Abstract
Accurate tRNA 3′ end maturation is essential for aminoacylation and thus for protein synthesis in all organisms. Here we report the first identification of protein and DNA sequences for tRNA 3′-processing endonucleases (RNase Z). Purification of RNase Z from wheat identified a 43 kDa protein correlated with the activity. Peptide sequences obtained from the purified protein were used to identify the corresponding gene. In vitro expression of the homologous proteins from Arabidopsis thaliana and Methano coccus janaschii confirmed their tRNA 3′-processing activities. These RNase Z proteins belong to the ELAC1/2 family of proteins and to the cluster of orthologous proteins COG 1234. The RNase Z enzymes from A.thaliana and M.janaschii are the first members of these families to which a function can now be assigned. Proteins with high sequence similarity to the RNase Z enzymes from A.thaliana and M.janaschii are present in all three kingdoms.
Keywords: archaea/endonuclease/plants/processing/tRNA
Introduction
Functional tRNA molecules are essential not only for protein synthesis but also for a number of other cellular processes in all organisms (Söll, 1993). Since tRNAs are transcribed as part of longer precursor molecules, several processing steps are required to yield functional tRNAs. Two of these processing steps are needed to release the tRNA from the precursor molecule. While the enzyme responsible for tRNA 5′ end maturation, RNase P, is well studied (for reviews see Altman et al., 1995; Frank and Pace, 1998), generation of the tRNA 3′ end is not as well characterized (for a review see Mörl and Marchfelder, 2001).
In bacteria, tRNA maturation has been analysed in detail in Escherichia coli. An endonuclease cleaves the tRNA precursor several nucleotides downstream of the tRNA 3′ end and some of the remaining nucleotides are removed by an exonuclease. After 5′ end maturation by RNase P, the residual nucleotides at the 3′ end are removed by further exonucleolytic activities (Deutscher, 1995). Although individual 3′ exoribonucleases play specific roles in 3′ processing with typical substrates specificities and specific mechanisms of action (Li and Deutscher, 1994), a single 3′ exoribonuclease can substitute several other RNases in mutant strains lacking these (Reuven and Deutscher, 1993).
In archaea, tRNA 3′ processing is performed by an endonuclease that cleaves the precursor 3′ to the discriminator (the discriminator is located 5′ to the CCA and serves as an identity element in many tRNAs) (Schierling et al., 2002). While in vitro 5′-extended pre-tRNAs are processed with lower efficiency than 5′-matured pre-tRNAs (Schierling et al., 2002), 5′ processing by RNase P has to precede 3′ processing in vivo (Palmer et al., 1994). tRNA 3′ processing can occur prior to intron splicing in vivo and in vitro (Palmer et al., 1994; Schierling et al., 2002).
In eukarya, generation of the mature tRNA 5′ end is catalysed by an RNase P activity as in all other organisms. For removal of the 3′ trailer sequence, two processing modes seem to exist in these organisms, one using an endonucleolytic cut at or close to the discriminator (Garber and Gage, 1979; Hagenbüchle et al., 1979; Castaño et al., 1985; Stange and Beier, 1987; Oommen et al., 1992; Franklin et al., 1995; Han and Kang, 1997; Nashimoto, 1997; Mayer et al., 2000) and the other taking advantage of exonucleolytic activities (Garber and Altman, 1979; Engelke et al., 1985). The majority of the eukaryotic species analysed use the first, the endonucleolytic pathway. In yeast, it has been shown that the presence of protein Lhp1p is required for endonucleolytic processing (Yoo and Wolin, 1997). Lhp1p, however, has no endonucleolytic activity by itself. Strains without the Lhp1p protein generate tRNA 3′ ends solely with exonucleases, while wild-type cells, containing the Lhp1p protein, use the endonucleolytic pathway. It has been suggested that Lhp1p stabilizes the tRNA structure and thus facilitates the endonucleolytic processing (Yoo and Wolin, 1997).
Studies of mitochondrial and plastid tRNA 3′-processing systems have shown that in these cellular compartments, endonucleases generate the tRNA 3′ ends (Manam and Van Tuyle, 1987; Chen and Martin, 1988; Hanic-Joyce and Gray, 1990; Gegenheimer, 1995; Kunzmann et al., 1998).
To date, no protein sequence and no gene has been assigned to a tRNA-specific 3′ endonuclease. tRNA 3′-processing activities have, however, been partially purified from several organisms. In Saccharomyces cerevisiae, two endonucleases of 45/60 and 55 kDa and three exonucleases of 33, 60 and 70 kDa were found to be competent in tRNA 3′ end maturation, confirming the observation that both exo- and endonucleolytic pathways are possible in this organism (Papadimitriou and Gross, 1996). Purification of the Aspergillus nidulans enzyme revealed an endonuclease of 160 kDa to be involved in tRNA 3′ processing (Han and Kang, 1997). In Xenopus laevis, a protein of 97 kDa was identified as tRNA 3′ endonuclease (Castaño et al., 1985). Analysis of tRNA 3′ processing in potato mitochondria has correlated a 43 kDa protein with the endonucleolytic activity (Kunzmann et al., 1998).
Here we report the isolation of the nuclear tRNA 3′-processing endonuclease, RNase Z, from wheat, resulting in the first protein and nucleic acid sequences for this class of enzymes, and the subsequent identification of RNase Z homologues in the plant Arabidopsis thaliana and the archaeon Methanococcus janaschii.
Results
Purification of the nuclear RNase Z from wheat germ
Earlier experiments showed that the tRNA 3′-processing activity in wheat germ is present in very low amounts, similar to other tRNA processing enzymes, such as RNase P and tRNA splicing endonuclease (Zimmerly et al., 1993; Kleman-Leyer et al., 1997). For the isolation of the wheat tRNA 3′ endonuclease, a lot of material was thus required, even though highly efficient fractionation steps had been worked out in earlier experiments (Mayer et al., 2000).
