Cajal bodies contain U3 precursors, suggesting that these compartments are directly involved in snoRNP biogenesis (see below).
It is remarkable that the yeast nucleolar body becomes visible only under particular conditions, i.e. growth on solid media. In higher eukaryotes, Cajal bodies are known to be highly dynamic structures that can vary greatly in size and number (Platani et al., 2000). Noticeably, some cells even lack detectable Cajal bodies (Young et al., 2001). Thus, they are likely to form in response to particular physiological conditions. In yeast, growth on liquid or solid media results in strikingly different conditions (Sherman, 1991). For instance, solid media provide high levels of oxygen, rapid removal of CO2 but poor access to soluble sources of amino acids, nitrogen and sugar. In contrast, liquid media provide high levels of solute, but low oxygen pressure and poor CO2 removal. This may result in different physiological states with specific requirements in snRNA and snoRNA metabolism, which eventually promote or inhibit formation of the nucleolar body. In any case, it is clear that the yeast nucleolus contains activities devoted to small RNP biogenesis, and that under certain conditions these activities can separate from those involved in rRNA production to give rise to a distinct domain. In higher eukaryotes, these activities may have become physically separated from nucleoli to form Cajal bodies. This may explain the close relationships between the two compartments (Ramón y Cajal, 1903; Bohmann et al., 1995), and the fact that in some cell lines Cajal bodies are found within nucleoli (Ochs et al., 1994).
Assembly and maturation of U3 in specific nuclear structures
In yeast, we observed that early, intron-containing precursors in the U3 maturation pathway were restricted to the nucleoplasm. In contrast, the later 3′-extended intermediates (U3-I and U3-II) were present in the nucleolus (see Figure 2). These results were obtained using both a U3 mutant that accumulated these 3′-extended intermediates (U3del) and an overexpressed wild-type U3 gene. U3del precursors were not bound by Nop58p, confirming that this mutant follows a maturation pathway similar if not identical to that of wild-type U3. Importantly, U3-I and U3-II are not only devoid of the core snoRNP proteins Nop1p, Nop56p and Nop58p, but also have an m7G cap (Kufel et al., 2000). Since the enzyme responsible for cap hypermethylation is localized in the nucleolus and requires prior binding of Nop58p and/or Nop56p (Mouaikel et al., 2002), our results collectively suggest that assembly with the core box C/D snoRNP proteins, final 3′ end trimming and cap hypermethylation are all occurring in the nucleolus. Interestingly, when cells were grown on solid media, both Tgs1 and U3del precursors accumulated in the nucleolar body, suggesting that in this case U3 is matured in this subnucleolar compartment.
Other RNAs like tRNAs or SRP RNA are also thought to be matured in the nucleolus (Bertrand et al., 1998; Politz et al., 2000; Grosshans et al., 2001). Interestingly, in Archaea, some RNA precursors generate both snoRNA and either rRNA or tRNA (Ome et al., 2000; d’Orval et al., 2001; Tang et al., 2002). Thus, rRNA, tRNA and snoRNA processing were physically connected in ancient organisms, and this spatial link may have been conserved during the evolution of the nucleolus, allowing distinct classes of RNA to be processed in the same compartment and use common processing factors.
In vertebrates, U3 maturation also involves the transient production of 3′-extended intermediates. These precursors have an m7G cap and are not assembled with the snoRNP proteins fibrillarin and hNop58 (see Figure 3), indicating that U3 follows a very similar maturation pathway in yeast and mammals. Whereas we could not determine whether hNop56 is actually bound to U3 precursors, this is unlikely since in yeast Nop56p binds snoRNAs only following Nop1p (Lafontaine and Tollervey, 2000). Most importantly, we show that these precursors are targeted to Cajal bodies. Mature U3, all core box C/D proteins and a U3 maturation enzyme, the human homolog of Tgs1p, were also detected in Cajal bodies. Furthermore, hTgs1p is likely to bind the snoRNA only following its assembly with hNop56 and hNop58, supporting the idea that not only the precursors but also the fully assembled snoRNPs are present in Cajal bodies. Taken together, these data strongly support the view that final 3′ end trimming, cap hypermethylation and snoRNP assembly all occur in Cajal bodies. The observation that U3 and other box C/D genes often cluster next to Cajal bodies would be consistent with this possibility (Gao et al., 1997), as this would facilitate transport of snoRNA precursors from the site of transcription to the site of snoRNP assembly. It is unclear at present if U3 snoRNP maturation in Cajal bodies is an obligatory event, or if it merely represents the preferred and most frequent situation. That some cells lack Cajal bodies would argue for the latter possibility, although it is also possible that the Cajal bodies in these cells are simply too small to be detected.
