Abstract
During each spliceosome cycle, the U6 snRNA undergoes extensive structural rearrangements, alternating between singular, U4–U6 and U6–U2 base-paired forms. In Saccharomyces cerevisiae, Prp24 functions as an snRNP recycling factor, reannealing U4 and U6 snRNAs. By database searching, we have identified a Prp24-related human protein previously described as p110nrb or SART3. p110 contains in its C-terminal region two RNA recognition motifs (RRMs). The N-terminal two-thirds of p110, for which there is no counterpart in the S.cerevisiae Prp24, carries seven tetratricopeptide repeat (TPR) domains. p110 homologs sharing the same domain structure also exist in several other eukaryotes. p110 is associated with the mammalian U6 and U4/U6 snRNPs, but not with U4/U5/U6 tri-snRNPs nor with spliceosomes. Recom binant p110 binds in vitro specifically to human U6 snRNA, requiring an internal U6 region. Using an in vitro recycling assay, we demonstrate that p110 functions in the reassembly of the U4/U6 snRNP. In summary, p110 represents the human ortholog of Prp24, and associates only transiently with U6 and U4/U6 snRNPs during the recycling phase of the spliceosome cycle.
Keywords: Prp24/snRNA/spliceosome/splicing/U6 snRNP
Introduction
mRNA splicing requires the assembly of the pre-mRNA into a large, dynamic RNA–protein complex, the spliceosome, in which five small nuclear RNAs (U1, U2, U4, U5 and U6) and >50 protein components are incorporated (reviewed by Will and Lührmann, 1997; Burge et al., 1999). After the ordered assembly of the spliceosome and the two catalytic steps, the complex dissociates, followed by recycling of its components and initiation of a new cycle. Although most of the components of the spliceosome are known by now, we still understand only in part the dynamic interactions occuring during each splicing cycle.
The spliceosome is made up of an extensive and dynamic RNA network that involves interactions of the snRNA components among themselves and with the pre-mRNA. The conformational changes during each spliceosome cycle are thought to be governed by protein factors with annealing or RNA helicase activities. U6 is the most highly conserved snRNA component of the spliceosome and occurs in at least three different conformations: the singular U6 form (U6 snRNP in the recycling phase), the U4–U6 base-paired form (in the U4/U6 and U4/U5/U6 snRNPs) and the U6–U2 form, which is generated in the active spliceosome after disruption of the U4–U6 base pairing. The latter isomerization of mutually exclusive RNA structures is likely to be directed by a specific RNA helicase of the DEXH type, called Prp44 in yeast and U5-200 kDa in the mammalian system (reviewed by Raghunathan and Guthrie, 1998a).
After the catalytic steps, U6 leaves the post-splicing complex in the singular form and has to reassociate with U4 snRNP to regenerate the U4/U6 snRNP. In contrast to spliceosome assembly and splicing catalysis, we still know very little about this recycling phase of the spliceosome cycle, about possible auxiliary factors that may associate only transiently with recycling snRNPs and how their interactions are regulated. In addition to recycling of U6 snRNPs, newly synthesized U6 snRNA has to assemble into an RNP and undergo the U4–U6 annealing step, a process known as snRNP biogenesis.
Here we focus on the transition from the singular U6 snRNP to the U4/U6 snRNP in the mammalian system. Due to its low abundance in nuclear extract, free U6 snRNP is not very well characterized. Regarding protein components, seven polypeptides, the LSm2–LSm8 proteins, are associated specifically with U6 snRNA (Achsel et al., 1999). In addition, the La protein is bound by a fraction of U6 snRNA (Rinke and Steitz, 1985). In contrast, the U4/U6 snRNP is relatively well characterized: besides the seven U6-associated LSm proteins, it contains the seven U4-bound Sm proteins (SmB, D1, D2, D3, E, F and G), as well as U4/U6- specific polypeptides of 90, 60, 20 and 15.5 kDa (Horowitz et al., 1997; Vidovic et al., 2000; and references therein).
Based on genetic suppression studies in yeast, the splicing factor Prp24 has been identified as a protein required for the U4–U6 annealing step (Shannon and Guthrie, 1991). Subsequent studies have characterized the U6 binding specificity of yeast Prp24 (Ghetti et al., 1995; Jandrositz and Guthrie, 1995) and characterized its function as a recycling factor (Raghunathan and Guthrie, 1998b). Recombinant yeast Prp24 was found to be active, although at a low efficiency, in annealing of yeast U4 and U6 snRNAs (Ghetti et al., 1995). In addition, there is genetic evidence that Prp24 may play a role in U4–U6 dissociation within the active spliceosome (Vidaver et al., 1999).
Although Prp24 was identified 10 years ago in the yeast system, no mammalian counterpart had been identified so far. Using U4/U6-depleted nuclear extract and an in vitro splicing complementation system (Wolff and Bindereif, 1992), we have established an assay for studying the interaction between purified U4 snRNP and U6 RNA. In this way, we obtained initial evidence that the interaction requires a protein factor separate from the U4 snRNP and appears to be ATP independent (Wolff and Bindereif, 1993). However, the responsible protein factor could not be identified at that time.
Here we report on the identification of a new human snRNP protein, p110, which is distantly related in its C-terminal third to the Saccharomyces cerevisiae protein Prp24. The N-terminal two-thirds of p110, however, which have no yeast counterpart in Prp24 itself, carry seven tetratricopeptide repeat (TPR) domains that also exist in other RNA processing factors. We demonstrate that p110 is associated with the U6 and U4/U6 snRNPs, but not with U4/U5/U6 tri-snRNPs nor with spliceosomes. Recom binant p110 binds in vitro specifically to an internal region of U6 snRNA and is responsible for post-spliceosomal U4/U6 snRNP recycling. Taken together, our data demonstrate that p110 associates only transiently with U6 during the recycling phase of the spliceosome cycle, mediating the U4–U6 interaction.
Results
Identification and domain structure of human p110: a protein distantly related to S.cerevisiae Prp24
Since no mammalian homolog of the yeast Prp24 has been identified so far, we searched databases for related sequences, using only subregions of the Prp24 protein sequence. This search revealed a human cDNA sequence, which had been reported previously as KIAA0156 (Nagase et al., 1995), coding for p110nrb (= p110 nuclear RNA-binding protein; Gu et al., 1998), also referred to as SART3 (Yang et al., 1999). Interestingly, this protein appears to be expressed specifically at high levels in various tumor tissues and cell lines (see Discussion).
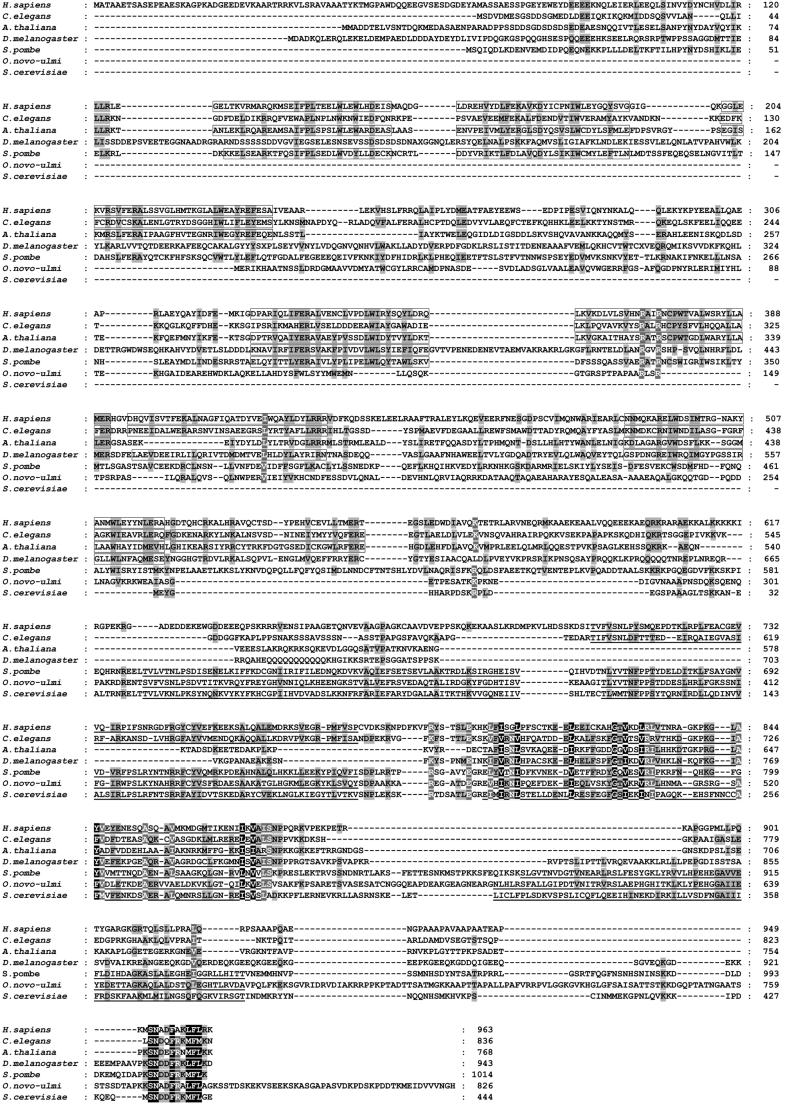
The human p110nrb/SART3 protein (called U6-p110, or p110 in the following) is much larger than Prp24 (963 compared with 444 amino acids) and only distantly related to Prp24 (see Figures 1 and 2). In its C-terminal region, the human protein shares only 22% sequence identity with Prp24 and contains two RNA recognition motifs (RRMs), compared with four RRMs in Prp24 (note that the fourth RRM conforms less well to the RRM consensus than the others and had not been reported previously). Comparing the RRMs between both proteins, the two RRMs of human p110 correspond to RRMs 2 and 3 in the yeast Prp24 protein, which are functionally important, based on mapping of suppressor mutations (Shannon and Guthrie, 1991; Vidaver et al., 1999). Significantly, the second RRM of p110 is the most highly conserved region of the human protein and also occurs in all other homologs (see below). The N-terminal two-thirds of the human p110 protein, which have no counterpart in Prp24 itself, carry seven TPR domains that also exist in other RNA processing factors and are thought to be involved in protein–protein interactions (Blatch and Lassle, 1999). Since the TPR motif present in human p110 deviates slightly from the TPR consensus, it has also been referred to as HAT (half a TPR; Preker and Keller, 1998) or cl- (crooked-neck-like) TPR motif (Zhang et al., 1991; McLean and Rymond, 1998).