Briefly, a soluble protein fraction (S30) was extracted from 2.9 kg of wheat germ and purified through six purification steps (Figure 1), the most efficient purification step being a tRNA affinity column to which the RNase Z bound tightly. Analysis of protein patterns from RNase Z active fractions showed that after the last purification step, only a single protein with an apparent mol. wt of 43 kDa remains detectable (Figure 1D). Gel filtration analysis correlates the tRNA 3′-processing activity with an enzyme of ∼64 kDa, suggesting the active enzyme to be present as a homodimer. Such potential dimer formation has to be investigated with additional experiments, e.g. two-hybrid assays.
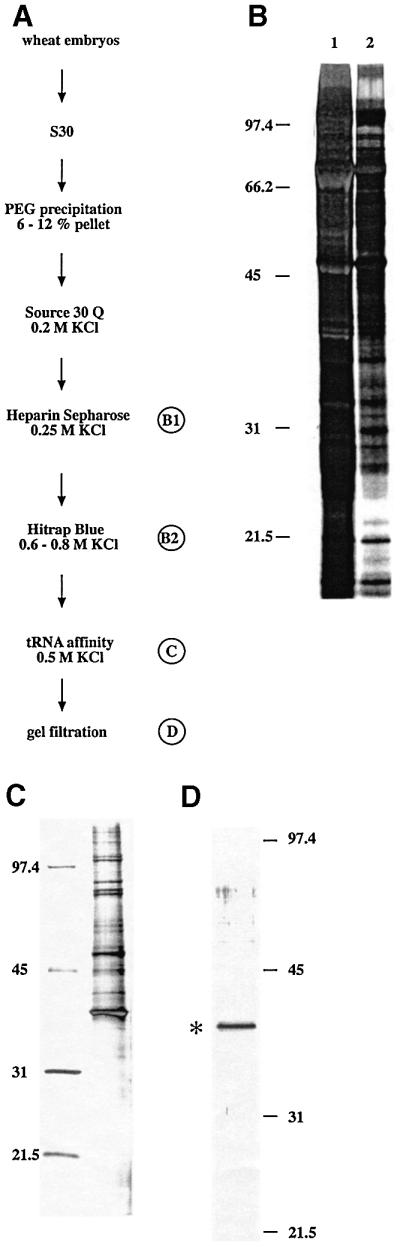
Fig. 1. Isolation of the nuclear RNase Z. (A) Purification scheme. RNase Z was purified from the soluble protein fraction from wheat germ (S30) in six fractionation steps. (B) SDS–PAGE of RNase Z active fractions from two purification steps. Aliquots of RNase Z active fractions from the two purification steps B1 and B2 were loaded onto SDS gels: lane 1, 57 µg of the 0.25 M heparin fraction; lane 2, 10 µg of the 0.6 M KCl Blue fraction. (C) Since little protein was left after the last purification step, protein concentrations could not be determined and, therefore, 10% (by volume) of the tRNA affinity fraction was loaded. On the left, a protein size standard is given in kDa. (D) SDS–PAGE of the RNase Z active fraction after the last purification step. Again, 10% (by volume) of the RNase Z active gel filtration fraction was loaded onto the gel. A protein size standard is given on the right in kDa.
The 43 kDa protein was isolated from an SDS– polyacrylamide gel and subjected to tryptic digestion and mass spectrometry (MS)/MS analysis. Subsequent database searches using the algorithm SEQUEST (Eng et al., 1994) and programs developed at Harvard Microchem Facility (Chittum et al., 1998) did not identify the corresponding gene. Therefore, peptides were separated by HPLC and sequenced using Edman degradation. Four peptide sequences were obtained and database searches revealed a wheat cDNA sequence (accession No. BE403456) and two open reading frames (ORFs) in the A.thaliana genome with high sequence similarity to these peptides. The wheat cDNA sequence translates into a protein 100% identical to one of the sequenced wheat peptides. Since the wheat sequence is only a partial cDNA sequence, the other three peptides are outside of this sequence and consequently show no match to the cDNA. The corresponding wheat genomic sequence could not be identified in the few wheat sequences available in public databases.
Characterization of homologous ORFs in A.thaliana
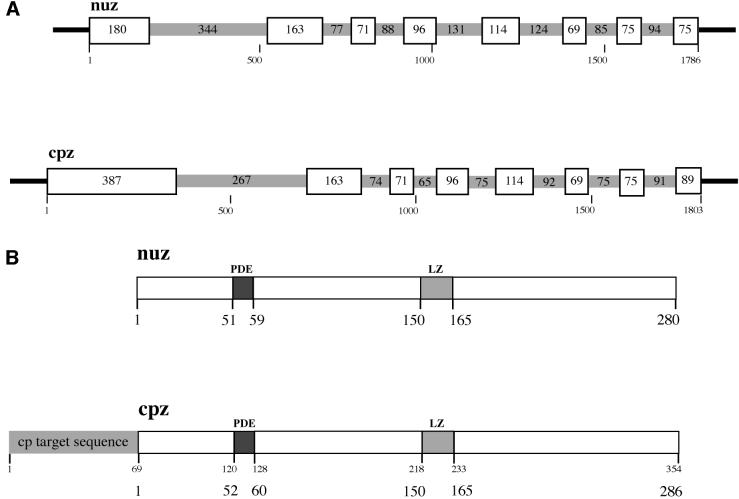
The two identified ORFs from A.thaliana differ in the length of the 5′ end encoding the N-terminus. The shorter ORF was named NUZ (for nuclear RNase Z) (accession No. for the gene, ac011765; for the protein, aag52354) and the longer ORF was termed CPZ (for chloroplast RNase Z) (accession No. for the gene, ac006951; for the protein, aad25827), since the longer ORF contains an N-terminal extension predicted to be a signal sequence potentially routing the protein into chloroplasts (see below).
