Abstract
The crystal structure of the motor domain of Dictyostelium discoideum myosin-IE, a monomeric unconventional myosin, was determined. The crystallographic asymmetric unit contains four independently resolved molecules, highlighting regions that undergo large conformational changes. Differences are particularly pronounced in the actin binding region and the converter domain. The changes in position of the converter domain reflect movements both parallel to and perpendicular to the actin axis. The orientation of the converter domain is ∼30° further up than in other myosin structures, indicating that MyoE can produce a larger power stroke by rotating its lever arm through a larger angle. The role of extended loops near the actin-binding site is discussed in the context of cellular localization. The core regions of the motor domain are similar, and the structure reveals how that core is stabilized in the absence of an N-terminal SH3-like domain.
Keywords: crystal structure/Dictyostelium discoideum/motor protein/myosin-I/unconventional myosin
Introduction
Myosins are molecular motors that use the energy of ATP hydrolysis to power movement along actin filaments. They power not only muscle contraction, but also function in cell division, cellular locomotion and vesicle transport. Since the first myosin was identified from skeletal muscle tissue, a large number of related myosins and myosin isoforms have been discovered in eukaryotic cells. To date, 17 classes have been identified (Hodge and Cope, 2000), with the myosin-II subfamily forming the largest class. Myosin-IIs have historically been referred to as ‘conventional’ myosins, with all other classes referred to as ‘unconventional’ myosins. Among these, the monomeric class-I myosins, which were the first class of unconventional myosins to be identified (Pollard and Korn, 1973), have the most members, followed by class-XI myosins, which are only found in plants, and class-V myosins. Based on the amino acid sequences of the motor domain, the class-I myosins have been grouped into four subclasses (Berg et al., 2001).
The role of the myosin-Is in cellular motility has been explored using Dictyostelium as a model system (reviewed in Uyeda and Titus, 1997). This simple organism expresses at least seven different myosin-Is: MyoA–F and MyoK. The myoB, myoC and myoD genes (Jung et al., 1989, 1993; Peterson et al., 1995) have heavy chains of ∼130 kDa. In contrast, the myoA, myoE and myoF genes (Titus et al., 1989, 1995; Urrutia et al., 1993) have shorter heavy chains of ∼115 kDa. The heavy chain of each of the latter myosin-Is can be divided into three distinct domains. The N-terminal region consists of a highly conserved motor domain and is followed by an α-helical light chain binding domain and a C-terminal region, rich in basic residues, that binds directly via electrostatic interactions to membranes in vitro (Adams and Pollard, 1989; Neuhaus and Soldati, 2000). The cellular function of MyoA–D and MyoK has been deduced primarily from double or triple knockout mutants, because the deletion of a single myosin-I gene results in subtle phenotypes. Mutant strains show defects in pseudopod extensions, fluid phase endocytosis, streaming and terminal development. Furthermore, they oversecrete lysosomal enzymes and cannot maintain cortical tension (Novak et al., 1995; Jung et al., 1996; Temesvari et al., 1996; Schwarz et al., 2000). The cellular role of MyoE is still under investigation.
In contrast to class-II myosins, which are regulated by phosphorylation of the myosin light chain or by binding of Ca2+, some of the myosin-I ATPases (the amoeboid, yeast and Aspergillus class-I myosins) as well as the ATPases of the class-VI myosins are activated by phosphorylation of the heavy chain at a single site in the cardiomyopathy (CM) loop. The site of phosphorylation, which is in the upper 50 kDa domain and thought to be part of the actomyosin interface, is called the TEDS site (Bement and Mooseker, 1995). Most myosins contain either a glutamate or an aspartate at this position, resulting in a constitutively activated form. In contrast, some of the myosin-Is contain a serine or a threonine at this position, which can be phosphorylated by specific myosin heavy chain kinases in order to regulate activity (Wang et al., 1998).
High-resolution crystal structures exist for a number of class-II myosins, which have been crystallized in a variety of nucleotide states (Rayment et al., 1993; Dominguez et al., 1998; Houdusse et al., 1999). All of the crystal structures assume one of three, presumably weak actin binding, conformations: (i) near rigor, in which the nucleotide binding pocket is open and the lever arm is down; (ii) transition, in which the nucleotide pocket is closed and the lever arm is up; or (iii) detached, which is thought to be non-actin binding and in which the lever arm is very far down. In order to understand the structural basis for the differences observed among myosin classes, we have solved the crystal structure of a recombinant motor domain of Dictyostelium myosin-IE. MyoE was crystallized with MgADP·VO4 in the active site, resulting in a closed structure. The overall fold is quite similar to the smooth muscle myosin and Dictyostelium myosin-II motors, except that it lacks the N-terminal SH3-like domain. A docking to the F-actin model (Holmes et al., 1990; Lorenz et al., 1995) shows that the lever arm of MyoE moves ∼30° further up than that of myosin-II, which could result in a larger power stroke. The four crystallographically independent molecules show different movements of the converter domain and surrounding structures about the actin long axis, demonstrating that this region of myosin has additional degrees of freedom beyond those described previously for myosin-IIs. The actin-binding site of MyoE is considerably different from that of myosin-II, with all loops involved in actin binding being different in length and conformation.
Results
Kinetic characterization
The head fragment of MyoE was cloned from genomic DNA into a Dictyostelium expression vector containing a C-terminal His8 tag. The protein was expressed in Dictyostelium cells and purified using ATP-dependent actin binding, Ni affinity and ion-exchange chromatography. In order to analyze the catalytic competence of our construct, we performed transient kinetic and steady-state experiments on the MyoE motor domain. The results summarized in Table I show that binding to nucleotide and actin is similar for MyoE, chicken smooth muscle myosin-II (GgSmS1) and rabbit skeletal muscle myosin-II (OcSkS1). In the presence of actin, the ADP affinity of MyoE is ∼100-fold greater than that of DdMyoII and OcSkS1. As indicated by the ratio of the dissociation equilibrium constants KAD/KD (1.5), coupling between nucleotide binding and actin binding is weak. In this respect, MyoE resembles GgSmS1, for which a KAD/KD ratio of 4.2 has been determined (Cremo and Geeves, 1998). In the presence of saturating amounts of actin, the steady-state ATPase activity of MyoE is activated from a basal level of 0.04/s to 1.3/s (kcat). Additionally, we demonstrated the motile competence of MyoE by generating a construct in which the motor domain was fused to an artificial lever arm consisting of two α-actinin repeats (Anson et al., 1996; Kliche et al., 2001) and showed that it was able to move actin filaments in vitro. The specific kinetic and mechanical properties of this construct, in its phosphorylated and unphosphorylated form, will be described in detail elsewhere (U.Dürrwang, S.Fujita-Becker and D.J.Manstein, manuscript in preparation).