Fig. 1. Sequence alignment of the human p110 protein with homologous proteins. The human p110 sequence (Homo sapiens; NP_055521) has been aligned with the following sequences: Caenorhabditis elegans (CAA97405); Arabidopsis thaliana (CAB45062); Drosophila melanogaster (CAA75535); Schizosaccharomyces pombe (CAB52740); Ophiostoma novo-ulmi (AAA76605); and Saccharomyces cerevisiae Prp24 (P49960). The total numbers of amino acids are given on the right. TPR (HAT) domains are boxed, RRM domains are underlined. Amino acid identity in the alignment is shown by the black background, shading indicates conservation of the physicochemical property of the residues.
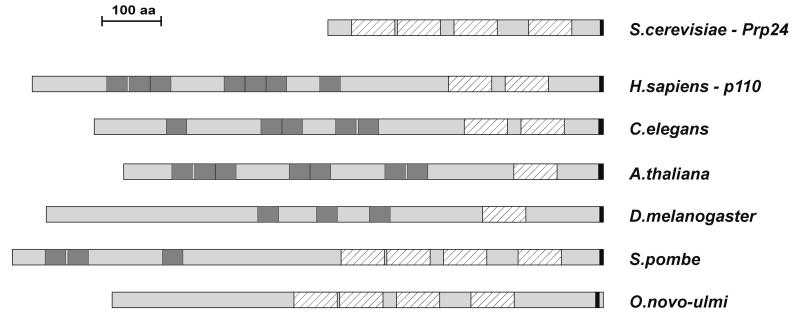
Fig. 2. Conserved domain structures of the S.cerevisiae Prp24 protein, human p110 and related proteins from other species. The proteins are aligned relative to their C-terminal ends. The RRM (striped boxes) and TPR (HAT) motifs (in dark gray) as well as the short C-terminal region (in black) are indicated.
Using the human p110 sequence for database searches, p110 orthologs were identified from the following species, carrying between 3 and 7 TPR domains in the N-terminal and 1–4 RRMs in the C-terminal region (see Figures 1 and 2): Caenorhabditis elegans (836 amino acids), Arabidopsis thaliana (768 amino acids), Schizo saccharomyces pombe (1014 amino acids) and Drosophila melanogaster (943 amino acids; Petschek et al., 1997). There is an additional ortholog from Ophiostoma novo-ulmi (826 amino acids; Pereira et al., 2000), with four RRMs and apparently no TPR motifs. All these orthologs are between 17 and 27% identical to human p110, with similarity ranging from 33 to 45%. With the exception of the D.melanogaster and O.novo-ulmi proteins, which are significantly conserved only in their C-terminal half, the homology is distributed equally throughout the entire protein. In addition to the common general organization of TPR and RRM motifs, all of these sequences carry a short region of 10 amino acids that is highly conserved and resides at the very C-terminus of the proteins, except for the O.novo-ulmi protein, where this motif is separated from the C-terminal end by 47 amino acids.
p110 protein is present in U6 and U4/U6 snRNP, but not in U4/U5/U6 tri-snRNPs
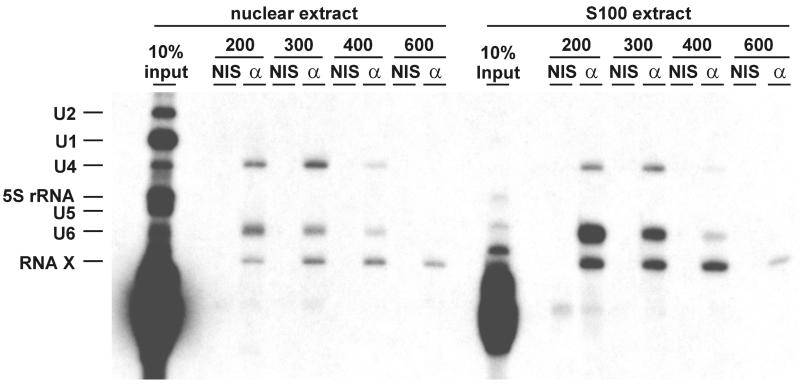
To search for p110-associated RNAs, we carried out immunoprecipitations from nuclear and S100 extracts derived from HeLa cells, using anti-p110 antibodies. Co-precipitated RNAs were detected by 3′ end labeling with [32P]pCp (Figure 3). In both nuclear and S100 extracts, a complex mixture of RNAs can be labeled (see input lanes), predominantly tRNAs and 5S rRNA as well as the spliceosomal snRNAs U1, U2, U4, U5 and U6 (in the S100 input, the known snRNAs can be detected only after longer exposure; data not shown). Immunoprecipitations were done at different stringencies, by varying the concentrations of NaCl between 200 and 600 mM. In each case, a control reaction was included with the corresponding non-immune serum (compare lanes NIS and α). As Figure 3 shows, at 200 and 300 mM NaCl, three major RNA bands were co-immunoprecipitated specifically by the anti-p110 antibody, both from nuclear extract and S100, the upper two corresponding in size to U4 and U6 snRNAs; the identities of these two snRNAs were confirmed by northern blot hybridization (data not shown). In addition, a third RNA species of ∼85 nucleotides was detected to be associated specifically with p110 (see band labeled RNA X). Based on direct RNA sequencing of this band, northern blot hybridization and cDNA cloning, RNA X most probably represents a 3′-truncated U6 snRNA fragment, which can be 3′ end labeled efficiently by [32P]pCp (A.Damianov, J.Medenbach and A.Bindereif, unpublished results). Under conditions of higher stringency, U4 and U6 snRNAs were reduced (400 mM NaCl) or undetectable (600 mM NaCl), while RNA X remained associated with p110. We conclude that the two spliceosomal snRNAs, U4 and U6, are associated specifically with p110.
Fig. 3. Human p110 protein is associated with U4 and U6 snRNAs: immunoprecipitation from cell extracts and [32P]pCp labeling. Immunoprecipitations were done from HeLa nuclear extract and S100 extract, using anti-p110 antibodies (lanes α) or non-immune antiserum (lanes NIS). The reactions were carried out in parallel at different stringencies, varying from 200 to 600 mM NaCl, as indicated above the lanes. Co-precipitated RNAs were purified, [32P]pCp labeled and analyzed by denaturing gel electrophoresis. For comparison, RNA from 10% of the input is shown (lanes 10% input). The identities of the [32P]pCp-labeled RNA bands are given on the left.