The NUZ gene is located on chromosome I and covers 1786 nucleotides. The combined exons of this gene translate into protein nuz, which is 280 amino acids long. The nuz polypeptide has clear sequence similarity (88–97%) to three of the peptide sequences from the isolated wheat protein and somewhat less sequence similarity (47%) to the fourth peptide. The predicted molecular weight of nuz is 31.3 kDa, with a pI of 6.1. Two expressed sequence tag (EST) clones were identified for ORF NUZ (accession Nos ai992894 and aa067482), confirming that this ORF is indeed actively expressed in Arabidopsis. An additional cDNA clone was obtained using RT–PCR and total RNA from Arabidopsis. Sequencing of the RT–PCR clone (these sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession No. aj428988) revealed that the genomic prediction of this ORF differs only slightly from the cDNA sequence. The boundary between the second exon and the second intron as found in the cDNA is three nucleotides downstream of the predicted exon–intron boundary.
The gene CPZ is located on chromosome II with 1803 nucleotides, of similar size to the NUZ gene, while the encoded protein cpz has 354 amino acids, 74 amino acids longer than nuz. The two sorting servers TargetP (Emanuelsson et al., 2000) and Predotar (http://www. inra.fr/Internet/Produits/Predotar, version 0.5) predict that the N-terminus of cpz contains a signal sequence of ∼68 amino acids, which routes the protein to chloroplasts with a probability of 97 and 98%, respectively. This prediction is supported by the observation that the sequence similarity of cpz to nuz begins at around amino acid 68, the potential leader cleavage site predicted by TargetP (Nielsen et al., 1997). The calculated molecular weight of cpz (without the 68 N-terminal amino acids) is 32.1 kDa, with a pI of 6.8. Thus nuclear and processed chloroplast versions of RNase Z have similar molecular weights and pIs. An EST clone was identified for ORF CPZ (accession Nos: 5′ sequence, av441601; 3′ sequence, av439700), confirming that cpz is expressed. Sequence analysis of the complete cpz EST clone (Asamizu et al., 2000) (these sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession No. aj428988) revealed the structure of the gene. While only seven exons are predicted, the cDNA sequence reveals eight exons with 75 nucleotides between exons 6 and 7, and 91 nucleotides between exons 7 and 8 (Figure 2A).
Fig. 2. RNase Z gene structures from A.thaliana. (A) Gene structures for NUZ and CPZ. Exons are boxed and introns are shown in grey. The sizes of exons and introns are indicated in base pairs. (B) Protein structures of cpz and nuz. Lengths of proteins and locations of domains are given in amino acids. The predicted import sequence of cpz, shown in grey, is predicted to be cleaved off after amino acid 68. The locations of several predicted domains in cpz are given with (smaller letters in upper line) and without the signal sequence. The positions of the PDE and the potential leucine zipper (LZ) are indicated.
The NUZ and the CPZ genes contain seven introns each, located at identical positions (Figure 2A). The proteins nuz and cpz show 85% sequence similarity. Analysis of the two Arabidopsis protein sequences reveals a conserved phosphodiesterase (PDE) motif between amino acids 51 and 59 in nuz (cpz: 52 and 60) and a potential leucine zipper between amino acids 150 and 165 (cpz: 150 and 165). Overlapping with the PDE motif is a Zn2+-binding motif (Figure 4).
Fig. 4. Alignment of RNase Z protein sequences. Alignment of Arabidopsis nuz with similar proteins from representatives of the eukarya (H.sapiens and S.cerevisiae), archaea (M.janaschii) and bacteria (E.coli). The PDE domain, the zinc-binding motif (HX2HX20–120E, where H and E are marked with *; in H.sapiens and S.cerevisiae, the third conserved amino acid E is marked with *hs and *sc, respectively, since it is not at the same position as in the other proteins) and the leucine zipper (LZ) are shown. Identical amino acids are shaded in grey and similar amino acids are shaded in light grey. For all aligned proteins, only relevant portions are shown (positions where amino acids were left out are marked with a black triangle). Accession Nos: H.sapiens, ak024822; S.cerevisiae, s38156; E.coli, E85867.
In vitro expressed Arabidopsis ORF NUZ cDNA has RNase Z activity
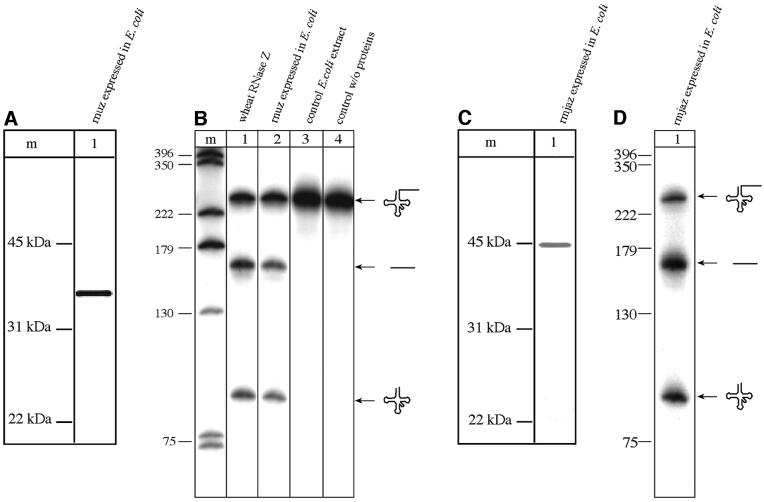
Proof for the correct identification of the NUZ gene as the RNase Z-encoding gene is the assay for catalytic activity. For this purpose, the Arabidopsis cDNA encoding nuz was cloned into the E.coli expression vector pET32a to incorporate a thioredoxin tag, a histidine tag and an S tag at the N-terminus of the resulting fusion protein. Although the majority of the recombinant nuclear RNase Z (rnuz) was found in inclusion bodies, enough protein was present in the soluble fraction to isolate the recombinant protein for activity assays. After purification of rnuz with S-protein–agarose, the N-terminal tags were removed with enterokinase. The apparent molecular weight of the Arabidopsis rnuz without the tags is ∼41 kDa (Figure 3A), very similar to the RNase Z isolated from wheat germ (Figure 1D) and somewhat larger than the calculated mol. wt of 31.4 kDa. Gel filtration analysis of the recombinant RNase Z revealed that an enzyme of ∼79 kDa correlates with the activity, suggesting that the Arabidopsis nuz, like its wheat homologue, might be active as a homodimer. Additional experiments will show whether this enzyme indeed forms a homodimer.