Table I. Transient kinetic analysis.
| Rate constant | MyoE | DdMyoII | GgSmS1a | OcSkS1a | |
|---|---|---|---|---|---|
| Nucleotide binding to myosin | |||||
| ATP | K1k2 (per µM/s) | 1.0 | 0.6 | 2.1 | 2.3 |
| ADP | KD (µM) | 7.1 | 14 | 1.2 | 2.0 |
| Nucleotide binding to actomyosin | |||||
| ATP | K1k+2 (per µM/s) | 0.40 | 0.16 | 0.47 | 2.1 |
| ADP | KAD (µM) | 11 | 253 | 5 | 200 |
| KAD/KD | 1.5 | 18 | 4.2 | 100 | |
| Actin binding | k+A (per µM/s) | 2.4 | 0.8 | 1.24 | 6.7 |
| k–A (per s) | 0.006 | 0.002 | 0.004 | 0.187 | |
| KA (nM) | 2.5 | 2.5 | 3.5 | 28.2 |
KA, KAD and KD are defined as dissociation equilibrium constants.
aThe data for nucleotide-binding of the chicken smooth muscle (GgSmS1) and the rabbit skeletal muscle myosins were taken from Cremo and Geeves (1998). Values for actin binding were taken from Van Dijk et al. (1999b).
Overall structure
Purified MyoE was crystallized in complex with MgADP·VO4, and a data set was collected to 3.0 Å (Table II). The structure of the MyoE motor domain was solved by molecular replacement using a polyalanine model of amino acids 80–759 of the closed myosin-II structure in complex with MgADP·BeF3 (F.J.Kull and K.C.Holmes, manuscript in preparation). The search process revealed four molecules in the asymmetric unit. Because some subdomains of all four molecules were in the wrong position, they were deleted and the remaining model was refined using strict non-crystallographic symmetry. After several rounds of model building and refinement, these subdomains were re-included and the structure was refined further, excluding regions that were not visible or differed significantly among the four molecules.
Table II. Data collection, refinement and model quality statistics.
| Data collection | |
| Temperature of data collection (K) | 100 |
| Crystal space group | P21 |
| Unit cell parameters | a = 51.82 Å, b = 143.67 Å, c = 236.05 Å; β = 94.86° |
| Resolution range (Å)a | 50.0–3.0 (3.2–3.0) |
| Data completeness (%) | 79.7 (57.4) |
| Unique reflections | 54 968 |
| Data redundancy | 3.8 (2.0) |
| Rsym (%)b | 19.5 (28.0) |
| I/σ | 7.02 (2.87) |
| Model refinement statistics | |
| Resolution range (Å) | 50.0–3.0 |
| Reflections (work set/test set) | 49 376/5579 |
| Number of protein/ligandc atoms | 21 069/132 |
| Sigma cutoff | None |
| Model quality | |
| Rwork (%)d | 22.8 |
| Rfree (%)e | 27.3 |
| Average B factor (Å2) | 57.4 |
| Ramachandran outliers | None |
| R.m.s. bond length (Å) | 0.008 |
| R.m.s. bond angles (°) | 1.376 |
| R.m.s. dihedrals (°) | 21.220 |
| R.m.s. improper (°) | 0.863 |
| Cross-validated σ coordinate error (Å) | 0.57 |
aValues in parentheses correspond to the highest resolution shell.
bRsym = ΣhΣi|I(h) – Ii(h)|/ΣhΣiIi(h), where Ii(h) and I(h) are the ith and mean measurements of the intensity of reflection, h.
cThe ligand atoms include four molecules of ADP, four Mg2+ ions and a vanadate group.
dRwork = Σh|Fo – Fc|/ΣhFo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection, h.
eRfree is the same as Rwork, but calculated on the ∼9% of the data excluded from refinement.
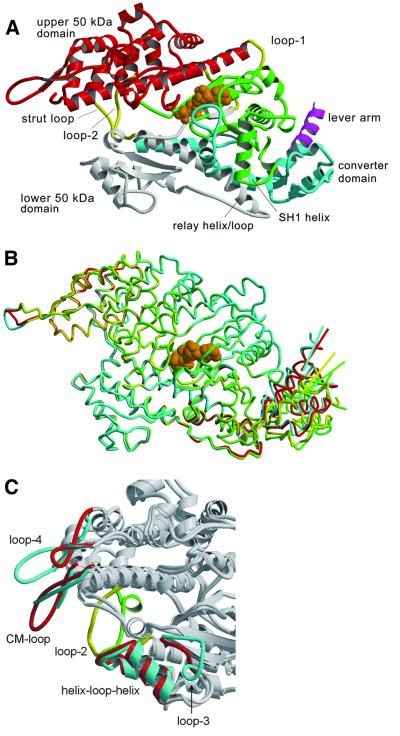
The final MyoE model consists of four molecules in complex with MgADP·VO4, containing a total of 2641 residues and 64 water molecules. The four molecules in the asymmetric unit are grouped around a solvent pore, which extends through the crystal. The overall fold of the structure is similar to that of the conventional myosins (Figure 1A). Superposition of the four crystallographically independent molecules shows that the positions of the converter domain, SH1 helix and the relay-loop/helix differ, while the core structures are very similar, with a root mean squared deviation (r.m.s.d.) of 1.2 Å for 574 common α-carbon atoms (Figure 1B). The converter domains and adjacent relay region in the four molecules vary substantially more, due to a rigid body rotation of ∼10° directed both parallel to and perpendicular to the long axis of the actin filament (Figure 1B).
