Abstract
A large group of hydrolytic enzymes, which contain a conserved stretch of ∼130 amino acids designated the amidase signature (AS) sequence, constitutes a super family that is distinct from any other known hydrolase family. AS family enzymes are widespread in nature, ranging from bacteria to humans, and exhibit a variety of biological functions. Here we report the first structure of an AS family enzyme provided by the crystal structure of malonamidase E2 from Bradyrhizobium japonicum. The structure, representing a new protein fold, reveals a previously unidentified Ser-cisSer-Lys catalytic machinery that is absolutely conserved throughout the family. This family of enzymes appears to be evolutionarily distinct but has diverged to acquire a wide spectrum of individual substrate specificities, while maintaining a core structure that supports the catalytic function of the unique triad. Based of the structures of the enzyme in two different inhibited states, an unusual action mechanism of the triad is proposed that accounts for the role of the cis conformation in the triad.
Keywords: amidase signature family/catalytic mechanism/crystal structure/new protein fold/novel catalytic triad/serine hydrolase
Introduction
Enzymatic hydrolysis of amide and ester bonds is usually initiated by an amino acid nucleophile that is polarized by the catalytic base(s) in contact with it. The classical Ser-His-Asp triad is probably most widespread in nature, existing not only in several protease families but also in esterases and lipases. Amazingly, the triad’s essentially identical stereochemistry is maintained in several different groups of hydrolytic enzymes whose folding patterns are entirely dissimilar. Some variations in the chemical make-up of the classic triad are found in different hydrolases, but the three active residues usually occupy the same locations in their respective structures (Dodson and Wlodawer, 1998). A dramatic variation on the triad is the Ser-Lys dyad that has been found in a subset of proteases, including signal peptidases (Paetzel et al., 1998), a DNA polymerase V accessory protein UmuD (Peat et al., 1996), a D1 C-terminal processing protease (CtpA) (Liao et al., 2000) and the LexA family of self-cleaving proteases (Bell et al., 2000; Luo et al., 2001). A comparison of the structures of these proteases reveals that they all share the same protein fold in the central catalytic domain, except for CtpA (Paetzel and Strynadka, 1999; Paetzel et al., 2002). The role of the lysine residue in these proteases is replaced by the α-amino group in another class of hydrolases, the so-called N-terminal nucleophile hydrolases (Ntn-hydrolases), which include penicillin acylase, glutamine PRPP amidotransferase, and the catalytic subunit of proteosomes (Oinonen and Rouvinen, 2000). In these enzymes, an N-terminal residue (serine, threonine or cysteine) utilizes the deprotonated α-amino group as the general base for the polarization of its own side chain. The Ntn-hydrolases share a common catalytic core framework despite the absence of any recognizable sequence similarity (Brannigan et al., 1995).
Perhaps the amidase signature (AS) super family is the largest group of enzymes that is distinct from any of the representative serine hydrolase families described above. The AS sequence was first identified in a group of proteins as a highly conserved stretch that is rich in glycine and serine residues (Mayaux et al., 1991; Chebrou et al., 1996). Knowledge of the AS family has now expanded to include more than 140 different members, which exist throughout the hierarchy of living organisms. The common biochemical nature of AS family enzymes has been characterized by the hydrolysis of the amide bond (CO-NH2). However, there are many predicted members with unknown biochemical functions in sequence databases, suggesting that some of them may hydrolyze peptide or ester bonds of substrates. The AS sequence contains several invariant residues including serine and lysine but not aspartic acid and histidine, indicating that the AS family enzymes are non-classical serine hydrolases. A series of site-directed mutagenesis studies on a well studied rat fatty acid amide hydrolase (FAAH) led to the suggestion that AS family enzymes are Ser-Lys dyad hydrolases (Patricelli et al., 1999; Patricelli and Cravatt, 2000).