Fig. 3. Comparison of RNase Z enzymes from plants and archaea. (A) SDS–PAGE showing the recombinant Arabidopsis RNase Z (rnuz). Lane 1: 180 ng of Arabidopsis rnuz were loaded onto the gel. The rnuz monomer has an apparent molecular mass of 41 kDa. Lane m: protein marker sizes are given in kDa. (B) tRNA 3′-processing activity of recombinant Arabidopsis rnuz. Pre-tRNATyr was incubated in in vitro processing assays with different protein fractions. Lane 1: incubation of pre-tRNA with 1.7 µg of wheat RNase Z (0.25 M KCl heparin fraction). Lane 2: 180 ng of Arabidopsis rnuz expressed in E.coli were incubated with pre-tRNA. Lane 3: 180 ng of the control E.coli extract were incubated with pre-tRNA, to test whether the processing activities observed derive from the NUZ gene cloned into pET32a-nuz and not from any residual E.coli activities present in the S-protein–agarose fraction. Lane 4: incubation of pre-tRNA with processing buffer, but without proteins. Precursor and products are depicted schematically on the right. Lane m: DNA size marker; sizes are given in nucleotides on the left. Incubation with either the enriched wheat RNase Z or the recombinant nuz expressed in E.coli yields the same two processing products, the tRNA and the 3′ trailer. (C) SDS–PAGE showing the recombinant RNase Z from M.janaschii (rmjaz). Lane 1: the recombinant M.janaschii RNase Z (177 ng loaded) runs at an apparent mol. wt of 45 kDa. Lane m: protein marker sizes given in kDa. (D) The archaeal RNase Z homologue shows tRNA 3′-processing activity. In vitro processing experiment with rmjaz. Pre-tRNATyr was incubated with rmjaz (708 ng). Precursor and products are shown schematically on the right, DNA marker sizes are indicated in nucleotides on the left. The pre-tRNA is processed efficiently by the archaeal RNase Z, yielding the tRNA and the 3′ trailer.
In vitro processing assays with the recombinant RNase Z expressed in E.coli revealed that this protein indeed does have tRNA 3′-processing activity (Figure 3B). This observation confirms earlier experiments which had suggested that RNA subunits are not part of RNase Z (Mayer et al., 2000) and shows, furthermore, that no other protein subunits are essential for the activity. The observed catalytic activity of nuz in a bacterial expression system suggests that no eukaryotic-specific modifications are required for this enzyme.
Identification of the archaeal RNase Z
Database searches with the plant nuz sequence revealed similar proteins in several archaea. The gene for the similar protein from M.janaschii (55% similarity to nuz) (Figure 4) was cloned (MJAZ, accession No. U67591) and the protein (mjaz, accession No. E64487) was expressed in E.coli. The recombinant protein has an apparent mol. wt of 45 kDa according to SDS–PAGE (Figure 3C). To determine whether MJAZ encodes an archaeal RNase Z, recombinant mjaz protein was incubated with precursor tRNAs in in vitro processing experiments (Figure 3D). Two processing products, tRNA and 3′ trailer, are released upon incubation, showing that rmjaz indeed has endonucleolytic tRNA 3′-processing activity.
RNase Z is conserved between bacteria, archaea and eukarya
Proteins with high sequence similarity are identified not only in archaea but also in the other two kingdoms, in bacteria and eukarya (Figure 4). All related ORFs contain the Zn2+-binding motif and the PDE motif, which might be a candidate for the catalytic domain of this enzyme. The RNase Z-like proteins in E.coli and cyanobacterium Synechocystis (accession No. q55132) share 53 and 51% sequence similarity to the Arabidopsis nuclear RNase Z, respectively (Figure 4). Plant EST sequences with high similarity (72–87%) to the Arabidopsis nuz protein are identified in soybean, potato, barrel medic, maize and tomato.
In humans, a cDNA has been identified, which encodes a previously unassigned protein, ELAC1 (Figure 4) with 48% sequence similarity to nuz. The nomenclature ELAC1 was derived from the similarity to the likewise uncharacterized E.coli ORF ElaC (Tavtigian et al., 2001), which we find to be the only E.coli protein similar to RNase Z and which we accordingly renamed ecoz. In S.cerevisiae, a protein has been identified whose C-terminal half (amino acids 510–770) has 33% sequence similarity to nuz (Figure 4) and which, with 838 amino acids, is twice as long as nuz.
Discussion
The plant RNase Z
The RNase Z from wheat has been characterized previously in detail (Mayer et al., 2000; Schiffer et al., 2001). The wheat RNase Z is an endonuclease that cleaves pre-tRNAs 3′ to the discriminator. Analysis of the substrate specificity of this enzyme showed that pre-tRNA variants, which lack one or more arms of the tRNA, are processed with drastically reduced cleavage efficiency (Schiffer et al., 2001). Pre-tRNA variants missing the T arm are not cleaved at all (Schiffer et al., 2001), showing that this enzyme has a high preference for substrates with an intact tRNA structure. This report shows that the RNase Z from wheat has, according to SDS–PAGE, an apparent molecular mass of 43 kDa and, according to gel filtration, an apparant molecular mass of 64 kDa. This result is in contrast to the previous observation that the wheat RNase Z has an apparent mol. wt of 122 kDa (Mayer et al., 2000). In the latter experiments, we had included detergent in the early purification steps, which accumulated during the purification. The presence of detergent shifted the protein to a much higher apparent molecular weight in gel filtration experiments. Furthermore, in the earlier purification, we used a tRNA affinity column to which the tRNA molecules were bound via a spacer of only one C-atom, while we now used a spacer with six C-atoms, resulting in a considerable improvement in separating the protein from contaminants.