Fig. 1. Structure of the motor momain of the class-I myosin MyoE from D.discoideum. (A) Ribbon diagram of MyoE, colored according to structural domains. The N-terminus (green) lacks the SH3-like domain present in conventional myosin, but otherwise has a similar fold. It contains part of the nucleotide-binding site and leads into the upper 50 kDa domain (red). The lower 50 kDa domain (white) contains the main actin-binding motifs, switch-2 and the relay region. The C-terminal domain (cyan) consists of the converter domain, the beginning of the lever arm helix (magenta), and a long α-helix connecting the converter to loop-2 in the actin-binding site. The transition state analog MgADP·VO4 is shown in orange, and loop-1, loop-2 and the strut loop are shown in yellow. (B) Superposition of the four molecules in the asymmetric unit shows good overlay in the core regions. The relay helices, adjacent relay loops and converter domains are in different positions. Colored lines indicate the trajectory of the lever arm helix as it leaves the converter domain for each molecule. (C) Superposition of MyoE and the Dictyostelium myosin II structures shows the differences in the actin-binding loops (MyoE, cyan and yellow; DdMyoII, red and green). All figures were produced using Molscript (Kraulis, 1991) and Raster3d (Merritt and Bacon, 1997).
Comparison of MyoE to the myosin II structures
The MyoE structure is very similar to that of the Dictyostelium motor domain in complex with MgADP· VO4 (Smith and Rayment, 1996), MgADP·AlF4 (Fisher et al., 1995) or MgADP·BeF3 (F.J.Kull and K.C.Holmes, manuscript in preparation), and to the chicken smooth muscle motor domain/motor domain essential light chain complex with bound MgADP·AlF4 or MgADP·BeF3 (Dominguez et al., 1998). MyoE shares ∼49% sequence identity with DdMyoII. Because the C-terminal regions of the Dictyostelium structures in complex with MgADP· AlF4 or MgADP·VO4 are considerably disordered, our comparison will focus on the closed DdMyoII·MgADP· BeF3 and the SmMD·MgADP·AlF4 structures. These two structures are very similar, with an r.m.s.d. of 1.2 Å for α-carbon atoms in 659 residues.
N-terminus
Myosin-Is lack the N-terminal SH3-like subdomain found in myosin-II. A functional myosin-II construct lacking the N-terminal 80 amino acids can be expressed in Dictyostelium, but displays greatly reduced stability and motile activity. In contrast, a mutant lacking only the SH3-like domain, but containing the first 15 amino acids shows wild-type-like properties (S.Fujita-Becker, D.J.Manstein and K.Sutoh, manuscript in preparation). Although the sequence of the N-terminus is not conserved among myosin-IIs, the SH3-like domain is held in place by contacts between aromatic residues in the N-terminus and hydrophobic pockets on the core domain. In order to stabilize this region in the absence of the SH3-like domain, MyoE has polar or charged amino acids at positions equivalent to the hydrophobic pockets.
Actin-binding region
The actin-binding region of myosin consists of a number of structures and loops (Milligan, 1996) including the helix–loop–helix motif of the lower 50 kDa domain, loop-2, the CM loop, loop-3 and loop-4 (Figure 1C). The helix–loop–helix motif forms part of the primary actin binding site, and its structure in MyoE is similar to that observed in conventional myosin. A notable feature of this site is the presence of a number of hydrophobic residues flanked by potentially complementary ionic and polar groups. Loop-2, which has a completely different conformation in MyoE, has been shown to be involved in both weak and the strong binding interactions with actin. It also plays an important role in controlling the rate of product release (Furch et al., 1998; van Dijk et al., 1999a; Joel et al., 2001). Loop-3, which contains positively charged residues and is involved in the secondary binding actin site (Van Dijk et al., 1999b), is more extended and has a different conformation. Loop-4 is five residues longer in MyoE, and contains many charged amino acids (Figure 1C).
The CM loop lies at the front of the motor domain and is important for normal myosin activity. It was shown that a point mutation at position Arg403 in human β-cardiac myosin (the equivalent of Arg397 in Dictyostelium myosin-II) is associated with familial hypertrophic cardiomyopathy (Geisterfer-Lowrance et al., 1990). The CM loop in MyoE is of special interest as it contains a serine at the TEDS-site position (Ser334). Phosphorylation of this serine stimulates actin-activated ATPase and motor activity. In myosin-II, aspartic or glutamic acid residues are found at the equivalent position. Motors without a negative charge at this position display low motility (Wang et al., 1998; De La Cruz et al., 2001; Liu et al., 2001). In the MyoE structure, this loop is disordered and the phosphorylation site is not visible. Based on the purification procedure and kinetic data, it is unlikely that the TEDS-site serine is phosphorylated. Because this loop is well defined in most structures of myosin II motor domains, its disorder in MyoE suggests that phosphorylation may be required in order to constrain its orientation.
Nucleotide-binding site
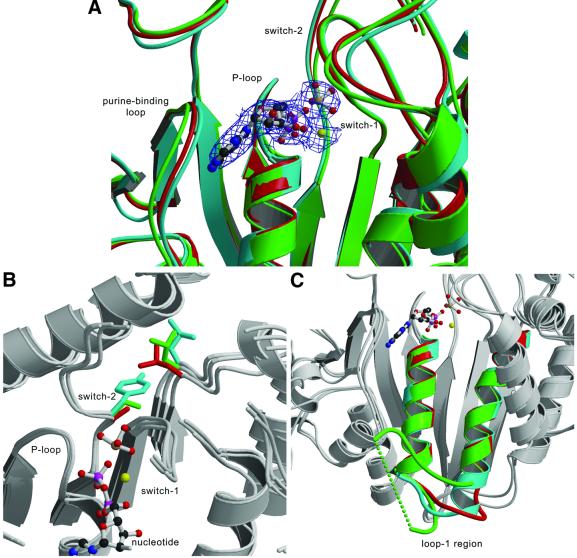
The nucleotide-binding site, which consists of the purine-binding loop (NPxxxxxxY), the P-loop (GESGAGKT), switch-1 [AKTxxN(N/D)NSSR] and switch-2 [DI(S/A/Y/F)GFE], is conserved among all myosins. It is therefore not surprising that only minor structural differences are observed in the polypeptide backbone atom positions of MyoE and myosin-II (Figure 2A). Differences, therefore, are restricted to the size and nature of non-conserved residues in the binding motifs. A noticeable difference between conventional and unconventional myosins is the presence of aromatic side chains at position three of the switch-2 motif (Figure 2B). It has been speculated that having a small side chain at this position is important for efficient release of phosphate (Yount et al., 1995). In a mutational study of this region (Murphy et al., 2001), DdSer456 was mutated to leucine, resulting in the ATPase being uncoupled from motility and a shorter step size. Introduction of a larger side chain was predicted to sterically interfere with formation of the closed conformation. However, the kinetic properties and structure of MyoE, which has a tyrosine in this position, indicate this is not the case (Table I; Figure 2B).