The known substrate specificities and biological functions of the AS family enzymes vary significantly. In most bacterial species, heterotrimeric Glu-tRNAGln amidotransferase, whose subunit A belongs to the AS family, catalyzes the formation of Gln-tRNAGln by the transamidation of misacylated Glu-tRNAGln via amidolysis of glutamine (Schon et al., 1988; Curnow et al., 1997). This is an essential reaction providing the precursor for protein biosynthesis, and is therefore a point of therapeutic intervention (Decicco et al., 2001). In mammals, FAAH is responsible for the catabolism of neuromodulatory fatty acid amides (Cravatt et al., 1996), which display a number of activities including induction of sleep (Cravatt et al., 1995) and analgesia (Calignano et al., 1998). The other known activities of AS family enzymes include the formation of indole-3-acetic acid (Gaffney et al., 1990), which is a fundamental regulator of plant development, metabolic turnover of nitrile (Mayaux et al., 1991) or other carbon-nitrogen-containing compounds (Gomi et al., 1991), and degradation of nylon oligomers (Okada et al., 1983). Amidases involved in the microbial nitrile metabolism are used in the industrial scale production of acrylamide and nicotinamide (Yamada and Kobayashi, 1996).
The functional variety and the wide evolutionary distribution of the AS family enzymes underscores the importance of information regarding the three-dimensional structure so that the structure–function relationship can begin to be understood. In order to provide a canonical structure, to identify catalytic components and to provide a structural basis for the widely varying individual substrate specificities of the AS family enzymes, we determined the crystal structure of malonamidase E2 (MAE2) from Bradyrhizobium japonicum, a member of the AS family. The enzyme catalyzes the hydrolysis of malonamate (–OOCCH2CONH2) into malonate and ammonia (Koo et al., 2000), and is involved in the transport of fixed nitrogen from bacteroids to plant cells in symbiotic nitrogen metabolism (Kim and Chae, 1990). This study led to an identification of a novel catalytic triad in a new folding architecture.
Results and discussion
Structure of MAE2
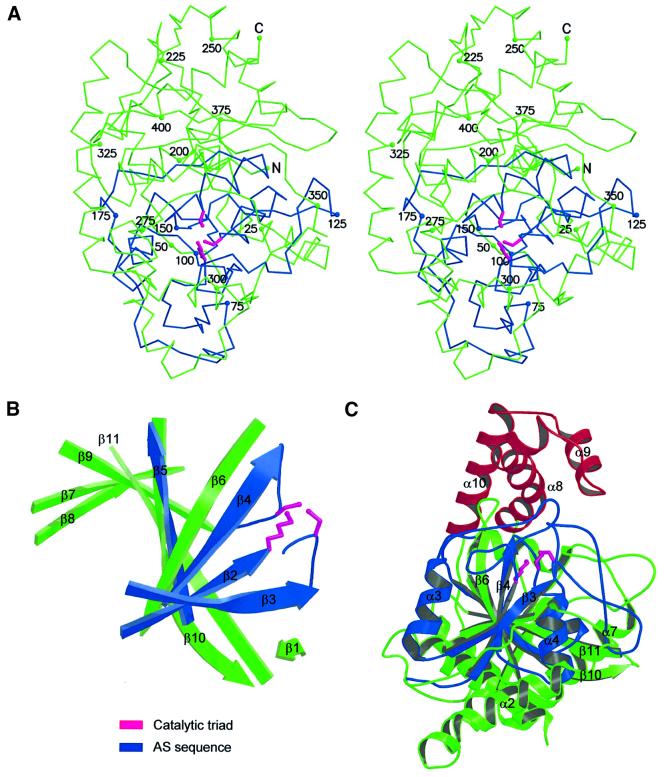
The crystal structure of MAE2 was solved with multiple-wavelength anomalous dispersion (MAD) phasing, by using one tungsten derivative crystal of the enzyme. The final model, refined against data to 1.8 Å resolution, consists of all the residues except for the last two C-terminal ones. The structure is a compact mixed α/β fold, containing 12 α-helices and one central and highly irregular β-sheet composed of 11 β-strands (Figures 1 and 2). Each β-strand is sequentially rotated in the same direction with respect to an adjacent β-strand, such that the β-strand at one end of the molecule is rotated by ∼180° relative to the β-strand at the other end of the molecule (Figure 2B). Most α-helices are intricately intermingled with β-strands and/or loops. The conserved AS sequence forms roughly the core of the molecule and interacts with a substantial part of the remainder of the sequence (Figure 2C). For example, β-strand 6, which is outside the AS sequence, lies between β-strands 4 and 5, which are part of the AS sequence composed of AS sequence residues (Figure 2B). Therefore, it is highly likely that the core structures of other AS family members are similar to that of MAE2. The enzyme forms a dimer, consistent with the dimer formation in solution (data not shown). Many non-conserved residues present at the dimer interface indicate that dimer formation is not a common feature of AS family enzymes. A search for analogous folding architectures in the Protein Data Bank (PDB) using the program DALI (Holm and Sanders, 1995) revealed that the structure of MAE2 is distinct from that of any other known protein fold. The structure of inositol monophosphatase (PDB code 1IMB), listed as the closest match (Z-score = 4.8, r.m.s.d. = 3.8), is an interleaved α/β/α/β/α fold and is obviously different to that of MAE2.
