Abstract
Attachment and invasion of host cells by apicomplexan parasites involve the exocytosis of the micronemal proteins (MICs). Most MICs are adhesins, which show homology with adhesive domains from higher eukaryote proteins and undergo proteolytic processing of unknown biological significance during their transport to micronemes. In Toxoplasma gondii, the micronemal homodimeric protein MIC3 is a potent adhesin that displays features shared by most Apicomplexa MICs. We have developed an original MIC3-binding assay by transfection of mammalian cells with complete or truncated MIC3 gene sequences and demonstrated that the receptor binding site of MIC3 is located in the N-terminal chitin-binding-like domain, which remains poorly accessible until the adjacent pro-peptide has been cleaved, and that binding requires dimerization. We have localized the dimerization domain in the C-terminal end of the protein and shown that it is able to convert MIC8, a monomeric micronemal protein sharing the MIC3 lectin-like domain, into a dimer able to interact with host cell receptors. These findings shed new light on molecular mechanisms that control functional maturation of MICs.
Keywords: adhesion/Apicomplexa/dimerization/microneme/pro-peptide
Introduction
The phylum Apicomplexa is a large group of mostly intracellular protozoan parasites of considerable medical and veterinary importance, responsible for diseases such as malaria, toxoplasmosis, neosporosis, coccidiosis and cryptosporidiosis. A key step in infection is host cell invasion. The sophisticated invasion apparatus of the Apicomplexa involves the sequential exocytosis of two types of apical secretory organelles, i.e. micronemes and rhoptries. Recent investigations have highlighted the central role of micronemes in the recognition–adhesion to host cells (for reviews see Menard, 2001; Soldati et al., 2001). Immunofluorescence observations of invading Apicomplexa parasites have shown that micronemal proteins (MICs) are rapidly redistributed onto the apical end of the parasite at the attachment site and then progressively capped, in an actin-dependent manner, toward the posterior end of the parasite, where they are finally released. This surface capping and posterior relocalization during penetration superimposes exactly with the timing of invasion. Moreover, in being able to bind to host cell surface receptors, MICs function as adhesins (Soldati et al., 2001). The recruitment of a high density of secretory adhesins on the surface of parasites at the time of invasion and their ability to interact with host cell receptors are likely to induce a tight attachment of the parasites to the host cell surface, which could then initiate invasion.
Many MICs are processed extensively either before storage in the microneme compartment or after release from micronemes (Soldati et al., 2001). This latter event may be responsible for disengaging receptors prior to completing invasion. The biological significance of the processing event occurring before sorting in micronemes and the proteases involved are still unknown. MICs contain structural domains that are morphologically and functionally conserved at the molecular level (Kappe et al., 1999; Di Cristina et al., 2000). Some of these domains present homology with known adhesive domains from higher eukaryote proteins (for a review see Tomley and Soldati, 2001). They include: (i) the type I repeat of thrombospondin; (ii) the I-domains (an integrin-like adhesive domain); (iii) epidermal growth factor (EGF)-like domains; (iv) the Apple domains; and (v) chitin-binding domains. These motifs are present in one or multiple copies and a growing number of possible combinations of these modules has been identified, so that every MIC is structurally unique.
Among Apicomplexa parasites, Toxoplasma gondii has the broadest host range. It is able to invade and infect virtually any nucleated cell from warm-blooded vertebrates. The molecular basis controlling host cell-type specificity has not yet been elucidated, but might involve receptor-mediated adhesive interactions and therefore the repertoire of MICs. This repertoire is currently expanding. It contains membrane-spanning or soluble MICs (Soldati et al., 2001). Recent studies have indicated that soluble and transmembrane MICs assemble into multimolecular complexes, and that transmembrane proteins function as escorts, targeting soluble adhesins to the micronemes (Meissner et al., 2002; Rabenau et al., 2001; Reiss et al., 2001). We have characterized, in T.gondii, a soluble micronemal protein, MIC3 (Achbarou et al., 1991). MIC3 is a dimeric 90 kDa protein, synthesized as 40 kDa precursors that are proteolytically processed to 38 kDa final products during their trafficking through the secretory pathway and before storage in mature organelles (J.F.Dubremetz, unpublished). It is escorted to micronemes by MIC8 (Meissner et al., 2002) and binds to all nucleated cells tested to date, and does so only in non-reducing conditions, indicating that an appropriate conformation of the protein is required for binding (Garcia-Reguet et al., 2000). MIC3 contains five partially overlapping EGF-like domains and a chitin-binding-like domain found in lectins and plant chitinases, which can be involved in protein–protein or protein–carbohydrate interactions, respectively. In this study we have carried out a structure–function analysis of MIC3, which has demonstrated that the lectin-like domain is responsible for the adhesion and that N-terminal pro-peptide cleavage and C-terminal dimer formation are needed to allow the expression of this binding property.
Results
In mammalian cells, MIC3 is synthesized as a dimeric pro-protein
In order to determine the functional domain(s) of MIC3, which promotes adhesion to cell surfaces, we have produced recombinant MIC3 proteins in eukaryotic cells and studied their interaction with host cells.
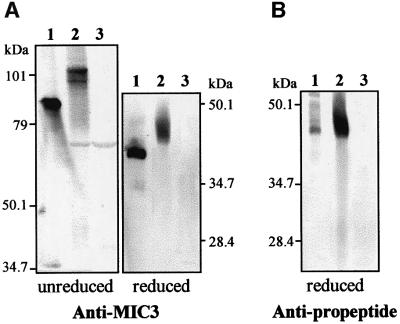
The entire MIC3 nucleotide-coding sequence was therefore cloned in the mammalian expression vector pcDNA3 and transiently expressed in baby hamster kidney (BHK)-21 cells. Lysates of transfected cells were then analyzed on immunoblots. As shown in Figure 1A, anti-MIC3 antibodies reacted specifically against cells transfected with the MIC3 gene. Recombinant MIC3 migrated at the size expected for a dimer (100 kDa) under unreducing conditions and for a monomer (42 kDa) in reducing conditions. This result showed that dimerization of recombinant MIC3 was obtained in mammalian cells. However, recombinant MIC3 migrated at a higher molecular weight than native MIC3 found in T.gondii tachyzoites. The difference was consistent with a failure to cleave the MIC3 pro-peptide in BHK-21 cells. This was confirmed by probing the immunoblots with a specific anti-pro-peptide serum. This serum strongly labeled recombinant MIC3 in BHK-21 cells and the small amount of immature MIC3 (proMIC3) found in tachyzoite lysate, providing direct evidence that the pro-peptide was not cleaved (Figure 1B). Identical results were obtained with 293 T or Vero cells transfected with the MIC3 gene (data not shown). This recombinant MIC3 protein was thereafter named R-proMIC3. Taken together, these results indicate that the natural processing of MIC3 involves a protease that does not traffic with MIC3 in mammalian cells, or is specific to T.gondii.
Fig. 1. In mammalian cells, the complete ORF of MIC3 is expressed as a dimeric pro-protein (R-proMIC3). Western blot analysis of cells transfected with MIC3 gene. Lane 1, T.gondii lysate; lane 2, lysate of BHK-21 cells transfected with plasmid p-SS-PRO-MIC3; lane 3, control (BHK-21 cells transfected with empty plasmid pcDNA3). Molecular weight standards are indicated. Anti-MIC3 mAb (A) and anti-pro-peptide serum (B) labeled the same band in immunoblot of cells transfected with the entire MIC3 gene. In contrast, anti- pro-peptide labeled only faintly the small amount of the proMIC3 in tachyzoite lysate, which co-migrated with recombinant MIC3.