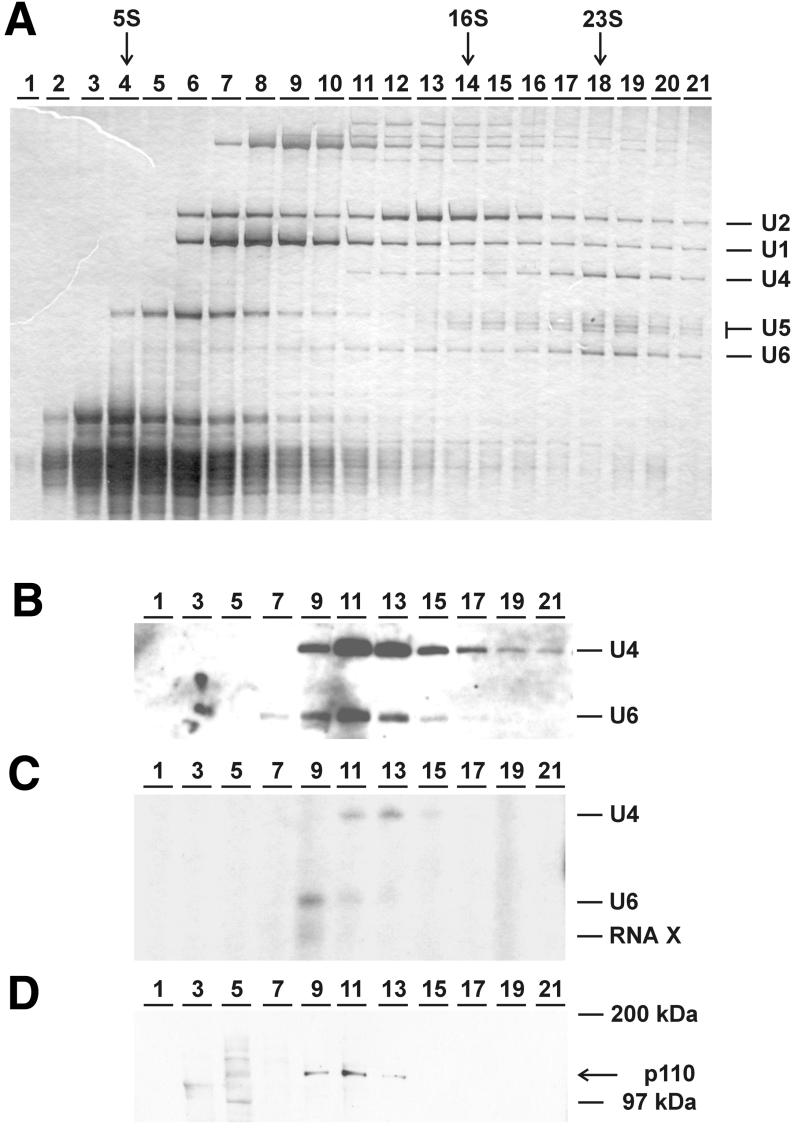
This initial result indicated that p110 is a component of the U6 and/or the U4/U6 snRNPs; U5 snRNA, however, could not be detected in the anti-p110 immunoprecipitate, suggesting that p110 is absent from the 25S tri-snRNP. To demonstrate this conclusively, spliceosomal snRNPs of HeLa cell nuclear extract were fractionated by glycerol gradient sedimentation (Figure 4). RNA was prepared from each gradient fraction and analyzed by denaturing gel electrophoresis and silver staining (Figure 4A). The identities and distribution of U4, U5 and U6 snRNAs were also confirmed by northern blot hybridization (data not shown). Figure 4A shows that U6 snRNPs (free of U4) fractionated heterogenously in the 5–10S region (fractions 4–10), and that U4/U6 snRNPs (∼15S; fractions 12/13) and U4/U5/U6 tri-snRNPs (25S; fractions 18/19) could be separated from each other.
Fig. 4. p110 protein is present in U6 and U4/U6 snRNPs, but not in U4/U5/U6 tri-snRNPs: glycerol gradient sedimentation of nuclear extract. HeLa nuclear extract was fractionated through glycerol gradient sedimentation, and RNA was prepared from each fraction (1–21, as indicated above). (A) The snRNA distribution was analyzed by denaturing gel electrophoresis and silver staining. The sedimentation profile of the 5S, 16S and 23S rRNA markers is given on the top and the identity of the snRNAs U1, U2, U4, U5 and U6 on the right. (B and C) Anti-p110 immunoprecipitations were carried out from the same gradient fractions, and co-precipitated RNA was prepared and analyzed by both northern blot hybridization with U4- and U6-specific probes (B) and [32P]pCp labeling (C). (D) The gradient fractions were analyzed for p110 protein by western blotting. The mobility of two marker proteins and p110 is marked on the right.
We looked for p110-associated RNAs, using anti-p110 immunoprecipitation from these gradient fractions, followed by northern blot hybridization with U4- and U6-specific probes (Figure 4B) or [32P]pCp labeling (Figure 4C). Consistent with our initial analysis, U4 and U6 snRNAs as well as the additional RNA X were immunoprecipitated specifically. Analyzing the distribution across the glycerol gradient, we conclude that both U6 snRNPs (peak in fraction 9) and U4/U6 snRNPs (peak in fraction 13) are associated with p110. Note that U6 snRNA fractionating in the U4/U6 snRNP region of the gradient (peaking around fraction 11) could not be labeled efficiently by [32P]pCp (compare fraction 11 in Figure 4B and C), most probably reflecting the 2′,3′-cyclic phosphate of mature U6 snRNA. Significantly, only a fraction of free U6 snRNPs can be immunoprecipitated by anti-p110 (peak at fraction 9), which does not coincide with the majority of U6 snRNPs (see broad peak in fractions 4–10; Figure 4A). Finally, no significant amounts of U6 could be immunoprecipitated from the 25S region of the gradient (fractions 18/19), indicating the absence of p110 from the U4/U5/U6 tri-snRNP.
In addition to this RNA analysis, we have also characterized the distribution of p110 across the gradient, using western blot analysis of total protein recovered from gradient fractions (Figure 4D). Most of the p110 protein was detected in fractions 9–13, corresponding to the regions where the U4/U6 and p110-associated U6 snRNPs migrate. This analysis of the p110 protein distribution was also consistent with the protein being absent from the U4/U5/U6 tri-snRNP, ruling out the possibility that p110 could not be detected due to epitope inaccessibility in this complex.
Similarly, we have characterized snRNP complexes containing p110 protein in HeLa cell S100 extract, which represents the cytoplasmic fraction as well as material leaked from the nuclei during the extract preparation (data not shown). S100 extract was fractionated by glycerol gradient sedimentation and RNA was prepared from each gradient fraction and analyzed by anti-p110 immunoprecipitation and [32P]pCp labeling; the p110 distribution was characterized by western blotting. One important difference from the snRNP distribution in nuclear extract was that in S100 extract, U6 snRNP complexes free of U4 represented the major form of U6. Accordingly, we have found in S100 extract more p110 protein, which co-sedimented with the free U6 form. In comparison with nuclear extract, there was also more of RNA X detectable in S100 extract (data not shown).
In sum, we conclude that p110 protein is associated specifically with a subfraction of the free U6 snRNP and with the U4/U6 snRNP, but is absent from the 25S U4/U5/U6 tri-snRNP. The U6–p110 snRNA complex exists in both HeLa nuclear and S100 extract; it is enriched in the S100 fraction, most probably due to its leakage during nuclear extract preparation. Based on these experiments, p110 appears to associate only transiently with U6 in the spliceosome cycle, in particular during recycling of the U4/U6 snRNP.
p110 cannot not be detected in spliceosomes
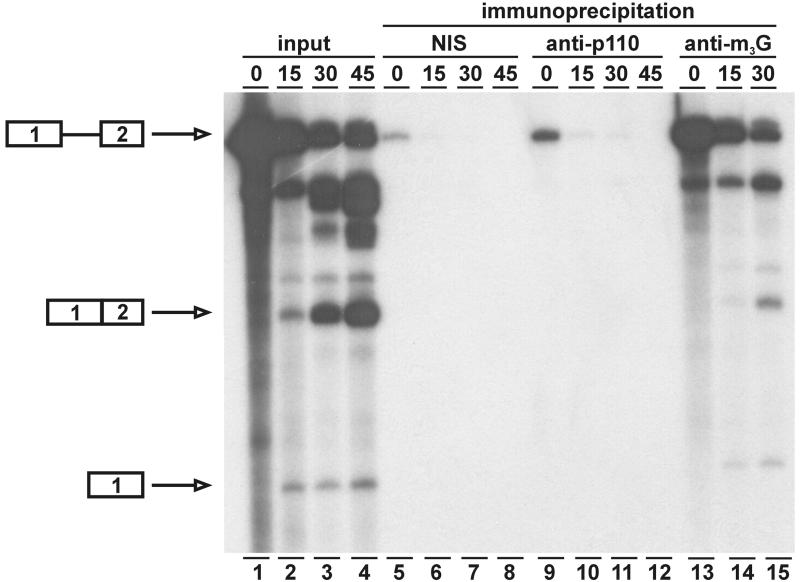
Since we were unable to detect p110 in U4/U5/U6 tri-snRNPs, we next asked whether it can be found in spliceosomes assembled during in vitro splicing (Figure 5). 32P-labeled MINX pre-mRNA was spliced in vitro in HeLa cell nuclear extract for 0, 15, 30 and 45 min. At each time point, aliquots were taken and immunoprecipitated with anti-p110 antiserum; separate control reactions were included with the corresponding non-immune serum and with anti-m3G cap antiserum. RNA was prepared from the immunoprecipitates and analyzed together with the total input samples. Splicing efficiency after 45 min reached ∼50%, and pre-mRNA, splicing intermediates and splicing products could be detected (see lanes 1–4). In the anti-m3G cap immunoprecipitates, pre-mRNA, both splicing intermediates and excised intron lariat could all be recovered efficiently, as expected from the snRNP association of the spliceosome; spliced mRNA was precipitated at lower efficiency (lanes 13–15). In contrast, anti-p110 antibodies (as well as the corresponding non-immune serum) did not precipitate significant levels of pre-mRNA, intermediates or products after 15, 30 and 45 min of in vitro splicing (lanes 10–12 and 6–8); the low level of pre-mRNA precipitated at time 0 is due to non-specific aggregates (compare lanes 5 and 9), and has not been observed under more stringent conditions (data not shown). Because of this negative result, we have also analyzed the anti-p110 and non-immune precipitates for their U4 and U6 snRNA content. Using direct [32P]pCp labeling, both U4 and U6 snRNAs could be readily detected in anti-p110, but not in the non-immune precipitate (data not shown).