Fig. 2. Comparison of the nucleotide-binding site of MyoE and myosin-II from Dictyostelium and smooth muscle. (A) The nucleotide-binding sites of MyoE (cyan), DdM754 (red) and Gg smII (green) are shown with the MgADP·VO4 from MyoE. (B) Although the sequences of the nucleotide binding regions, including the P-loop, switch-1 and switch-2 are almost entirely conserved, a significant difference occurs in switch-2, where a conserved alanine (red) or serine in amoeboid class-II myosins (green), is replaced by tyrosine (cyan). The introduction of a bulkier side chain pushes a conserved leucine away. (C) Major differences are seen in the length and conformation of loop-1 in MyoE (cyan), DdM754 (red) and Gg smII (green). The green dotted line connects the part of loop-1 that is not visible in the crystal structure of Gg smII.
Loop-1 has been implicated in influencing access to the nucleotide-binding site. It has been speculated that the mobility of this loop correlates to the rate of ADP release (Kurzawa-Goertz et al., 1998; Sweeney et al., 1998), and our results lend support to this view. MyoE has a short, conformationally restrained loop and high ADP affinity (Figure 2C; Table I). The higher ADP affinity of MyoE when compared with that of DdMyoII is caused by differences in the rate of ADP release, as nucleotide binding to both types of myosin occurs at similar rates.
Relay region and converter domain
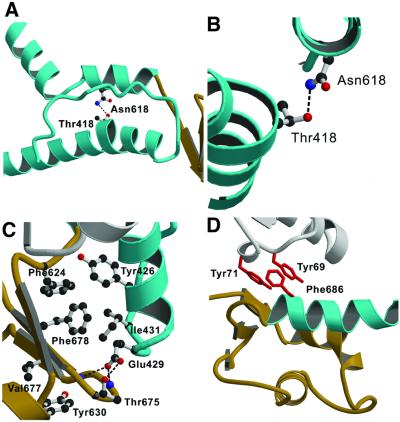
As its name implies, the relay region functions in communicating conformational information between the actin binding site, nucleotide binding site and converter domain. Major sequence differences between myosin-IIs and myosin-Is in the relay region occur at the point in the relay helix where a kink forms when switch-2 is in the ‘closed’ position (Figure 3A). Myosin is in the ‘closed’ state when ATP or transition state analogues are bound, and in the ‘open’ state when ADP is bound or nucleotide is absent from the binding pocket. In MyoE, this kink is slightly more pronounced due to Asn616 forming a hydrogen bond with Thr418 at the position of the kink of the relay helix, which allows the Cα of Thr418 to move 1.5 Å further into the core of myosin (Figure 3B).
Fig. 3. Contacts between the relay region, converter domain and lever arm helix allow these structural elements to move together. (A) The relay region and SH1 helix of MyoE are shown in cyan. In MyoE and other class-I myosins, there is a hydrogen bond between Thr418 in the relay helix and Asn618 from the SH1 helix. (B) Close-up view of this region, viewed along the relay helix. The kink forms at Thr418. (C) Highly conserved residues form a hydrophobic core, and polar residues further stabilize the link via conserved hydrogen bonds (dashed lines). This core interaction is further supported by a small, hydrophobic, highly conserved extension into the converter formed by residues Tyr630 and Val677 (DdTyr699 and DdIle744). At the tip of the relay loop (cyan), conserved Glu429 (DdGlu497) forms hydrogen bonds to residue Thr675 of the converter domain (brown) (DdThr742; at this position there is always a threonine or a serine) and to the backbone nitrogen atoms of converter residues Lys674 and Lys676. (D) Hydrophobic interactions between the lever arm helix (cyan) and core domain (white). All class-I myosins contain an aromatic residue at the positions of Tyr69 and/or Tyr71 (red) in close contact with the conserved Phe686 (red) in the lever arm helix. Either a glycine or an alanine is found at the equivalent position to Phe686 in class-II myosins.
As the C-terminal end of the relay helix is tightly coupled to the converter domain, this kinking leads to a rotational movement of the converter domain. Highly conserved hydrophobic residues are responsible for the coupling, which occurs primarily in the area between the tip of the relay loop and the β-core of the converter domain (Figure 3C). Hydrophobic contacts between the first turn of the lever arm helix and the N-terminal domain further stabilize the ‘up’ conformation. In MyoE, contacts are formed between Phe686, Tyr69 and Tyr71 (Figure 3D). Although these residues are completely conserved in myosin-Is, similar hydrophobic contacts are absent in myosin-IIs.
Discussion
Our results show the core structural elements and topology of MyoE to be essentially identical to those in myosin-II. Given the high sequence conservation between the two motors, this is not unexpected. The differences that are observed are subtle, and occur in loop regions and residues mediating contacts between elements essential for motor function. Spudich has proposed that motor activity of myosin is fine-tuned by changes in the length and flexibility of surface loops (Spudich, 1994), and the MyoE structure certainly lends support to this paradigm. The greatest variation in the loops occurs in the actin binding site and the loop-1 region, and is reflected in differences in the kinetics of actin and nucleotide binding.
An important difference between myosin-I and conventional myosin is that myosin-I has its motor activity regulated by phosphorylation at the TEDS site on the CM loop. Kinetics show that phosphorylation stabilizes the actomyosin complex by reducing the koff rate without greatly affecting kon (U.Dürrwang and D.J.Manstein, manuscript in preparation) and consistent effects have been observed after changes in a single negatively charged residue at the actin binding site of myosin-II (Furch et al., 2000). These data indicate that the CM loop is primarily involved in stabilizing electrostatic interactions with actin.