The homologous protein from A.thaliana has an apparent molecular mass of 41 kDa according to SDS–PAGE and an apparent molecular mass of ∼79 kDa in gel filtration experiments. The slight differences in size between the two plant RNase Z enzymes might be due to sequence differences. In addition, the wheat enzyme was isolated from germ cells while the Arabidopsis protein was analysed as a recombinant protein expressed in a bacterial system (E.coli). The latter may be lacking some post-translational modification(s) that modify its behaviour. Nevertheless, both proteins seem to be similar enough, since database searches with the wheat peptides readily identified the Arabidopsis homologues, and the protein sequence encoded by the partial wheat cDNA shows 83% similarity to the respective region of the Arabidopsis nuz ORF.
tRNA 3′ endonucleases of similar sizes have been enriched previously from yeast cells, where a 45/60 kDa protein is associated with the endonucleolytic activity (Papadimitriou and Gross, 1996). This protein can be found in complexes of either 90 or 450 kDa in gel filtration analyses. In contrast, nuclear tRNA 3′-processing endonucleases from Aspergillus and Xenopus have been reported to be much larger polypeptides of 160 (Han and Kang, 1997) and 97 kDa (Castaño et al., 1985), respectively. Both these latter 3′ endonucleases have been reported to be monomeric enzymes. These size variations suggest that there may be different, additional tRNA 3′-processing enzymes in eukaryotes, although we expect from the general conservation of the here identified RNase Z through all kingdoms that Aspergillus and Xenopus will also contain proteins similar to the here defined RNase Z group.
Chloroplast and nuclear enzyme are encoded by different genes
Since database searches revealed two different genes in Arabidopsis coding for a nuclear and a predicted chloroplast RNase Z, these two compartments seem to be served by two separate nuclear genes for their tRNA 3′ endonucleases. The high similarity (85%) between the proteins cpz and nuz suggests a common evolutionary origin. The identical positions of all of the introns in both genes strongly support the occurence of a duplication event resulting in these two genes.
The identified similarities to both archaeal and bacterial RNase Z-like proteins do not allow a clear decision on the evolutionary origin of the ancestor of the two plant genes. The rather low sequence identity between chloroplast RNase Z and the Synechocystis ORF (26%) suggests, however, that the RNase Z genes in plants are not derived directly from a common ancestor with cyanobacteria and supports the scenario of the cpz gene being derived from the nuclear RNase Z gene by duplication.
The plant mitochondrial and nuclear enzymes have been reported to be of similar sizes, of ∼43 kDa (Kunzmann et al., 1998), while analysis of the substrate specificities of the two enzymes revealed considerable differences (Marchfelder and Brennicke, 1994; Kunzmann et al., 1998; Schiffer et al., 2001). In this context, it will be interesting to investigate whether the CPZ gene also encodes the mitochondrial RNase Z (mtz). A number of proteins, notably several involved in tRNA recognition and charging, have been found previously to be encoded by single nuclear genes, and to be routed to both organelles, chloroplast and mitochondrion (Small et al., 1998). This indicates that the signal sequences of these proteins are recognized by both organellar import systems. Experiments with green fluorescent protein (GFP) fusion proteins will show where the cpz is routed to in the cell.
Predicted domain structure of the nuclear RNase Z
Analysis of the two Arabidopsis protein sequences reveals a conserved PDE motif and a potential leucine zipper (Figure 2). The PDE motif was first described in cAMP PDE enzymes, which have been grouped into two subclasses (class I and class II) based on sequence similarities (Francis et al., 2001). The PDE motif found in nuz is 78% identical to the PDE domain in the class II cAMP PDE from Vibrio fischeri (Dunlap and Callahan, 1993). In evolutionary terms, this could indicate that tRNA 3′ endonucleases and cAMP PDEs are derived from a common ancestor and that the substrate-binding domains have evolved differently. Another option is that the PDE domain was acquired through exon shuffling and proved to be more efficient than other catalytic domains acting on phosphodiester bonds (such as, for example, the catalytic domain in RNase T2). Comparing the primary structure of nuz with that of Zn2+-dependent enzymes, a motif closely resembling the Zn2+-binding motif is detectable, overlapping with the PDE domain (Figure 4), suggesting that a metal ion is an important cofactor in the catalytic reaction. Zn2+-metalloproteases as well as class I cAMP PDEs contain the Zn2+-binding motif: HX3HX20–120E (Francis et al., 1994), the corresponding motif in nuz is HX2HX29E or HX2HX32E (Figure 4). Of the three histidine residues in the potential catalytic domain of nuz, one could act as a base, the second as an acid and the third histidine could be used for positioning the substrate in the catalytic centre, analogously to the catalytic reaction of RNase T2 (Irie, 1997). In addition, COGnitor (http://www.ncbi.nlm.nih. gov/COG/xognitor.html) places nuz into the protein cluster COG 1234, which is a group of metal-dependent hydrolases.
The RNase Z homologue in the archaeal kingdom
tRNA 3′ processing in the archaeon Haloferax volcanii is generated by an endonuclease cleaving the pre-tRNA 3′ to the discriminator (Schierling et al., 2002), similar to the respective processing reaction in eukaryotes. A protein with high sequence similarity to the plant nuclear RNase Z is identified in several members of the archaeal kingdom, e.g. Halobacterium salinarum, Methanobacterium thermoautotrophicum, Pyrococcus horikoshii and Sulfolobus solfataricus. The here analysed recombinant protein rmjaz from M.janaschii processes pre-tRNAs at the same site as the H.volcanii RNase Z and the plant RNase Z (see Supplementary data available at The EMBO Journal Online). This experimental result shows functionally that the archaeal protein has the same enzymatic activity and is a true homologue of the eukaryotic plant RNase Z.
Similar proteins in the bacterial kingdom
The discovery of a similar protein (ecoz) in E.coli is surprising since the pathway and the mechanistics of the tRNA maturation processes seem to differ between E.coli and eukarya/archaea. Nuclear and archaeal RNase Z enzymes are endonucleases cleaving the precursor close to or at the discriminator (Mörl and Marchfelder, 2001). In contrast, in E.coli, the final maturation steps at the tRNA 3′ end are performed by exonucleases (Deutscher, 1995). An E.coli strain carrying a null mutation of the RNase Z-like ECOZ gene is not viable, showing that this gene is essential (A.Jellen-Ritter and A.Marchfelder, unpublished data). It will be interesting to analyse the in vivo function of the E.coli RNase Z using mutant strains.
