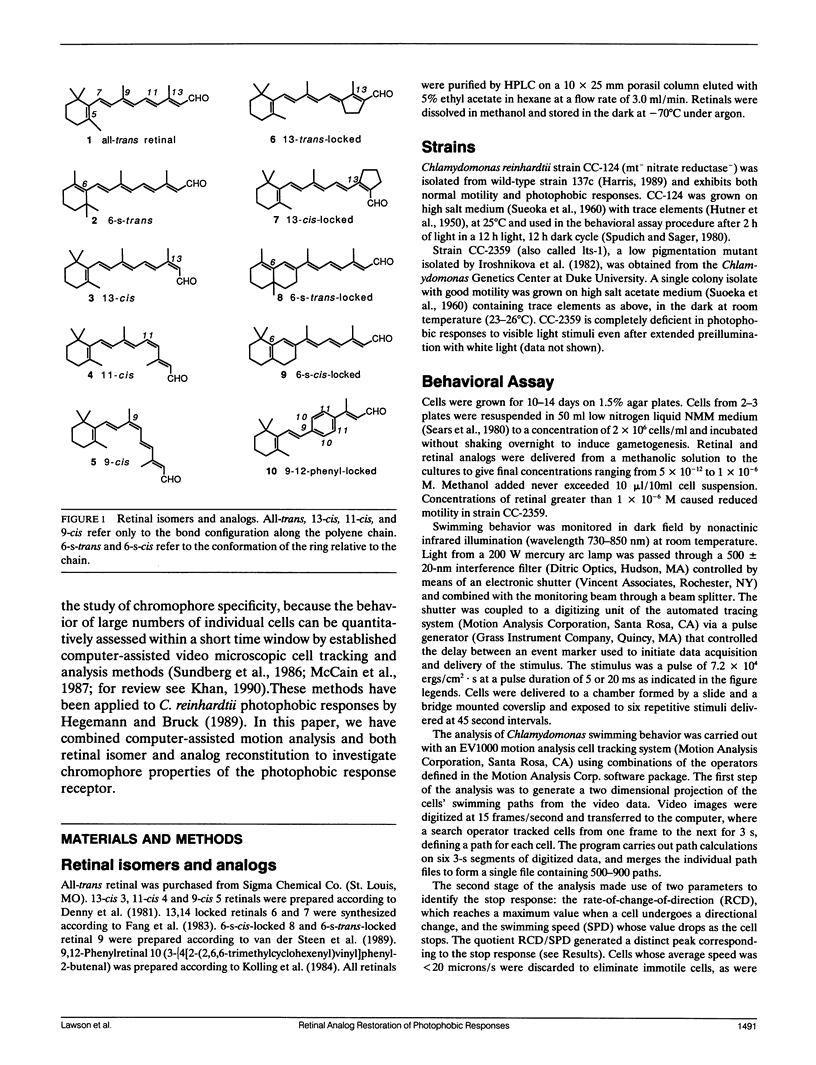
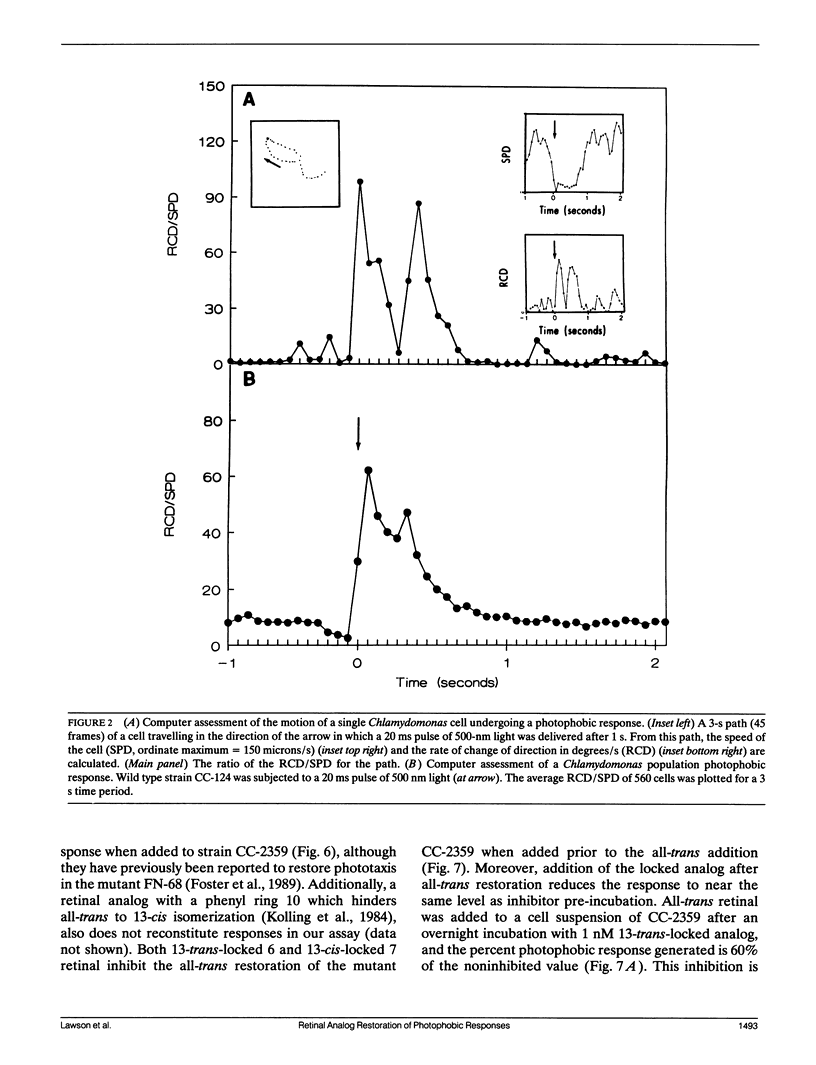
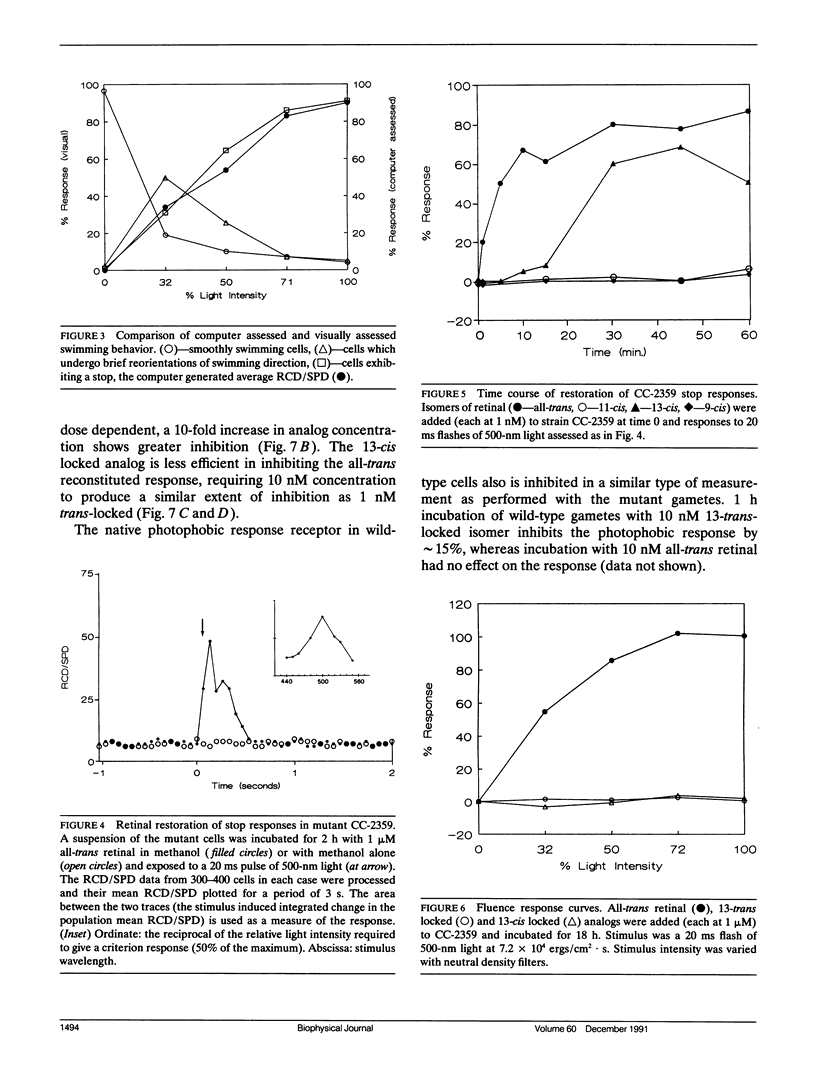
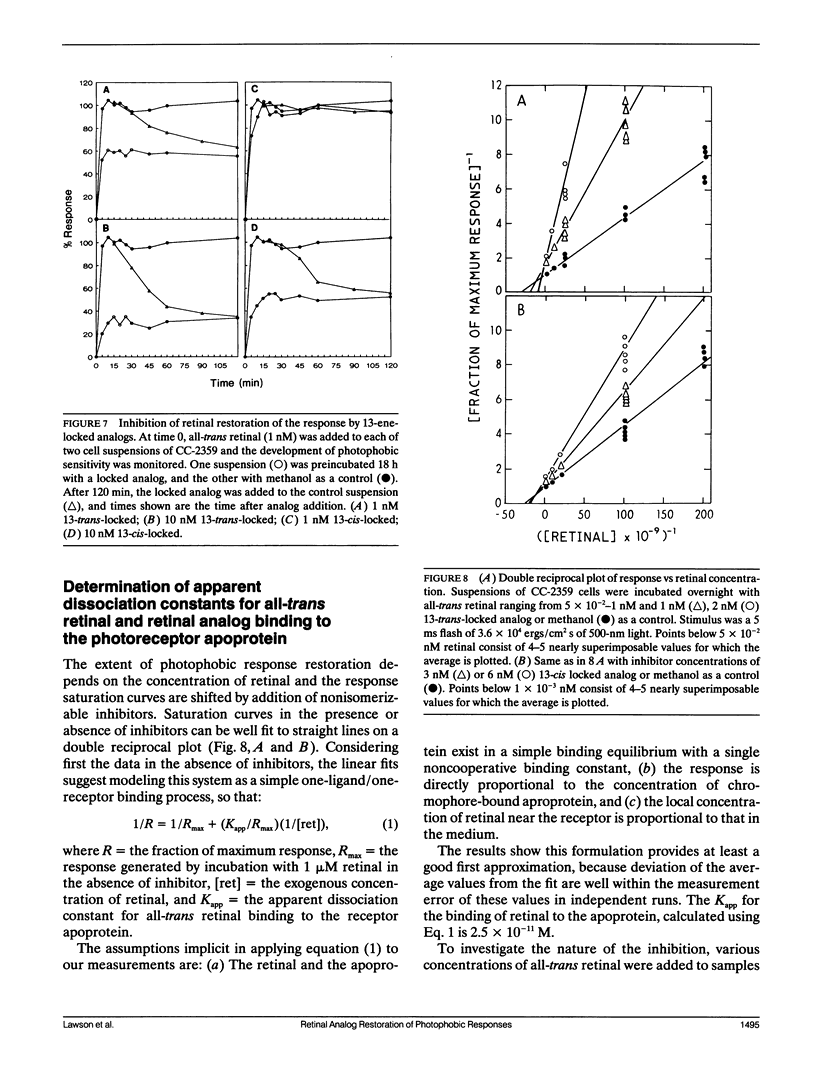
Abstract
The strain CC-2359 of the unicellular eukaryotic alga Chlamydomonas reinhardtii originally described as a low pigmentation mutant is found to be devoid of photophobic stop responses to photostimuli over a wide range of light intensities. Photophobic responses of the mutant are restored by exogenous addition of all-trans retinal. We have combined computer-based cell-tracking and motion analysis with retinal isomer and retinal analog reconstitution of CC-2359 to investigate properties of the photophobic response receptor. Most rapid and most complete reconstitution is obtained with all-trans retinal compared to 13-cis, 11-cis, and 9-cis retinal. An analog locked by a carbon bridge in a 6-s-trans conformation reconstitutes whereas the corresponding 6-s-cis locked analog does not. Retinal analogs prevented from isomerization around the 13-14 double bond by a five-membered ring in the polyene chain (locked in either the 13-trans or 13-cis configuration) do not restore the response, but enter the chromophore binding pocket as evidenced by their inhibition of all-trans retinal regeneration of the response. Results of competition experiments between all-trans and each of the 13-locked analogs fit a model in which each chromophore