Processing of the MIC3 pro-peptide is a prerequisite to the expression of the binding function of the protein
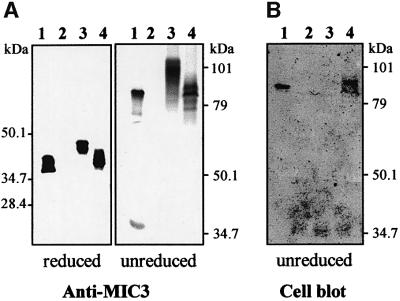
To determine whether R-proMIC3 possessed adhesin function, its interaction with putative host cells was tested in ‘cell blot’ experiments. Extracts of cells transfected with the MIC3 gene (p-SS-PRO-MIC3 plasmid) were separated by SDS–PAGE and transferred onto a nitrocellulose sheet, which was then incubated with Vero cell suspension. As expected, the cells bound strongly to native MIC3 (Figure 2B, lane 1). In contrast, cells were unable to bind to R-proMIC3 (lane 3). The possibility that the presence of the pro-sequence was inhibiting the adhesive function of the protein was then considered. We thus constructed a plasmid encoding the MIC3 signal sequence fused directly to the mature MIC3 coding sequence, and the resulting p-SS-MIC3 plasmid was transfected into BHK-21 cells. After 24 h of expression, cells were analyzed by western and cell blots. Anti-MIC3 antibodies reacted with protein bands migrating with molecular weights expected for MIC3 dimers in unreducing conditions and monomers in reducing conditions, indicating that the pro-sequence is not essential for dimerization of MIC3 (Figure 2A). This recombinant protein was thereafter named R-MIC3. It appeared as two closely migrating bands co-migrating with MIC3 from the T.gondii lysate. These two bands showed affinity to host cell surfaces in cell blot experiments (Figure 2B, lane 4), contrasting with the absence of binding of R-proMIC3 (lane 3) and showing that processing of the pro-peptide is a prerequisite for the expression of MIC3 binding properties.
Fig. 2. Recombinant R-MIC3 has a strong affinity to host cell surfaces. Western blot and cell blot analysis of cells transfected with MIC3 constructs. Lane 1, T.gondii lysate; lane 2, control (BHK-21 cells transfected with empty plasmid pcDNA3); lane 3, lysate of BHK-21 cells transfected with plasmid p-SS-PRO-MIC3; lane 4, lysate of BHK-21 cells transfected with plasmid p-SS-MIC3. Molecular weight standards are indicated. (A) The nitrocellulose membrane was probed with anti-MIC3 mAb. (B) A duplicate nitrocellulose membrane was incubated with BHK-21 cells, washed, and bound cells were stained with amidoblack (cell blot). Cells bind to native MIC3 (lane 1) and R-MIC3 (lane 4), but not to R-proMIC3 (lane 3).
R-MIC3 is strongly expressed at the cell surface of transfected mammalian cells, although it contains no transmembrane sequence
The presence of the recombinant proteins in the supernatant of transfected cells and their localization in cells were then investigated using western blotting and immunofluorescence microscopy. Sequences allowing expression of R-proMIC3 and R-MIC3 were first subcloned in-frame with a sequence coding for the V5 epitope, in pTRACER-A vector, which allows co-expression of the sequence of interest and of green fluorescent protein (GFP). GFP is expressed in the cytoplasm and its intrinsic fluorescence allows the direct visualization of transfected cells.
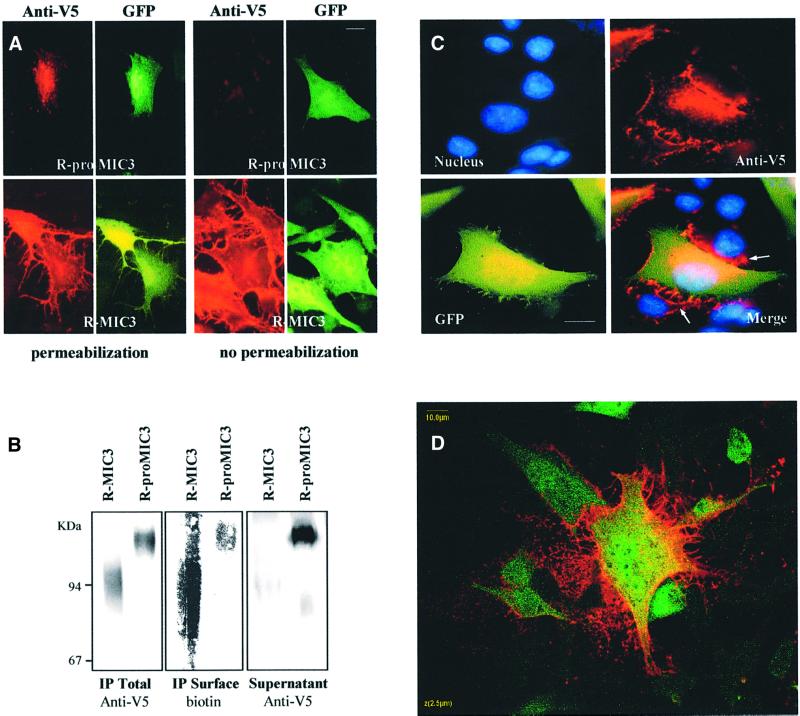
In transfected cells expressing R-proMIC3 and permeabilized after fixation, anti-MIC3 (data not shown) and anti-V5 antibodies (Figure 3A, left panel) gave a perinuclear and vesicular labeling pattern, suggesting that R-proMIC3 was present in the secretory pathway. Western blots of supernatant showed abundant secretion in the cell culture media (Figure 3B). GFP was not detected in supernatants (data not shown), confirming that the presence of R-proMIC3 in supernatants was not due to cell lysis. When cells were not permeabilized, a few fluorescent dots were found on some transfected cells (Figure 3A, right panel). In contrast, in cells expressing R-MIC3, strong anti-MIC3 (data not shown) or anti-V5 labeling was detected on transfected cells, permeabilized (Figure 3A, left panel) or not (right panel), indicating that R-MIC3 was bound to the transfected cell surface. The surface expression of R-MIC3 was essentially observed in transfected cells (GFP fluorescence), but also in some cases in non-transfected adjacent cells (absence of GFP; Figure 3C). Apart from a putative signal peptide, MIC3 has no other hydrophobic region suggestive of a transmembrane domain. Therefore, these results indicate that surface localization of mature MIC3 occurs by direct binding with its cell surface receptor. Single optical sections in confocal analysis (Figure 3D) confirmed that, in contrast to GFP, which was homogeneously distributed in the cytosol, R-MIC3 was found essentially associated with the plasma membrane, with barely detectable cytosol staining. Remarkably, the labeling extended beyond the edge of the cell surface. To confirm the difference between R-proMIC3 and R-MIC3 in their affinity for the host cell surface, we biotinylated the surface proteins of transfected cells, immunopurified recombinant MIC3 proteins from the cells and compared the amount of R-proMIC3 and R-MIC3 at the cell surface (streptavidin probing), and analyzed protein secretion in supernatants. When comparing similar amounts of immunopurified proteins, more R-MIC3 than R-proMIC3 was found to be biotinylated, showing a higher expression on the surface, whereas comparison of similar volumes of supernatant showed a large amount of R-proMIC3 and little R-MIC3, demonstrating that the absence of R-proMIC3 on the surface was not due to a defect in secretion (Figure 3B). Taken together, these results showed that the signal peptide of MIC3 allowed correct sorting of recombinant proteins into the secretory pathway of mammalian cells and that recombinant R-MIC3 bound to the surface of transfected cells, whereas R-proMIC3 had a much lower affinity for the host cell surface.