RNase Z proteins in eukarya
In the eukaryotic kingdom, database searches reveal similar proteins in several plant species. The proteins identified in human and yeast have 48 and 33% less sequence similarity to nuz, but they might still have the same function as nuz, namely tRNA 3′ processing. Experiments with yeast mutant strains will show whether the yeast sequence indeed encodes a tRNA 3′-processing endonuclease.
Assigning a function to a conserved group of proteins
The proteins with sequence similarity to the plant RNase Z belong to two overlapping conserved groups of proteins. Mjaz and the E.coli protein ecoz are members of the protein cluster COG 1234, and COGnitor (http://www.ncbi.nlm.nih.gov/COG/xognitor.html) places nuz into the same protein cluster. The COG database contains information of 42 prokaryotic and four eukaryotic genomes, of which 39 prokaryotic and four eukaryotic proteins are grouped into the protein cluster COG 1234. This cluster includes a protein with similarity to RNase Z from almost all of the analysed prokaryotic genomes. So far, no member of COG 1234 had been assigned a function, the only general prediction for these enzymes having been that they are metal-dependent hydrolases of the β-lactamase superfamily III. Now we can tentatively assign a function to two members of this family, although our in vitro data will have to be complemented by in vivo analyses.
In the second classification, the RNase Z-like proteins had been grouped into the so-called ELAC1/2 family of proteins (Tavtigian et al., 2001). The ELAC1 group of proteins consists of polypeptides of 300–400 amino acids, while the ELAC2 group comprises larger proteins with 800–900 amino acids. Whereas ELAC2 proteins are only found in the eukaryotic domain, representatives of ELAC1 are present in eukarya, bacteria and archaea. The C-terminal region of ELAC2 proteins aligns with ELAC1 proteins, both containing the PDE domain. The N-terminal region of ELAC2 covers a Ψ-histidine motif (a motif similar to the PDE domain but without the conserved histidines), and it has been suggested that ELAC2 proteins have been derived from a duplication event of ELAC1 with subsequent degeneration of its N-terminal region (Tavtigian et al., 2001).
Mutations in the human ELAC2 gene have been associated with the occurrence of prostate cancer, but the precise function of the encoded protein has not been clarified yet. The in vivo functional importance of these genes is emphasized by the observation that the yeast ELAC2 protein YKR079C is essential, since yeast cells with the respective gene disruption are not viable (Tavtigian et al., 2001). Some of the ELAC1/2 members have been annotated as sulfatase homologues because of sequence similarities (Tavtigian et al., 2001). However, to date, the function of the ELAC1/2 proteins was not clear. The two RNase Z enzymes identified here are the first members of the ELAC1/2 family to which a function can be assigned, and may now provide clues for the search for functions for the other members.
Outside of the ELAC1/2 family, the RNase Z proteins show similarity to the PSO2 (also named SNM1) (Haase et al., 1989; Niegemann and Brendel, 1994) proteins and to the group of 3′ mRNA cleavage and polyadenylation specificity factors (CPSFs) (Jenny et al., 1994, 1996; Chanfreau et al., 1996). CPSF and ELAC proteins originally had been grouped into one COG cluster (Tatusov et al., 1997) and the alignment of the human cpsf73 and nuz reveals that both proteins are 22% identical to each other. The primary structure similarity is reflected in the functional similarity between the here identified RNase Z enzymes of the ELAC1 group and the CPSF proteins. This suggests an intriguing possibility for the evolution of these enzymes from common ancestors, possibly with a monomeric structure as retained in the RNase Z family.
Materials and methods
Purification of the nuclear RNase Z from wheat
The wheat nuclear tRNA 3′-processing activity was isolated from wheat germ as described (Mayer et al., 2000), with the following modifications. All buffers were prepared without detergent. The tRNA affinity column was made using wheat tRNA (Sigma), which was coupled to HiTrap NHS-activated Sepharose (Amersham Biosciences). The column was equilibrated with buffer A [40 mM Tris–HCl pH 8, 5 mM MgCl2, 5% glycerol, 2 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] and loaded with the RNase Z active fraction (0.6–0.8 M KCl) from the preceding Blue column. Proteins were eluted with a step gradient (0.2, 0.5, 1.0 and 2.0 M KCl), and RNase Z activity was recovered in the 0.5 M KCl fraction. RNase Z purified with this tRNA affinity column showed an apparent molecular mass of 64 kDa on the subsequent gel filtration column. After the gel filtration column, RNase Z active fractions were loaded onto an SDS–polyacrylamide gel, where only a 43 kDa protein correlated with the activity. Approximately 0.5 µg of the 43 kDa protein was excised from the gel and submitted to the MicroChemFacilty at Harvard University for protein sequence analysis.
Gel filtration analysis
To determine the molecular mass of nuz from wheat and rnuz from Arabidopsis, gel filtration was performed as described earlier (Mayer et al., 2000).
Isolation of the Arabidopsis cDNA for NUZ
Total RNA was isolated from leaves (2 weeks old) from A.thaliana var. Columbia, using the RNeasy Plant Mini Kit (Qiagen) and reverse transcribed into cDNA with oligo(dT) primers (primer sequences are available upon request). The resulting cDNA was amplified with PCR using primers AT-Z-2-5 and AT-Z-2-3. The PCR product was digested with NcoI and XhoI and the fragment was cloned into pET32a (digested with NcoI and XhoI), yielding pET32a-nuz.
Isolation of the gene for mjaz
Chromosomal DNA from M.janaschii was isolated using the sarcosyl method (Hofman et al., 1979). The gene for mjaz was amplified with primers Ja3 and Ja4 and cloned into pET32a, yielding pET32a-mjaz.