Fig. 5. p110 protein is not detectable in spliceosomes. 32P-labeled MINX pre-mRNA was spliced in vitro in HeLa cell nuclear extract for 0, 15, 30 and 45 min, followed by immunoprecipitation by non-immune control serum (NIS; lanes 5–8), anti-p110 antibodies (lanes 9–12) or anti-m3G antibodies (lanes 13–15). Aliquots of the total reactions were also analyzed (lanes 1–4). The mobilities of pre-mRNA, spliced mRNA and exon 1 intermediate are marked on the left.
We conclude that p110 cannot be detected in spliceosomes assembled and processed in vitro; this represents further evidence for a transient association of p110 with snRNPs during the recycling phase of the spliceosome cycle.
p110 binds specifically to U6 snRNA in vitro
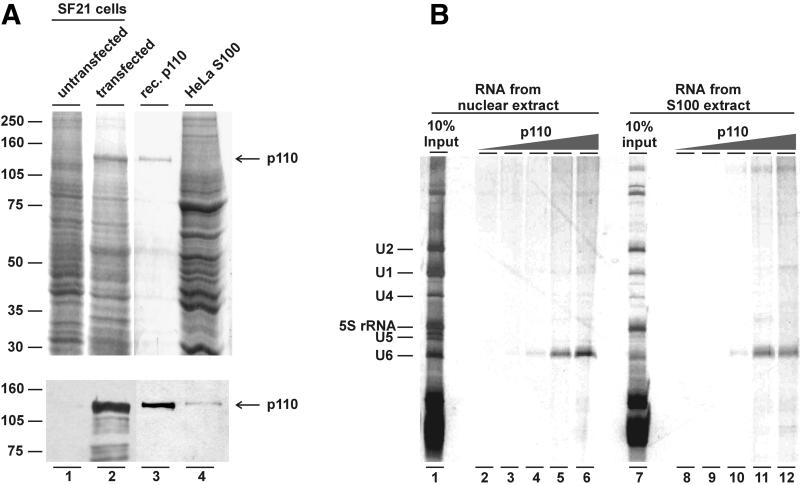
The presence of p110 in U6 and U4/U6 snRNPs suggested that it may recognize U6 snRNA specifically. To characterize its RNA-binding properties, we first expressed and purified recombinant p110 protein carrying an N-terminal His6 tag from baculovirus-transfected SF21 cells. Figure 6A demonstrates the purification and purity of the recombinant p110 protein, as detected by Coomassie Blue staining and western blot analysis; note that recombinant p110 co-migrated with p110 protein present in HeLa S100 extract (compare lanes 3 and 4). Total RNA was prepared from either nuclear extract or S100 (Figure 6B, lanes 1 and 7, respectively) and was incubated with recombinant p110 protein (50 ng to 6.25 µg per assay, as indicated above lanes 3–6 and 9–12); in addition, control reactions were done in the absence of added protein (lanes 2 and 8). Subsequently, anti-p110 immunoprecipitations were carried out, and co-selected RNAs were analyzed directly by denaturing gel electrophoresis and silver staining (Figure 6B). As this titration of p110 shows, U6 snRNA was bound by recombinant p110 from the complex RNA mixture with high specificity and with an efficiency of 10–20% (the identity of U6 snRNA was confirmed by northern blotting; data not shown). Only at the highest protein concentrations, some low background of high molecular weight RNA and some very minor discrete RNA species appeared. We conclude that recombinant p110 is able to recognize specifically and efficiently endogenous U6 snRNA present in total RNA from extracts.
Fig. 6. Recombinant p110 protein specifically binds U6 snRNA. (A) Expression and purification of recombinant p110 protein from baculovirus- transfected SF21 cells. Aliquots of extract prepared from untransfected SF21 cells (lane 1) or from SF21 cells transfected with the p110 expression construct (lane 2), purified His-tagged p110 protein (lane 3) and HeLa S100 extract (lane 4) were analyzed by SDS–PAGE and Coomassie Blue staining (top panel) and western blotting with anti-p110 antibodies (bottom panel). The arrow marks the position of p110 protein. (B) U6-specific RNA binding of p110 protein. Purified His-tagged p110 protein (0, 0.05, 0.25, 1.25 and 6.25 µg per assay; see lanes 2–6 and 8–12, respectively) was incubated with total RNA prepared from 125 µl of HeLa nuclear extract or S100 extract, respectively. After anti-p110 immunoprecipitation, co-precipitating RNAs were purified and analyzed by denaturing gel electrophoresis and silver staining (nuclear extract, lanes 2–6; S100 extract, lanes 8–12). The RNA composition of the total nuclear and S100 extracts is also shown (lanes 1 and 7, respectively). The positions of U1, U2, U4, U5, U6 and 5S rRNA are marked on the left.
U6 snRNA sequence requirements for p110 binding
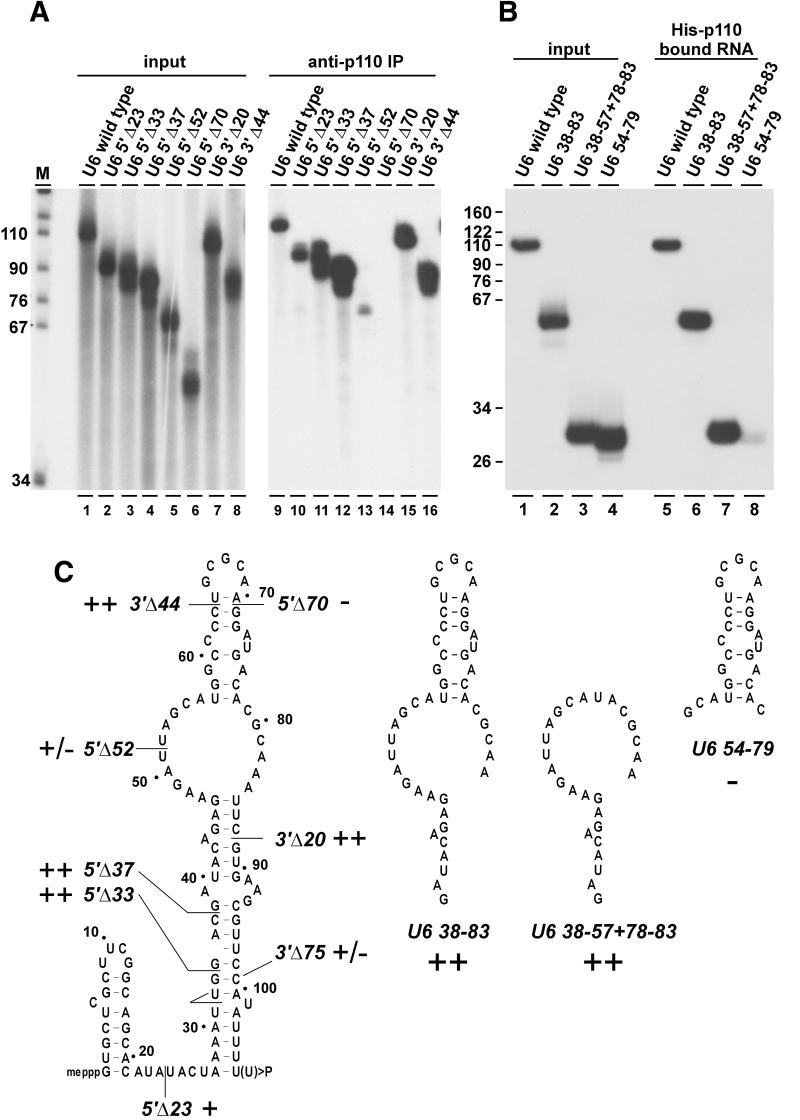
To determine where on U6 snRNA p110 binds, we first used a series of 5′- and 3′-truncated mutant derivatives of U6 snRNA (Figure 7A; see Figure 7C for a schematic representation). Each of these 32P-labeled mutant U6 RNAs was incubated with recombinant p110 protein, and binding was assayed by immunoprecipitation with anti-p110 antibodies (Figure 7A) or by selection on Ni-NTA–agarose (Figure 7B). Twenty-three, 33 and 37 nucleotides could be deleted from the 5′ end of U6 snRNA without affecting p110 binding; the efficiency of binding was even increased for the U6 5′ Δ37 RNA, when compared with wild-type U6 (lanes 1–4 and 9–12). However, if the 5′-terminal 52 or 70 nucleotides were removed, p110 binding was reduced to very low or undetectable levels, respectively (lanes 5–6 and 13–14). Deleting 20 or 44 nucleotides from the 3′ end still allowed p110 to bind, even at an increased efficiency compared with full-length U6 (lanes 7–8 and 15–16). A 3′ deletion of 75 nucleotides, however, abolished p110 binding (data not shown).