Fig. 3. Immunofluorescence assay (IFA) and biotinylation demonstrate the difference in affinity to host cell surface between R-MIC3 and R-proMIC3. (A) Localization of recombinant MIC3 proteins in transfected cells by IFA. BHK-21 cells were transfected with pOC1 or pOC2 (see Figure 4A), which encode, respectively, R-proMIC3 and R-MIC3. Intracellular staining was performed on permeabilized cells with mAb anti-V5 followed by an anti-mouse antibody conjugated to TRITC (left panel). Surface expression of recombinant proteins was performed on unpermeabilized cells (right panel). GFP fluorescence (green) monitored transfected cells. In contrast to R-proMIC3, which is barely detectable in the absence of cell permeabilization, mature R-MIC3 covers the entire cell surface of transfected cells. Scale bar = 10 µm. (B) Comparative distribution of R-MIC3 and R-proMIC3 on cell surface and in supernatants. Transfected cells expressing R-proMIC3 or R-MIC3 were surface biotinylated and recombinant MIC3 proteins were immunopurified from cell lysates with anti-MIC3 agarose. The western blot was revealed by anti-V5–alkaline phosphatase (IP Total) and streptavidin–alkaline phosphatase conjugates (IP Surface). Note that equal amounts of R-proMIC3 and R-MIC3 were immunoprecipitated, in contrast to unequal amounts that were biotinylated, R-MIC3 being strongly biotinylated. Analysis of supernatants obtained from the same number of cells shows that R-proMIC3 is highly enriched, whereas R-MIC3 is almost absent. (C) Three-color overlay (green, GFP; red, anti-V5; blue, Hoechst DNA stain) of a cell monolayer transfected with pOC2, showing surface binding of R-MIC3 on neighboring untransfected BHK-21 cells (arrows). Scale bar = 10 µm. (D) Single confocal section through a BHK-21 cell expressing R-MIC3 (on the surface) and GFP (in cytoplasm). Scale bar = 10 µm.
Dimerization plays a crucial role in the expression of the binding properties of the lectin-like motif
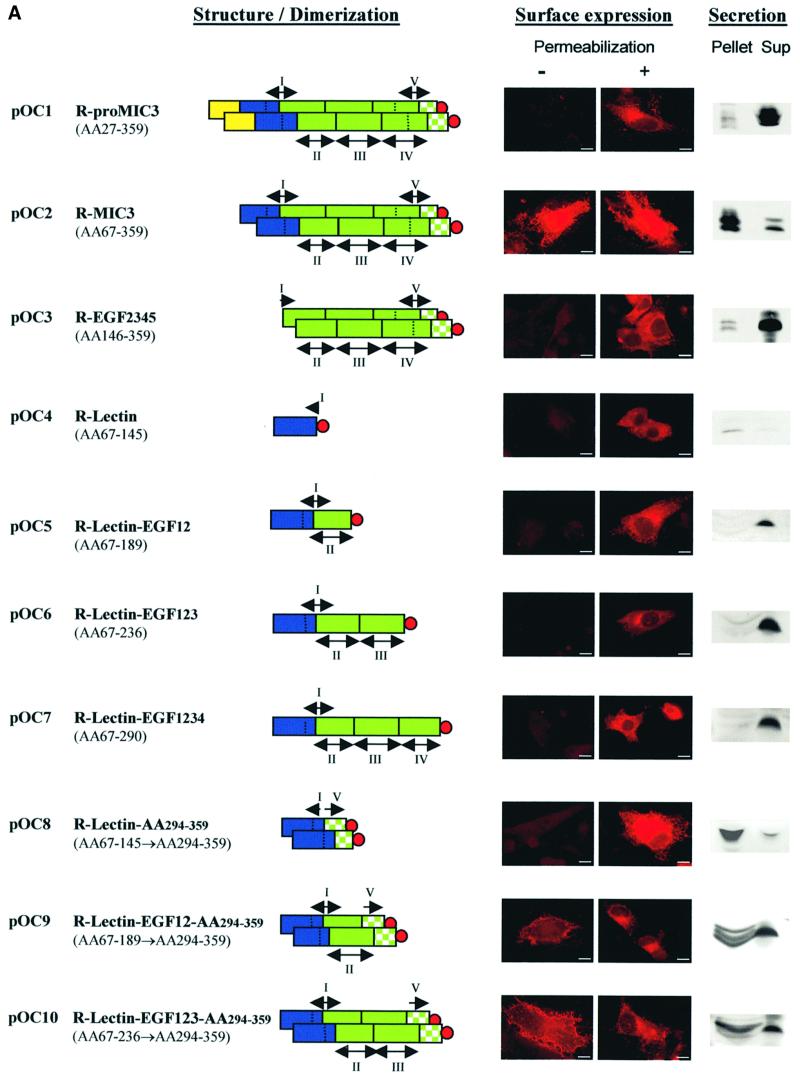
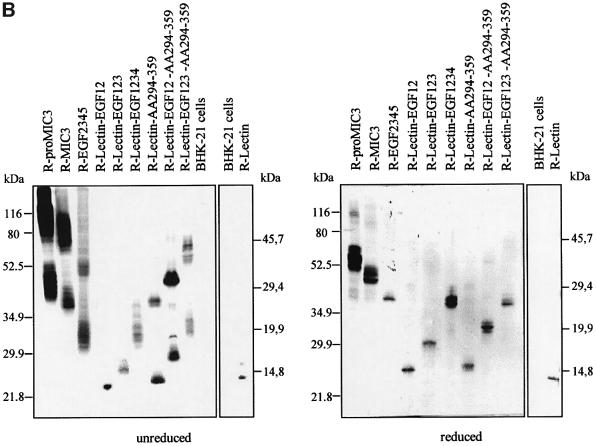
The deduced protein sequence of MIC3 contains a lectin-like domain (Wright et al., 1991) and five EGF-like domains, of which three are found in tandem (EGF234) whereas two other less conserved domains overlap with the others (EGF1 and EGF5; Figure 4A).
Fig. 4. Analysis of the binding domain of MIC3. (A) Schematic drawings of the MIC3 constructs produced. The color code is yellow for pro-peptide, blue for lectin and green for tandemly repeated EGF domains. EGF domains are numbered as indicated. The two overlapping EGF domains are I and V. The C-terminal stretch (amino acids 294–359) is represented by a checkered green area. The V5 epitope tag is indicated by a red circle. BHK-21 cells were transfected with plasmids pOC1–pOC10. Intracellular or surface detection were performed with mAb anti-V5 (red) on cells permeabilized or not before incubation, respectively. GFP fluorescence monitored transfected cells. Surface staining was considered as an indicator of the binding property of the recombinant protein. Scale bar = 10 µm. Secretion was monitored by western blotting: identical fractions of cell monolayers and corresponding supernatants after 18 h of expression were analyzed in reduced conditions and probed with anti-V5. Note that, except for the chimera R-lectin–AA294–359, all non-adhesive recombinant proteins were well secreted (amount in supernatant greater than amount in pellet), in contrast to adhesive proteins, which were present in supernatant and pellet, most likely resulting from their surface association. (B) Dimerization status of MIC3 constructs expressed in BHK-21 cells. Cell lysates (pellet) of BHK-21 cells expressing the different constructs were separated by SDS–PAGE in reduced or unreduced conditions, western blotted and probed with anti-V5 antibodies. Molecular weight standards are indicated.