The greatest changes in loop length are observed in loop-4 (Figure 1C). Based on actomyosin models derived from electron microscopy, loop-4 is the loop that is furthest removed from the actin surface. Its extended length in myosin-Is suggests it may be involved in either interactions with actin or regulatory proteins that are bound to actin. It has been shown for rat myr-1 that a head fragment localizes to the same actin structures as the full-length construct (Tang and Ostap, 2001). According to this study, the head of myr-1 localizes to the highly dynamic actin structures at the cell cortex and not to the actin filaments that are regulated by tropomyosin. An extended loop-4 might be responsible for this localization due to steric interference with tropomyosin, which is bound to less dynamic actin filaments as they occur in stress fibers.
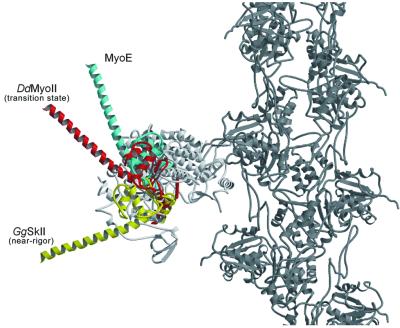
While changes in the loop structures are mainly involved in regulating kinetics and interactions with binding partners, changes in amino acid residues mediating contacts between elements essential for motor function act to fine-tune the mechanical properties of myosin-Is. Examples of this fine-tuning include the more highly kinked relay helix and the hydrophobic interactions between the lever arm and N-terminal domain. These changes result in MyoE being able to undergo a greater lever arm rotation than conventional myosin-IIs. As the overall shapes of the catalytic domains are so similar and image reconstructions of actin filaments decorated with myosin-I in the rigor state showed the geometry of actin binding by the catalytic domain to be virtually indistinguishable from that of myosin-IIs (Jontes et al., 1995, 1998; Jontes and Milligan, 1997; Carragher et al., 1998), we superposed the MyoE structure with a model of myosin S1 attached to an actin filament in order to visualize how MyoE is oriented when bound to an actin filament (Figure 4). We found that the MyoE converter extends ∼30° further up than the converter of myosin-II in the closed conformation. Based on this, we predict that MyoE could produce a power stroke 20–30° greater than the 60–70° rotation of the myosin-II lever arm. Recent single molecule step size measurements support the presence of a larger power stroke in myosin-I, and indicate its lever arm undergoes a rotation of at least 100° during force generation (D.Köhler, C.Ruff, E.Meyhöfer and M.Bähler, manuscript in preparation).
Fig. 4. A model of chicken skeletal muscle myosin motor core (white), converter domain and lever arm (yellow) in the near-rigor state attached to the actin filament (dark gray). Dictyostelium myosin-II in complex with ADP-BeF3 with a modeled extended lever arm in the ‘up’ or transition state position is shown in red. The MyoE converter domain and modeled extended lever arm (cyan) is in an ∼30° higher position.
The structure also reveals detailed information about the contacts that persist during movement of the converter domain and relay region. Tight coupling of these structural elements is an essential part of the pathway linking small conformational changes in the nucleotide binding site to larger movements of the converter domain and lever arm. As described above, the converter domains and adjacent relay region in the four molecules in the asymmetric unit vary substantially in their orientation due to a rigid body rotation of up to 10°. In contrast, myosin-II crystal structures have indicated only translations along the actin long axis. Thus, the four independent molecules observed in the MyoE crystal visualize three important structural features at atomic resolution: (i) the converter domain and relay regions are rigidly linked; (ii) the identity of residues that are essential for effective coupling of these domains; and (iii) the converter domain is able to move not only along the actin axis, but also with a component perpendicular to it. Similar movements of the converter domain and lever arm about the actin long axis have been described in electron microscopy (EM) studies of actin/myosin-II complexes (Volkmann et al., 2000). Our results and those of the EM study suggest that the pathway of converter movement is not a simple translation, but rather a combination of a major translational component along the actin axis with a minor component perpendicular to it, and may describe an arc as it moves between up and down positions. Biochemical evidence supporting the tight coupling between the relay helix and the converter domain throughout the catalytic cycle has been found using a ‘cysteine-light’ myosin construct (Shih and Spudich, 2001) in which two cysteines were introduced, one into the relay loop and one into the converter. Covalent cross-linking of the cysteines resulted in only minimal changes in the actin-binding properties, ATP-induced actin release and actin-activated ATPase.
The myosin-I crystal structure described in this paper visualizes how subtle evolutionary changes in myosin sequence fine-tune motor function. Building on a core containing activities necessary for molecular motor function, alterations in the MyoE motor domain result in distinct kinetic and mechanical properties. Although the sequence changes that produce these altered activities are minor, taken together their effects are additive and result in the distinct properties of class-I myosins. In order to understand more fully the relationship between structure and function in myosin-Is and other myosin classes, it will be necessary to solve additional atomic-level structures of other myosin family members, as well as to determine the structure of class-I myosins in other nucleotide states.
Materials and methods
Cloning and expression
General methods for molecular biology were as described by Sambrook et al. (1989). Genomic DNA was isolated from AX2 cells according to Bain and Tsang (1991). A degenerate primer was used to amplify the head fragment (amino acids 2–698) of the myoE gene. The fragment was ligated into the pDXA-3H expression vector (Manstein et al., 1995), which contains a C-terminal His8 tag. The derived sequence of MyoE was different from the published sequence (Urrutia et al., 1993) (DDBJ/EMBL/GenBank accession number L06805) in 25 places: while some changes concerned non-conserved residues, especially in the regions where additional residues or deletions were found, the derived sequence was in better agreement with the amino acid alignment of the Dictyostelium myosins. In one region, a 10-residue surface loop was found to be completely different. In detail, the sequence contained the following modifications compared with the published one: D26E, R48T, I77M, I137L, R138D, F139 absent, Q140 absent, N215D, L371I, S372I, I373N, V374C, H375T, R376T, G378K, T379G, P380 added, V427 added, R428 added, K429E, N440 added, N498I, D604V, I681N, R683T. Our sequence completely agrees with the fragment obtained from the genome sequencing project (amino acids 270–1003).
