Abstract
Ssc1, a molecular chaperone of the Hsp70 family, drives preprotein import into the mitochondrial matrix by a specific interaction with the translocase component Tim44. Two other mitochondrial Hsp70s, Ssc3 (Ecm10) and Ssq1, show high sequence homology to Ssc1 but fail to replace Ssc1 in vivo, possibly due to their inability to interact with Tim44. We analyzed the structural basis of the Tim44 interaction by the construction of chimeric Hsp70 proteins. The ATPase domains of all three mitochondrial Hsp70s were shown to bind to Tim44, supporting the active motor model for the Hsp70 mechanism during preprotein translocation. The peptide-binding domain of Ssc1 sustained binding of Tim44, while the peptide-binding domains of Ssc3 and Ssq1 exerted a negative effect on the interaction of the ATPase domains with Tim44. A mutation in the peptide-binding domain of Ssc1 resulted in a similar negative effect not only on the ATPase domain of Ssc1, but also of Ssq1 and Ssc3. Hence, the determination of a crucial Hsp70 function via the peptide-binding domain suggests a new regulatory principle for Hsp70 domain cooperation.
Keywords: Hsp70/protein interaction/mitochondria/Saccharomyces cerevisiae/Tim44
Introduction
Molecular chaperones of the 70 kDa class (Hsp70) perform critical functions in many cellular processes (Craig et al., 1994; Hartl, 1996). Hsp70 chaperones promote protein folding and assembly by binding to unfolded or partially folded polypeptide chains. In Saccharomyces cerevisiae, 14 Hsp70 homologs have been identified. Despite their functional diversity, proteins of the Hsp70 family have retained a remarkably high sequence conservation and exhibit a well-conserved structural composition (Bukau and Horwich, 1998). The N-terminal part forms a 44 kDa domain exhibiting ATPase activity. Hsp70 proteins can switch between two main conformational states depending on the nucleotide bound (McCarthy et al., 1995). The nucleotide state of the ATPase domain controls the affinity of the C-terminal part for substrate polypeptides. The C-terminal part can be divided into the 18 kDa peptide-binding domain (PBD) and a small 10 kDa variable domain at the extreme C-terminus. In some types of eukaryotic Hsp70 proteins the variable domain has been shown to act as a binding site for co-factors that modulate their activity (Demand et al., 1998; Horton et al., 2001). When ATP is bound the affinity of the PBD for the substrate polypeptide is low, with a high on and off rate. Hydrolysis of the nucleotide leads to conformational changes in both domains and stabilizes the interaction with the substrate. Two types of co-factors stimulate the cellular activity of Hsp70 family members (Cyr et al., 1994; Rassow et al., 1995). Proteins of the DnaJ family mainly regulate substrate interaction while proteins of the GrpE family catalyze nucleotide exchange.
Specific interactions with specialized partner proteins but also the subcellular localization determine the functional specificity of the different Hsp70s. In mitochondria from S.cerevisiae three different Hsp70 molecules have been identified, Ssc1, Ecm10 and Ssq1, all located as soluble proteins in the matrix (Schilke et al., 1996; Baumann et al., 2000). The essential protein Ssc1, the most abundant mitochondrial Hsp70, plays a major role in the membrane translocation of cytosolic preproteins through the mitochondrial inner membrane into the matrix and in subsequent folding reactions. Together with the proteins Tim44 and Mge1, Ssc1 forms an essential import motor complex that is responsible for the coupling of ATP hydrolysis to polypeptide translocation (Matouschek et al., 2000; Strub et al., 2000). By the ATP-regulated interaction with Tim44, Ssc1 is recruited to the import site at the inner membrane pore, thus favoring binding of Ssc1 to the incoming preprotein. A functional interaction between Ssc1 and Tim44 is indispensable for the full efficiency of the preprotein translocation reaction. The introduction of mutations in both Ssc1 and Tim44 that destabilize or abolish their interaction result in significant preprotein import defects (Voos et al., 1996; Merlin et al., 1999; Milisav et al., 2001). In particular, the import of preproteins containing stable folded domains is affected by a defective Ssc1–Tim44 interaction. Here, Ssc1 is not able to exert an inward-directed translocation force on the polypeptide in transit that would lead to the active unfolding of preprotein domains exposed on the cytosolic face of the mitochondrial membranes (Voisine et al., 1999; Lim et al., 2001).
In contrast to Ssc1, Ssq1 and Ecm10 seem to be less important for the mitochondrial metabolism. Mutations in Ssq1 have only minor effects on cellular growth, and no preprotein import defect has been observed (Schilke et al., 1996); however, the maturation of the yeast frataxin homolog is delayed and mitochondria exhibit a significantly increased iron content (Knight et al., 1998). Both genetic and biochemical data seem to indicate that Ssq1 plays a role in the mitochondrial iron metabolism and Fe/S cluster assembly (Craig et al., 1999; Lutz et al., 2001), although its direct cellular role is not fully understood. Although there is extensive sequence overlap Ssq1 is not able to take over Ssc1 function (Schilke et al., 1996). A possible reason is the inability of Ssq1 to bind to the preprotein translocase component Tim44 (Schmidt et al., 2001). The protein Ecm10, encoded by the open reading frame YEL030w, was initially classified in a group together with the mitochondrial Hsp70 Ssc1 due to the extremely high sequence similarity and a potential mitochondrial targeting sequence (Rassow et al., 1997). Although ECM10 was also described as being involved in cell wall formation (Lussier et al., 1997), recent evidence demonstrated clearly a mitochondrial localization rendering a function in cell wall biogenesis highly unlikely (Baumann et al., 2000). Based on these results we recommend using the name Ssc3 instead of Ecm10. Despite their functional differences, both Ssq1 and Ssc3 (Ecm10) show typical chaperone properties and also interact specifically with the Hsp70 nucleotide exchange factor Mge1 (Baumann et al., 2000; Schmidt et al., 2001). However, the functional relevance of Ssc3 (Ecm10) under normal growth conditions is unclear, since it is expressed at extremely low levels and no phenotype was observed in a deletion mutant.
These observations raised the question of whether the closely related SSC3 is capable of complementing a ssc1Δ mutation when its expression levels were raised considerably. Since we found that Ssc3 could not replace Ssc1, we asked which structural properties are responsible for the distinct functional differences between the mitochondrial Hsp70s. We investigated the relative influence of the Hsp70 domains on the interaction with the essential partner protein Tim44 by the construction of chimeric proteins consisting of different combinations of ATPase, peptide-binding and variable C-terminal domains derived from the three mitochondrial Hsp70s. We found that the ATPase domains of all three Hsp70s are principally able to bind to Tim44, while the PBDs of Ssq1 and Ssc3 can exert a negative influence on the ATPase domain, which results in the inability of the respective full-length proteins to interact with Tim44.