In order to identify domains of MIC3 essential for the binding process, we cloned the corresponding sequences in pTRACER-A, transfected mammalian cells, and analyzed the recombinant proteins by immunofluorescence, taking advantage of the capacity of recombinant mature MIC3 to relocalize at the surface of transfected cells and using this parameter as an indicator of the binding properties. All constructs contained the MIC3 signal peptide, were deleted of the pro-peptide and fused at their C-terminal end to the V5 epitope. The efficiency of secretion of the different constructs was analyzed by western blotting of equivalent amounts of cells and supernatants after 18 h of expression. Dimerization status was analyzed by western blotting in reduced and unreduced conditions. The nomenclature of recombinant proteins, their dimerization, their binding properties and their secretion are summarized in Figure 4. Consistent with the strong binding properties of R-MIC3, a large amount of this protein was present in pellets compared with the supernatant (Figure 4A). Conversely, in the absence of binding, R-proMIC3 was essentially secreted in the supernatant. The mobility shift observed for R-proMIC3 and R-MIC3 compared with that in Figure 2B is consistent with the addition of V5 epitope followed by His6 residues. Two recombinant proteins were then generated that comprised either the EGF-like motifs or the lectin-like motif. The recombinant EGF2345 protein was expressed as variable amounts of protein bands migrating with molecular weights expected for dimers in non-reducing conditions and monomers in reducing conditions (Figure 4B). They were all recognized by anti-MIC3 antibodies (data not shown). The recombinant EGF2345 protein was dimeric and abundantly secreted, but did not bind to the surface of transfected cells (Figure 4A) and failed to bind cells in cell blot experiments (data not shown). When expressed alone, the lectin-like domain was expressed as a monomer (Figure 4B). This construct was always weakly expressed and secreted, and no transfected cell expressed the protein at the surface. These results indicate that the lectin-like domain is probably essential for binding, and has no potential for dimerization. New constructs were generated in which EGF-like motifs were successively added to the lectin-like domain. In summary, R-lectin–EGF12, R-lectin–EGF123 and R-lectin–EGF1234 were all monomerics, secreted, and not adhesins (Figure 4). These results suggest that the C-terminal part of MIC3 is involved in dimerization and that the dimerization may be essential for the adhesive function.
Finally, the last amino acids (294–359) of MIC3 were fused to R-lectin, R-lectin–EGF12 or R-lectin–EGF123 to create R-lectin–AA294–359, R-lectin–EGF12–AA294–359 and R-lectin–EGF123–AA294–359 chimeras. The three chimeras were dimerics (Figure 4B), and R-lectin– EGF12–AA294–359 and R-lectin–EGF123–AA294–359 chimeras exhibited surface relocalization in transfected cells (Figure 4A). R-lectin–AA294–359 was poorly secreted and accumulated in wide perinuclear vesicules. The bind ing properties of this construct were, therefore, difficult to analyze. A gradual increase in the intensity of sur face detection was observed (R-lectin–EGF12–AA294–359 < R-lectin–EGF123–AA294–359 < R-MIC3), parallel to an increasing retention of the proteins in pellets. Taken together, these results showed that the amino acid stretch 294–359 is sufficient for dimerization and that dimerization plays a crucial role in the expression of the binding properties of the lectin-like motif, which may itself be modulated by the addition of the EGF motifs.
Dimerization of another lectin-like-containing protein, MIC8, leads to acquisition of adhesive function
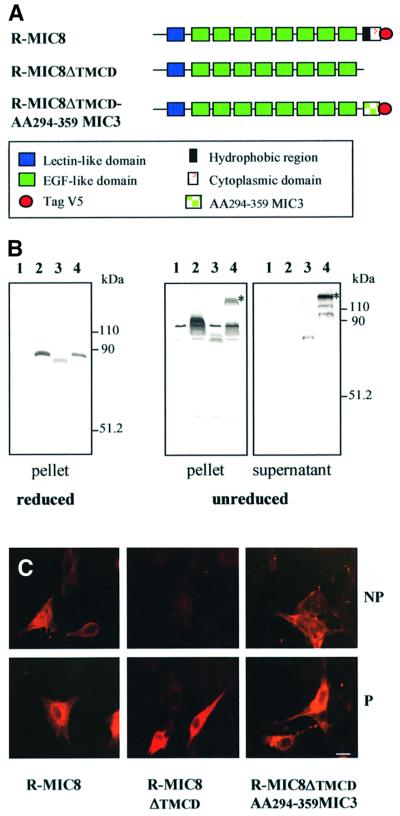
MIC3 shares a significant degree of homology with the recently described T.gondii microneme protein MIC8 (Meissner et al., 2002). The two proteins contain a peptide signal, a chitin-binding-like domain and several EGF-like domains (Figure 5A). In MIC8, the last EGF motif is followed by a hydrophobic region, likely to promote transmembrane insertion, and a short cytoplasmic domain. MIC8 lacks the dimerization domain of MIC3, and is detectable in immunoblots of tachyzoites as a monomer of uncharacterized adhesive properties. Homologous to what we had observed with MIC3, we suspected that dimerization of MIC8 could reveal adhesive properties.
Fig. 5. Replacement of the transmembrane insertion domain of MIC8 by the C-terminal end of MIC3 led to dimerization of MIC8 and acquisition of adhesive function. (A) Schematic drawing of the domains of MIC8 and recombinant MIC8 constructs produced. (B) Western blot analysis of MIC8 constructs. Supernatant and corresponding cell lysates (pellet) of BHK-21 cells expressing full-length MIC8 (lane 2), MIC8ΔTMCD (lane 3) and the chimera MIC8ΔTMCD–AA294–359-MIC3 (lane 4) were western blotted and probed with rabbit anti-MIC8 serum. Lane 1 shows cells transfected by empty vector. Molecular weight standards are indicated. The chimera is secreted as a dimer also present in the pellet. The asterisk indicates the band corresponding to the dimer. (C) Intracellular detection and surface expression of MIC8 constructs were performed on cells permeabilized (P) or not (NP) by incubation with rabbit anti-MIC8 serum. The chimera is detected on the cell surface. Scale bar = 10 µm.
New constructs were generated in pTRACER-A to transiently express: (i) R-MIC8; (ii) R-MIC8ΔTMCD (deletion of transmembrane and cytoplasmic domains); and (iii) the chimera R-MIC8ΔTMCD–AA294–359MIC3 in which the dimerization stretch of MIC3 was fused in-frame with the C-terminal end of MIC8ΔTMCD (Figure 5A). A V5 epitope sequence was fused at the end of R-MIC8 and of the chimera. Dimerization and binding properties were analyzed as described above, using anti-V5 or anti-MIC8 serum (this serum was raised against EGF domains and does not recognize the last 66 amino acids of MIC3).
R-MIC8 was strongly expressed at the plasma membrane of transfected cells (Figure 5C). Under permeabilization, both polyclonal anti-MIC8 and anti-V5 (data not shown) strongly labeled R-MIC8, while in the absence of permeabilization, only anti-MIC8 serum revealed R-MIC8 at the surface of transfected cells. The inaccessibility of the C-terminal V5 epitope suggested that the C-terminal hydrophobic region of MIC8 promoted efficient type 1 transmembrane insertion in mammalian cells. Consist ently, R-MIC8 was not found in culture media (Figure 5B, lane 2). As expected, deletion of the transmembrane domain (R-MIC8ΔTMCD) led to secretion of the protein in supernatants (Figure 5B, lane 3). R-MIC8ΔTMCD was not found on the surface of transfected cells (Figure 5C). While R-MIC8 and R-MIC8ΔTMCD were exclusively detected as monomers in immunoblots, R-MIC8ΔTMCD– AA294–359MIC3 was detected as a monomer and a dimer with both anti-MIC8 (Figure 5B, lane 4) and anti-V5 (data not shown) antibodies. The recombinant chimera was detected in supernatant essentially as a dimer. It was detected by IFA at the surface of transfected cells, with both anti-MIC8 (Figure 5C) and anti-V5 (data not shown) antibodies, showing that unlike R-MIC8, the surface localization of R-MIC8ΔTMCD–AA294–359MIC3 was not due to transmembrane insertion and, like R-MIC3, probably resulted from direct binding of the chimera with cell surface molecules. Taken together, these results confirmed that amino acids 294–359 of MIC3 allow dimerization, and that such dimerization could induce binding of a MIC3 homolog.