Fig. 1. Sequence alignments and secondary structure assignment. The AS sequences of selected AS family enzymes from low to high organisms are aligned: GluAT, Glu-tRNAGln amidotransferase, Bacillus subtilis (gi: 2589195); Yeast Amd2p, a putative amidase, Saccharomyces cerevisiae (gi: 6320448); Celeg, a putative amidase, Caenorhabditis elegans (gi: 6425411); VDHAP, vitamin D3 hydroxylase-associated protein, Gallus domesticus (gi: 1079452); FAAH, fatty acid amide hydrolase, Homo sapiens (gi: 4557575). The secondary structure assignment is shown at the top of the sequence. The solid-line boxes indicate the three invariant residues of the catalytic triad and the dotted-line box indicates the highly conserved residue that provides the augmenting catalytic role (see text). Asterisks indicate the residues comprising the oxyanion hole.
Fig. 2. Crystal structure of the MAE2 monomer. (A) Stereoview of Cα tracings of the MAE2 monomer. Every 25th residue, positions are numbered. (B) Ribbon diagram of the β-sheet of MAE2, showing the extreme curvature of the sheet and the varying lengths of the β-strands. (C) Ribbon diagram of the MAE2 monomer with elements of secondary structures numbered in the order of appearance in the primary sequence. For clarity, some of them are not labeled. The three α-helices, highlighted in red, are noticeably separated from the core structure. Helices α8 and α9 restrict the size of the entrance to the active site cavity. The AS sequence is in blue and the catalytic triad is in magenta in (A–C). This figure was prepared using BobScript and rendered using Raster3D.
The Ser-cisSer-Lys triad
The active site is located in the middle of the core structure. Many conserved residues in the AS sequence decorate the bottom of the active site. Among these, the absolutely conserved residues Ser155, Ser131 and Lys62 (Figure 1) form a hydrogen-bonded network (Figure 3A). Substitution of each of the residues in MAE2 (Koo et al., 2000; this study) or the corresponding residues in FAAH (Patricelli and Cravatt, 2000) to alanine resulted in virtually no remaining catalytic activity. Ser155 must be a reactive nucleophile, initiating the catalysis because it forms a covalent pyrophospho-serine as described below. Ser131 is in the middle of the most highly conserved glycine- and serine-rich segment, G129-G130-S131-S132-S133-G134 (Figure 1). Remarkably, Ser131 is in the unusual cis conformation, which places both the amide nitrogen and the side chain of the residue at ideal positions to polarize Ser155 together. The conservation of the sequence of the segment indicates that the residues corresponding to Ser131 in other AS family members also adopt the cis conformation. The amino group of Lys62 is correctly positioned by Ser132 to interact with cisSer131 Oγ. Ser132 is a nearly invariant residue, and the substitution of the corresponding residue in FAAH with alanine decreased the catalytic activity by ∼100-fold (Patricelli and Cravatt, 2000). Thr150, which is also highly conserved as threonine or serine, interacts with the amino group of Lys62 via its Oγ atom. The role of the interaction may also lie in the positioning of the side chain of Lys62. The MAE2 structure, adequately reflecting the previous mutagenesis and kinetic studies, identifies the core catalytic component of the AS family enzymes as the Ser155-cisSer131-Lys62 triad, with an additional supporting role of Ser132. Since this triad or a similar variation has not been found in other hydrolase families, the structure presented here defines the AS family as a new class of serine hydrolases that has a novel catalytic triad in a unique protein fold.
Fig. 3. Ser-cisSer-Lys catalytic triad and oxyanion hole. (A) Spatial arrangement of the catalytic triad in the structure of uninhibited MAE2. Ser132 and Thr150, involved in hydrogen bonds with Lys62, are shown. Wat1 is the one of multiple water molecules on a hydrogen-bonded network, which is extended to the bulk solvent. (B) View of catalytic triads and oxyanion holes for MAE2 (left) and thrombin (right). Covalently modified serine nucleophiles in MAE2 and thrombin (PDB code 1AWF) are shown in an orientation that roughly superposes the serine residues in the two structures. For clarity, a portion of the inhibitor boned to thrombin is not shown beyond the || mark. This figure was prepared using BobScript and rendered using Raster3D.