Purification
The protein was purified as described previously for myosin-II head fragments (Manstein and Hunt, 1995). Before crystallization, the protein was further purified by cation-exchange chromatography (SP 650; Supelco), dialyzed against 50 mM HEPES–KOH pH 7.3, 0.5 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM MgCl2, 1 mM benzamidine, 0.02% NaN3 and 3% sucrose, and concentrated to 5.5 mg/ml.
Kinetics
Stopped-flow measurements were performed at 20°C with a Hi-tech Scientific SF-61 DX2 double mixing stopped-flow system using a 100 W Xe/Hg lamp and a monochromator for wavelength selection. Pyrene and mantATP fluorescences were excited at 365 nm, and emission was detected after passing through a Schott (Mainz, Germany) KV 389-nm cutoff filter. Tryptophan fluorescence, excited at 295 nm, was monitored through a WG 320 nm cutoff filter. Data were stored and analyzed using software provided by Hi-Tech. The experimental buffer used was 25 mM MOPS, 5 mM MgCl2, 100 mM KCl, pH 7.0. Data interpretation was performed as described previously (Batra et al., 1999). Steady-state ATPase activities were measured at 25°C with the NADH-coupled assay (Furch et al., 1998).
Crystallization
For crystallization, the protein solution was used immediately after purification or stored at –78°C. Prior to crystallization, MgADP VO4 (Goodno, 1982) was added to a final concentration of 0.2 mM ADP and 0.18 mM vanadate, and incubated on ice for 1 h. Crystals were grown by the hanging drop method either at 16 or 7°C. The mother liquor contained 11% (w/v) PEG 8K, 170 mM NaCl, 50 mM HEPES–NaOH pH 7.2, 5 mM MgCl2, 5 mM DTT, 0.5 mM EGTA and 2% 2-methyl-1,3-propanediol. Crystals appeared after 3–7 days at 16°C and 5–6 weeks at 7°C, and reached maximum dimensions of 0.06 × 0.06 × 0.06 mm.
Crystallography and structure refinement
Crystals of MyoE-MgADP·VO4 diffracted to 2.5 Å, but due to radiation damage complete data could only be collected to 3 Å. A low-resolution data set of a single crystal was collected at DESY beamline BW7B (λ = 0.8424 Å) on a MarCCD detector at 80 K to 4 Å. High-resolution X-ray data was collected from the same crystal using an ESRF beamline ID-13 (λ = 0.9600 Å) on a MarCCD detector at 80 K to 3.0 Å. Both data sets were individually indexed and integrated, and then scaled and merged using the program XDS (Kabsch, 1993), producing a data set 79.7% complete to 3.0 Å, with 3.8-fold redundancy and an Rsym of 19.5%. The MyoE crystals belonged to spacegroup P21 (a = 51.818 Å, b = 143.667 Å, c = 236.050 Å, β = 94.858°) and contain four independent molecules in the asymmetric unit. Molecular replacement was performed with the program CNS Version 1.0 (Brünger et al., 1998), using the polyalanine model of the crystal structure of Dictyostelium myosin residues 80–754 complexed with MgADP-BeF3 (F.J.Kull and K.C.Holmes, manuscript in preparation) as a starting model (the nucleotide and all solvent molecules were excluded). The converter domains (last 64 amino acids) were removed and molecule A was refined using strict non-crystallographic symmetry, resulting in initial R-values of R = 38.1% and Rfree = 43.2%. Following several rounds of rigid body refinement, minimization and simulated annealing refinement, side chains were placed using the program O Version 7.0 (Jones et al., 1991). Subsequent rounds of model building and refinement (including bulk solvent correction) improved the map. The converters were built and the structure refined applying NCS restraints (weights: 200 kcal/mol/Å) to amino acids Gly9-Gln287, Thr292-Ser324, Val335-Thr418, Lys450-Pro548 and Glu560-Gly612. Subsequent rounds of model building and simulated annealing refinement using torsional dynamics and a maximum likelihood target produced the final structure of four MyoE molecules, four molecules of MgADP VO4 and 64 water molecules (R-factor = 22.8, Rfree = 27.3). Ramachandran analysis with the program PROCHECK (Laskowski et al., 1993) revealed that 81.5% of the residues adopt the most favorable conformational angles. Atomic coordinates have been deposited in the Protein Data Bank under accession code 1LKX.
Acknowledgments
Acknowledgements
We would like to thank P.Tucker at DESY beamline BW7B and M.Burghammer at ESRF beamline ID-13 for their help with data collection, W.Kabsch, S.Eschenburg and K.Scheffzek for crystallographic advice, M.L.Knetsch for help with the Dictyostelium cells; S.Zimmermann and A.Scherer for technical assistance, J.Wray for helpful comments and discussion, and K.C.Holmes for discussions and continuous support. This work was supported by Deutsche Forschungs gemeinschaft grants KU1288/2-2 (to F.J.K.) and MA1081/5-2 (to D.J.M.), and the Max-Planck Society.