Thus, it appears that in both yeast and mammals the same critical steps of U3 biogenesis are organized spatially, and occur in specific and related nuclear compartments, the nucleolus/nucleolar body and Cajal bodies, respectively (Figure 6). One obvious reason that might favor a spatial confinement of RNA maturation is to increase considerably the rates of the reactions by concentrating substrates and enzymes. We observed such a precise localization for Tgs1p in yeast and mammals. Another example could be the SMN complex, which localizes in Cajal bodies and was shown recently to bind unassembled fibrillarin (Jones et al., 2001; Pellizzoni et al., 2001), thus possibly facilitating its incorporation into the snoRNP. Likewise, Tgs1p binds free Nop58p and probably free Nop56p (Mouaikel et al., 2002), and could thus provide an anchor to concentrate these proteins in Cajal bodies.
Fig. 6. Schematic of box C/D snoRNA biogenesis in yeast and mammalian cells. In yeast, U3 becomes associated with the complete set of the core box C/D proteins in the nucleolus and/or the nucleolar body, and this triggers 3′ end maturation and cap hypermethylation. In mammals, these events take place in Cajal bodies.
Intranuclear RNA transporters
In yeast, the 3′-extended U3 precursors are stabilized by Lhp1 (the yeast La protein). Being the only protein currently known to be associated with these precursors (Kufel et al., 2000), Lhp1 could play a role in targeting U3 to the nucleolus and the nucleolar body. Interestingly, a number of other RNA precursors bound by Lhp1 are found at least transiently in the nucleolus, including pre-tRNA, pre-U1 and pre-U6 snRNA (Bertrand et al., 1998; Ganot et al., 1999; Xue et al., 2000; Mouaikel et al., 2002). From this, it would be expected that Lhp1p encodes an essential function. This is, however, not the case (Yoo and Wolin, 1994), but a functional redundancy with the nuclear Lsm complex, which binds similar sequences, is possible (Pannone et al., 2001). While identification of the molecules that transport U3 precursors will require additional studies, it is already clear that they are distinct from those involved in the localization of the mature molecules. Indeed, the precursors are not assembled with the complete set of core snoRNP proteins, which we showed previously to be required for the nucleolar localization of the mature snoRNP (Verheggen et al., 2001).
A function for Cajal bodies in the biogenesis of small RNPs
Since the discovery that snRNAs and their associated proteins concentrate in Cajal bodies, these nuclear compartments have attracted much attention, but no particular functions have yet been assigned to them. In this work, our findings indicate that specific and important steps of U3 biogenesis are likely to occur in Cajal bodies. This is consistent with the recent proposal that snoRNA-mediated modification of snRNAs bases is also occurring there (Darzacq et al., 2002), and collectively supports the view that Cajal bodies are devoted to the biogenesis of small RNPs, similar to the role of the nucleolus for ribosome assembly.
Materials and methods
Cell culture
HeLa and 293T cells were grown at 37°C in 5% CO2, and in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS). Cells were transfected by the calcium phosphate co-precipitation procedure as described previously (Samarsky et al., 1998). The precipitate was left overnight on the cells, removed by resolubilizing it in an isotonic solution without calcium and phosphate, and cells were then split. For in situ hybridization and immunofluorescence, cells were plated on gelatin-coated glass coverslips.
Strains
The Saccharomyces cerevisiae strains used in this study are derived from W303, except the RRP6 deletion strain that was in a BMA38 background (TRP1::RRP6), the ZZ-Nop58 strain (Lafontaine and Tollervey, 1999) and the tgs1::KAN/TGS1 strain (Mouaikel et al., 2002). When needed, plasmids were introduced into these strains by the PEG–lithium procedure, and transformants were selected on appropriate drop-out media. For imaging or biochemical analyses, cells were grown at 30°C in drop-out minimal media containing 2% glucose.
Plasmids
DNA manipulation was performed by standard techniques, and with the Gateway™ system (Invitrogen). Fibrillarin–GFP was described previously (Dundr et al., 2000). The 15.5 kDa–GFP and hNop58–GFP constructs were generated by first amplifying their cDNA by PCR, and then fusing it to the C-terminus of eGFP, downstream of the mouse ribosomal L30 promoter. The yeast U3del plasmid was described previously (Samarsky and Fournier, 1998), and the rat U3B.7 gene was cloned following PCR amplification. To place the GFP–Tgs1 fusion under its own Tgs1 promoter, an 180 bp 5′-untranslated region of TGS1 was PCR-amplified using oligonucleotides containing SacI and EcoRI sites. The SacI–EcoRI fragment containing the promoter and the EcoRI–HindIII fragment containing the GFP–Tgs1 DNA were cloned into the pGFP-Nfus vector previously cut with SacI and HindIII. The obtained plasmid was named pTgs1-GFP.