Fig. 7. U6 snRNA sequence requirements for p110 binding. (A) p110 binding to 5′ and 3′ truncated mutant derivatives of U6 snRNA. 32P-labeled wild-type U6 and U6 derivatives (as indicated above the lanes; see C) were incubated with recombinant p110 protein, followed by immunoprecipi tation with anti-p110 antibodies. In lanes 1–8, 20% of the input RNAs were analyzed and in lanes 9–16 the total immunoprecipitated material (anti-p110 IP). (B) p110 binding to short internal U6 fragments. 32P-labeled wild-type U6 and three derivatives containing internal fragments of U6 (as indicated above the lanes; see C) were incubated with recombinant p110 protein, followed by binding to Ni-NTA–agarose and recovery of bound material. In lanes 1–4, 20% of the input RNAs were analyzed and in lanes 5–8 the total precipitated material. (C) U6 singular secondary structure model (Rinke and Steitz, 1985). The 5′ and 3′ truncations and the short internal fragments of U6 are represented schematically as well as their p110 binding properties (in comparison with full-length wild-type U6 snRNA: ++, >50%; +, 10–50%; +/–, <10%, but above background level; –, undetectable).
Based on these results, we generated three shorter U6 RNA derivatives that contained various internal U6 regions (Figure 7C): U6/38–83 containing U6 nucleotides G38 to A83; U6/38–57 + 78–83, with most of the internal loop region, but not the intramolecular stem–loop; and U6/54–79 consisting of the intramolecular stem–loop (nucleotides G54 to C79). Binding assays with His-tagged p110 revealed that, with the exception of the intramolecular stem–loop RNA, these short RNAs still bound p110 efficiently (Figure 7B). We conclude that an internal region of U6, G38 to U57, which includes the highly conserved ACAGAG hexanucleotide and part of the internal loop sequence in the singular U6 structure, suffices for efficient p110 binding.
p110 is required for U4/U6 snRNP recycling in vitro
Since p110 associates only transiently with U6 in the U6 and U4/U6 snRNPs, but apparently not during the subsequent stages, we next tested for a function in U4/U6 snRNP recycling. Using purified U4 snRNPs and U6 snRNA, we were able to detect only low levels of a U4 snRNP–U6 snRNA interaction that was dependent on p110 protein; preliminary U4 RNA–U6 RNA annealing experiments with recombinant p110 protein were unsuccessful (data not shown). Therefore, we developed an assay system to assess the base-pairing status of U4 and U6 in the extract and to measure quantitatively the relative levels of U4/U6 as well as of free U4 and U6 snRNPs, using CsCl density gradient centrifugation. Under these highly stringent conditions, core complexes of snRNPs are characteristically stable and fractionate according to their density, which is determined by their RNA:protein ratio (Lelay-Taha et al., 1986).
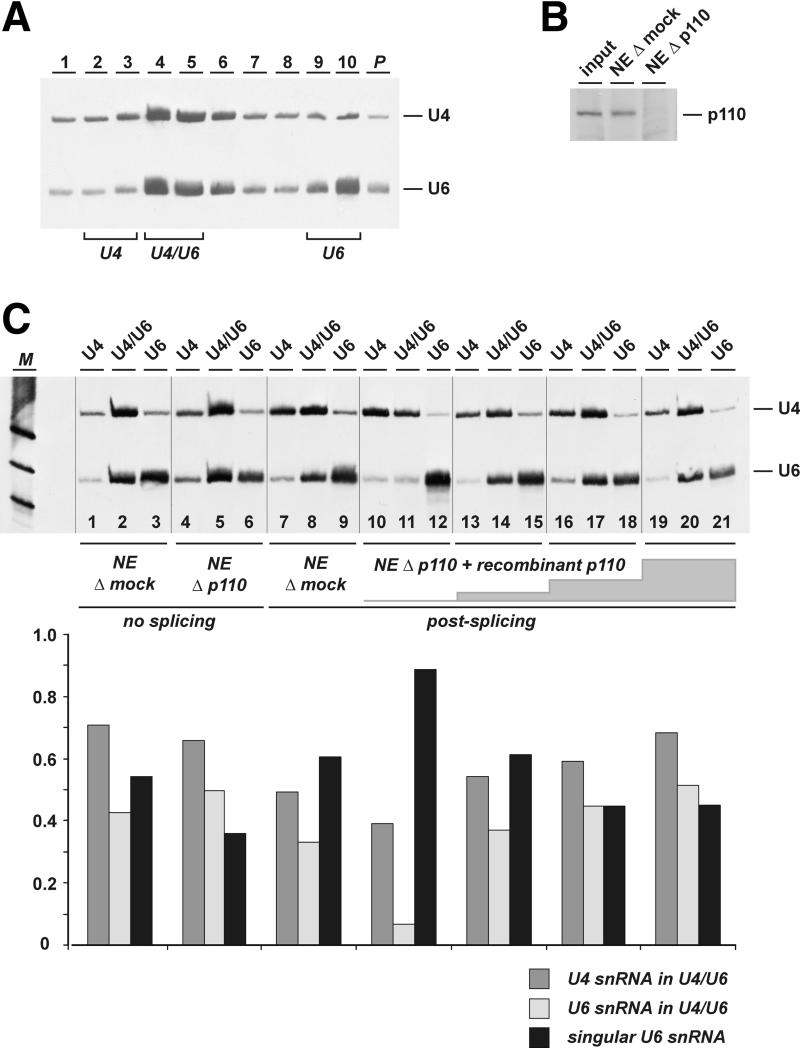
When normal nuclear extract was fractionated by CsCl density gradient centrifugation and U4 and U6 snRNAs were detected by northern blotting, we found a peak for both U4 and U6 at fractions 4–5, representing the U4/U6 core snRNP (Figure 7A). Approximately half of the U6 snRNA fractionated at the bottom of the CsCl gradient (fractions 9–10), representing free U6 snRNA, which is derived from U6 snRNPs unstable under these high ionic strength conditions. When nuclear extract was heat treated at 45°C for 10 min, which should disrupt the U4–U6 base-pairing interaction, we detected a shift of the U4 peak from fractions 4–5 (U4/U6 snRNP) to 2–3, which most probably represents free U4 snRNPs; in parallel, most of U6 snRNA shifted from fractions 4–5 (U4/U6 snRNP) to 9–10 (free U6) (data not shown). In sum, this approach allowed us to monitor quantitatively the relative levels of base-paired U4/U6, free U4 and free U6 snRNPs. For each assay, the extract or splicing reaction was fractionated on a CsCl gradient, followed by northern blot analysis of RNA from pooled fractions 2–3, 4–5 and 9–10, indicative of the abundance of free U4, base-paired U4/U6 and free U6, respectively.
We first determined the U4/U6 distribution in mock-depleted and p110-immunodepleted nuclear extract. Immunodepletion with anti-p110 antibodies routinely resulted in the selective removal of at least 95% of p110, as determined by western blot analysis (Figure 8B). Quantitation of the relative distribution of U6 in the U4/U6 and free forms gave the following results: for mock-depleted extract (Figure 8C, lanes 1–3), 42% of U6 as U4/U6 and 54% as free U6; for p110-immunodepleted nuclear extract (lanes 4–6), 50% of U6 as U4/U6 and 35% as free U6.
Fig. 8. p110 is required for U4/U6 snRNP recycling in vitro. (A) Nuclear extract was fractionated by CsCl gradient centrifugation, and RNA from fractions 1–10 (top to bottom; P, pellet fraction) was analyzed by denaturing gel electrophoresis and northern blotting with U4 and U6 probes. The distribution of U4/U6 and U4 snRNPs as well as U6 snRNA is indicated below. (B) p110 immunodepletion of nuclear extract. Aliquots of normal nuclear extract (input), and nuclear extract after mock depletion (NE Δ mock) and p110 depletion (NE Δ p110) were analyzed by western blotting with anti-p110 antibodies. (C) Analysis of the distribution of U4/U6, free U4 and free U6 snRNPs. Mock-depleted (lanes 1–3) and p110-depleted nuclear extracts (lanes 4–6) were fractionated by CsCl gradient centrifugation as shown in (A). RNA was purified from pooled fractions 2/3 (free U4), 4/5 (U4/U6) and 9/10 (free U6) (as indicated above the lanes) and was analyzed by northern blotting with U4 and U6 probes. In addition, splicing reactions were performed in mock-depleted extract (lanes 7–9), and in p110-depleted extract, without p110 protein (lanes 10–12) and after complementation with recombinant p110 protein (200 ng per 25 µl reaction, lanes 13–15; 500 ng, lanes 16–18; 1000 ng, lanes 19–21). For each assay, the three lanes derived from one CsCl gradient are separated by black lines. Below the northern blot, the U4–U6 distribution is represented quantitatively for each extract or reaction (dark gray bars, fraction of U4 in the U4/U6 snRNP; light gray bars, fraction of U6 in the U4/U6 snRNP; black bars, free U6 fraction). M, DIG molecular weight marker V (Roche; 122, 110 and 89 nucleotides).