References
- Adams R.J. and Pollard,R.D. (1989) Binding of myosin I to membrane lipids. Nature, 340, 565–568. [DOI] [PubMed] [Google Scholar]
- Anson M., Geeves,M.A., Kurzawa,S.E. and Manstein,D.J. (1996) Myosin motors with artificial lever arms. EMBO J., 15, 6069–6074. [PMC free article] [PubMed] [Google Scholar]
- Bain G. and Tsang,A. (1991) Disruption of the gene encoding the p34/31 polypeptides affects growth and development of Dictyostelium discoideum. Mol. Gen. Genet., 226, 59–64. [DOI] [PubMed] [Google Scholar]
- Batra R., Geeves,M.A. and Manstein,D.J. (1999) Kinetic analysis of Dictyostelium discoideum myosin motor domains with glycine-to-alanine mutations in the reactive thiol region. Biochemistry, 38, 6126–6134. [DOI] [PubMed] [Google Scholar]
- Bement W.M. and Mooseker,M.S. (1995) TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil. Cytoskeleton, 31, 87–92. [DOI] [PubMed] [Google Scholar]
- Berg J.S., Powell,B.C. and Cheney,R.E. (2001) A millennial myosin census. Mol. Biol. Cell, 12, 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr., 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carragher B.O., Cheng,N., Wang,Z.-Y., Korn,E.D., Reilein,A., Belnap,D.M., Hammer,J.A.,III and Steven,A.C. (1998) Structural invariance of constitutively active and inactive mutants of Acanthamoeba myosin IC bound to F-actin in the rigor and ADP-bound states. Proc. Natl Acad. Sci. USA, 95, 15206–15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremo C.R. and Geeves,M.A. (1998) Interaction of actin and ADP with the head domain of smooth muscle myosin: implications for strain-dependent ADP release in smooth muscle. Biochemistry, 37, 1969–1978. [DOI] [PubMed] [Google Scholar]
- De La Cruz E.M., Ostap,E.M. and Sweeney,H.L. (2001) Kinetic mechanism and regulation of myosin VI. J. Biol. Chem., 276, 32373–32381. [DOI] [PubMed] [Google Scholar]
- Dominguez R., Freyzon,Y., Trybus,K. and Cohen,C. (1998) Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell, 94, 559–571. [DOI] [PubMed] [Google Scholar]
- Fisher A.J., Smith,C.A., Thoden,J.B., Smith,R., Sutoh,K., Holden,H.M. and Rayment,I. (1995) X-ray structures of myosin motor domain of Dictyostelium discoideum complexed with MgADP·BeFx and MgADP·Al4–. Biochemistry, 34, 8960–8972. [DOI] [PubMed] [Google Scholar]
- Furch M., Geeves,J.A. and Manstein,D.J. (1998) Modulation of actin affinity and actomyosin adenosin triphosphatase by charge changes in the myosin motor domain. Biochemistry, 37, 6317–6326. [DOI] [PubMed] [Google Scholar]
- Furch M., Remmel,B., Geeves,J.A. and Manstein,D.J. (2000) Stabilization of the actomyosin complex by negative charges on myosin. Biochemistry, 39, 11602–11608. [DOI] [PubMed] [Google Scholar]
- Geisterfer-Lowrance A.A., Kass,S., Tanigawa,G., Vosberg,H.P., McKenna,W., Seidman,C.E. and Seidman,J.G. (1990) A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell, 62, 999–1006. [DOI] [PubMed] [Google Scholar]
- Goodno C.C. (1982) Myosin active-site trapping with vanadate ion. In Frederiksen,D. and Cunningham,L. (eds), Methods in Enzymology. Academic Press, NY, pp. 116–123. [DOI] [PubMed]
- Hodge T. and Cope,M.J.T.V. (2000) A myosin family tree. J. Cell Sci., 113, 3353–3354. [DOI] [PubMed] [Google Scholar]
- Holmes K.C., Popp,D., Gebhard,W. and Kabsch,W. (1990) Atomic model of the actin filament. Nature, 347, 44–49. [DOI] [PubMed] [Google Scholar]
- Houdusse A., Kalabokis,V.N., Himmel,D., Szent-Györgyi,A.G. and Cohen,C. (1999) Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell, 97, 459–470. [DOI] [PubMed] [Google Scholar]
- Joel P.B., Trybus,K.M. and Sweeney,H.L. (2001) Two conserved lysines at the 50/20 kDa junction of myosin are necessary for triggering actin-activation. J. Biol. Chem., 276, 2998–3003. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jontes J.D. and Milligan,R.A. (1997) Brush border myosin-I structure and ADP-dependent conformational changes revealed by cryoelectron microscopy and image analysis. J. Cell Biol., 139, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jontes J.D., Wilson-Kubalek,W.M. and Milligan,R.A. (1995) A 32° tail swing in brush border myosin I on ADP release. Nature, 378, 751–753. [DOI] [PubMed] [Google Scholar]
- Jontes J.D., Ostap,E.M., Pollard,T.D. and Milligan,R.A. (1998) Three-dimensional structure of Acanthamoeba castellanii myosin-IB (MIB) determined by cryoelectron microscopy of decorated actin filaments. J. Cell Biol., 141, 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Saxe,C.L.,III, Kimmel,A.R. and Hammer,J.A.,III (1989) Dictyostelium discoideum contains a gene encoding a myosin I heavy chain isoform. Proc. Natl Acad. Sci. USA, 86, 6186–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Fukui,Y., Martin,B. and Hammer,J.A.,III (1993) Sequence, expression pattern, extracellular localization, and targeted disruption of the Dictyostelium myosin ID heavy chain isoform. J. Biol. Chem., 268, 14981–14990. [PubMed] [Google Scholar]
- Jung G., Wu,X. and Hammer,J.A.,III (1996) Dictyostelium mutants lacking multiple callsi myosin I isoforms reveal combinations of shared and distinct functions. J. Cell Biol., 133, 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795–800. [Google Scholar]
- Kliche W., Fujita-Becker,S., Kollmar,M., Manstein,D.J. and Kull,F.J. (2001) Structure of a genetically engineered molecular motor. EMBO J., 20, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Kurzawa-Goertz S.E., Perreault-Micale,C.L., Trybus,K.M., Szent-Gyorgyi,A.G. and Geeves,M.A. (1998) Loop I can modulate ADP affinity, ATPase activity, and motility of different scallop myosins. Transient kinetic analysis of S1 isoforms. Biochemistry, 37, 7517–7525. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemistry quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Liu X., Osherov,N., Yamashita,R., Brzeska,H., Korn,E.D. and May,G.S. (2001) Myosin I mutants with only 1% of wild-type actin-activated MgATPase activity retain essential in vivo function(s). Proc. Natl Acad. Sci. USA, 98, 9122–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M., Poole,K.J., Popp,D., Rosenbaum,G. and Holmes,K.C. (1995) An atomic model of the unregulated thin filament obtained by X-ray fiber diffraction on oriented actin-tropomyosin gels. J. Mol. Biol., 246, 108–119. [DOI] [PubMed] [Google Scholar]