Covalent modification of Ser155
The structure of the enzyme reacted with phenylphosphorodiamidate (PPD) was determined at 1.9 Å. We used this inhibitor with the expectation that the enzyme would cleave off the phenyl group and form a covalent bond with the resulting diamidophosphate, (NH2)2PO2–. This was the actual compound that competitively inhibited a nickel ion-dependent bacterial urease in the presence of PPD (Benini et al., 1999). However, clear electron density connected to the side chain of Ser155 (Figure 4A) revealed the presence of a diphosphoryl group covalently bonded to Ser155. We concluded that the compound is a pyrophosphoryl group on the basis of its specific interactions with the enzyme, as shown in Figure 4C and explained in the legend. The formation of pyrophospho-serine is likely to be the result of multiple displacements of the leaving groups by the enzyme, including the amino groups, which finally led to the dead-end modification of Ser155 during the inhibitor soaking of the crystal. The covalent modification demonstrates that Ser155 acts as the nucleophile and forms a covalent acyl-enzyme intermediate in the course of the catalysis. Since pyrophospho-Ser155 is a mimic of the tetrahedral intermediate, the picture of the covalent complex also led to the identification of the oxyanion hole, which is known to stabilize a tetrahedral oxyanion intermediate in the mechanism for serine proteases. Thr152, Gly153, Gly154 and Ser155 form a hairpin loop structure, with their backbone nitrogen atoms all pointing to, and within hydrogen-bonding distances (<3.1 Å) of, the oxygen atom O2 of pyrophospho-Ser155 (Figure 4C). The direct interactions provided by the peptide segment conform to the general concept for the oxyanion hole. The two residues preceding the invariant Ser155 are highly conserved, mostly as glycine and infrequently as alanine. Note that the structure of the oxyanion hole and the relative positions of the triad residues are markedly different from those found in thrombin (Figure 3B).
Fig. 4. Irreversible and competitive inhibition of MAE2. (A) Stereoview of covalent pyrophospho-Ser155 shown along with a 2Fo – Fc map (1.5 σ) at 1.9 Å. Arg113 interacting directly with the phosphate group shown in (C) is omitted for clarity. (B) Mode of binding of malonate (labeled ‘Mal’) to the active site shown along with a 2Fo – Fc map (1.5 σ) at 2.0 Å. The interactions of three carboxyl oxygens of malonate with Arg158 and the backbone amide of Gly153 suggest that the position of O1 would be occupied by the amino group of the substrate when it binds to the active site. (C) Detailed interactions between pyrophospho-Ser155 and the active site. The three oxygen atoms of the phosphate group interact directly with the guanidine groups of Arg158 and Arg113, while O2 interacts directly with the four backbone nitrogen atoms. These specific interactions demonstrate that the covalently modified Ser155 is pyrophospho-serine. Bound malonate is shown in yellow. Distances are shown in angstroms. The residues consisting of the triad and oxyanion hole in the two structures of the inhibited enzyme are superposed. This figure was prepared using BobScript and rendered using Raster3D.