exhibits reversible binding to the photoreceptor apoprotein. A competitive inhibition scheme closely fits the data and permits calculation of apparent dissociation constants for the in vivo reconstitution process of 2.5 x 10(-11) M, 5.2 x 10(-10) M, and 5.4 x 10(-9) M, for all-trans, 13-trans-locked and 13-cis-locked analogs, respectively. The chromophore requirement for the trans configuration and 6-s-trans conformation, and the lack of signaling function from analogs locked at the 13 position, are characteristic of archaebacterial rhodopsins, rather than the previously studied eukaryotic rhodopsins (i.e., visual pigments).
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baselt D. R., Fodor S. P., van der Steen R., Lugtenburg J., Bogomolni R. A., Mathies R. A. Halorhodopsin and sensory rhodopsin contain a C6-C7 s-trans retinal chromophore. Biophys J. 1989 Jan;55(1):193–196. doi: 10.1016/S0006-3495(89)82791-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann M., Hegemann P. In vitro identification of rhodopsin in the green alga Chlamydomonas. Biochemistry. 1991 Apr 16;30(15):3692–3697. doi: 10.1021/bi00229a014. [DOI] [PubMed] [Google Scholar]
- Birge R. R. Photophysics of light transduction in rhodopsin and bacteriorhodopsin. Annu Rev Biophys Bioeng. 1981;10:315–354. doi: 10.1146/annurev.bb.10.060181.001531. [DOI] [PubMed] [Google Scholar]