Results
The mitochondrial Hsp70, Ssc3 (Ecm10), cannot replace the essential function of the related chaperone Ssc1
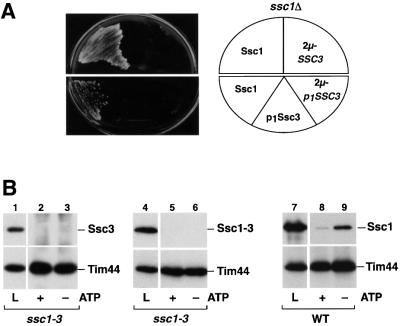
Of the three species of Hsp70 proteins in the mitochondrial matrix, Ssc1 and Ssc3 (Ecm10) exhibit an exceptionally high degree of sequence conservation. Ssc3 and Ssc1 share 82% identical amino acids, slightly more in the ATPase domain than in the PBD. The other mitochondrial Hsp70, Ssq1, is less well conserved and has an overall identity of 52%. It has been shown that Ssq1 is not able to take over the Ssc1 function in vivo even if expressed at similar levels as Ssc1, but the much higher sequence conservation between Ssc3 and Ssc1 indicated an overlapping function. To determine the functional relationship between Ssc3 and Ssc1 in vivo we tested whether Ssc3 might be able to complement the lethal effect of a ssc1Δ mutation. To increase the expression level of Ssc3, we inserted the SSC3 (ECM10) gene including the promotor and downstream region into a high copy number vector and performed an in vivo complementation assay with an ssc1Δ mutant strain. No colonies were detected with cells containing the high copy plasmid expressing Ssc3, whereas the control cells expressing Ssc1 grew normally (Figure 1A, upper panel). However, a biochemical characterization of mitochondria isolated from these cells revealed that the amount of Ssc3 was still lower than the amount of Ssc1 in wild-type mitochondria. When we tested the mitochondrial import efficiency in vitro it became apparent that a low intrinsic import rate of the Ssc3 precursor protein was the reason for the low mitochondrial abundance (data not shown). To further raise the mitochondrial levels of Ssc3 we exchanged the endogenous pre-sequence of Ssc3 for the highly efficient pre-sequence of Ssc1 without altering the cleavage site for the matrix processing peptidase (MPP) and additionally replaced the endogenous promotor by the promotor region of SSC1. The resulting protein construct, p1Ssc3, was efficiently imported and processed in isolated mitochondria. However, despite mitochondrial protein amounts similar to Ssc1 (data not shown), Ssc3 was not able to complement the lethal phenotype of the ssc1 null mutation (Figure 1A, lower panel). Even when expressed from a high copy 2µ vector, p1Ssc3 did not take over the function of Ssc1 (Figure 1A, lower panel). In summary, Ssc3 cannot complement the essential function of Ssc1 despite their high sequence similarity.
Fig. 1. Ssc3 (Ecm10) is not able to functionally replace Ssc1. (A) Overproduction of Ssc3 cannot rescue the lethal phenotype of a mutant lacking Ssc1. Plasmids expressing SSC1 (pASC07) or the SSC3 gene in an overexpression vector (pASE10) were tested for complementation of the ssc1Δ mutation (upper panel). In addition, the mature Ssc3 protein with the pre-sequence and the promotor of Ssc1 (p1Ssc3) was expressed in a low copy (pASE08) and high copy (pNZ46) vector. (B) Ssc3 does not interact with the essential Ssc1 partner protein Tim44. Interaction of Ssc3 with Tim44 was assayed by co-immunoprecipitation with antibodies against Tim44 from mitochondrial lysates overproducing Ssc3 in ssc1-3 mutant cells in the presence (+) or absence (–) of ATP as described in Materials and methods. Cells were pre-incubated for 15 min at 37°C to induce the mutant phenotype prior to lysis. Five percent of the lysed mitochondria were applied as a control (L). Precipitated proteins were separated by SDS–PAGE and the presence of Ssc3 (lanes 1–3) or Ssc1 (lanes 4–9) was detected by western blot.
Both Ssc3 and Ssq1 show no interaction with the essential partner protein Tim44
We wished to determine what property of Ssc3 might be responsible for the observed inability to take over the function of the related Ssc1. The import of cytosolic preproteins into the mitochondrial matrix is considered to be the essential function of Ssc1, requiring an efficient interaction with the inner membrane translocase component Tim44. Hence, we tested whether Ssc3 is able to interact specifically with Tim44 by performing co-immunoprecipitation experiments in organello. Detergent lysates of mitochondria isolated from the temperature-sensitive mutant strain ssc1-3 overexpressing p1SSC3 were prepared under native buffer conditions. The temperature-sensitive mutant protein Ssc1-3 is completely inactivated under non-permissive conditions, and unable to bind to Tim44 (Voos et al., 1996; Figure 1B, lanes 4–6); therefore, a possible competition between Ssc1 and Ssc3 for binding sites at Tim44 could be excluded. However, no signal for Ssc3 could be detected when Tim44 was precipitated from the mitochondrial lysates with affinity-purified antibodies in either the presence or absence of nucleotides (Figure 1B, lanes 1–3). As control, Ssc1 co-precipitated efficiently with Tim44 in an ATP-sensitive manner, indicating a specific and functional interaction (Figure 1B, lanes 7–9). In summary, Ssc3 failed to interact with the Hsp70 partner protein Tim44, and this is a possible reason for the inability of Ssc3 to complement the deletion of Ssc1.
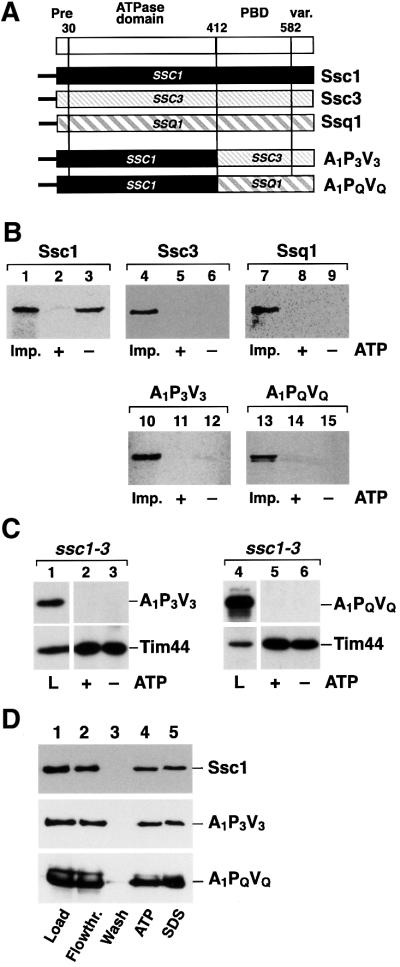
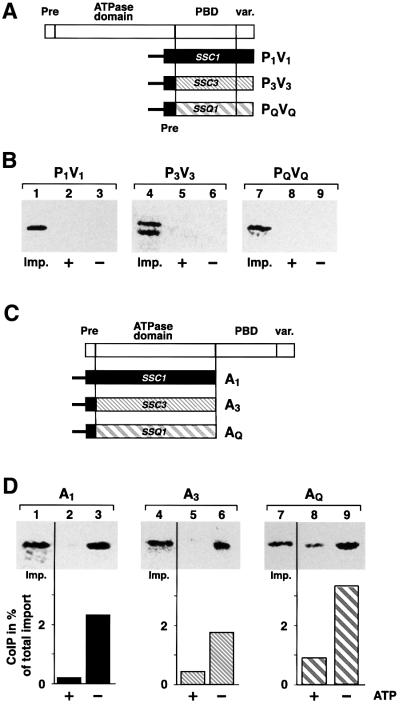
The conserved structural organization of Hsp70 family members allowed us to determine the properties of Hsp70s that are responsible for the interaction with Tim44 using a domain-swap approach. In previous experiments we were able to demonstrate that the ATPase domain of Ssc1 is crucial for interaction with Tim44 (Krimmer et al., 2000). We asked whether it is possible to convert the non-binding proteins Ssc3 and Ssq1 into proteins capable of an interaction with Tim44 by exchanging their ATPase domain for that of Ssc1. For this purpose we constructed chimeric proteins that contained the ATPase domain of Ssc1 (A1) fused to the PBD and variable domains of Ssc3 (P3V3) or Ssq1 (PQVQ) (Figure 2A). The resulting chimeric proteins A1P3V3 and A1PQVQ were tested for their ability to form a complex with Tim44. In order to be able to detect minor amounts of co-precipitated proteins we performed the analysis with radiolabeled precursor proteins that were imported into isolated mitochondria. After stopping the import reaction and removal of excess preprotein, mitochondria were lysed and the immunoprecipitation with Tim44 antibodies in the presence or absence of ATP was performed. Efficient binding to Tim44 was detected with the imported Ssc1. Up to 3.5% of the imported and processed preprotein was found bound to Tim44 (Figure 2B, lane 3), reflecting the relative amounts of Tim44 and Hsp70 in the matrix. Since endogenous Ssc1 is in high excess over Tim44, only a small part of the total amount of Ssc1 is found in a complex with Tim44 in mitochondria. Similar to the co-immunoprecipitation experiments in organello, the Tim44–Ssc1 complex was dissociated in the presence of ATP (Figure 2B, lane 2), demonstrating that the imported proteins are functional. Unexpectedly, neither A1P3V3 (Figure 2B, lanes 11 and 12) nor A1PQVQ (Figure 2B, lanes 14 and 15) was found in complex with Tim44, even though they contain the functional ATPase domain of Ssc1. Here the chimeric constructs behaved indistinguishably from wild-type Ssc3 and Ssq1 (Figure 2B, lanes 4–9).