Proposed action mechanism of the Ser-cisSer-Lys triad
The product malonate is different from the substrate malonamate only in that it has a carboxyl group in place of an amide group at one end of the molecule. Pyrophospho-Ser155 should mimic the tetrahedral adduct of the malonamate-Ser155 owing to their similar lengths. In fact, the side chain of Arg158 at similar positions interacts with malonate and pyrophospho-Ser155 in the two structures of the complexes (Figure 4). As shown in Figure 4B, one of the carboxyl carbons of the bound malonate is close to Oγ of Ser155 (3.39 Å), indicating that the corresponding carbon of the substrate will be similarly positioned and attacked by this nucleophile. The specific hydrogen-bonding interactions between malonate and the enzyme suggest that the amino group of malonamate, upon binding to the enzyme, would occupy the position of the oxygen atom O1 of malonate (Figure 4B), which is away from the oxyanion hole. This oxygen atom is 4.68 Å apart from Oγ of cisSer131. In comparison, O1 of pyrophospho-Ser155, which is also away from the oxyanion hole, is 2.74 Å apart from Oγ of cisSer131 (Figure 4C). The covalent bond formation, through the nucleophilic attack on the substrate by Ser155, is therefore expected to bring the amino group of the resulting tetrahedral adduct near the position of the O1 of pyrophospho-Ser155. Subsequently, the hydroxyl group of cisSer131 will be able to provide its proton (serving as the general acid) to the amino group of the tetrahedral adduct, which is stabilized by the oxyanion hole. Lys62 is likely to play a critical role in helping cisSer131 to donate its proton to the leaving group by stabilizing the resulting oxyanion of the serine residue. The product ammonia subsequently leaves the active site and a water molecule will occupy the vacated site near the O1 position of pyrophospho-Ser155. Activated by cisSer131 (serving as the general base), the water molecule will attack the carbonyl carbon atom of the acyl-enzyme intermediate to finally regenerate Ser155. Possibly, a solvent molecule (S840), close to cisSer131 in the uninhibited enzyme but displaced in the covalent complex, could be the catalytic water molecule.
The profile of reaction rate versus pH for FAAH indicated a single titrable group at pH 7.9 (Patricelli et al., 1999). In a similar experiment, MAE2 also exhibited a single titrable group at pH 6.8 (data not shown). The environment surrounding the amino group of Lys62 is polar, but is free of positive charges within 8 Å of the group. Therefore, Lys62 is most probably protonated at neutral pH and is unsuitable for initiating the polarization of Ser155, and is unlikely to be responsible for the observed pH titration. Ser155 Oγ, which is involved in the direct hydrogen-bonding interactions with cisSer131 and a water molecule, is oriented to and within 5.0 Å of the three backbone –NH groups comprising the oxyanion hole (Figure 3A) and is within 5.2 Å of the guanidine group of Arg158. In this environment, the pKa of Ser155 would be substantially lowered and is possibly responsible for the pH titration of the reaction rate of MAE2. We suggest that the catalysis starts with deprotonated Ser155, stabilized mainly by virtue of the two direct interactions provided by cisSer131. The proposed mechanism is different from the catalytic mechanisms proposed for the Ser-His-Asp triad (Fersht, 1985), which commonly start with proton transfer from the serine residue to the His-Asp pair. However, we do not rule out other possibilities for polarization of the serine nucleophile in other members of the AS superfamily, because individual pKa values of the triad residues are affected by the microenvironment surrounding them.
Substrate specificity and catalytic efficiency
The structure of MAE2 provides an insight into the structural basis for the substrate specificities of AS family members. The upper half of the active site cavity is lined mostly with non-conserved residues. Helices α8 and α9 in particular, which constitute a substantial portion and restrict the size of the active site cavity (Figure 2C), are composed of residues outside the AS sequence. We suggest that the active site of each member is tailored to recognize its substrate by the substitution of these non-conserved residues. Consistent with this, Arg158, which provides prominent interactions with the bound malonate (Figure 4B), is not a conserved residue in the AS sequence (Figure 1). It should be noted that α8, α9 and α10 are noticeably separated from the structural core (Figure 2C). Therefore, substantial variations in the number and composition of amino acids in this region for acquiring different substrate specificities would not affect the catalytic core framework in other AS family members.
Low acidity of the base in catalytic triads or dyads is often associated with a low enzymatic activity. For example, the cytomegalovirus protease contains a Ser-His- His triad and exhibits a kcat of 18/min, which is comparable to a kcat of 3.0/min for a mutant trypsin containing the Ser-His-Asn triad (Chen et al., 1996). It is conceivable that a low catalytic activity of the Ser-cisSer-Lys triad may be expected because both serine and lysine residues are weak acids. However, MAE2 exhibits a kcat of 2200/s (Koo et al., 2000), indicating that the novel triad can be a very efficient catalytic machinery. The data support our proposal that Lys62 in MAE2 does not serve as a base in the initiation of the catalysis.