- Chabre M. Trigger and amplification mechanisms in visual phototransduction. Annu Rev Biophys Biophys Chem. 1985;14:331–360. doi: 10.1146/annurev.bb.14.060185.001555. [DOI] [PubMed] [Google Scholar]
- Creuzet F., McDermott A., Gebhard R., van der Hoef K., Spijker-Assink M. B., Herzfeld J., Lugtenburg J., Levitt M. H., Griffin R. G. Determination of membrane protein structure by rotational resonance NMR: bacteriorhodopsin. Science. 1991 Feb 15;251(4995):783–786. doi: 10.1126/science.1990439. [DOI] [PubMed] [Google Scholar]
- Foster K. W., Saranak J., Derguini F., Zarrilli G. R., Johnson R., Okabe M., Nakanishi K. Activation of Chlamydomonas rhodopsin in vivo does not require isomerization of retinal. Biochemistry. 1989 Jan 24;28(2):819–824. doi: 10.1021/bi00428a061. [DOI] [PubMed] [Google Scholar]
- Foster K. W., Saranak J., Patel N., Zarilli G., Okabe M., Kline T., Nakanishi K. A rhodopsin is the functional photoreceptor for phototaxis in the unicellular eukaryote Chlamydomonas. Nature. 1984 Oct 25;311(5988):756–759. doi: 10.1038/311756a0. [DOI] [PubMed] [Google Scholar]
- Harbison G. S., Smith S. O., Pardoen J. A., Courtin J. M., Lugtenburg J., Herzfeld J., Mathies R. A., Griffin R. G. Solid-state 13C NMR detection of a perturbed 6-s-trans chromophore in bacteriorhodopsin. Biochemistry. 1985 Nov 19;24(24):6955–6962. doi: 10.1021/bi00345a031. [DOI] [PubMed] [Google Scholar]