Fig. 2. Chimeric Hsp70 constructs containing the ATPase domain of Ssc1 fused with the PBD and C-terminus of Ssc3 or Ssq1 do not interact with Tim44. (A) Scheme of the used Hsp70 constructs. Indicated are the N-terminal pre-sequence (Pre), the ATPase domain, the peptide-binding domain (PBD) and the C-terminal variable domain (var.). The numbers indicate the first amino acid of each Ssc1 subdomain. (B) The wild-type proteins or the chimeric constructs A1P3V3 and A1PQVQ were imported into isolated mitochondria as described in Materials and methods. Mitochondria were lysed in the presence (+) or absence (–) of ATP. Five percent of the lysis fraction was taken as import control (Imp.). Proteins interacting with Tim44 were isolated by co-immunoprecipitation and detected by SDS–PAGE and autoradiography. (C) The constructs A1P3V3 and A1PQVQ do not bind to Tim44 in vivo. A1P3V3 (pASC31) and A1PQVQ (pASC26) were expressed in ssc1-3 mutant cells and co-immunoprecipitation analysis was performed as described in (A). Proteins were detected by western blot using antibodies raised against the C-terminal parts of Ssc3 or Ssq1. Five percent of the mitochondrial lysates were included as control (L). (D) ATPase domains of A1P3V3 and A1PQVQ are functional. ATP-agarose binding assays were performed under non-permissive conditions with mitochondrial lysates expressing the constructs A1P3V3 and A1PQVQ in ssc1-3. Samples were split and bound proteins were eluted by incubation in lysis buffer that contained 1 mM ATP or in SDS sample buffer, respectively. Ten percent lysate (Load) and flowthrough (Flowthr.), and 25% of the final wash (Wash) fractions were included. Proteins were detected by western blot using antibodies recognizing Ssc1 (first panel), the C-terminus of Ssc3 (second panel) or Ssq1 (third panel).
To ascertain that the surprising inability to interact with Tim44 was not a consequence of the in vitro import assay procedure, we repeated the co-immunoprecipitation experiments with mitochondrial extracts from cells that expressed the chimeric proteins in vivo. Immunopre cipitations were performed in the ssc1-3 strain background in order to avoid an interference of endogenous Ssc1. As was observed with the radiolabeled imported versions the chimeric proteins A1P3V3 and A1PQVQ were not found in a complex with Tim44 under the in organello conditions (Figure 2C, lanes 1–6). To confirm that the ATPase domain of Ssc1 in the heterologous protein context was functional, we tested the ability of A1P3V3 and A1PQVQ to bind nucleotides. We performed ATP-agarose binding experiments with mitochondrial extracts of either wild-type Ssc1 or of A1P3V3 and A1PQVQ expressed in ssc1-3. Owing to its mutation in the ATPase domain, Ssc1-3 is not able to interact with nucleotides under non-permissive conditions (von Ahsen et al., 1995). The mitochondrial extracts were incubated with ATP-agarose and bound proteins were eluted either by an incubation in buffer containing ATP or in SDS sample buffer. Similar to the wild-type Ssc1 (Figure 2D, upper panel), both con structs, A1P3V3 (Figure 2D, middle panel) and A1PQVQ (Figure 2D, lower panel), showed efficient and specific binding to ATP (Figure 2D, lane 5) and could be released from the agarose matrix upon addition of excess ATP (Figure 2D, lane 4). In addition, all constructs also showed an efficient and specific interaction with the nucleotide exchange factor Mge1 (data not shown). The binding to ATP and the physical interaction with Mge1 are consistent with a functional ATPase domain in the chimeric protein background of the constructs A1P3V3 and A1PQVQ.
The Ssc1 PBD enables Ssc3 and Ssq1 to interact with Tim44
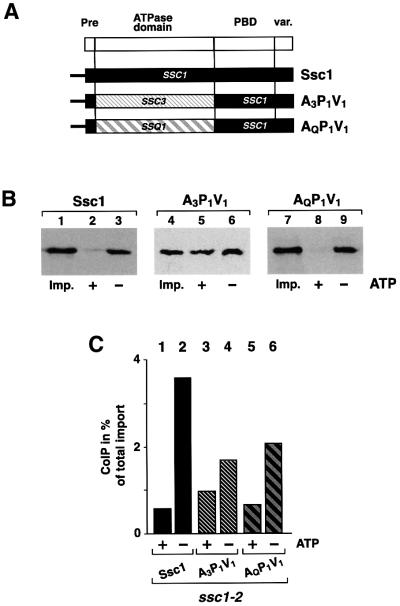
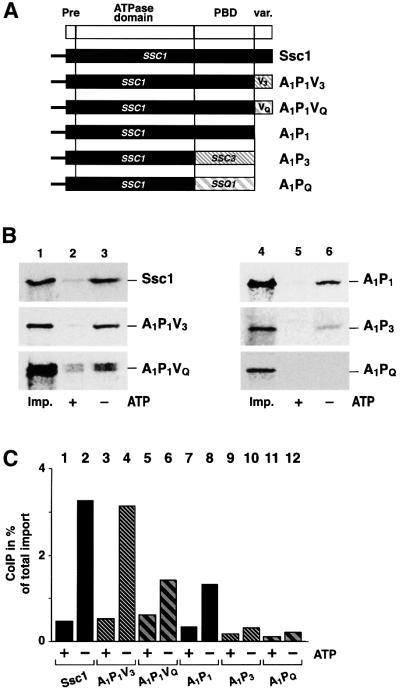
Since the presence of the ATPase domain of Ssc1 does not restore the ability to interact with Tim44, the possibility remained that the Tim44 interaction is governed by the PBD and variable domain. To determine the influence of these domains, we exchanged the ATPase domain of the Tim44-binding protein Ssc1 by the ATPase domains of the non-binding proteins Ssc3 or Ssq1. The resulting proteins A3P1V1 and AQP1V1 both contained the full PBD (P1) and variable domain (V1) of Ssc1, but the ATPase domains of Ssc3 (A3) or Ssq1 (AQ) (Figure 3A). To ensure efficient import in vitro, all constructs also contained the pre-sequence of Ssc1. Both chimeric constructs were efficiently imported into isolated mitochondria dependent on the inner membrane potential and showed correct maturation in the matrix (data not shown). We then analyzed the interaction of A3P1V1 and AQP1V1 with Tim44 by co-immunoprecipitation experiments after import. Surprisingly, both, A3P1V1 and AQP1V1 were detected in a complex with Tim44. The efficiency of complex formation of both A3P1V1 (Figure 3B, lane 6) and AQP1V1 (Figure 3B, lane 9) was similar to that observed with full-length Ssc1 (Figure 3B, lane 3). As in the case of Ssc1, the complex of AQP1V1 and Tim44 dissociated in the presence of ATP (Figure 3B, lane 8), demonstrating that the ATPase domain of Ssq1 is functional and the protein interaction is specific. Compared with wild-type Ssc1 (Figure 3B, lane 2) the complex of A3P1V1 with Tim44 showed a lower sensitivity to ATP (Figure 3B, lane 5), but a significant amount of the bound protein was still released by ATP.