Conclusions
The crystal structure of MAE2 reported here is the first for any member of the AS family. The precise contact between Lys62 and Ser155 via cisSer131 and the heavy contact between the AS sequence and other sequence regions suggest that the core structure should be highly conserved throughout the family to support the functional catalytic components. The catalytic function of the novel triad appears to be supported only by the new main chain framework revealed in this study, unlike the Ser-His-Asp catalytic triad and the Ser-Lys dyad, which exist in different folding scaffolds. However, the AS enzymes are widely distributed and therefore the newly identified Ser-cisSer-Lys catalytic triad is prevalent in nature. An action mechanism of the triad, incorporating the role of the cis conformation within it, explains how the cisSer-Lys pair replaces the functional role of the His-Asp pair of the Ser-His-Asp triad, despite their very different chemical nature. The set of structures presented here will undoubtedly serve as a framework for studying structure–function relationships of many other members of the AS family, including detailed studies of the new catalytic triad.
Materials and methods
Structure determination
MAE2 was expressed in Escherichia coli AD494 (DE3) (Novagen) and purified by a series of column purification steps as described previously (Koo et al., 2000). The purified enzyme was concentrated to 10 mg/ml in 20 mM Tris–HCl buffer pH 7.4 containing 15% glycerol. Crystals of MAE2 were grown by hanging-drop vapor diffusion against a reservoir containing 20% polyethyleneglycol 1000 and 100 mM Tris–HCl pH 7.0 as described previously (Shin et al., 2002). Crystals belonged to the space group P21212 with unit cell dimensions of a = 104.29, b = 95.58, c = 74.90 Å. Before cryocooling, crystals were briefly immersed in the same precipitant solution containing 10–15% glycerol. All diffraction data were collected at 100 K on the beamline 6B of the Pohang Accelerator Laboratory (Pohang, Korea) and were processed using HKL software (Otwinowski and Minor, 1997). A MAD data set at three different wavelengths was collected with a tungsten derivative crystal soaked in 10 mM Na2WO4 dissolved in the same precipitant solution for 32 h at 4°C. Two tungsten sites, coinciding with the active sites, were located and used for phase determination at 2.7 Å with the program SOLVE (Terwilliger and Berendzen, 1999) and phases were subsequently improved by density modification with the program RESOLVE (Terwilliger, 2000). The electron density was of excellent quality, showing nearly all features of the protein side chains. The program O (Jones et al., 1991) was used for model building, and refinement was performed against 1.8 Å native data with the CNS package (Brünger et al., 1998).
Structure determination of MAE2 in covalent complex with pyrophosphate and in complex with malonate
The MAE2 crystals were transferred to the mother liquor containing 10 mM PPD, and incubated for 62 h at 4°C before X-ray data collection. The crystals of MAE2 in complex with malonate were obtained using the co-crystallization method in the presence of 15 mM malonate in the same precipitant solution used for growing the native crystals. The structures of the complexes were solved by direct refinement of the dimeric structure of MAE2 against the diffraction data for PPD-soaked crystal (1.9 Å) and malonate-soaked crystal (2.0 Å). The 2Fo – Fc map for each crystal showed well defined electron density for the bound inhibitor. The resulting models fell within the limits of all the quality criteria of the program PROCHECK (CCP4, 1994). Crystallographic and refinement statistics of the three structures are provided in Table I.
Table I. Data collection and structure solution.
| Parameters | Peak | Edge | Remote |
|---|---|---|---|
| Wavelength (Å) | 1.21425 | 1.21459 | 1.20280 |
| Resolution range (Å) | 30.0–2.66 | 30.0–2.66 | 30.0–2.66 |
| Completenessa (%) | 96.9 (92.6) | 95.9 (90.7) | 96.1 (90.3) |
| Rsymb (%) | 5.5 (15.4) | 6.4 (18.8) | 6.1 (18.8) |
| I/σ | 21.87 (6.45) | 16.84 (4.41) | 17.81 (4.55) |
| FOM (centric/acentric)c |
0.42/0.63 |
|
|
| Refinement statistics | |||
| Parameters |
Native |
Pyrophosphate |
Malonate |
| Resolution (Å) | 30–1.8 | 30–1.9 | 20–2.0 |
| Rsym (%) | 3.7 (11.3) | 5.4 (23.9) | 6.1 (15.2) |
| Total reflections | 474 213 | 621 134 | 401 931 |
| Unique reflections | 68 327 | 55 720 | 48 851 |
| Completeness (%) | 97.4 (93.4) | 95.2 (87.5) | 96.4 (91.3) |
| Number of refined atoms | |||
| Protein/water | 6126/939 | 6126/927 | 6126/562 |
| R-factord (Rfree) (%) | 19.0 (22.6) | 18.1 (22.2) | 19.8 (24.3) |
| R.m.s.d. bond length (Å) | 0.0049 | 0.0053 | 0.0049 |
| R.m.s.d. bond angle (°) | 1.276 | 1.126 | 1.258 |
| Ramachandran plot (%) | |||
| Most favored region | 90.6 | 90.1 | 90.1 |
| Additionally allowed region | 9.4 | 9.8 | 9.8 |
| Generously allowed region | 0.0 | 0.1 | 0.1 |
The numbers in parentheses are statistics from the highest resolution shell except for Rfree values.