Dimerization of MIC3 occurs through protein–protein interaction
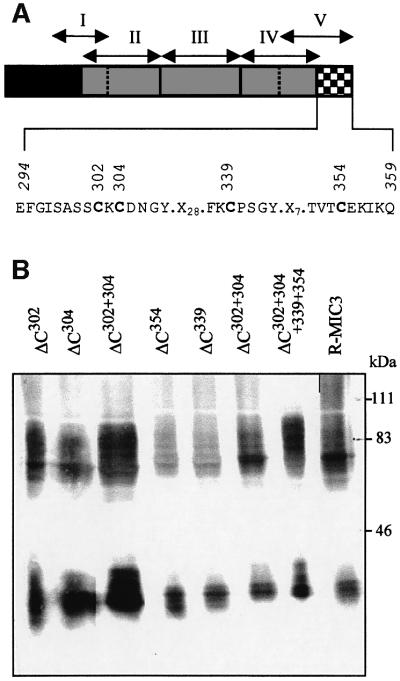
Dimerization of proteins may occur through inter-chain disulfide bonds or through protein–protein dimerization motifs like leucine zipper or helix–loop–helix motifs. No consensus for such motifs was found in the dimerization stretch of MIC3. However, four cysteines were present. All four cysteine residues were mutagenized individually to glycine, and the dimerization of recombinant mutated proteins was analyzed (Figure 6). No single cysteine mutation (Cys302, Cys304, Cys339 or Cys354) abolished dimerization (Figure 6B). We then performed double mutations (Cys302,304 and Cys339,354) and a quadruple mutation (Cys302,304,339,354), but all mutants dimerized (Figure 6B). These data clearly indicated that dimerization through the amino acid stretch 294–359 does not involve inter-chain disulfide links and is probably due to protein– protein interaction.
Fig. 6. Expression of site-directed mutagenized R-MIC3 proteins in BHK-21 cells. (A) Schematic drawing of the domains of MIC3 and position of substituted amino acids. The sequence of the dimerization stretch is given and the cysteine residues are indicated in bold letters. (B) BHK-21 cells were transfected with plasmids pOC302, pOC304, pOC339, pOC354, pOC302-304, pOC339-354 and pOC302-304-339-354 coding for R-MIC3 with cysteine mutations in the dimerization stretch. Cell lysates were separated by SDS–PAGE in unreduced conditions, western blotted and probed with anti-V5 antibody. Molecular weight standards are indicated. Dimerization occurs for all constructs.
Discussion
During their transport to micronemes, some of the MICs undergo proteolytic processing of unknown biological significance. The data reported here stress the importance of two post-translational events, N-terminal pro-peptide cleavage and C-terminal dimer formation, in the expression of the binding properties of the micronemal protein MIC3. We also show that EGF-like domains of MIC3 are not able to promote recognition of host cell receptor, a function that can be attributed essentially to a chitin-binding-like domain.
Development of a MIC3 functional binding assay by gene transfection of mammalian cells
Our data show convincingly that expression of mature MIC3 within the eukaryotic secretory pathway leads to a correctly folded, secreted and dimeric recombinant protein, which displays adhesive function. Binding of MIC3 to a cell surface receptor may occur either in the secretory pathway or after secretion. These two possibilities are not mutually exclusive, but the occurrence of the second one is supported by the presence of R-MIC3 on untransfected neighboring cells. To our knowledge, this is the first description of surface localization of a soluble protein expressed in a heterologous system. We then targeted different MIC3 domains into the BHK-21 cell secretory pathway and characterized their adhesive properties by surface localization. Except for the chimera R-lectin–AA294–359, all non-adhesive recombinant proteins were well secreted (amount in supernatant greater than amount in pellet), in contrast to the retention of the adhesive proteins in the pellet, most likely resulting from their surface association. The dimerization and binding observed in mammalian cells were not due to an artefact of the heterologous expression system used, since the truncated forms of MIC3, which were expressed in mammalian cells as (i) monomers (lectin–EGF12 or lectin–EGF123), (ii) dimers (lectin–AA294–359 chimera) or (iii) retained in the secretory pathway (R-MIC3-Cys107), behave the same way when expressed in parasites (data not shown). Moreover, as in mammalian cells, lectin–EGF123 expressed in tachyzoites did not display binding function in cell blot experiments (data not shown), although it was correctly targeted to micronemes. We observed a great deal of heterogeneity in the migration of the MIC3 constructs in western blots of pellets of transfected cells. We do not believe that this is due to the presence of the two N-glycosylation sites in the MIC3 sequence, since site-directed mutagenesis of asparagine residues present in the N-glycosylation consensus sites did not suppress the heterogeneity of the bands in R-proMIC3 (unpublished observations). Interestingly, such heterogeneity was significantly reduced in the supernatant (only one or two bands), indicating that the multiple bands correspond probably to incorrectly folded proteins that are not secreted.
We suggest that this new suface-binding assay could be used for identification of putative adhesins or characterization of adhesive properties of polypeptides, avoiding the use of specific antibodies and purified recombinant proteins.
Proteolytic cleavage is a critical step in the functional maturation of MIC3
Many MICs are processed extensively before storage in their target compartment (Soldati et al., 2001), and this is the case for MIC3. This processing occurs in the trans-Golgi network (J.F.Dubremetz, unpublished) and typically involves the removal of an N-terminal pro-peptide, but the significance of this process is still unknown. In mammalian proteins, the N-terminal pro-domain may be required for proper folding of the catalytic domain in proteases or cytokines, intracellular trafficking, secretion of the mature protein or in the control of proteolytic activity of proteinases by acting as a substrate-mimicking inhibitor (Gray and Mason, 1990; Taylor et al., 1995; Bauskin et al., 2000). Our study has shown that the pro-peptide is not required for secretion by mammalian cells or stabilization of MIC3 dimer, but also revealed the importance of the removal of the MIC3 pro-peptide to express efficient binding. The pro-peptide is located just upstream of the lectin-like domain and, therefore, it may be involved in preventing the interaction of MIC3 with a possible receptor until the protein is in its definitive storage compartment. Interestingly, N-terminal processing of the galactose-specific lectin of Sarcocystis muris also occurs before storage in the microneme compartment (Klein et al., 1996).
The chitin-binding-like sequence is the adhesive motif
By expression of a different segment of the MIC3 sequence, we showed that the chitin-binding domain, in a dimeric form, is required for binding. It is a disulfide-rich domain, first identified in wheat germ agglutinin, and then in a large number of other plant chitin-binding proteins (Wright et al., 1991). This domain has been implicated in the binding of N-acetylglucosamine and its polymers. Carbohydrate binding experiments were performed, but they did not reveal any interaction between MIC3 and GlcNAc, chitobiose or chitotriose (not shown), suggesting a different specificity between MIC3 and the plant lectins sharing sequence homologies. In our transfection experiments, the staining for R-MIC3 extended beyond the cell surface, suggesting that MIC3 also recognized extracellular matrix molecules. Analysis of the human genome shows that the EGF family is the fifth most common protein family, with 3% of all potential proteins containing EGF domains. The EGF module, which often occurs as multiple tandem repeats, is widely distributed among extracellular proteins involved in protein–protein interaction. This type of domain is being found in a growing number of MICs in Apicomplexa (Garcia-Reguet et al., 2000; Meissner et al., 2002; Tomley et al., 2001). In this work, we provide evidence that EGF-like domains alone are not able to promote the adhesive property of MIC3 to cells, but may contribute to correct exposure of the binding motif, and may then play a role in conformation. In addition, the EGF domains may be involved in the association between MIC3 and its escort MIC8 (Meissner et al., 2002), as has been shown for the third EGF-like domain of MIC6, which is a transmembrane micronemal protein that escorts the two soluble proteins, MIC1 and MIC4, to micronemes (Reiss et al., 2001).