Fig. 3. Chimeric constructs containing the ATPase domains of the different Hsp70 homologs fused to the PBD and C-terminus of Ssc1 can be efficiently co-precipitated with Tim44. (A) Scheme of the chimeric constructs A3P1V1 and AQP1V1. (B) Tim44 interaction of A3P1V1, AQP1V1 and Ssc1. Binding assays were performed after preprotein import by co-immunoprecipitation in the presence (+) or absence (–) of ATP as described. Five percent of total mitochondria was added as import control (Imp.). (C) Binding of A3P1V1 and AQP1V1 to Tim44 is direct and independent of functional Ssc1. Ssc1, A3P1V1 and AQP1V1 were imported into isolated ssc1-2 mutant mitochondria (PK81) under permissive conditions as described in Materials and methods. Mitochondria were heat-treated for 15 min at 37°C to induce the phenotype of Ssc1-2. Proteins bound to Tim44 were isolated by co-immunoprecipitation in the presence (+) or absence (–) of ATP. The efficiency of the co-immunoprecipitation is given as a percentage of the total imported protein.
The possibility remained that the occurrence of the chimeric proteins in the precipitation with Tim44 was due to a formation of heterodimers with endogenous Ssc1. In order to exclude this we performed the binding assays in mitochondria from the mutant strain ssc1-2. Under the chosen conditions Ssc1-2 is not able to form a stable complex with Tim44, while it retains the ability to import denatured preproteins (Gambill et al., 1993; von Ahsen et al., 1995). After import of A3P1V1 or AQP1V1 into ssc1-2 mitochondria both chimeric proteins (Figure 3C, lanes 4 and 6) were found in complex with Tim44. Essentially no difference from the situation in wild-type mitochondria was observed. Again, the interaction of A3P1V1 was less sensitive to ATP (Figure 3C, lane 3), whereas AQP1V1 showed the typical sensitivity of the Hsp70–Tim44 interaction to nucleotides (Figure 3C, lane 5). As a control, wild-type Ssc1 was imported into ssc1-2 mitochondria, showing a specific and efficient interaction with Tim44, as expected (Figure 3C, lanes 1 and 2). Taken together, it is highly unlikely that the observed interaction of the imported proteins with Tim44 did occur via a dimerization with endogenous Ssc1. The co-precipitation experiments therefore indicate clearly that chimeric proteins containing the ATPase domains of Ssc3 and Ssq1 and the C-terminal domains of Ssc1 can interact specifically with Tim44.
The interaction of chimeric Hsp70 constructs with Tim44 does not restore preprotein import
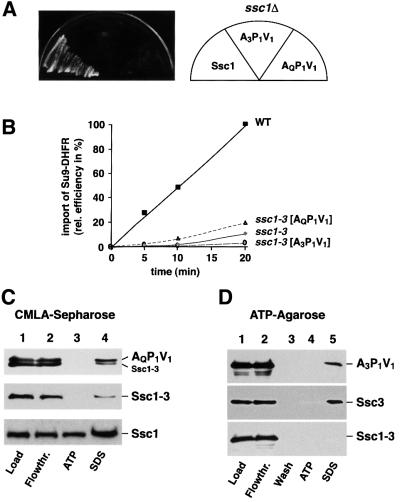
Since the chimeric proteins A3P1V1 and AQP1V1 were able to bind to the preprotein translocase component Tim44, we tested whether the constructs were able to complement the growth defect of ssc1Δ mutants in vivo. The constructs were expressed under control of the SSC1 promotor and growth in the absence of functional Ssc1 was tested as described above. Surprisingly, neither construct was able to restore growth of the ssc1Δ cells (Figure 4A). To analyze the phenotype in more detail, we expressed the chimeric constructs in the ssc1-3 temperature-sensitive mutant. Mitochondria were isolated under permissive conditions and preprotein import was tested after a short pre-incubation at 37°C to inactivate the endogenous Ssc1-3. Interestingly, both chimeric constructs exhibited a pronounced import defect of the reporter preprotein Su9-DHFR (Figure 4B). Mitochondria containing the chimeric constructs behaved virtually indistinguishably from those derived from the parent strain ssc1-3. The significant preprotein translocation defect shown by both chimeric proteins despite a specific interaction with Tim44 is the most likely reason for their inability to replace Ssc1.
Fig. 4. The chimeric constructs A3P1V1 and AQP1V1 are not able to replace Ssc1. (A) A3P1V1 and AQP1V1 cannot rescue the lethal phenotype of a mutant lacking Ssc1. Complementation assays with strain WVY37 expressing Ssc1 (pASC07), A3P1V1 (pASC25) and AQP1V1 (pASC11) were essentially performed as described in Figure 1A. (B) A3P1V1 and AQP1V1 cannot complement the import defect of a ssc1 mutant. Mitochondria isolated from the strains ASYC32 (WT) or ssc1-3 expressing A3P1V1 or AQP1V1 were incubated at 37°C for 15 min to induce the mutant phenotype and the model protein Su9(70)-DHFR was imported for the indicated times. The import reactions were quantified and the amount imported into wild-type after 20 min was set to 100%. (C) Substrate proteins bound to AQP1V1 are not released by addition of ATP. Mitochondria from strain ssc1-3 expressing the construct AQP1V1 (upper panel), a control plasmid (middle panel) and wild-type mitochondria (lower panel) were lysed under non-permissive conditions and binding assays with CMLA–Sepharose were performed as described. Samples were split and proteins were eluted with lysis buffer containing 3 mM ATP or SDS sample buffer. Load (10%), and flowthrough (Flowthr.; 10%) were included. Bound proteins were detected by western blot using antibodies recognizing the C-terminus of Ssc1. (D) Ssc3 and A3P1C1 are not able to release bound ATP. ssc1-3 mitochondria expressing the constructs p1Ssc3 (pASE08; upper panel), A3P1V1 (middle panel) or a control plasmid (lower panel) were lysed under non-permissive conditions and binding assays with ATP-agarose were performed as described. Samples were split and proteins were eluted with lysis buffer containing 1 mM ATP or SDS sample buffer. Load (10%), flowthrough (Flowthr.; 10%) and final wash (25%) were included. Bound proteins were detected by western blot using antibodies recognizing the C-terminus of Ssc3 (upper panel) or Ssc1 (middle and lower panels).
To elucidate which particular functional property of the chimeric proteins is responsible for the defect, we assayed their basic biochemical behavior, substrate binding and nucleotide interaction. As substrate we chose carboxy-methylated α-lactalbumin (CMLA), a typical unfolded model substrate for Hsp70 proteins. We incubated AQP1V1 that was expressed in ssc1-3 with immobilized CMLA and eluted the bound proteins either in the presence of ATP or with SDS sample buffer. Interestingly, the construct AQP1V1 showed significant binding to the substrate CMLA, indicating a functional PBD (Figure 4C, lane 4, upper panel). However, in contrast to wild-type Ssc1 (Figure 4C, lower panel), no release from the immobilized substrate in the presence of ATP was observed (Figure 4C, lane 3, upper panel), although the Tim44-binding assays showed full reactivity of the ATPase domain in AQP1V1 to ATP. Ssc1-3, with its inability to bind ATP, shows a comparable substrate binding behavior to the construct AQP1V1 (Figure 4C, middle panel), indicating that the information of the nucleotide state of the ATPase domain can not be propagated to the PBD and alter the substrate affinity. Since Ssc1-3 does not show any binding activity to ATP (Figure 4D, lanes 4 and 5, lower panel), we were able to analyze the nucleotide interaction of the construct A3P1V1 by binding assays with ATP-agarose. A3P1V1 was able to bind ATP-agarose but, in contrast to Ssc1, was not released in the presence of excess ATP (Figure 4D, lanes 4 and 5, upper panel). The chimeric construct behaved very similar to the full-length Ssc3 (Figure 4D, lanes 4 and 5, middle panel), indicating a possible lower nucleotide exchange rate of the A3 domain. Hence, both chimeric constructs A3P1V1 and AQP1V1 have severe biochemical defects concerning nucleotide reactivity or interdomain communication, resulting in a significant preprotein import defect and eventually in a pronounced growth inhibition in vivo.