Expression of nuz in E.coli
PET32a-nuz was transformed into strain AD494(DE3)pLys (Novagen) to express nuz, the recombinant protein was isolated using S-protein– agarose (Novagen), and N-terminal tags were removed using recombinant enterokinase (Novagen). All procedures were performed according to the manufacturer’s instructions. As a control, the expression vector pET32a (without insert) was transformed likewise into the strain AD494(DE3)pLys and proteins were isolated and purified as described above. This control purification was performed to show that the RNase Z activity was not due to any E.coli proteins.
Expression of mjaz using the RTS system
Protein synthesis was performed in vitro using the RTS 500 instrument and the RTS 500 E.coli circular template kit (Roche) following the manufacturer’s instructions. PET32a-mjaz was used as DNA template for the in vitro translation. Proteins were purified as described for expression in E.coli (above), yielding the recombinant protein rmjaz.
Preparation of RNA substrates
Precursor tRNATyr from Oenothera berteriana mitochondria was transcribed from template pTyrII as described (Kunzmann et al., 1998).
In vitro processing assays
In vitro processing assays were performed as described (Mayer et al., 2000), with the following modifications. For assays with the recombinant RNase Z from Arabidopsis, 180 ng of the protein was used, and with the recombinant RNase Z from M.janaschii, 708 ng of the protein were used. A 180 ng aliquot of protein was employed in the control reaction, using proteins isolated from the control purification as detailed above. A 1.7 µg aliquot of the purified wheat enzyme (RNase Z active fraction from the heparin column) was used as positive control, and all reactions were incubated for 15 min at 37°C.
Database analyses
Sequence comparisons were carried out using the BLAST program at NCBI (Altschul et al., 1990) or the FASTA program from the GCG package (Pearson and Lipman, 1988). Analyses of signal sequences were done with Predotar (http://www.inra.fr/Internet/ Produits/Predotar, version 0.5) and TargetP v1.01 (Nielsen et al., 1997).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We appreciate the expert protein sequence analysis done at the Harvard MicroChem Facility by Bill Lane and colleagues. We thank Elli Bruckbauer and Bärbel Weber for expert technical assistance, Karl O.Stetter and Harald Huber for M.janaschii cells, and Axel Brennicke and Stefan Binder for helpful discussion and critical reading of the manuscript. Work presented here was funded by the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie and the VolkswagenStiftung.
References
- Altman S., Kirsebom,L. and Talbot,S. (1995) Recent studies of RNase P. In Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. American Society for Microbiology, Washington, DC, pp. 67–78.
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Asamizu E., Nakamura,Y., Sato,S. and Tabata,S. (2000) A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res., 7, 175–180. [DOI] [PubMed] [Google Scholar]
- Castaño J.G., Tobian,J.A. and Zasloff,M. (1985) Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNA-Met(i) (3′ pre-tRNase). J. Biol. Chem., 260, 9002–9008. [PubMed] [Google Scholar]
- Chanfreau G., Noble,S.M. and Guthrie,C. (1996) Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF). Science, 274, 1511–1514. [DOI] [PubMed] [Google Scholar]
- Chen J.Y. and Martin,N.C. (1988) Biosynthesis of tRNA in yeast mitochondria. An endonuclease is responsible for the 3′-processing of tRNA precursors. J. Biol. Chem., 263, 13677–13682. [PubMed] [Google Scholar]
- Chittum H.S., Lane,W.S., Carlson,B.A., Roller,P.P., Lung,F.D., Lee,B.J. and Hatfield,D.L. (1998) Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry, 37, 10866–10870. [DOI] [PubMed] [Google Scholar]
- Deutscher M.P. (1995) tRNA processing nucleases. In Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. American Society for Microbiology, Washington, DC, pp. 51–65.
- Dunlap P.V. and Callahan,S.M. (1993) Characterization of a periplasmic 3′:5′-cyclic nucleotide phosphodiesterase gene, cpdP, from the marine symbiotic bacterium Vibrio fischeri. J. Bacteriol., 175, 4615–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen,H., Brunak,S. and von Heijne,G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol., 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Eng J.K., McCormick,A. and Yates,J.R.III. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom., 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Engelke D.R., Gegenheimer,P. and Abelson,J. (1985) Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J. Biol. Chem., 260, 1271–1279. [PubMed] [Google Scholar]
- Francis S.H., Colbran,J.L., McAllister-Lucas,L.M. and Corbin,J.D. (1994) Zinc interactions and conserved motifs of the cGMP-binding cGMP-specific phosphodiesterase suggest that it is a zinc hydrolase. J. Biol. Chem., 269, 22477–22480. [PubMed] [Google Scholar]
- Francis S.H., Turko,I.V. and Corbin,J.D. (2001) Cyclic nucleotide phosphodiesterases: relating structure and function. Prog. Nucleic Acid Res. Mol. Biol., 65, 1–52. [DOI] [PubMed] [Google Scholar]
- Frank D.N. and Pace,N.R. (1998) Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem., 67, 153–180. [DOI] [PubMed] [Google Scholar]
- Franklin S.E., Zwick,M.G. and Johnson,J.D. (1995) Characterization and partial purification of two pre-tRNA 5′-processing activities from Daucus carrota (carrot) suspension cells. Plant J., 7, 553–563. [DOI] [PubMed] [Google Scholar]
- Garber R.L. and Altman,S. (1979) In vitro processing of B.mori transfer RNA precursor molecules. Cell, 17, 389–397. [DOI] [PubMed] [Google Scholar]
- Garber R.L. and Gage,L.P. (1979) Transcription of a cloned Bombyx mori tRNA2Ala gene: nucleotide sequence of the tRNA precursor and its processing in vitro. Cell, 18, 817–828. [DOI] [PubMed] [Google Scholar]
- Gegenheimer P. (1995) Structure, mechanism and evolution of chloroplast transfer RNA processing systems. Mol. Biol. Rep., 22, 147–150. [DOI] [PubMed] [Google Scholar]
- Haase E., Riehl,D., Mack,M. and Brendel,M. (1989) Molecular cloning of SNM1, a yeast gene responsible for a specific step in the repair of cross-linked DNA. Mol. Gen. Genet., 218, 64–71. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Larson,D., Hall,G.I. and Sprague,K.U. (1979) The primary transcription product of a silkworm alanine tRNA gene: identification of in vitro sites of initiation, termination and processing. Cell, 18, 1217–1229. [DOI] [PubMed] [Google Scholar]
- Han S.J. and Kang,H.S. (1997) Purification and characterization of the precursor tRNA 3′-end processing nuclease from Aspergillus nidulans. Biochem. Biophys. Res. Commun., 233, 354–358. [DOI] [PubMed] [Google Scholar]
- Hanic-Joyce P.J. and Gray,M.W. (1990) Processing of transfer RNA precursors in a wheat mitochondrial extract. J. Biol. Chem., 265, 13782–13791. [PubMed] [Google Scholar]
- Hofman J.D., Lau,R.H. and Doolittle,W.F. (1979) The number, physical organization and transcription of ribosomal RNA cistrons in an archaebacterium: Halobacterium halobium. Nucleic Acids Res., 7, 1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie M. (1997) RNase T1/RNase T2 family RNases. In D’Alessio,G. and Riordan,J.F. (eds), Ribonucleases: Structures and Functions. Academic Press, New York, NY, pp. 101–130.