- McCain D. A., Amici L. A., Spudich J. L. Kinetically resolved states of the Halobacterium halobium flagellar motor switch and modulation of the switch by sensory rhodopsin I. J Bacteriol. 1987 Oct;169(10):4750–4758. doi: 10.1128/jb.169.10.4750-4758.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollevanger L. C., Kentgens A. P., Pardoen J. A., Courtin J. M., Veeman W. S., Lugtenburg J., de Grip W. J. High-resolution solid-state 13C-NMR study of carbons C-5 and C-12 of the chromophore of bovine rhodopsin. Evidence for a 6-S-cis conformation with negative-charge perturbation near C-12. Eur J Biochem. 1987 Feb 16;163(1):9–14. doi: 10.1111/j.1432-1033.1987.tb10729.x. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971 Sep 29;233(39):149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- Smith S. O., Palings I., Miley M. E., Courtin J., de Groot H., Lugtenburg J., Mathies R. A., Griffin R. G. Solid-state NMR studies of the mechanism of the opsin shift in the visual pigment rhodopsin. Biochemistry. 1990 Sep 4;29(35):8158–8164. doi: 10.1021/bi00487a025. [DOI] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Sensory rhodopsins of halobacteria. Annu Rev Biophys Biophys Chem. 1988;17:193–215. doi: 10.1146/annurev.bb.17.060188.001205. [DOI] [PubMed] [Google Scholar]