Next, we measured how the U4 and U6 snRNP dis tribution changed after pre-mRNA splicing. Preliminary experiments had shown that at the normal, low pre-mRNA concentration (10 ng of MINX pre-mRNA per 25 µl reaction) we observed no significant difference of splicing efficiency between mock- and p110-depleted extract; only at high pre-mRNA concentrations (100 ng per 25 µl reaction) was the splicing efficiency reduced at least 3-fold in p110-depleted splicing extract in comparison with mock-depleted extract (data not shown). Most probably this reflects that p110 is important only under conditions where splicing factors have to be reutilized after a single round of the spliceosome cycle, i.e. under recycling conditions. Therefore, MINX pre-mRNA was spliced in vitro at the high concentration for 60 min, using mock-depleted or p110-immunodepleted nuclear extract (Figure 8C, lanes 7–9 and 10–12, respectively). After splicing in vitro in mock-depleted extract, the U6 levels in the U4/U6 snRNPs decreased moderately from 42 to 36% and, in parallel, levels of free U6 increased from 54 to 60% (compare lanes 1–3 and 7–9). We interpret these changes to reflect the consumption of U4/U6 snRNPs during splicing and the corresponding accumulation of post-spliceosomal free U4 and U6 snRNPs.
In p110-depleted extract, however, these differences in the U4 and U6 status before and after splicing were much more pronounced: The corresponding U4/U6 levels decreased dramatically from 45 to 6% and, in parallel, the free U6 level increased from 36 to 86% (compare lanes 4–6 and 10–12). This behavior strongly suggests that p110 is required for the efficient regeneration of U4/U6 snRNPs following splicing catalysis.
Finally, we used complementation with recombinant p110 protein to show that this effect is due to the specific depletion of p110. Increasing quantities of recombinant p110 protein (200, 500 and 1000 ng per 25 µl reaction) were added back to splicing reactions in p110-depleted extract, and the U4 and U6 status was determined (lanes 13–21), in comparison with the situation in the absence of p110 protein (lanes 10–12). Clearly, the addition of p110 protein reversed—in a concentration-dependent manner—the imbalance of the U4/U6 distribution. The abundance of U6 in the U4/U6 snRNP increased to 38, 42 and 50%, respectively, compared with 6% without complementation; in parallel, complementation decreased the relative level of free U6 to 60, 45 and 42%, respectively, compared with 86% without added p110 protein. In sum, this demonstrates that p110 functions as a U6-specific recycling factor and is responsible for the regeneration of base-paired U4/U6 snRNPs from post-spliceosomal free U4 and U6 snRNPs.
Discussion
Conserved domain structure of human p110/SART3 and related proteins
We identified the p110 protein sequence during extensive database searches for mammalian proteins that are related to Prp24 of S.cerevisiae and possibly involved in U4/U6 snRNP assembly and/or recycling. The human p110 protein is only distantly related in its sequence and domain structure to the yeast splicing factor Prp24, sharing only RRM domains and the short C-terminal sequence motif (see Figures 1 and 2). In addition, the human p110 protein carries seven TPR domains in its N-terminal half. On the basis of the human p110 sequence, putative orthologs have been identified from several other species (see Figures 1 and 2). Except for the O.novo-ulmi protein, which apparently lacks the TPR motifs in its N-terminal half, all these orthologs share the same overall domain structure: a flexible number of TPR domains in the N-terminal half, between one and four RRMs in the C-terminal half of the protein and a highly conserved 10 amino acid segment at the C-terminus. In contrast, the S.cerevisiae Prp24 protein deviates from this pattern in that it lacks the entire TPR domain. Therefore, it will be important to investigate what additional function, such as specific protein interactions, the TPR domain mediates.
In the course of our search, we found that the human p110 protein and its gene had been identified and characterized initially by two different groups:
First, Reddy and co-workers identified a nuclear protein called p110nrb that exhibited general RNA-binding activity and co-purified with U6 snRNA capping activity (Shimba and Reddy, 1994; Gu et al., 1998). Based on the peptide sequence of the purified p110nrb protein, they found that the corresponding cDNA coding for a protein of 963 amino acids previously had been isolated in a random cDNA cloning project (KIAA0156; Nagase et al., 1995). The human gene for p110 (KIAA0156/SART3) maps to chromosome 12q23–24, extends over 38 kb and contains 19 exons within a 3.8 kb cDNA sequence (genomic contig NT_009660; Nagase et al., 1995; Yang et al., 1999).
Second, Itoh and co-workers isolated a human cDNA coding for a protein called SART3, based on a search for tumor epitope-encoding genes (Yang et al., 1999). This sequence turned out to be almost identical to KIAA0156 (see above). Interestingly, although present in almost all tissues analyzed, the SART3 protein is overexpressed in many different tumor cell lines and patient tissues (Yang et al., 1999; Niiya et al., 2000; Tanaka et al., 2000; Suefuji et al., 2001); in addition, certain SART3 epitopes are capable of inducing cytotoxic T lymphocytes (CTLs), making this protein a promising candidate as a tumor rejection antigen to be used for immunotherapy of cancer (Yang et al., 1999; Ito et al., 2000). More recently, Harada et al. (2000) isolated a mouse SART3 cDNA, which is highly homologous to its human counterpart (AF172722; 86% identity on the protein level).
Function of human p110 in U4/U6 snRNP recycling: implications for splicing regulation
What is the in vivo function of p110? The human p110 protein originally had been isolated in the search for the U6 capping activity; recombinant p110nrb protein, however, had no U6 capping activity, indicating that it may not be sufficient for or not be involved in U6 snRNA processing (Gu et al., 1998). In addition, general RNA-binding activity had been reported under the conditions of northwestern blotting (Gu et al., 1998). We have characterized native p110 protein for its RNA-binding activity in solution, both with total RNA and with in vitro transcribed U6 RNA (see Figures 6 and 7), and clearly demonstrated that p110 is able to recognize U6 snRNA by itself with high specificity. Based on the original co-purification with the U6 capping activity, p110 may constitute a component of a U6 modification and maturation complex.
Regarding the U6 snRNA binding specificity, we have mapped an internal region of U6 that is sufficient for specific p110 binding (see Figure 7). Interestingly, this region includes the highly conserved ACAGAG hexanucleotide and the stem I region. The ACAGAG sequence is involved in 5′ splice site recognition by U6 and in splicing catalysis; the stem I region participates in both U4–U6 and U6–U2 RNA–RNA interactions. Our results therefore suggest that these sequences play an additional role during snRNP assembly, serving as a binding site for p110.
We have observed that only a small fraction of ∼10% of free U6 snRNP is p110 associated (see Figure 3 and data not shown). Another known U6 snRNP-specific protein, La, also occurs in only 10% of U6 RNPs and binds only to U6 with a 3′ hydroxyl terminus, but not to U6 with the mature 2′,3′-cyclic phosphate (Terns et al., 1992). Both La and p110 proteins associate transiently, but differentially with U6 snRNA in the spliceosome cycle: whereas La protein occurs only in U6 snRNPs, p110 is found in both the singular U6 and the U4/U6 snRNP. Taken together, U6 snRNPs appear to be heterogenous in composition. Since U6 snRNA undergoes extensive modifications, it will be important in the future to dissect further this complex pathway of U6 snRNA maturation and snRNP assembly.
We have shown here that p110 functions only transiently during the spliceosome cycle; we could detect it neither in the U4/U5/U6 tri-snRNP nor in spliceosomes. This behavior is very similar to that of yeast Prp24, and therefore we wanted to obtain direct evidence for a role in U4/U6 snRNP recycling. Using CsCl gradient fractionation, we were able to demonstrate (i) that free forms of U4 and U6 accumulate in p110-depleted extract as a consequence of splicing; and (ii) that recombinant p110 protein can greatly enhance the reassociation of these separate, post-spliceosomal U4 and U6 snRNPs (Figure 8). Only little is known so far about the recycling phase of the spliceosome cycle, and this work provides the first experimental evidence in the mammalian system for an snRNP recycling factor. Two other factors, the human U4/U5/U6 triple snRNP-specific 61K protein (Makarova et al., 2002) and Aar2 from yeast (Gottschalk et al., 2001), recently have been suggested to function during recycling of the triple snRNP.