- Manstein D.J. and Hunt,D.M. (1995) Overexpression of myosin motor domains in Dictyostelium: screening of transformants and purification of the affinity tagged protein. J. Muscle Res. Cell Motil., 16, 325–332. [DOI] [PubMed] [Google Scholar]
- Manstein D.J., Schuster,H.-P., Morandini,P. and Hunt,D.M. (1995) Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene, 162, 129–134. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Milligan R.A. (1996) Protein–protein interactions in the rigor actomyosin complex. Proc. Natl Acad. Sci. USA, 93, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C.T., Rock,R.S. and Spudich,J.A. (2001) A myosin II mutation uncouples ATPase activity from motility and shortens step size. Nature Cell Biol., 3, 311–315. [DOI] [PubMed] [Google Scholar]
- Neuhaus E.M. and Soldati,T. (2000) A myosin I is involved in membrane recycling from early endosomes. J. Cell Biol., 150, 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak K.D., Peterson,M.D., Reedy,M.C. and Titus,M.A. (1995) Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J. Cell Biol., 131, 1205–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M.D., Novak,K.D., Reedy,M.C., Ruman,J.I. and Titus,M.A. (1995) Molecular genetic analysis of myoC, a Dictyostelium myosin I. J. Cell Sci., 108, 1093–1103. [DOI] [PubMed] [Google Scholar]
- Pollard T.D. and Korn,E.D. (1973) Acanthamoeba myosin. I. Isolation from Acanthamoeba castellanii of an enzyme similar to muscle myosin. J. Biol. Chem., 248, 4682–4690. [PubMed] [Google Scholar]
- Rayment I., Rypniewski,W.R., Schmidt-Bäse,K., Smith,R., Tomchick,D.R., Benning,M.M., Winkelmann,D.A., Wesenberg,G. and Holden,H.M. (1993) Three-dimensional structure of myosin subfragment-1: a molecular motor. Science, 261, 50–58. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schwarz E.C., Neuhaus,E.M., Kistler,C., Henkel,A.W. and Soldati,T. (2000) Dictyostelium myosin IK is involved in the maintenance of cortical tension and affects motility and phagocytosis. J. Cell Sci., 113, 621–633. [DOI] [PubMed] [Google Scholar]
- Shih W.M. and Spudich,J.A. (2001) The myosin relay helix to converter interface remains intact throughout the actomyosin ATPase cycle. J. Biol. Chem., 276, 19491–19494. [DOI] [PubMed] [Google Scholar]
- Smith C.A. and Rayment,I. (1996) X-ray structure of the magnesium(II)·ADP·vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry, 35, 5404–5417. [DOI] [PubMed] [Google Scholar]
- Spudich J.A. (1994) How molecular motors work. Nature, 372, 515–518. [DOI] [PubMed] [Google Scholar]
- Sweeney H.L., Rosenfeld,S.S., Brown,F., Faust,L., Smith,J., Xing,J., Stein,L.A. and Sellers,J.R. (1998) Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket. J. Biol. Chem., 273, 6262–6270. [DOI] [PubMed] [Google Scholar]
- Tang N. and Ostap,E.M. (2001) Motor domain-dependent localization of myo1b (myr-1). Curr. Biol., 11, 1131–1135. [DOI] [PubMed] [Google Scholar]
- Temesvari L.A., Bush,J.M., Peterson,M.D., Novak,K.D., Titus,M.A. and Cardelli,J.A. (1996) Examination of the endosomal and lysosomal pathways in Dictyostelium discoideum myosin I mutants. J. Cell Sci., 109, 663–673. [DOI] [PubMed] [Google Scholar]
- Titus M.A., Warrick,H.M. and Spudich,J.A. (1989) Multiple actin-based motor genes in Dictyostelium. Cell Regul., 1, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus M.A., Novak,K.D., Hanes.G.P. and Urioste,A.S. (1995) Molecular genetic analysis of myoF, a new Dictyostelium unconventional myosin gene. Biophys. J., 68, 152S–157S. [PMC free article] [PubMed] [Google Scholar]
- Urrutia R., Jung,G. and Hammer,J.A.,III (1993) The Dictyostelium myosin IE heavy chain gene encodes a truncated isoform that lacks sequences corresponding to the actin binding site in the tail. Biochim. Biophys. Acta, 1173, 225–229. [DOI] [PubMed] [Google Scholar]
- Uyeda T.Q.P. and Titus,M.A. (1997) The myosins in Dictyostelium. In Maeda,Y., Inouye,K. and Takeuchi,I. (eds), Dictyostelium: A Model System for Cell and Developmental Biology. Universal Academy Press, Tokyo, Japan, pp. 43–64.
- Van Dijk J., Furch,M., Derancourt,J., Batra,R., Knetsch,M.L., Manstein,D.J. and Chaussepied,P. (1999a) Differences in the ionic interaction of actin with the motor domains of nonmuscle and muscle myosin II. Eur. J. Biochem., 260, 672–683. [DOI] [PubMed] [Google Scholar]
- Van Dijk J., Furch,M., Lafont,C., Manstein,D.J. and Chaussepied,P. (1999b) Functional characterization of the secondary actin binding site of myosin II. Biochemistry, 38, 15078–15085. [DOI] [PubMed] [Google Scholar]
- Volkmann N., Hanein,D., Ouyang,G., Trybus,K.M., DeRosier,D.J. and Lowey,S. (2000) Evidence for cleft closure in actomyosin upon ADP release. Nature Struct. Biol., 7, 1147–1155. [DOI] [PubMed] [Google Scholar]
- Wang Z.-H., Wang,F., Sellers,J.R., Korn,E.D. and Hammer,J.A.,III (1998) Analysis of the regulatory phosphorylation site in Acanthamoeba myosin IC by using site-directed mutagenesis. Proc. Natl Acad. Sci. USA, 95, 15200–15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo C.M., Chrin,L., Rovner,A.S. and Berger,C.L. (1999) Intrinsic tryptophan fluorescence identifies specific conformational changes at the actomyosin interface upon actin binding and ADP release. Biochemistry, 38, 14515–14523. [DOI] [PubMed] [Google Scholar]
- Yount R.G., Lawson,D. and Rayment,I. (1995) Is myosin a ‘back door’ enzyme? Biophys. J., 68, 44S–49S. [PMC free article] [PubMed] [Google Scholar]