Dimerization of MIC3 does not depend on disulfide bonds and involves a previously undescribed dimerization sequence
The dimerization stretch has been mapped in the C-terminal part of the protein (amino acids 294–359). The dimerization does not involve intermolecular disulfide bonds, since site-directed mutagenesis of all the cysteine residues present in the dimerization stretch did not impair the formation of dimers, and, in addition, a small amount of the dimeric form of MIC3 or recombinant MIC3 proteins is resistant to reduction when boiled in the presence of SDS and dithiothreitol (DTT), even when using iodoacetamide to alkylate sulfhydryl groups. Intramolecular disulfide bridges in other domains of the molecule are apparently necessary and probably act in stabilizing the dimerization of the protein. A similar observation has been reported in the case of mixed lineage kinase-3 (MLK-3) homodimerization form, where the leucine zipper motif is responsible for dimerization (Leung and Lassam, 1998). No dimerization consensus such as helix–turn–helix, leucine zipper motifs or short peptide motifs (i.e. KXXXTVXXXE of cytosolic transferases, GVXXGVXXA of neurotransmitter transporters) (Hastrup et al., 2001; Petrotchenko et al., 2001), known to mediate strong homodimerization, are found in the MIC3 sequence. Except for Ncp38, which is the homolog of MIC3 in Neospora caninum, we failed to detect homologous sequences in databases. Interestingly, NcMIC3 is dimeric. In contrast, MIC8 does not possess this sequence and appears monomeric when analyzed in western blots of T.gondii tachyzoites. Here, we show that replacement of the C-terminal transmembrane domain of the monomeric MIC8 with the dimerization stretch of MIC3 leads to a dimeric chimer, which is also partially resistant to reduction by boiling in the presence of SDS and DTT, confirming that the dimerization motif of MIC3 promotes strong protein–protein interaction. Further deletions or site-directed mutagenesis analysis will be necessary to precisely delineate this new dimerization motif.
Oligomerization is necessary for adhesion
In this study, we demonstrated that the dimeric form of MIC3 displays adhesive function that monomers are unable to perform. Interestingly, dimerization was also shown to be a prerequisite for other micronemal proteins to bind sugars, i.e. for a recombinant circumsporozoite protein of Plasmodium falciparum (Cerami et al., 1992) and for the major micronemal protein of S.muris (Klein et al., 1998). Dimerization and oligomerization are one of the strategies for enhancing both the affinity and specificity of lectins toward carbohydrate ligands (Rini, 1995). The process of dimer formation may juxtapose two lectin-like domains and increase affinity through proximity. Multimer formation may also cause a conformational change of the lectin-like binding motif in each MIC3 subunit.
MICs assemble into multimolecular complexes, the transmembrane proteins functioning as escorts of the soluble proteins to micronemes (Reiss et al., 2001). MICs are ligands, which, when secreted onto the parasite surface, allow a direct or indirect link between the actomyosin motor of the parasite (Kappe et al., 1999) and receptors present on the surface of target host cells. We have previously shown that MIC3 is rapidly redistributed towards the posterior pole of the parasite during invasion (Garcia-Reguet et al., 2000), presumably via its interaction with the transmembrane micronemal protein MIC8 (Meissner et al., 2002). A close association of the two proteins, which both contain a lectin-like domain, can lead to juxtaposition of binding sites that may induce (for MIC8) or increase (for MIC3) affinity for receptor. However, we cannot exclude the possibility that MIC8 self-associates in the membrane of parasites to form oligomers, which may bind cells. In Apicomplexa, oligomerization may be a crucial step to enhance avidity of adhesive domains present in MICs, which, when secreted at the surface of the parasite, allow a tight and local attachment to the host cell and initiation of invasion. Consistent with this model, MIC3 is probably a fundamental component of the T.gondii attachment/invasion process, and therefore an important goal of future investigation will be to dissect the role of MIC3 in this process.
Materials and methods
Reagents, plasmids and antibodies
Sulfosuccinimidobiotin was purchased from Sigma. Monoclonal antibodies (mAbs) T4 2F3 and T8 2C10, which react to the micronemal protein MIC3, were as described elsewhere (Garcia-Reguet et al., 2000). Monoclonal anti-V5, anti-V5 covalently linked to alkaline phosphatase and GFP antiserum were purchased from Invitrogen. Anti-MIC8 serum was kindly provided by M.Meissner and D.Soldati (Imperial College, London, UK).
Host cells and parasite cultures
RH strain T.gondii tachyzoites were maintained by serial passage in the peritoneal cavity of Swiss mice. Vero cells (ATCC CCL 81) were grown in Dulbecco’s modified Eagle’s medium (Bio Whittaker) supplemented with 5% fetal calf serum (FCS), 2 mM glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin. BHK-21 cells (ATCC CCL 10) were grown in BHK-21 medium (Gibco-BRL) supplemented with 5% FCS, 2 mM tryptose, 100 U/ml penicillin and 100 µg/ml streptomycin.
Transient transfection experiments and immunofluorescence
Plasmids were purified using the Qiagen kit and transfections were carried out using LipofectAMINE reagent (Gibco-BRL) as instructed by the manufacturer, with 3 × 105 BHK-21 cells grown on coverslips for 24 h in 6-well plates. Cells were grown for an additional 24 h before analysis. Some coverslips and corresponding supernatants were processed for western blot analysis (see below). For immunofluorescence analysis, cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min, washed and permeabilized with 0.1% Triton X-100 in PBS for 10 min. Coverslips were subsequently washed in PBS, 0.5% BSA, incubated in the same buffer containing mAb T8 2C10 (1:200) or anti-V5 (1:500) for 1 h and then with affinity-purified goat anti-mouse immunoglobulin G (IgG, whole molecule) conjugated to TRITC (1:400) (Sigma) for 1 h, with several washings in PBS between each incubation. Finally, the coverslips were washed and mounted onto microscope slides using Immunomount (Calbiochem). Confocal images were collected with an Olympus laser scanning confocal microscope Fluoview FV500-IX70 using a Planapo 60× oil NA 1.40 objective. All other micrographs were obtained with a Zeiss Axiophot microscope equipped for epifluorescence. Adobe Photoshop (Adobe Systems, Mountain View, CA) was used for image processing. Matching pairs of images were recorded with the same exposure time and processed identically.
Preparation of anti-pro-peptide serum
Antibodies specific for the MIC3 pro-peptide were raised against a fusion between glutathione S-transferase (GST) and peptide 17–62 of MIC3. The DNA sequence coding for amino acids 17–62 was amplified by PCR from a pBluescript II®SK plasmid containing the genomic MIC3 sequence (namely pBlueMIC3) (Garcia-Reguet et al., 2000) using primers 5′-TTCAGGAATTCCGTGTGGATGTGCACCCCAGCG-3′ and 5′-TTGCACACTCGAGTGAGTCTCAGTCACCGC-3′, and cloning into EcoRI and XhoI sites of pGEX-4T3 vector (Pharmacia). Expression of the recombinant pro-peptide fragment fused to GST was achieved in Escherichia coli strain BL21 after 4 h induction with isopropyl-β-d-thiogalactopyranoside. The protein was purified under native conditions according to the manufacturer. Antisera to purified GST–pro-peptide fusion were obtained from OF1 mice immunized with 25 µg of protein in complete Freund’s adjuvant and boosted twice at an interval of 2 weeks. Blood was collected 3 weeks after the final boost. Antiserum to the fusion protein is referred to as anti-pro-peptide. The specificity of the serum to proMIC3 was demonstrated by differential immunoprecipitation of radiolabeled proMIC3 and MIC3.
DNA constructs for mammalian expression
The primers used for constructions are listed in Table I.