We are unable as yet to demonstrate direct U4–U6 RNA annealing activity with recombinant p110 (data not shown). Therefore, additional factors such as the LSm complex may be required, consistent with a recent study on the human U6 snRNP-specific LSm proteins that found that they can promote RNA–RNA annealing of U4 and U6 (Achsel et al., 1999). Interactions between yeast Prp24 and several of the U6-specific Lsm proteins (Fromont-Racine et al., 2000; S.Rader and C.Guthrie, in preparation) are likely to be relevant in this regard.
Is there a role for p110 in splicing regulation? Recent work by Harada et al. (2001) found an association of p110 with RNPS1, a known splicing activator protein, suggesting that p110 might participate in some way in splicing regulation. It is plausible that mRNA splicing may also be controlled at the level of the recycling phase. In this context, it should be revealing to investigate the expression, cellular localization and function of p110 in different cell lines and during development. As Hamm and Mattaj (1989) have reported, the distribution of free U6 versus U4/U6 snRNPs is shifted dramatically during Xenopus embryonic development; furthermore, they discovered differences in the U6 status between embryonic and other cell lines. These differences in the U6 particle status may be caused by different levels or activities of p110.
Materials and methods
Extracts; glycerol gradient sedimentation
S100 and nuclear extracts (4C Biotec, Belgium) were prepared from HeLa cells by the method of Dignam et al. (1983). For fractionation of snRNP complexes, 500 µl of HeLa nuclear extract dialyzed against buffer G (10 mM HEPES pH 8.0, 150 mM KCl, 1.5 mM MgCl2) were loaded onto an 11 ml glycerol gradient (10–30% glycerol in buffer G). After ultracentrifugation (16 h, 4°C, 32 000 r.p.m., Beckman SW 40 Ti rotor), 500 µl fractions were taken from the top to the bottom of the gradient. RNA from 50 µl of each fraction was isolated and analyzed by denaturing PAGE and silver staining. For analysis of p110 complexes, equal amounts of two fractions were pooled, and aliquots corresponding to 15 or 75 µl of one fraction were subjected to western blot analysis or immunoprecipitation, respectively.
RNA analysis
RNA was separated by electrophoresis in denaturing polyacrylamide– urea gels (8% acrylamide, 0.42% bisacrylamide, 50% urea, 1× TBE buffer). For 3′-terminal labeling, RNA was incubated with 10 µCi of 5′ [32P]pCp and 50 U of T4 RNA ligase (Roche) in the provided buffer, containing in addition 10% dimethylsulfoxide (5 h, 18°C). After phenolization and precipitation, the labeled RNA was separated by denaturing PAGE. Northern blot analysis of RNA was carried out as described previously (Bell and Bindereif, 1999). Digoxigenin-labeled probes directed against human U4 and U6 snRNA were obtained by PCR including the PCR DIG labeling mix (Roche), M13 forward and reverse primers and SP6-U4 (Wersig and Bindereif, 1990) or SP6-U6 (Bindereif et al., 1990), respectively, as templates.
Expression and purification of recombinant p110 protein
The open reading frame of p110 (NM_014706) was cloned into the vector pFASTa (Life Technologies) replacing the first methionine with the N-terminal His tag of the vector. Recombinant baculovirus for protein expression in SF21 cells was obtained using the Bac-to-Bac® baculovirus expression system (Life Technologies). For purification of recombinant p110, cytoplasmic and nuclear extracts from infected SF21 cells were isolated (Dignam et al., 1983), and the protein was affinity selected on Ni-NTA–agarose (Qiagen) under native conditions, eluted with 250 mM imidazole and dialyzed against buffer D (Dignam et al., 1983).
Antibodies, western blot analysis and immunoprecipitations
Polyclonal antibodies against recombinant p110 protein were either provided by R.Reddy (Gu et al., 1998) or raised against gel-purified recombinant p110 protein expressed in baculovirus-infected SF21 cells (see above; Eurogentec). For protein analysis, cell extracts, gradient fractions or recombinant p110 were mixed with protein sample buffer, separated on an 8% SDS–polyacrylamide gel and analyzed by Coomassie Blue staining or western blot. Therefore, the proteins were transferred to a Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech) and immunostained with p110 antisera (1:2000 dilution) and anti-rabbit-POD (Roche; 1:20 000 dilution), followed by chemiluminescence detection (ECL, Amersham).
For immunoprecipitation experiments, 50 µl packed volume of protein A–Sepharose CL-4B (Amersham Pharmacia Biotech) in 220 µl of N100 [50 mM Tris–HCl pH 8.0, 100 mM NaCl, 0.05% NP-40, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF)] were incubated with 30 µl of serum, rotating for 4 h at 4°C, and washed five times with 1 ml of N100. Then the beads were incubated for 3 h together with 300 µl of N200, N250, N300, N400 or N600 (as N100, with the number indicating the concentration of NaCl in mM) and 50 µl of HeLa S100, nuclear extract or a gradient fraction for 3 h (4°C, rotating), followed by washing five times with 1 ml each of the corresponding buffer. Co-selected RNAs were eluted in 300 µl of PK buffer (100 mM Tris pH 8.0, 12.5 mM EDTA, 15 mM NaCl, 1% SDS) for 10 min at 80°C, phenolized and precipitated. Immunopurified RNA was isolated from denaturing polyacrylamide gels after staining with Sybr Green (FMC BioProducts) or analyzed by [32P]pCp labeling.
In vitro pre-mRNA splicing was performed as described (Bindereif and Green, 1987), using in vitro transcribed 32P-labeled MINX RNA (Zillmann et al., 1988) as a template. At different time points, aliquots of the reaction were taken, diluted 10-fold with N200, centrifuged to remove aggregates and subjected to immunoprecipitation. Selected RNA was analyzed by denaturing PAGE.
RNA binding in vitro; mutational analysis
Total RNA was purified from 125 µl of HeLa S100 or nuclear extract and incubated with different amounts of recombinant p110 for 30 min at 30°C in a 200 µl reaction [20 mM HEPES pH 8.0, 100 mM KCl, 15 mM MgCl2, 0.2 mM EDTA, 625 µM ATP, 25 mM creatine phosphate, 100 U of RNasin (Promega), 16% glycerol]. After addition of 200 µl of N100, p110 complexes were immunoprecipitated with immobilized p110 antibodies. Co-selected RNAs were analyzed by denaturing PAGE and silver staining.
5′- or 3′-truncated mutant derivatives of U6 snRNA (Bindereif et al., 1990) were transcribed in vitro in the presence of [α-32P]UTP. A 50 ng aliquot of each mutant RNA was incubated for 30 min at 30°C with 300 ng of His-tagged p110 (an ∼2-fold molar excess of the protein) in a 30 µl reaction containing buffer D50 (20 mM HEPES–KCl pH 7.5, 50 mM KCl, 15 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT), 0.5 mM ATP and 40 U of RNasin. RNA–protein complexes were immunoprecipitated as described above, and co-precipitated RNA was analyzed by denaturing PAGE and autoradiography.
U6 snRNA derivatives containing short internal regions of U6 (see Figure 7B for details) were transcribed in vitro from double-stranded oligonucleotide templates and 3′ end-labeled by [32P]pCp. A 50 ng aliquot of each RNA sample was incubated for 30 min at 30°C with 500 ng of His-tagged p110 in a 30 µl reaction containing buffer D50, 0.5 mM ATP and 40 U of RNasin. After the addition of 240 µl of buffer D50, p110 complexes were bound to Ni-NTA–agarose beads, and the co-precipitated RNAs were analyzed as described above.
p110 immunodepletion and U4/U6 snRNP recycling in vitro
For p110 immunodepletion, protein A–Sepharose beads were incubated overnight at 4°C with polyclonal anti-p110 antiserum. Beads were then washed 10 times with 1 ml each of N100 buffer, followed by the addition of nuclear extract (4C Biotec, Belgium) and incubation at 4°C for 2 h (400 µl of extract per 100 µl of packed beads). As control, mock-depleted extract was prepared in parallel without antiserum.
In vitro pre-mRNA splicing was done as described (Krainer et al., 1984), using 100 ng of MINX pre-mRNA per 25 µl reaction, and normal, mock-depleted or p110-depleted nuclear extract. For complementation assays, recombinant p110 protein was added to the depleted extract (200–1000 ng per 25 µl reaction). After splicing for 60 min at 30°C, the entire reaction (75 µl) was fractionated by CsCl gradient centrifugation, as described (Lücke et al., 1997), using a CsCl–buffer D solution with a density of 1.55 g/ml and containing 15 mM MgCl2. Ten fractions were obtained, and fractions 2–3 (free U4), 4–5 (U4/U6) and 9–10 (free U6) were pooled. RNA was purified and analyzed on an 8% denaturing polyacrylamide gel, followed by northern hybridization with digoxigenin-labeled U4 and U6 probes.