- Spudich J. L., Sager R. Regulation of the Chlamydomonas cell cycle by light and dark. J Cell Biol. 1980 Apr;85(1):136–145. doi: 10.1083/jcb.85.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. MITOTIC REPLICATION OF DEOXYRIBONUCLEIC ACID IN CHLAMYDOMONAS REINHARDI. Proc Natl Acad Sci U S A. 1960 Jan;46(1):83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. A., Alam M., Spudich J. L. Excitation signal processing times in Halobacterium halobium phototaxis. Biophys J. 1986 Nov;50(5):895–900. doi: 10.1016/S0006-3495(86)83530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Yan B., Mazur P., Derguini F., Nakanishi K., Spudich J. L. Color regulation in the archaebacterial phototaxis receptor phoborhodopsin (sensory rhodopsin II). Biochemistry. 1990 Sep 11;29(36):8467–8474. doi: 10.1021/bi00488a038. [DOI] [PubMed] [Google Scholar]
- Uhl R., Hegemann P. Probing visual transduction in a plant cell: Optical recording of rhodopsin-induced structural changes from Chlamydomonas reinhardtii. Biophys J. 1990 Nov;58(5):1295–1302. doi: 10.1016/S0006-3495(90)82469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G. The molecular basis of visual excitation. Nature. 1968 Aug 24;219(5156):800–807. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- Yan B., Takahashi T., Johnson R., Derguini F., Nakanishi K., Spudich J. L. All-trans/13-cis isomerization of retinal is required for phototaxis signaling by sensory rhodopsins in Halobacterium halobium. Biophys J. 1990 Apr;57(4):807–814. doi: 10.1016/S0006-3495(90)82600-X. [DOI] [PMC free article] [PubMed] [Google Scholar]