Tim44 is not able to bind to the C-terminal part of any mitochondrial Hsp70
The observed interaction of A3P1V1 and AQP1V1 with Tim44 raised the question of whether the PBD derived from Ssc1 rather than the ATPase domain would be able to interact with Tim44. The pre-sequence of Ssc1 was fused to the PBD and the adjacent variable domain of Ssc1, Ssc3 or Ssq1 (Figure 5A). The constructs P1V1, P3V3 and PQVQ were imported into isolated mitochondria. Although the processing of P3V3 was not as efficient as P1V1 and PQVQ (Figure 5B, lane 4), import into mitochondria was specific, as shown by its full protection against protease digestion. When the interaction with Tim44 was analyzed by co-immunoprecipitation, no binding was observed for P1V1, P3V3 and PQVQ (Figure 5B), either in the presence or absence of ATP. Similar results were obtained with the PBD and variable domain of Ssc1 that were expressed in vivo (data not shown). Since the C-terminal domains did not bind to Tim44, we tested the ATPase domains for their binding behavior. We constructed truncated versions of Ssc1, Ssc3 and Ssq1 that contained the pre-sequence of Ssc1 fused to the respective ATPase domains, resulting in the constructs A1, A3 and AQ (Figure 5C). After import into isolated mitochondria, we observed that the ATPase domain of Ssc1 (A1) bound to Tim44 in the absence of ATP, as expected (Figure 5D, lane 3). Interestingly, we found that the ATPase domains of the other two mitochondrial Hsp70s, A3 and AQ, were also able to interact efficiently with Tim44 (Figure 5D, lanes 6 and 9). These results confirmed that the observed binding of the chimeric constructs A3P1V1 and AQP1V1 (Figure 3B) indeed occurred via the ATPase domain. For all three ATPase domains, between 2 and 3.5% of the imported protein was found in complex with Tim44 (Figure 5D, lanes 3, 6 and 9), a similar value as was found for full-length Ssc1 (Figure 3B). The sensitivity of the Tim44 complex of all three proteins to the presence of ATP indicated that the interaction is specific and functional (Figure 5D, lanes 2, 5 and 8). In summary, the ATPase domains of all three mitochondrial Hsp70s were sufficient to bind to Tim44 with characteristics comparable to the full-length Ssc1. The interaction was also observed in the context of the full-length protein as long it contains the PBD and variable domains of Ssc1. In contrast, no interaction was found for the wild-type or chimeric proteins that contained the C-terminal domains of either Ssq1 or Ssc3. We conclude that the C-terminal domains of Ssc3 and Ssq1 exert a negative influence on the binding behavior of their corresponding ATPase domains.
Fig. 5. The PBDs of all three Hsp70s do not interact with Tim44, while the ATPase domains bind specifically to Tim44. (A) Scheme of the constructs P1V1, P3V3 and PQVQ. The pre-sequence of Ssc1 was fused to the PBDs of either Ssc1, Ssc3 or Ssq1. (B) In vitro import and Tim44 co-immunoprecipitation of P1V1, P3V3, PQVQ and Ssc1. After import into mitochondria, binding to Tim44 was analyzed in the presence (+) or absence (–) of ATP. Five percent of mitochondria were added as control (Imp.). (C) Scheme of the ATPase domain constructs A1, A3 and AQ. (D) Co-immunoprecipitation analysis of the single ATPase domains A1, A3 and AQ with Tim44. Immunoprecipitations with Tim44 antibodies in the presence (+) or absence (–) of ATP after import into mitochondria were performed as described. As a control, 2% of the mitochondrial extracts after import were applied (Imp.). Precipitation efficiency is given as a percentage of the total imported protein.
The variable domain of Ssc1, but not of Ssc3 and Ssq1, has a stabilizing influence on the Tim44 interaction
We then tried to dissect the relative influence of the PBD and the variable domain on the Tim44 interaction. We exchanged the variable domain of Ssc1 by the corresponding segments of Ssc3 or Ssq1, resulting in the proteins A1P1V3 and A1P1VQ (Figure 6A). The constructs were imported into isolated mitochondria and processed to the mature protein (data not shown). Both A1P1V3 and A1P1VQ were able to bind to Tim44 in an ATP-sensitive fashion (Figure 6B, lanes 1–3, middle and lower panel). In the construct A1P1V3 the C-terminus of Ssc3 had no influence on Tim44 binding efficiency compared with the full-length Ssc1 (Figure 6C, column 2 and 4). The construct A1P1VQ also showed significant and ATP-sensitive binding to Tim44 (Figure 6B, lanes 2 and 3, lower panel), but its interaction was less efficient than wild-type Ssc1 (Figure 6C, columns 2 and 6). The truncated protein A1P1, which did not contain the variable domain, was able to bind to Tim44 (Figure 6B, lane 6, upper panel), but with decreased efficiency. The interaction of the shortened construct A1PQ was completely abolished, while the construct A1P3 was reduced to almost background levels (Figure 6B, lane 6, middle and lower panels). Interestingly, the binding efficiency of A1P1 was lower than wild type, but almost identical to the value obtained with A1P1VQ. This indicates that the variable domain of Ssc1 could contribute to the overall efficiency of the Tim44 interaction. The variable domain of Ssc3 was able to exercise this stabilizing effect, but the corresponding domain of Ssq1 was not. This effect of the variable domain was qualitatively different from the influence of the PBDs on the Tim44 interaction. In the absence of a stabilizing effect exerted by variable domains the residual binding of the truncated proteins was again abrogated by the negative influence of the PBD derived from Ssc3 or Ssq1.
Fig. 6. The variable C-terminal domains of Ssc1 and Ssc3 have a stabilizing effect on the interaction with Tim44. (A) Scheme of the Hsp70 constructs. The variable C-terminal domain of Ssc1 was replaced by the domains of Ssc3 or Ssq1 (A1P1V3 and A1P1VQ). The constructs A1P3 and A1PQ were obtained by replacing the PBD and C-terminus of Ssc1 with the PBD of Ssc3 or Ssq1. (B) Interaction with Tim44. Import into isolated mitochondria and co-immunoprecipitation with Tim44 antibodies in the presence (+) or absence (–) of ATP was performed as described. As a control, 5% of the mitochondria after the import reactions were applied (Imp.). (C) Relative efficiencies of the interaction of Tim44 with the chimeric constructs. The co-immunoprecipitation experiments shown in (B) were quantified and binding efficiencies are given as a percentage of the total imported preprotein.
A mutation in the PBD of Ssc1 abolishes Tim44 interaction
The negative influence of the PBD of Ssq1 and Ssc3 was reminiscent of the behavior of the mutant Ssc1-2 containing a point mutation in the PBD (substitution of Pro442 by serine) that alters its reactivity towards Tim44. We asked whether the same amino acid exchange would be able to exert a similar negative influence on the binding of the ATPase domains derived from Ssc3 and Ssq1. We fused the PBD of Ssc1-2 with the ATPase domains of Ssc3 and Ssq1, resulting in the constructs A3P1-2V1 and AQP1-2V1 (Figure 7A). After import of the radiolabeled Ssc1, A3P1-2V1, AQP1-2V1 and Ssc1-2, mitochondria were either lysed directly (Figure 7B, lanes 1–3) or incubated at 37°C for 15 min before lysis (Figure 7B, lanes 4–6), and the complex formation with Tim44 was analyzed. Similar to the original mutant protein Ssc1-2, the binding of AQP1-2V1 and A3P1-2V1 to Tim44 was already decreased at permissive temperature (Figure 7B, lanes 2 and 3, second and third panels) compared with Ssc1 (Figure 7B, lane 3, first panel). Compared with wild-type Ssc1, all constructs carrying the ssc1-2 mutation showed <20% binding efficiency. Under non-permissive conditions the complex formation between Tim44 and A3P1-2V1 or AQP1-2V1 was completely abolished (Figure 7B, lanes 5 and 6, second and third panels). In contrast, binding of wild-type Ssc1 to Tim44 was not affected (Figure 7B, lane 6, first panel). The binding behavior of the chimeric constructs containing the PBD derived from Ssc1-2 was identical to the binding defect of the original Ssc1-2 mutant protein (Figure 7B, lanes 1–6, fourth panel). Thus, the destabilizing effect of the ssc1-2 mutation on the interaction with Tim44 can also be observed in the case of the ATPase domains of Ssc3 and Ssq1. The point mutation Pro442→Ser essentially converted the stabilizing effect of the PBD into a destabilizing effect, as was found for the PBDs of Ssc3 and Ssq1.