The intensities of the U4 and U6 signals obtained by northern analysis were determined by AIDA 1D quantitation software (Raytest, Strauchenhardt, Germany). The amounts of U4 and U6 snRNAs present in the U4/U6 or the free snRNPs (Figure 8C) are expressed as ratios of the intensities of the corresponding peak bands to the integrated intensities of all fractions of the CsCl gradient shown.
Sequence analysis
For database searches, BLAST programs at NCBI were used. Protein pattern predictions were done with SMART (Schultz et al., 2000), and sequence alignments with Clustal_W (Thompson et al., 1994).
Acknowledgments
Acknowledgements
We thank Christine Guthrie, Stephen Rader and Karla Neugebauer for communicating unpublished results and discussions, John Abelson and Brian Rymond for discussions, and Assen Roguev for the quantitation of northern hybridization. We acknowledge Jan Medenbach for investigating the nature of RNA X. This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
References
- Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M. and Bindereif,A. (1999) Cloning and mutational analysis of the Leptomonas seymouri U5 snRNA gene: function of the Sm site as a conserved sequence element in core RNP formation and nuclear localization. Nucleic Acids Res., 27, 3986–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindereif A. and Green,M.R. (1987) An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J., 6, 2415–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindereif A., Wolff,T. and Green,M.R. (1990) Discrete domains of human U6 snRNA required for the assembly of U4/U6 snRNP and splicing complexes. EMBO J., 9, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch G.L. and Lassle,M. (1999) The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. BioEssays, 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M. et al. (2000) Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast, 17, 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti A., Company,M. and Abelson,J. (1995) Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA, 1, 132–145. [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A., Kastner,B., Lührmann,R. and Fabrizio,P. (2001) The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA, 7, 1554–1565. [PMC free article] [PubMed] [Google Scholar]
- Gu J., Shimba,S., Nomura,N. and Reddy,R. (1998) Isolation and characterization of a new 110 kDa human nuclear RNA-binding protein (p110nrb). Biochim. Biophys. Acta, 1399, 1–9. [DOI] [PubMed] [Google Scholar]
- Hamm J. and Mattaj,I.W. (1989) An abundant U6 snRNP found in germ cells and embryos of Xenopus laevis. EMBO J., 8, 4179–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K., Yamada,A., Mine,T., Kawagoe,N., Takasu,H. and Itoh,K. (2000) Mouse homologue of the human SART3 gene encoding tumor-rejection antigen. Jap. J. Cancer Res., 9, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K., Yamada,A., Yang,D., Itoh,K. and Shichijo,S. (2001) Binding of a SART3 tumor-rejection antigen to a pre-mRNA splicing factor RNPS1: a possible regulation of splicing by a complex formation. Int. J. Cancer, 93, 623–628. [DOI] [PubMed] [Google Scholar]
- Horowitz D.S., Kobayashi,R. and Krainer,A.R. (1997) A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA, 3, 1374–1387. [PMC free article] [PubMed] [Google Scholar]
- Ito M., Shichijo,S., Miyagi,Y., Kobayashi,T., Tsuda,N., Yamada,A., Saito,N. and Itoh,K. (2000) Identification of SART3-derived peptides capable of inducing HLA-A2-restricted and tumor-specific CTLs in cancer patients with different HLA-A2 subtypes. Int. J. Cancer, 88, 633–639. [DOI] [PubMed] [Google Scholar]
- Jandrositz A. and Guthrie,C. (1995) Evidence for a Prp24 binding site in U6 snRNA and in a putative intermediate in the annealing of U6 and U4 snRNAs. EMBO J., 14, 820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A.R., Maniatis,T., Ruskin,B. and Green,M.R. (1984) Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell, 36, 993–1005. [DOI] [PubMed] [Google Scholar]
- Lelay-Taha M.N., Reveillaud,I., Sri-Widada,J., Brunel,C. and Jeanteur,P. (1986) RNA–protein organization of U1, U5 and U4–U6 small nuclear ribonucleoproteins in HeLa cells. J. Mol. Biol., 189, 519–532. [DOI] [PubMed] [Google Scholar]
- Lücke S., Klöckner,T., Palfi,Z., Boshart,M. and Bindereif,A. (1997) Trans mRNA splicing in trypanosomes: cloning and analysis of a PRP8-homologous gene from Trypanosoma brucei provides evidence for a U5-analogous RNP. EMBO J., 16, 4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O.V., Makarov,E.M., Liu,S., Vornlocher,H.P. and Lührmann,R. (2002) Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6·U5 tri-snRNP formation and pre-mRNA splicing. EMBO J., 21, 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean M.R. and Rymond,B.C. (1998) Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol. Cell. Biol. 18, 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T., Seki,N., Tanaka,A., Ishikawa,K. and Nomura,N. (1995) Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121–KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res., 2, 167–174. [DOI] [PubMed] [Google Scholar]
- Niiya F., Nishizaka,S., Matsunaga,K., Koufuji,K., Mori,M., Katai,H., Yamana,H. and Itoh,K. (2000) Expression of SART3 tumor-rejection antigen in gastric cancers. Jap. J. Cancer Res., 91, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira V., Royer,J.C., Hintz,W.E., Field,D., Bowden,C., Kokurewicz,K., Hubbes,M. and Horgen,P.A. (2000) A gene associated with filamentous growth in Ophiostoma novo-ulmi has RNA-binding motifs and is similar to a yeast gene involved in mRNA splicing. Curr. Genet., 37, 94–103. [DOI] [PubMed] [Google Scholar]
- Petschek J.P., Scheckelhoff,M.R., Mermer,M.J. and Vaughn,J.C. (1997) RNA editing and alternative splicing generate mRNA transcript diversity from the Drosophila 4f-rnp locus. Gene, 204, 267–276. [DOI] [PubMed] [Google Scholar]
- Preker P.J. and Keller,W. (1998) The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci., 23, 15–16. [DOI] [PubMed] [Google Scholar]
- Raghunathan P.L. and Guthrie,C. (1998a) RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol., 8, 847–855. [DOI] [PubMed] [Google Scholar]
- Raghunathan P.L. and Guthrie,C. (1998b) A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science, 279, 857–860. [DOI] [PubMed] [Google Scholar]
- Rinke J. and Steitz,J.A. (1985) Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 13, 2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res., 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K.W. and Guthrie,C. (1991) Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev., 5, 773–785. [DOI] [PubMed] [Google Scholar]
- Shimba S. and Reddy,R. (1994) Purification of human U6 small nuclear RNA capping enzyme. Evidence for a common capping enzyme for γ-monomethyl-capped small RNAs. J. Biol. Chem., 269, 12419–12423. [PubMed] [Google Scholar]
- Suefuji Y., Sasatomi,T., Shichijo,S., Nakagawa,S., Deguchi,H., Koga,T., Kameyama,T. and Itoh,K. (2001) Expression of SART3 antigen and induction of CTLs by SART3-derived peptides in breast cancer patients. Br. J. Cancer, 84, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Tsuda,N., Kawano,K., Sakamoto,M., Nishida,T., Hashimoto,T., Shichijo,S., Kamura,T. and Itoh,K. (2000) Expression of tumor-rejection antigens in gynecologic cancers. Jap. J. Cancer Res., 91, 1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns M.P., Lund,E. and Dahlberg,J.E. (1992) 3′-end-dependent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevis oocyte nuclei. Mol. Cell. Biol., 12, 3032–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Clustal_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaver R.M., Fortner,D.M., Loos-Austin,L.S. and Brow,D.A. (1999) Multiple functions of Saccharomyces cerevisiae splicing protein Prp24 in U6 RNA structural rearrangements. Genetics, 153, 1205–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic I., Nottrott,S., Hartmuth,K., Lührmann,R. and Ficner,R. (2000) Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell, 6, 1331–1342. [DOI] [PubMed] [Google Scholar]
- Wersig C. and Bindereif,A. (1990) Conserved domains of human U4 snRNA required for snRNP and spliceosome assembly. Nucleic Acids Res., 18, 6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]
- Wolff T. and Bindereif,A. (1992) Reconstituted mammalian U4/U6 snRNP complements splicing: a mutational analysis. EMBO J., 11, 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T. and Bindereif,A. (1993) Conformational changes of U6 RNA during the spliceosome cycle: an intramolecular helix is essential both for initiating the U4–U6 interaction and for the first step of splicing. Genes Dev., 7, 1377–1389. [DOI] [PubMed] [Google Scholar]
- Yang D. et al. (1999) Identification of a gene coding for a protein possessing shared tumor epitopes capable of inducing HLA-A24-restricted cytotoxic T lymphocytes in cancer patients. Cancer Res., 59, 4056–4063. [PubMed] [Google Scholar]
- Zhang K., Smouse,D. and Perrimon,N. (1991) The crooked neck gene of Drosophila contains a motif found in a family of yeast cell cycle genes. Genes Dev., 5, 1080–1091. [DOI] [PubMed] [Google Scholar]
- Zillmann M, Zapp,M.L. and Berget,S.M. (1988) Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol., 8, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]