aCompleteness for I/σ (I) > 1.0.
bRsym = Σ |Iobs – Iavg|/Σ Iobs, where Iobs is the observed intensity of individual reflection and Iavg is average over symmetry equivalents.
cFigure of merit, defined as < |Σ P(α) e iα/Σ P(α)| >, where α is the phase and P(α) is the phase probability distribution.
dR-factor = Σ ||Fo| – |Fc||/Σ |Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with 5% of the data.
Coordinates
The coordinates of the MAE2 structures have been deposited in the Protein Data Bank under accession codes 1GR8 (native), 1GRK (the covalent complex) and 1GR9 (malonate complex) (with the condition of immediate release upon publication).
Acknowledgments
Acknowledgements
This study used the beamline 6B at the Pohang Accelerator Laboratory and was supported by Creative Research Initiatives of the Korean Ministry of Science and Technology. S.S., T.-H.L. and N.-C.H. were supported by the Brain Korea 21 Project. H.M.K. and S.-y.K. were supported by a grant from the Korea Science and Engineering Foundation through the Protein Network Research Center at Yonsei University.
References
- Bell C.E., Frescura,P., Hochschild,A. and Lewis,M. (2000) Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding. Cell, 101, 801–811. [DOI] [PubMed] [Google Scholar]
- Benini S., Rypniewski,W.R., Wilson,K.S., Miletti,S., Ciurli,S. and Mangani,S. (1999) A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels. Structure Fold. Des., 7, 205–216. [DOI] [PubMed] [Google Scholar]
- Brannigan J.A., Dodson,G., Duggleby,H.J., Moody,P.C., Smith,J.L., Tomchick,D.R. and Murzin,A.G. (1995) A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature, 378, 416–419. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Calignano A., La Rana,G., Giuffrida,A. and Piomelli,D. (1998) Control of pain initiation by endogenous cannabinoids. Nature, 394, 277–281. [DOI] [PubMed] [Google Scholar]
- CCP 4 (1994) Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chebrou H., Bigey,F., Arnaud,A. and Galzy,P. (1996) Study of the amidase signature group. Biochim. Biophys. Acta, 1298, 285–293. [DOI] [PubMed] [Google Scholar]
- Chen P. et al. (1996) Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad. Cell, 86, 835–843. [DOI] [PubMed] [Google Scholar]
- Cravatt B.F., Prospero-Garcia,O., Siuzdak,G., Gilula,N.B., Henriksen, S.J., Boger,D.L. and Lerner,R.A. (1995) Chemical characterization of a family of brain lipids that induce sleep. Science, 268, 1506–1509. [DOI] [PubMed] [Google Scholar]
- Cravatt B.F., Giang,D.K., Mayfield,S.P., Boger,D.L., Lerner,R.A. and Gilula,N.B. (1996) Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature, 384, 83–87. [DOI] [PubMed] [Google Scholar]
- Curnow A.W., Hong,K., Yuan,R., Kim,S., Martins,O., Winkler,W., Henkin,T.M. and Soll,D. (1997) Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl Acad. Sci. USA, 94, 11819–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decicco C.P. et al. (2001) Glutamyl-γ-boronate inhibitors of bacterial Glu-tRNA(Gln) amidotransferase. Bioorg. Med. Chem. Lett., 11, 2561–2564. [DOI] [PubMed] [Google Scholar]
- Dodson G. and Wlodawer,A. (1998) Catalytic triads and their relatives. Trends Biochem. Sci., 23, 347–352. [DOI] [PubMed] [Google Scholar]
- Fersht A.R. (1985) Enzyme Structure and Mechanism. Freeman, New York, NY.