In situ hybridization and immunofluorescence
In situ hybridization of yeast cells and mammalian cells was performed as described previously (Samarsky et al., 1998), with Cy3- or Cy5-conjugated oligonucleotide probes. Briefly, mammalian cells were fixed for 30 min with 4% paraformaldehyde in phosphate-buffered saline (PBS; 100 mM Na2HPO4, 20 mM KH2PO4, 137 mM NaCl, 27 mM KCl pH 7.4) and permeabilized overnight in 70% ethanol. After rehydration in 2× SSC (300 mM NaCl, 30 mM sodium citrate pH 7.0), yeast and mammalian cells were hybridized overnight at 37°C in 40 µl of a mixture containing 10% dextran sulfate, 2 mM vanadyl-ribonucleoside complex, 0.02% RNase-free bovine serum albumin (BSA), 40 µg of E.coli tRNA, 2× SSC, 10% formamide and 30 ng of probe. Cells were washed twice for 30 min in 1× SSC, 10% formamide, and at 37°C. The probes used were the following: yeast U3 intron: AT*GTTTTCAAATCCT*GGTTTTTTGTGGGT*AAAATGTTTGGGT*A; yeast U3-3′-extended precursors: AT*GGAAAAAAGTGGTT*AACTTGTCAGACT*A; yeast U3- mature: TT*CTATAGAAATGATCCT*ATGAAGTACGTCGACT*A; mature rU3B.7: TT*GCACAGAAGCAGCACCT*AGAGCCGGCTTCACGCTT*T; rU3B.7 3′-extended precursors: TT*CATAAAGGTTAGACCGAAGT*CCACTCAGACTGT*T; plasmid: T*CCTCGCTCACT* GACTCGCTGCGCT*CGGTCGTTCGGCT*GCGGCGAGCGGT*T. T* denotes the modified bases.
Immunofluorescence was performed either following in situ hybridization, or directly with cells fixed in paraformaldehyde and permeabilized in acetone (3 min at –20°C). Anti-coilin and anti-hTGS1 rabbit polyclonal antibodies were diluted 1/400, and fibrillarin mouse monoclonal antibody 1/10 (antibody 72B9; Reimer et al., 1987). Incubations were for 1 h at 37°C in PBS containing 1% BSA, and then slides were washed three times for 10 min in PBS. Secondary antibodies were diluted 1/400 and utilized in the same conditions. Anti-hTgs1 antibodies were raised by Eurogentech against peptides NH2-CAYFGDLIRRPASET and NH2-CRFDDGIKLDREGWFS, and sequentially immunopurified against each peptide. Two rabbits were immunized and their sera gave identical results. Image acquisition and deconvolution were carried out as described previously (Verheggen et al., 2001).
Co-immunoprecipitations
For yeast cells, we followed the procedure described in Lafontaine and Tollervey (1999). For mammalian cells, experiments were done as described in Jady and Kiss (2001). Briefly, transfected cells were resuspended in extraction buffer (1–3 × 106 cells for 0.5 ml), and sonicated three times for 3 s. Extracts were centrifuged further for 10 min at 10 000 r.p.m. and pre-cleared for 30 min with protein A–Sepharose beads (CL-4B, Sigma Aldrich). For each immunoprecipitation assay, 5 mg of hydrated beads were incubated for 2 h in 400 µl of extraction buffer with either 4 µg of GFP antibody (Roche Diagnostic) or 4 µl of 9E10 anti-myc antibody (mouse ascites fluid, Sigma). The beads coupled to the antibodies were then incubated with 400 µl of extract for 2 h at 4°C, and washed six times with 1 ml of extraction buffer. Bound RNAs were purified by proteinase K–SDS treatment, phenol extraction and precipitation with isopropanol. RNAs were then analyzed by RNase protection assays (RPA) with a probe covering nucleotides +98 to +236 of rU3B.7 (+1 is first nucleotide of mature U3) with the RPAIII kit (Ambion).
Extraction buffers were NET1 for fibrillarin [50 mM Tris–HCl pH 7.4, 0.1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 150 mM NaCl], NET2 for the 15.5 kDa protein (identical to NET1 except that it contained 250 mM NaCl) and NET3 for hNop58 [20 mM Tris–HCl pH 8, 5 mM MgCl2, 300 mM potassium acetate, 1 mM dithiothreitol, 0.2% Triton X-100, 0.1% NP-40, 0.5 mM PMSF].