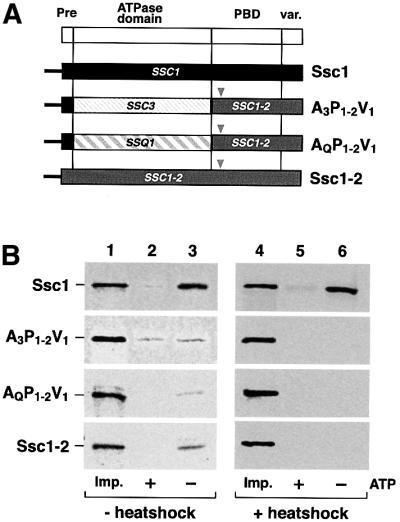
Fig. 7. A point mutation in the PBD of Ssc1 exerts a negative influence on the Tim44 interaction of all three Hsp70s. (A) Scheme of the Hsp70 constructs. The point mutation of the ssc1-2 mutant (substitution of Pro442 by serine, arrowhead) was introduced into the constructs A3P1V1 and AQP1V1. (B) Co-immunoprecipitation of constructs carrying the ssc1-2 point mutation in the PBD. After import, mitochondria were divided into two fractions and either directly lysed and subjected to co-immunoprecipitation analysis (lanes 1–3) or resuspended in import buffer and incubated for 15 min at 37°C to induce the phenotype of the mutation (lanes 4–6). Samples were divided into two fractions and mitochondria were isolated and lysed in the presence (+) or absence (–) of ATP as described. As a control, 5% of the mitochondrial lysates were applied (Imp.).
Discussion
In this study we addressed the question of the functional correlation between the three mitochondrial Hsp70s. We were able to show by in vivo complementation assays that Ssc3 cannot functionally replace the major mitochondrial Hsp70, Ssc1, despite an extremely high sequence conservation. It was previously observed that overproduction of Ssc3 could partially rescue the temperature-sensitive mutant phenotype of the ssc1-3 mutation (Baumann et al., 2000). Hence, some functional overlap between both proteins must exist, but this is not sufficient to rescue the lethal effect of a ssc1 null mutation. As was revealed by database analysis, the ECM10 gene, encoding Ssc3, most probably arose from an ancient gene duplication of the SSC1 locus (Wolfe and Shields, 1997). A block of several genes from chromosome X, containing SSC1, is found repeated on chromosome V, resulting in the ECM10 gene, a situation similar to the genes for two cytosolic Hsp70s, SSB1 and SSB2. The other mitochondrial Hsp70, Ssq1, is also unable to complement an ssc1 deletion strain (Schilke et al., 1996), despite showing typical chaperone properties (Schmidt et al., 2001). All three mitochondrial chaperones have previously been shown to interact with the Hsp70 nucleotide exchange factor Mge1 (Baumann et al., 2000; Schmidt et al., 2001). Thus, the deficiency of both Ssc3 and Ssq1 to bind to the inner membrane translocase component Tim44 is the most likely reason for the inability of Ssc3 and Ssq1 to take over the essential cellular function of the related Ssc1. Efficient binding to Tim44 has been shown to be indispensable for the successful membrane translocation of nuclear-encoded precursor proteins into the mitochondrial matrix, especially under in vivo conditions (Merlin et al., 1999; Voisine et al., 1999; Milisav et al., 2001). This indicates that the differential interaction of the mitochondrial Hsp70s with Tim44 is a very specific property, with functional relevance.
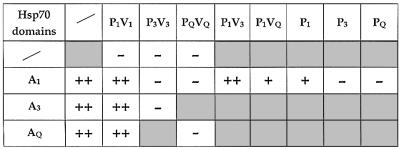
The conserved domain structure of Hsp70 proteins provided the opportunity to investigate the structural determinants of the Tim44 interaction by examining domain exchange constructs for their ability to bind to Tim44. The results of the interaction assays are summarized in Table I. Interestingly, the constructs that contained the ATPase domain derived from Ssc1 (A1P3V3 and A1PQVQ) were not able to interact with Tim44, while chimeric proteins with the C-terminal domains of Ssc1 (A3P1V1 and AQP1V1) showed an efficient interaction. Since the ATPase domains seemed to be functional in the foreign protein context these results seemed contradictory to our previous work in which the ATPase domain of Ssc1 was determined to be the principal Tim44 binding site (Krimmer et al., 2000). Very recently, a report was published that also indicated a possible interaction of Tim44 with the C-terminal domain of Ssc1 (Moro et al., 2002). However, our experiments demonstrate that a specific interaction of the C-terminal domains of the mitochondrial Hsp70s (PBD and variable domain) with Tim44 is very unlikely, since the single domains (P1V1, P3V3 and PQVQ) showed no binding to Tim44 at all. Most interestingly, when we analyzed the ATPase domains separately for their interaction, the ATPase domain of Ssc1 (A1), as well as A3 and AQ, showed specific and efficient binding to Tim44. This interaction was not been noted by Moro et al. (2002), probably due to the use of non-physiological buffer conditions in the binding assays that did not include potassium ions. Potassium ions have been shown to be indispensable for the full enzymatic activity of the Hsp70 ATPase domain (O’Brien and McKay, 1995; von Ahsen et al., 1995).
Table I. Tim44 interaction of mitochondrial Hsp70 constructs.
A co-precipitation of the chimeric proteins with Tim44 that has been caused by a formation of heterodimers (Azem et al., 1997) between endogenous Ssc1 and the imported constructs can be excluded by two arguments. Binding assays using ssc1-2 mutant mitochondria that do not show binding of endogenous Ssc1-2 to Tim44 did not affect the behavior of the chimeric proteins. In addition, only trace amounts of A1 could be precipitated by an antibody predominantly recognizing the C-terminal domains of Ssc1, while almost all of the imported full-length Ssc1 was precipitated (data not shown). Taken together, the results demonstrate that the ATPase domains of all three mitochondrial Hsp70s are principal binding sites for Tim44. The PBDs of Ssc3 and Ssq1 exert a negative influence on their ATPase domains that results in the inability to interact with Tim44 and to perform the translocation function of Ssc1. Interestingly, the interaction of the ATPase domains of Ssc3 or Ssq1 with Tim44 was destabilized by the ssc1-2 mutation in the PBD, a defect that was indistinguishable from the effect of the mutation in the full-length Ssc1-2. This indicates that a slight alteration of the PBD can abolish interaction with Tim44 even when the ATPase domain is unaltered. In contrast to the PBD the variable domain at the C-terminus of Ssc1 exerted a stabilizing effect on the Tim44 interaction, while the variable domains of Ssq1 and Ssc3 did not contribute to the negative effect of the PBD.