- Gaffney T.D., da Costa e Silva,O., Yamada,T. and Kosuge,T. (1990) Indoleacetic acid operon of Pseudomonas syringae subsp. savastanoi: transcription analysis and promoter identification. J. Bacteriol., 172, 5593–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K., Kitamoto,K. and Kumagai,C. (1991) Cloning and molecular characterization of the acetamidase-encoding gene (amdS) from Aspergillus oryzae. Gene, 108, 91–98. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sanders,C. (1995) Dali: a network tool for protein structure comparison. Trends Biochem. Sci., 20, 478–480. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kim Y.S. and Chae,H.Z. (1990) A model of nitrogen flow by malonamate in Rhizobium japonicum—soybean symbiosis. Biochem. Biophys. Res. Commun., 169, 692–699. [DOI] [PubMed] [Google Scholar]
- Koo H.M., Choi,S.O., Kim,H.M. and Kim,Y.S. (2000) Identification of active-site residues in Bradyrhizobium japonicum malonamidase E2. Biochem. J., 349, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D.I., Qian,J., Chisholm,D.A., Jordan,D.B. and Diner,B.A. (2000) Crystal structures of the photosystem II D1 C-terminal processing protease. Nature Struct. Biol., 7, 749–753. [DOI] [PubMed] [Google Scholar]
- Luo Y., Pfuetzner,R.A., Mosimann,S., Paetzel,M., Frey,E.A., Cherney, M., Kim,B., Little,J.W. and Strynadka,N.C. (2001) Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell, 106, 585–594. [DOI] [PubMed] [Google Scholar]
- Mayaux J.F., Cerbelaud,E., Soubrier,F., Yeh,P., Blanche,F. and Petre,D. (1991) Purification, cloning and primary structure of a new enantiomer-selective amidase from a Rhodococcus strain: structural evidence for a conserved genetic coupling with nitrile hydratase. J. Bacteriol., 173, 6694–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinonen C. and Rouvinen,J. (2000) Structural comparison of Ntn-hydrolases. Protein Sci., 9, 2329–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Negoro,S., Kimura,H. and Nakamura,S. (1983) Evolutionary adaptation of plasmid-encoded enzymes for degrading nylon oligomers. Nature, 306, 203–206. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Paetzel M. and Strynadka,N.C. (1999) Common protein architecture and binding sites in proteases utilizing a Ser/Lys dyad mechanism. Protein Sci., 8, 2533–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzel M., Dalbey,R.E. and Strynadka,N.C. (1998) Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature, 396, 186–190. [DOI] [PubMed] [Google Scholar]
- Paetzel M., Dalbey,R.E. and Strynadka,N.C. (2002) Crystal structure of a bacterial signal peptidase apo-enzyme. Implications for signal peptide binding and the ser-lys dyad mechanism. J. Biol. Chem., 277, 9512–9519. [DOI] [PubMed] [Google Scholar]
- Patricelli M.P. and Cravatt,B.F. (2000) Clarifying the catalytic roles of conserved residues in the amidase signature family. J. Biol. Chem., 275, 19177–19184. [DOI] [PubMed] [Google Scholar]
- Patricelli M.P., Lovato,M.A. and Cravatt,B.F. (1999) Chemical and mutagenic investigations of fatty acid amide hydrolase: evidence for a family of serine hydrolases with distinct catalytic properties. Biochemistry, 38, 9804–9812. [DOI] [PubMed] [Google Scholar]
- Peat T.S., Frank,E.G., McDonald,J.P., Levine,A.S., Woodgate,R. and Hendrickson,W.A. (1996) Structure of the UmuD′ protein and its regulation in response to DNA damage. Nature, 380, 727–730. [DOI] [PubMed] [Google Scholar]
- Schon A., Kannangara,C.G., Gough,S. and Soll,D. (1988) Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature, 331, 187–190. [DOI] [PubMed] [Google Scholar]
- Shin S., Lee,T.-H., Koo,H.M., Kim,S.-y., Lee,H.-S., Kim,Y.S. and Oh,B.-H. (2002) Crystallization and preliminary X-ray crystallographic analysis of malonamidase E2, an amidase signature family member. Acta Crystallogr. D, 58, 562–563. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D, 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H. and Kobayashi,M. (1996) Nitrile hydratase and its application to industrial production of acrylamide. Biosci. Biotechnol. Biochem., 60, 1391–1400. [DOI] [PubMed] [Google Scholar]