The cap structure of U3 was analyzed by incubating 20 µg of purified RNA with H20 or K121 antibody immobilized on protein A–Sepharose beads. The beads (5 mg) were incubated with 10 µl of antibody in 400 µl of buffer NET1 for 2 h at 4°C. After washing, the bound RNAs were analyzed by RPA as described above.
Acknowledgments
Acknowledgements
We wish to thank M.L.Violet and G.Cabal for their help with immuno precipitations, M.Fournier for the gift of numerous U3 plasmids, R.Lührmann for the gifts of anti-cap antibodies, A.Lamond for the gift of coilin antibodies, M.Dundr for the gift of fibrillarin–GFP, D.Tollervey for the gift of the ΔRRP6 strain, and M.Caizergues-Ferrer for helpful advice and critical reading of the manuscript. D.L.J.L. is a Chercheur qualifié du Fonds National de la Recherche Scientifique Belge (FNRS). This work was supported by grants from the AFM to R.B., 9043 of the ARC and ACI of the MNRT to E.B. C.V. was supported by a fellowship from the ARC.
References
- Bataille N., Helser,T. and Fried,H. (1990) Cytoplasmic transport of ribosomal subunits microinjected into the Xenopus laevis oocyte nucleus: a generalized, facilitated process. J. Cell Biol., 111, 1571–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E., Houser-Scott,F., Kendall,A., Singer,R. and Engelke,D. (1998) Nucleolar localization of early tRNA processing. Genes Dev., 12, 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K., Ferreira,J. and Lamond,A. (1995) Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J. Cell Biol., 131, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E., Fatica,A., Prislei,S., De Gregorio,E., Fragapane,P. and Bozzoni,I. (1996) Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J., 15, 1121–1131. [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Jády,B.E., Verheggen,C., Kiss,A.M., Bertrand,E. and Kiss,T. (2002) Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J., 21, 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Orval B., Bortolin,M., Gaspin,C. and Bachellerie,J. (2001) Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res., 29, 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Misteli,T. and Olson,M.O. (2000) The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol., 150, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Morlando,M. and Bozzoni,I. (2000) Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J., 19, 6218–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Pelczar,P., Pogacic,V. and Dragon,F. (1999) Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim. Pol., 46, 377–389. [PubMed] [Google Scholar]
- Frey M., Bailey,A., Weiner,A. and Matera,A. (1999) Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol., 9, 126–135. [DOI] [PubMed] [Google Scholar]
- Gall J. (2000) Cajal bodies: the first 100 years. Annu. Rev. Cell. Dev. Biol., 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Ganot P., Jady,B., Bortolin,M., Darzacq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Frey,M. and Matera,A. (1997) Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res., 25, 4740–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi S. and Borovjagin,A. (1997) U3 snoRNA may recycle through different compartments of the nucleolus. Chromosoma, 105, 401–406. [DOI] [PubMed] [Google Scholar]
- Grosshans H., Deinert,K., Hurt,E. and Simos,G. (2001) Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA and Xpo1p-mediated export. J. Cell Biol., 153, 745–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady B. and Kiss,T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia L., Segura-Valdez,M., Ochs,R., Rothblum,L., Hannan,R. and Spector,D. (1994) Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell, 9, 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K., Gorzynski,K., Hales,C., Fischer,U., Badbanchi,F., Terns,R. and Terns,M. (2001) Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem., 276, 38645–38651. [DOI] [PubMed] [Google Scholar]
- Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Allmang,C., Chanfreau,G., Petfalski,E., Lafontaine,D. and Tollervey,D. (2000) Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol., 20, 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D. and Tollervey,D. (1999) Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D. and Tollervey,D. (2000) Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. (1999) Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol., 9, 302–309. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. (1986) Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell, 46, 905–911. [DOI] [PubMed] [Google Scholar]
- Mouaikel J., Verheggen,C., Bertrand,E., Tazi,J. and Bordonné,R. (2002) Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell, 9, 891–901. [DOI] [PubMed] [Google Scholar]
- Narayanan A., Speckmann,W., Terns,R. and Terns,M. (1999) Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell, 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs R., Stein,T.J. and Tan,E. (1994) Coiled bodies in the nucleolus of breast cancer cells. J. Cell Sci., 107, 385–399. [DOI] [PubMed] [Google Scholar]