- Jenny A., Hauri,H.P. and Keller,W. (1994) Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol. Cell. Biol., 14, 8183–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Minvielle-Sebastia,L., Preker,P.J. and Keller,W. (1996) Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science, 274, 1514–1517. [DOI] [PubMed] [Google Scholar]
- Kleman-Leyer K., Armbruster,D.W. and Daniels,C.J. (1997) Properties of H.volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell, 89, 839–847. [DOI] [PubMed] [Google Scholar]
- Kunzmann A., Brennicke,A. and Marchfelder,A. (1998) 5′ end maturation and RNA editing have to precede tRNA 3′ processing in plant mitochondria. Proc. Natl Acad. Sci. USA, 95, 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. and Deutscher,M.P. (1994) The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J. Biol. Chem., 269, 6064–6071. [PubMed] [Google Scholar]
- Manam S. and Van Tuyle,G.C. (1987) Separation and characterization of 5′- and 3′-tRNA processing nucleases from rat liver mitochondria. J. Biol. Chem., 262, 10272–10279. [PubMed] [Google Scholar]
- Marchfelder A. and Brennicke,A. (1994) Characterization and partial purification of tRNA processing activities from potato mitochondria. Plant Physiol., 105, 1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M., Schiffer,S. and Marchfelder,A. (2000) tRNA 3′ processing in plants: nuclear and mitochondrial activities differ. Biochemistry, 39, 2096–2105. [DOI] [PubMed] [Google Scholar]
- Mörl M. and Marchfelder,A. (2001) The final cut. The importance of tRNA 3′-processing. EMBO rep., 2, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M. (1997) Distribution of both lengths and 5′ terminal nucleotides of mammalian pre-tRNA 3′ trailers reflects properties of 3′ processing endoribonuclease. Nucleic Acids Res., 25, 1148–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niegemann E. and Brendel,M. (1994) A single amino acid change in SNM1-encoded protein leads to thermoconditional deficiency for DNA cross-link repair in Saccharomyces cerevisiae. Mutat. Res., 315, 275–279. [DOI] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Oommen A., Li,X.Q. and Gegenheimer,P. (1992) Cleavage specificity of chloroplast and nuclear tRNA 3′-processing nucleases. Mol. Cell. Biol., 12, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J.R., Nieuwlandt,D.T. and Daniels,C.J. (1994) Expression of a yeast intron-containing tRNA in the archaeon Haloferax volcanii. J. Bacteriol., 176, 3820–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou A. and Gross,H.J. (1996) Pre-tRNA 3′-processing in Saccharomyces cerevisiae. Purification and characterization of exo- and endoribonucleases. Eur. J. Biochem., 242, 747–759. [DOI] [PubMed] [Google Scholar]
- Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven N.B. and Deutscher,M.P. (1993) Multiple exoribonucleases are required for the 3′ processing of Escherichia coli tRNA precursors in vivo. FASEB J., 7, 143–148. [DOI] [PubMed] [Google Scholar]
- Schierling K., Rösch,S., Rupprecht,R., Schiffer,S. and Marchfelder,A. (2002) tRNA 3′ end maturation in archaea has eukaryotic features: the RNase Z from Haloferax volcanii. J. Mol. Biol., 316, 895–902. [DOI] [PubMed] [Google Scholar]
- Schiffer S., Helm,M., Théobald-Dietrich,A., Giegé,R. and Marchfelder,A. (2001) The plant tRNA 3′ processing enzyme has a broad substrate spectrum. Biochemistry, 40, 8264–8272. [DOI] [PubMed] [Google Scholar]
- Small I., Wintz,H., Akashi,K. and Mireau,H. (1998) Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol. Biol., 38, 265–277. [PubMed] [Google Scholar]
- Söll D. (1993) Transfer RNA: an RNA for all seasons. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 157–183.
- Stange N. and Beier,H. (1987) A cell-free plant extract for accurate pre-tRNA processing, splicing and modification. EMBO J., 6, 2811–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. [DOI] [PubMed] [Google Scholar]
- Tavtigian S.V. et al. (2001) A candidate prostate cancer susceptibility gene at chromosome 17p. Nature Genet., 27, 172–180. [DOI] [PubMed] [Google Scholar]
- Yoo C.J. and Wolin,S.L. (1997) The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell, 89, 393–402. [DOI] [PubMed] [Google Scholar]
- Zimmerly S., Drainas,D., Sylvers,L.A. and Söll,D. (1993) Identification of a 100-kDa protein associated with nuclear ribonuclease P activity in Schizosaccharomyces pombe. Eur. J. Biochem., 217, 501–507. [DOI] [PubMed] [Google Scholar]