Table I. Primers used for DNA constructs.
| Primer | Sequence |
|---|---|
| MNM1 | 5′-GTGTAAGCTTCTTGTCCAACACTGGTA-3′ |
| MNM2 | 5′-CACGGATATCTGCGAATGGGCG-3′ |
| ML1 | 5′-GCTCTAGATCCCCCAGCAAGCAGGAGA-3′ |
| ML2 | 5′-ATTAAGCTTTCAGCAGTCTCCTCCATAGCTTTTG-3′ |
| ML3 | 5′-GCTCTAGAAGCTGTGAAAAGCAGGGCCAT-3′ |
| ML4 | 5′-ATTAAGCTTTCACTGCTTAATTTTCTCACACGTCAC-3′ |
| ML5 | 5′-GATGAGCTCGCTAGCCTTGTCCAACACTGGTAAA-3′ |
| ML6 | 5′-GCTCTAGAAGCCTCCGCTGGGGTGCACAT-3′ |
| ML11 | 5′-GCACAATTGAGATCTAAAATGCGAGGCGGGACGTCC-3′ |
| ML13 | 5′-GCTCTAGATGCGGAGAATTCTGCTCCAGC-3′ |
| ML15 | 5′-TGCTATGCATTCCTAGGCTGCTTAATTTTCTCACACGTCAC-3′ |
| ML17 | 5′-CGGGATATCTCCTAGGGCAGTCTCCTCCATAGCTTTTG-3′ |
| ML21 | 5′-GCACAATTCCCTAGGTCCAGTCCTCTTGCATCCTTG-3′ |
| ML22 | 5′-GCACAATTCCCTAGGTTTCTCAGCCAGCGTGACTTC-3′ |
| ML25 | 5′-GCACAATTGCCTAGGGCAGTCTCCTCCATAGCTTTTGTC-3′ |
| ML26 | 5′-GCACAATTGCCTAGGAGGATCCTCGGAGCAAGTCAA-3′ |
| ML27 | 5′-GCACAATTGCCTAGGTCCAGTCCTCTTGCATCCTTG-3′ |
| ML28 | 5′-GGGGTACCAGATCTAAAATGCGAGGCGGGACGTCC-3′ |
| ML32 | 5′-TAGGATATCTTATTTCGAATATCGACCTTTGTT-3′ |
| ML33 | 5′-TAGGCTAGCGACAGAATGAAGGCCAATCGA-3′ |
| ML40 | 5′-GCTCTAGAGGACCAGATACCGCCCGAAGG-3′ |
| ML41 | 5′-CGGAATTCTTTCGAATATCGACCTTTGTTGTC-3′ |
Plasmid pSS-PRO-MIC3 is designed to transfect mammalian cells with the complete MIC3 ORF. It was constructed by PCR amplification of the MIC3 gene from pBlueMIC3 with primers MNM1/2, and cloning into NotI and HindIII sites of pcDNA3. Plasmid pSS-MIC3 was designed to express the mature MIC3 protein spanning amino acids 67–359, such as the one stored in Toxoplasma micronemes. It was obtained by deletion of the pro-peptide sequence by PCR ligation–PCR amplification. The signal peptide sequence and the mature sequence of MIC3 were PCR-amplified independently with primers ML5/6 and ML1/4, respectively. Both XbaI-digested PCR fragments were ligated together. The ligation product was PCR-amplified with primers ML5/4. The PCR fragment was cloned into NheI and HindIII sites of pcDNA3.1. Plasmid pOC1 was constructed by PCR amplification of the MIC3 gene from pBlueMIC3 with primers ML11/15 and cloning into EcoRI and XbaI sites of pTRACER-A. Plasmid pOC2 was constructed by PCR amplification of MIC3 mature sequence from pSS-MIC3 with primers ML11/15, and cloning into EcoRI and XbaI sites of p-TRACER-A. Plasmid pOC3 was constructed in pTRACER-A by the same PCR ligation–PCR approach described for pSS-MIC3, using primers ML11/6 and primers ML3/15 for the first PCR amplifications and ML11/15 for the second PCR amplification. The second PCR fragment was cloned into plasmid pTRACER-A digested by EcoRI–XbaI. Plasmids pOC4, pOC5, pOC6 and pOC7 were constructed in pTRACER-A by PCR amplification of the sequences from pSS-MIC3 with primers ML11/17, ML28/26, ML11/21 and ML11/22, respectively, digestion of the PCR fragments by MfeI–EcoRV for pOC4, KpnI–MfeI for pOC5 and MfeI–AvrII for pOC6 and pOC7, and cloning into pTRACER-A digested by EcoRI–EcoRV for pOC4, KpnI–EcoRI for pOC5 and EcoRI–XbaI for pOC6 and pOC7. Plasmids pOC8, pOC9 and pOC10 were constructed by PCR amplification from pSS-MIC3 with primers ML28/25, ML28/26 and ML28/27, respectively, digestion of the PCR fragments by KpnI and MfeI, and cloning into pOC2 digested by KpnI and EcoRI. Plasmid pOC11 was designed to express the complete ORF of MIC8. It was constructed by PCR amplification of a plasmid containing the MIC8 sequence with primers ML33/40, digestion by NheI and XbaI, and cloning into pTRACER-A digested by NheI and XbaI. Plasmid pOC12 was designed to express MIC8ΔTMCD (transmembrane and cytoplasmic tail deletions). It was constructed by PCR amplification of the MIC8 gene with primers ML32/33, digestion of the PCR fragment by NheI and EcoRV, and cloning into pTRACER-A digested by NheI and SmaI. Plasmid pOC13 was designed to express a chimeric protein containing MIC8ΔTMCD fused to the C-terminal dimerization stretch of MIC3 (MIC8ΔTMCD–AA294–359MIC3) (amino acids 1–599 of MIC8 fused to amino acids 294–359 of MIC3). It was constructed by PCR amplification of MIC8ΔTMCD with primers ML33/41, digestion of the PCR fragment by NheI and EcoRI, and cloning into pOC2 digested by NheI and EcoRI.
Except for pOC12, all the constructs obtained in pTRACER-A were designed to express recombinant proteins with a V5 epitope followed by six histidine residues at the C-terminal end. All constructs were verified by sequencing.
SDS–PAGE, western blotting and cell blot assays
SDS–PAGE was performed according to Laemmli (1970). Freshly released tachyzoites or transfected cells were boiled in SDS sample buffer with (reduced) or without (unreduced) 0.1 M DTT and separated on 8–15% polyacrylamide gels. Proteins were transferred to nitrocellulose membranes. Western blots were probed as described previously (Garcia-Reguet et al., 2000) with mAbs anti-MIC3 T4 2F3 or T8 2C10 (1:400), mouse anti-pro-peptide (1:200), rabbit anti-GFP (1:2000), mouse anti-V5 (1:5000) or rabbit anti-MIC8 (1:1000), followed by goat anti-mouse or goat anti-rabbit IgG–alkaline phosphatase conjugates (1:1000). For biotin detection, streptavidin–alkaline phosphatase (Amersham) was used at 1:1000. Proteins from transfected cell supernatants were precipitated with 5% TCA prior to western blotting analysis. When comparing relative amounts of recombinant proteins in cells and supernatants, identical fractions of the total cell pellet or supernatant from the same dish were processed for western blotting. Cell blot assays were performed as described previously (Garcia-Reguet et al., 2000), with Toxoplasma or transfected cells solubilized in non-reducing conditions.
Surface biotinylation of transfected cells and purification of recombinant MIC3 and pro-MIC3 proteins
To detect specifically surface recombinant MIC3 proteins, BHK-21 cells were transfected with pOC1 or pOC2 and grown for 18 h prior to surface labeling. Cells were loosened by incubation in PBS containing 2 mM EDTA, and after washings in PBS containing 1 mM MgCl2 and 0.5 mM CaCl2, were resuspended at 1.5 × 107/ml in PBS. The GFP fluorescence of 105 transfected cells was analyzed using a FACScan (Becton Dickinson) equipped with CELLQUEST sofware in order to quantify the rate of transfection. Biotinylation and extraction of surface proteins were performed as described in Isberg and Leong (1990) using the non-permeant amine-reactive compound sulfosuccinimidobiotin. The extract from the same number of transfected cells was immediately incubated with 100 µl of anti-MIC3 immunosorbent for immunopurification of recombinant MIC3 proteins as described in Garcia-Reguet et al. (2000). After extensive washes, immunosorbent beads were pelleted and proteins were solubilized in SDS–PAGE sample buffer. The culture supernatant corresponding to the same amount of both transfected cell cultures was TCA-precipitated and analyzed simultaneously. Biotinylated recombinant MIC3 proteins were detected by probing filters with streptavidin covalently linked to alkaline phosphatase, and total and supernatant recombinant MIC3 proteins were detected by probing filters with anti-V5 covalently linked to alkaline phosphatase.