The determination of the Ssc1 interaction site with Tim44 has direct consequences for the definition of the molecular mechanism of Ssc1 during mitochondrial preprotein translocation (Jensen and Johnson, 1999; Matouschek et al., 2000). A spatial separation between the substrate-binding site and the interaction site with the membrane anchor Tim44 would be a prerequisite to allow the generation of a leverage on the preprotein in transit by Ssc1. A functional interaction with Tim44 is required but not sufficient for the full translocation function of Ssc1, as described by the active motor model (Voisine et al., 1999; Strub et al., 2000). This fact is reflected especially by the behavior of the chimeric construct AQP1V1, which showed an ATP-dependent interaction with Tim44 and also an efficient binding of substrate proteins. However, in this construct the conformational change in the ATPase domain induced by nucleotides can not be propagated to the PBD, resulting in a strong translocation defect and a lethal phenotype in vivo. Since in contrast to Ssc1 substrate binding of AQP1V1 was not influenced by the presence of ATP, the observed interaction with Tim44 correlates with the behavior of the ATPase domain but not with that of the PBD. Although it has been proposed that Tim44 functions in analogy to members of the conserved DnaJ protein family (Merlin et al., 1999), the interaction properties of the chimeric constructs indicate that the binding between Hsp70 and Tim44 is structurally different from the binding of DnaJ homologs. For the bacterial Hsp70 DnaK it was proposed that DnaJ has two binding sites, one in the ATPase domain and one near the substrate-binding pocket (Gässler et al., 1998). In contrast to Ssc1, neither the single ATPase domain nor the PBD of DnaK can be found in complex with DnaJ (Suh et al., 1999). Not only the complex formation but also the stimulation of ATPase activity and substrate binding of DnaK requires at least the presence of the PBD adjacent to the ATPase domain. A definition of the molecular mechanism of both substrate-ATPase coupling in the case of DnaJ proteins or the negative effect of the PBD on the Tim44 interaction requires an understanding of the structural cooperation between Hsp70 ATPase and the C-terminal domains. The negative influence on the ATPase domains of the mitochondrial Hsp70s could be either caused by a direct steric hindrance of Tim44 interaction by the PBD or indirectly by the alteration of the properties of the ATPase domains. The characterization of cytosolic members of the Hsp70 family established that an important determinant of functional specificity is largely based on specific interactions between the ATPase and the substrate-binding domain (James et al., 1997). The substrate specificity set by the particular molecular environment of the peptide-binding pocket is only of minor influence. The activity of Hsp70 proteins like substrate-binding affinity is mostly determined by the nucleotide-state of the ATPase domains. In the case of mitochondrial Hsp70s a functionally important property, i.e. the interaction with Tim44, is determined by a reverse control of the PBD on the ATPase domain. This destabilizing effect of the PBD on the Tim44 interaction observed in Ssc3 and Ssq1 has significant consequences for the cellular function of structurally very closely related proteins.
Materials and methods
Construction of plasmids and yeast strains
Table II contains brief descriptions of all yeast strains and plasmids used in this study. Details of their construction are listed as Supplementary data (available at The EMBO Journal Online).
Table II. Yeast strains and plasmids.
| Strain or plasmid | Genotype | Source |
|---|---|---|
| WVY37 | trp1-1; ura3-1; leu2-3,112; his3-11, 15; ade2-1; can1-100; met2-Δ1; lys2-Δ2; ssc1ΔClaI::LEU2 [pNZ05] | This study |
| ASYC32 | trp1-1; ura3-1; leu2-3,112; his3-11, 15; ade2-1; can1-100; met2-Δ1; lys2-Δ2; ssc1ΔClaI::LEU2 [pASC07] | This study |
| PK81 | MATα; ade2-101; lys2; ura3-52; trp1; leu2-3,112; ssc1-2::LEU2 | Gambill et al. (1993) |
| PK82 | MATα; his4-713; lys2; ura3-52; trp1; leu2-3 | Gambill et al. (1993) |
| PK83 | MATα; ade2-101; lys2; ura3-52; trp1; leu2-3,112; ssc1-3::LEU2 | Gambill et al. (1993) |
| pASC07 | AmpR CEN6 TRP1 ARSH4 SSC1 | This study |
| pASC08 | KanaR AQ | This study |
| pASC10 | KanaR AQP1V1 | This study |
| pASC11 | AmpR CEN6 TRP1 ARSH4 AQP1V1 | This study |
| pASC24 | AmpR CEN6 TRP1 ARSH4 A1P1V3 | This study |
| pASC25 | AmpR CEN6 TRP1 ARSH4 A3P1V1 | This study |
| pASC26 | AmpR CEN6 TRP1 ARSH4 A1PQVQ | This study |
| pASC27 | AmpR CEN6 TRP1 ARSH4 A1P1VQ | This study |
| pASC31 | AmpR CEN6 TRP1 ARSH4 A1P3V3 | This study |
| pASC33 | AmpR CEN6 TRP1 ARSH4 A1 | This study |
| pASC35 | AmpR CEN6 TRP1 ARSH4 AQ | This study |
| pASC36 | AmpR CEN6 TRP1 ARSH4 A3 | This study |
| pASC37 | AmpR CEN6 TRP1 ARSH4 A3P1-2V1 | This study |
| pASC38 | AmpR CEN6 TRP1 ARSH4 AQP1-2V1 | This study |
| pASC40 | AmpR CEN6 TRP1 ARSH4 P1V1 | This study |
| pASC41 | AmpR CEN6 TRP1 ARSH4 P3V3 | This study |
| pASC42 | AmpR CEN6 TRP1 ARSH4 PQVQ | This study |
| pASE02 | AmpR CEN6 TRP1 ARSH4 SSC3 | This study |
| pASE06 | KanaR p1SSC3 | This study |
| pASE08 | AmpR CEN6 TRP1 ARSH4 p1SSC3 | This study |
| pASE10 | AmpR 2µ TRP1 ARSH4 SSC3 | This study |
| pASQ04 | AmpR SSQ1 | This study |
| pNZ05 | AmpR CEN6 URA3 ARSH4 SSC1 | This study |
| pNZ12 | AmpR SSC3 | This study |
| pNZ46 | AmpR 2µ TRP1 ARSH4 p1SSC3 | This study |
| pRS314 | AmpR CEN6 TRP1 ARSH4 | Sikorski and Hieter (1989) |
| pRS316 | AmpR CEN6 URA3 ARSH4 | Sikorski and Hieter (1989) |
| pRS424 | AmpR 2µ TRP1 ARSH4 | Sikorski and Hieter (1989) |
Complementation assays
Plasmids carrying the gene of interest were transformed into strain WVY37 (ssc1Δ::LEU2 [pNZ5]) containing the wild-type gene of SSC1 on a centromeric plasmid with the URA3 marker gene following standard procedures. Transformants were plated on solid YPD medium containing 5-fluoro-orotic acid (5-FOA), which only allowed growth of cells without the wild-type SSC1 gene.
Protein import into mitochondria and co-immunoprecipitation
Mitochondria were isolated from yeast cells according to published pro cedures (Ryan et al., 2001). Preproteins were synthesized and imported in vitro into yeast mitochondria essentially as described previously (Ryan et al., 2001). Mitochondria were resuspended in lysis buffer [0.3% (v/v) Triton X-100, 200 mM KCl, 30 mM Tris–HCl, pH 7.4, 5% (v/v) glycerol, 0.5 mM phenylmethylsulfonyl fluoride, protease inhibitor mix (Roche) and either 5 mM MgCl2/2 mM ATP or 5 mM EDTA]. After a clarifying spin (20 000 g, 5 min) the supernatant was transferred to Tim44 antibodies that were bound to protein A–Sepharose (Amersham-Pharmacia) in lysis buffer. The sepharose was incubated for 1 h at 4°C and washed three times with lysis buffer. Bound proteins were eluted by addition of electrophoresis sample buffer, applied to SDS–PAGE, and detected and quantified using a storage PhosphorImaging system (Molecular Dynamics).
Co-immunoprecipitation of Tim44–Hsp70 complexes
The co-immunoprecipitation experiments using affinity-purified Tim44 antibodies were performed essentially as described previously (Schmidt et al., 2001).