- Olson M., Dundr,M. and Szebeni,A. (2000) The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol., 10, 189–196. [DOI] [PubMed] [Google Scholar]
- Ome R.A., Lowe,T., Russell,A., Ebhardt,H., Eddy,S. and Dennis,P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- Pannone B., Kim,S., Noe,D. and Wolin,S. (2001) Multiple functional interactions between components of the Lsm2–Lsm8 complex, U6 snRNA and the yeast La protein. Genetics, 158, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. (1998) The plurifunctional nucleolus. Nucleic Acids Res., 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L., Baccon,J., Charroux,B. and Dreyfuss,G. (2001) The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol., 11, 1079–1088. [DOI] [PubMed] [Google Scholar]
- Platani M., Goldberg,I., Swedlow,J. and Lamond,A. (2000) In vivo analysis of Cajal body movement, separation and joining in live human cells. J. Cell Biol., 151, 1561–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessel G., Fischer,U. and Lührmann,R. (1994) m3G cap hyper methylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol. Cell. Biol., 14, 4160–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J., Yarovoi,S., Kilroy,S., Gowda,K., Zwieb,C. and Pederson,T. (2000) Signal recognition particle components in the nucleolus. Proc. Natl Acad. Sci. USA, 97, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón y Cajal S. (1903) Un sencillo método de coloración selectiva del retículo protoplasmático y sus efectos en los diversos órganos nerviosos de vertebrados e invertebrados. Trab. Lab. Invest. Biol. (Madrid), 2, 129–221. [Google Scholar]
- Raska I., Ochs,R., Andrade,L., Chan,E., Burlingame,R., Peebles,C., Gruol,D. and Tan,E. (1990) Association between the nucleolus and the coiled body. J. Struct. Biol., 104, 120–127. [DOI] [PubMed] [Google Scholar]
- Reimer G., Pollard,K., Penning,C., Ochs,R., Lische,M., Bush,H. and Tan,E. (1987) Monoclonal antibody from a (New Zealand black × New Zealand White) F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 snoRNP particle. Arthritis Rheum., 30, 793–800. [DOI] [PubMed] [Google Scholar]
- Samarsky D. and Fournier,M. (1998) Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky D., Fournier,M., Singer,R. and Bertrand,E. (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J., 17, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U. and Hock,R. (1999) Structure and function of the nucleolus. Curr. Opin. Cell Biol., 11, 385–390. [DOI] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Stroke I. and Weiner,A. (1985) Genes and pseudogenes for rat U3A and U3B small nuclear RNA. J. Mol. Biol., 184, 183–193. [DOI] [PubMed] [Google Scholar]
- Tang T.H. et al. (2002) RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res., 30, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry M., Cheutin,T., O’Donohue,M., Kaplan,H. and Ploton,D. (2000) Dynamics and three-dimensional localization of ribosomal RNA within the nucleolus. RNA, 6, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumtel S., Leger-Silvestre,I., Gleizes,P., Teulieres,F. and Gas,N. (2000) Assembly and functional organization of the nucleolus: ultrastructural analysis of Saccharomyces cerevisiae mutants. Mol. Biol. Cell, 11, 2175–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C., Mouaikel,J., Thiry,M., Blanchard,J., Tollervey,D., Bordonne,R., Lafontaine,D. and Bertrand,E. (2001) Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J., 20, 5480–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Ceradini,F. and Bozzoni,I. (2000) Identification of a novel element required for processing of intron-encoded box C/D small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein L. and Steitz,J. (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- Will C. and Lührmann,R. (2001) Spliceosomal U snRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- Woolford J.L.J. (1991) The structure and biogenesis of yeast ribosomes. Adv. Genet., 29, 63–118. [DOI] [PubMed] [Google Scholar]
- Xue D., Rubinson,D., Pannone,B., Yoo,C. and Wolin,S. (2000) U snRNP assembly in yeast involves the La protein. EMBO J., 19, 1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C. and Wolin,S. (1994) La proteins from Drosophila melanogaster and Saccharomyces cerevisiae: a yeast homolog of the La autoantigen is dispensable for growth. Mol. Cell. Biol., 14, 5412–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P., Le,T., Dunckley,M., Nguyen,T., Burghes,A. and Morris,G. (2001) Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp. Cell Res., 265, 252–261. [DOI] [PubMed] [Google Scholar]
- Zhang G., Taneja,K., Singer,R. and Green,M. (1994) Localization of pre-mRNA splicing in mammalian nuclei. Nature, 372, 809–812. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qi,C., Cao,W., Yeldandi,A., Rao,M. and Reddy,J. (2001) Cloning and characterization of PIMT, a protein with a methyl transferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc. Natl Acad. Sci. USA, 98, 10380–10385. [DOI] [PMC free article] [PubMed] [Google Scholar]