Site-directed mutagenesis
QuikChange, a PCR-based site-directed mutagenesis system, was used to introduce mutations in the MIC3 gene (Stratagene). The reactions were performed as described by the manufacturer. Plasmid pOC2 was used as template for constructions of plasmids pC302 (mutant MIC3-C302), pC304 (mutant MIC3-C304), pC339 (mutant MIC3-C339), pC354 (mutant MIC3-C354) and pC302-304 (mutant MIC3-C302,304). Plasmid pC339 was used as template for construction of plasmid pC339-354 (mutant MIC3-C339,354). Plasmid pC339-354 was used as template for construction of plasmid pC302-304-339-354 (mutant MIC3-C302,304,339,354). The presence of expected mutations was verified by DNA sequencing.
Acknowledgments
Acknowledgements
We thank Markus Meissner for generous gifts of MIC8 plasmid and anti-MIC8 antibodies, and communication of results before publication. We are grateful to Pascale Cossart, Shaynoor Dramsi and Dominique Soldati for critical review of the manuscript. Special thanks are due to Nathalie Garciat-Reguet for providing the anti-pro-peptide serum. O.C. is a recipient of a fellowship from VIHPAL and M.L. was supported by Sidaction. Grants from Sidaction and Fondation pour la Recherche Médicale to J.F.D. are gratefully acknowledged.
References
- Achbarou A., Mercereau-Puijalon,O., Autheman,J.M., Fortier,B., Camus,D. and Dubremetz,J.F. (1991) Characterization of microneme proteins of Toxoplasma gondii. Mol. Biochem. Parasitol., 47, 223–233. [DOI] [PubMed] [Google Scholar]
- Bauskin A.R., Zhang,H.P., Fairlie,W.D., He,X.Y., Russell,P.K., Moore,A.G., Brown,D.A., Stanley,K.K. and Breit,S.N. (2000) The propeptide of macrophage inhibitory cytokine (MIC-1), a TGF-β superfamily member, acts as a quality control determinant for correctly folded MIC-1. EMBO J., 19, 2212–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami C., Frevert,U., Sinnis,P., Takacs,B., Clavijo,P., Santos,M.J. and Nussenzweig,V. (1992) The basolateral domain of the hepatocyte plasma membrane bears receptors for the circumsporozoite protein of Plasmodium falciparum sporozoites. Cell, 70, 1021–1033. [DOI] [PubMed] [Google Scholar]
- Di Cristina M., Spaccapelo,R., Soldati,D., Bistoni,F. and Crisanti,A. (2000) Two conserved amino acid motifs mediate protein targeting to the micronemes of the apicomplexan parasite Toxoplasma gondii. Mol. Cell. Biol., 20, 7332–7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reguet N. et al. (2000) The microneme protein MIC3 of Toxoplasma gondii is a secretory adhesin that binds to both the surface of the host cells and the surface of the parasite. Cell. Microbiol., 2, 353–364. [DOI] [PubMed] [Google Scholar]
- Gray A.M. and Mason,A.J. (1990) Requirement for activin A and transforming growth factor-β 1 pro-regions in homodimer assembly. Science, 247, 1328–1330. [DOI] [PubMed] [Google Scholar]
- Hastrup H., Karlin,A. and Javitch,J.A. (2001) Symmetrical dimer of the human dopamine transporter revealed by cross- linking Cys-306 at the extracellular end of the sixth transmembrane segment. Proc. Natl Acad. Sci. USA, 98, 10055–10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg R.R. and Leong,J.M. (1990) Multiple β1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell, 60, 861–871. [DOI] [PubMed] [Google Scholar]
- Kappe S., Bruderer,T., Gantt,S., Fujioka,H., Nussenzweig,V. and Menard,R. (1999) Conservation of a gliding motility and cell invasion machinery in Apicomplexan parasites. J. Cell Biol., 147, 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H., Mehlhorn,H. and Ruger,W. (1996) In vitro biosynthesis and in vivo processing of the major microneme antigen of Sarcocystis muris cyst merozoites. Parasitol. Res., 82, 468–474. [DOI] [PubMed] [Google Scholar]
- Klein H., Loschner,B., Zyto,N., Portner,M. and Montag,T. (1998) Expression, purification and biochemical characterization of a recombinant lectin of Sarcocystis muris (Apicomplexa) cyst merozoites. Glycoconj. J., 15, 147–153. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Leung I.W. and Lassam,N. (1998) Dimerization via tandem leucine zippers is essential for the activation of the mitogen-activated protein kinase kinase kinase, MLK-3. J. Biol. Chem., 273, 32408–32415. [DOI] [PubMed] [Google Scholar]
- Meissner M., Reiss,M., Viebig,N., Carruthers,V., Trousel,C., Tomavo,S., Ajioka,J.W. and Soldati,D. (2002) A family of transmembrane microneme proteins of Toxoplasma gondii contain EGF-like domains and function as escorters. J. Cell Sci., 115, 563–574. [DOI] [PubMed] [Google Scholar]
- Menard R. (2001) Gliding motility and cell invasion by Apicomplexa: insights from the Plasmodium sporozoite. Cell. Microbiol., 3, 63–73. [DOI] [PubMed] [Google Scholar]
- Petrotchenko E.V., Pedersen,L.C., Borchers,C.H., Tomer,K.B. and Negishi,M. (2001) The dimerization motif of cytosolic sulfo transferases. FEBS Lett., 490, 39–43. [DOI] [PubMed] [Google Scholar]
- Rabenau K.E., Sohrabi,A., Tripathy,A., Reitter,C., Ajioka,J.W., Tomley,F.M. and Carruthers,V.B. (2001) TgM2AP participates in Toxoplasma gondii invasion of host cells and is tightly associated with the adhesive protein MIC2. Mol. Microbiol., 41, 537–547. [DOI] [PubMed] [Google Scholar]
- Reiss M., Viebig,N., Brecht,S., Fourmaux,M.N., Soete,M., Di Cristina, M., Dubremetz,J.F. and Soldati,D. (2001) Identification and characterization of an escorter for two secretory adhesins in Toxoplasma gondii. J. Cell Biol., 152, 563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini J.M. (1995) Lectin structure. Annu. Rev. Biophys. Biomol. Struct., 24, 551–577. [DOI] [PubMed] [Google Scholar]
- Soldati D., Dubremetz,J.F. and Lebrun,M. (2001) Microneme proteins: structural and functional requirements to promote adhesion and invasion by the apicomplexan parasite Toxoplasma gondii. Int. J. Parasitol., 31, 1293–1302. [DOI] [PubMed] [Google Scholar]
- Taylor M.A., Baker,K.C., Briggs,G.S., Connerton,I.F., Cummings,N.J., Pratt,K.A., Revell,D.F., Freedman,R.B. and Goodenough,P.W. (1995) Recombinant pro-regions from papain and papaya proteinase IV are selective high affinity inhibitors of the mature papaya enzymes. Protein Eng., 8, 59–62. [DOI] [PubMed] [Google Scholar]
- Tomley F.M. and Soldati,D.S. (2001) Mix and match modules: structure and function of microneme proteins in apicomplexan parasites. Trends Parasitol., 17, 81–88. [DOI] [PubMed] [Google Scholar]
- Tomley F.M., Billington,K.J., Bumstead,J.M., Clark,J.D. and Monaghan,P. (2001) EtMIC4: a microneme protein from Eimeria tenella that contains tandem arrays of epidermal growth factor-like repeats and thrombospondin type-I repeats. Int. J. Parasitol., 31, 1303–1310. [DOI] [PubMed] [Google Scholar]
- Wright H.T., Sandrasegaram,G. and Wright,C.S. (1991) Evolution of a family of N-acetylglucosamine binding proteins containing the disulfide-rich domain of wheat germ agglutinin. J. Mol. Evol., 33, 283–294. [DOI] [PubMed] [Google Scholar]