Binding assays with ATP-agarose and CMLA–Sepharose
The binding assays to ATP-agarose and CMLA–Sepharose were performed as described previously (Schmidt et al., 2001).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr E.A.Craig for the ssc1-2, ssc1-3 and ssc1Δ mutants, N.Zufall for expert technical assistance and Dr A.Chacinska for critically reading the manuscript. This work was supported by the Deutsche Forschungs gemeinschaft (Sonderforschungsbereich 388).
References
- Azem A., Oppliger,W., Lustig,A., Jenö,P., Feifel,B., Schatz,G. and Horst,M. (1997) The mitochondrial hsp70 chaperone system. Effect of adenine nucleotides, peptide substrate and mGrpE on the oligomeric state of mhsp70. J. Biol. Chem., 272, 20901–20906. [DOI] [PubMed] [Google Scholar]
- Baumann F., Milisav,I., Neupert,W. and Herrmann,J.M. (2000) Ecm10, a novel Hsp70 homolog in the mitochondrial matrix of the yeast Saccharomyces cerevisiae. FEBS Lett., 487, 307–312. [DOI] [PubMed] [Google Scholar]
- Bukau B. and Horwich,A.L. (1998) The Hsp70 and Hsp60 chaperone machines. Cell, 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Craig E.A., Weissman,J.S. and Horwich,A.L. (1994) Heat shock proteins and molecular chaperones: mediators of protein conformation and turnover in the cell. Cell, 78, 365–372. [DOI] [PubMed] [Google Scholar]
- Craig E.A., Voisine,C. and Schilke,B. (1999) Mitochondrial iron metabolism in the yeast Saccharomyces cerevisiae. Biol. Chem., 380, 1167–1173. [DOI] [PubMed] [Google Scholar]
- Cyr D.M., Langer,T. and Douglas,M.G. (1994) DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci., 19, 176–181. [DOI] [PubMed] [Google Scholar]
- Demand J., Luders,J. and Höhfeld,J. (1998) The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol. Cell. Biol., 18, 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambill B.D., Voos,W., Kang,P.J., Miao,B., Langer,T., Craig,E.A. and Pfanner,N. (1993) A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol., 123, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gässler C.S., Buchberger,A., Laufen,T., Mayer,M.P., Schröder,H., Valencia,A. and Bukau,B. (1998) Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl Acad. Sci. USA, 95, 15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U. (1996) Molecular chaperones in cellular protein folding. Nature, 381, 571–579. [DOI] [PubMed] [Google Scholar]
- Horton L.E., James,P., Craig,E.A. and Hensold,J.O. (2001) The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J. Biol. Chem., 276, 14426–14433. [DOI] [PubMed] [Google Scholar]
- James P., Pfund,C. and Craig,E.A. (1997) Functional specificity among Hsp70 molecular chaperones. Science, 275, 387–389. [DOI] [PubMed] [Google Scholar]
- Jensen R.E. and Johnson,A.E. (1999) Protein translocation: is hsp70 pulling my chain? Curr. Biol., 9, R779–R782. [DOI] [PubMed] [Google Scholar]
- Knight S.A.B., Sepuri,N.B.V., Pain,D. and Dancis,A. (1998) Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J. Biol. Chem., 273, 18389–18393. [DOI] [PubMed] [Google Scholar]
- Krimmer T., Rassow,J., Kunau,W.H., Voos,W. and Pfanner,N. (2000) Mitochondrial protein import motor: the ATPase domain of matrix Hsp70 is crucial for binding to Tim44, while the peptide binding domain and the carboxy-terminal segment play a stimulatory role. Mol. Cell. Biol., 20, 5879–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.H., Martin,F., Guiard,B., Pfanner,N. and Voos,W. (2001) The mitochondrial Hsp70-dependent import system actively unfolds preproteins and shortens the lag phase of translocation. EMBO J., 20, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M. et al. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics, 147, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz T., Westermann,B., Neupert,W. and Herrmann,J.M. (2001) The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J. Mol. Biol., 307, 815–825. [DOI] [PubMed] [Google Scholar]
- Matouschek A., Pfanner,N. and Voos,W. (2000) Protein unfolding by mitochondria. The Hsp70 import motor. EMBO Rep., 1, 404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.S., Buchberger,A., Reinstein,J. and Bukau,B. (1995) The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol., 249, 126–137. [DOI] [PubMed] [Google Scholar]
- Merlin A., Voos,W., Maarse,A.C., Meijer,M., Pfanner,N. and Rassow,J. (1999) The J-related segment of Tim44 is essential for cell viability: a mutant Tim44 remains in the mitochondrial import site, but inefficiently recruits mtHsp70 and impairs protein translocation. J. Cell Biol., 145, 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milisav I., Moro,F., Neupert,W. and Brunner,M. (2001) Modular structure of the TIM23 preprotein translocase of mitochondria. J. Biol. Chem., 276, 25856–25861. [DOI] [PubMed] [Google Scholar]
- Moro F., Okamoto,K., Donzeau,M., Neupert,W. and Brunner,M. (2002) Mitochondrial protein import: molecular basis of the ATP-dependent Interaction of MtHsp70 with Tim44. J. Biol. Chem., 277, 6874–6880. [DOI] [PubMed] [Google Scholar]
- O’Brien M.C. and McKay,D.B. (1995) How potassium affects the activity of the molecular chaperone Hsc70. I. Potassium is required for optimal ATPase activity. J. Biol. Chem., 270, 2247–2250. [DOI] [PubMed] [Google Scholar]
- Rassow J., Voos,W. and Pfanner,N. (1995) Partner proteins determine multiple functions of hsp 70s. Trends Cell Biol., 5, 207–212. [DOI] [PubMed] [Google Scholar]
- Rassow J., von Ahsen,O., Bömer,U. and Pfanner,N. (1997) Molecular chaperones: towards a characterization of the heat-shock protein 70 family. Trends Cell Biol., 7, 129–133. [DOI] [PubMed] [Google Scholar]
- Ryan M.T., Voos,W. and Pfanner,N. (2001) Assaying protein import into mitochondria. Methods Cell Biol., 65, 189–215. [DOI] [PubMed] [Google Scholar]
- Schilke B., Forster,J., Davis,J., James,P., Walter,W., Laloraya,S., Johnson,J., Miao,B. and Craig,E.A. (1996) The cold sensitivity of a mutant of Saccharomyces cerevisiae lacking a mitochondrial heat shock protein 70 is suppressed by loss of mitochondrial DNA. J. Cell Biol., 134, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S., Strub,A., Röttgers,K., Zufall,N. and Voos,W. (2001) The two mitochondrial heat shock proteins 70, Ssc1 and Ssq1, compete for the cochaperone Mge1. J. Mol. Biol., 313, 13–26. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub A., Lim,J.H., Pfanner,N. and Voos,W. (2000) The mitochondrial protein import motor. Biol. Chem., 381, 943–949. [DOI] [PubMed] [Google Scholar]
- Suh W.C., Lu,C.Z. and Gross,C.A. (1999) Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J. Biol. Chem., 274, 30534–30539. [DOI] [PubMed] [Google Scholar]
- Voisine C., Craig,E.A., Zufall,N., von Ahsen,O., Pfanner,N. and Voos,W. (1999) The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell, 97, 565–574. [DOI] [PubMed] [Google Scholar]
- von Ahsen O., Voos,W., Henninger,H. and Pfanner,N. (1995) The mitochondrial protein import machinery. Role of ATP in dissociation of the Hsp70·Mim44 complex. J. Biol. Chem., 270, 29848–29853. [DOI] [PubMed] [Google Scholar]
- Voos W., von Ahsen,O., Müller,H., Guiard,B., Rassow,J. and Pfanner,N. (1996) Differential requirement for the mitochondrial Hsp70–Tim44 complex in unfolding and translocation of preproteins. EMBO J., 15, 2668–2677. [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H. and Shields,D.C. (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature, 387, 708–713. [DOI] [PubMed] [Google Scholar]