Abstract
During the course of meiotic prophase, intrinsic double-strand breaks (DSBs) must be repaired before the cell can engage in meiotic nuclear division. Here we investigate the mechanism that controls the meiotic progression in Schizosaccharomyces pombe that have accumulated excess meiotic DSBs. A meiotic recombination-defective mutant, meu13Δ, shows a delay in meiotic progression. This delay is dependent on rec12+, namely on DSB formation. Pulsed-field gel electrophoresis analysis revealed that meiotic DSB repair in meu13Δ was retarded. We also found that the delay in entering nuclear division was dependent on the checkpoint rad+, cds1+ and mek1+ (the meiotic paralog of Cds1/Chk2). This implies that these genes are involved in a checkpoint that provides time to repair DSBs. Consistently, the induction of an excess of extrinsic DSBs by ionizing radiation delayed meiotic progression in a rad17+-dependent manner. dmc1Δ also shows meiotic delay, however, this delay is independent of rec12+ and checkpoint rad+. We propose that checkpoint monitoring of the status of meiotic DSB repair exists in fission yeast and that defects other than DSB accumulation can cause delays in meiotic progression.
Keywords: DNA damage checkpoint/double-strand break/homologous recombination/meiosis/pachytene checkpoint
Introduction
Meiosis is a special type of cell division that produces haploid gametes from diploid parental cells. During meiosis, homologous recombination occurs at a high frequency and can generate new allelic combinations in the resulting gametes, thereby increasing the genetic diversity of the offspring and promoting the survival of the species during critical environmental changes. Homologous recombination is also essential for ensuring that chromosome segregation occurs correctly during meiosis. In the absence of recombination, homologs mis-segregate and the resulting anneuploid gametes produce defective or inviable progeny. Thus, homologous recombination is vital for the generation of viable gametes (Kleckner, 1996; Roeder, 1997).
When there are defects in critical cell cycle events, checkpoint mechanisms delay the cell cycle progression so as to prevent aneuploidy and cell lethality (Hartwell and Weinert, 1989). This arrest or delay in cell cycle progression occurs in eukaryotic cells when they are exposed to DNA-damaging agents or when DNA synthesis is blocked. The checkpoints require the function of the checkpoint rad+ genes (rad1+, rad3+, rad9+, rad17+, rad26+ and hus1+), crb2+/rhp9+, cds1+ and chk1+ (Caspari and Carr, 1999) and also inhibit Tyr15 dephosphorylation of Cdc2 (Rhind et al., 1997; Rhind and Russell, 1998). It has been found that a pre-meiotic replication checkpoint also operates in both budding and fission yeasts (Stuart and Wittenberg, 1998; Murakami and Nurse, 1999).
There are several meiotic mutants such as hop2Δ (Saccharomyces cerevisiae homolog of meu13), dmc1Δ and zip1Δ that exhibit an arrest at the pachytene stage of meiotic prophase in S.cerevisiae (Roeder and Bailis, 2000). Mammalian germ cells exhibit meiotic arrest and apoptosis in response to the defects of recombination and/or synapsis, suggesting the conservation throughout evolution of a checkpoint that prevents entry into meiosis I (Roeder and Bailis, 2000). Thus, to ensure the generation of viable meiotic products, the progression through meiosis must be tightly regulated by mechanisms that monitor the status of recombination repair, synaptonemal complex (SC) formation and proper chromosome segregation.
In S.cerevisiae, several proteins (Mec1, Rad24 and Rad17) involved in the DNA damage checkpoint also participate in the pachytene checkpoint (Lydall et al., 1996). In mammals, Atm, Atr, Rad1 and Chk1 proteins, which are involved in the damage checkpoint, also localize to the meiotic chromosomal cores and are thought to play important roles in meiotic recombination, meiotic arrest and apoptosis (Roeder and Bailis, 2000). In S.cerevisiae and probably also in mammals, all mutants that arrest at the pachytene stage exhibit defects in SC formation as well as in recombination. For this reason, it is not yet clear whether the defects in recombination or synapsis are responsible for the arrest in cell cycle progression that is mediated by the pachytene checkpoint. As Schizo saccharomyces pombe does not form an SC, meiotic recombination in S.pombe is a good model for studying the meiotic checkpoint during prophase.
In fission yeast, no meiotic recombination-deficient mutants show meiotic arrest and the precise meiotic progression in meiotic recombination-deficient mutants has not been fully investigated so far. To examine these aspects, we previously isolated and analyzed meu13+ (Nabeshima et al., 2001), a homolog to budding yeast HOP2, and dmc1+ (Fukushima et al., 2000), a meiotic RecA homolog that is conserved among a variety of organisms. Here, we report that meu13Δ cells show a delay in entering into meiosis I. The checkpoint Rad proteins (Rad17, Rad3, Rad1 and Rad9), Cds1 and the meiotic-specific Cds1 paralog, Mek1, are required for this control. These regulatory genes, which are found to be required for normal levels of recombination, delay the onset into meiosis I in meu13Δ because they recognize the presence of unrepaired double-strand breaks (DSBs). Introducing excess DSBs in normal cells by γ-ray irradiation similarly delayed the onset of meiosis. Furthermore, radΔ meu13Δ, cds1Δ meu13Δ and mek1Δ meu13Δ double mutants exhibited decreased recombination frequencies and reduced spore viability, indicating that rad+, cds1+ and mek1+ act to delay the cell’s progression into meiosis I until the DSBs are repaired. Thus, we propose that a meiotic recombination checkpoint exists in S.pombe to ensure the production of viable spores and recombination at the proper frequencies.
Results
Meiotic progression is delayed in the meu13Δ and dmc1Δ mutants
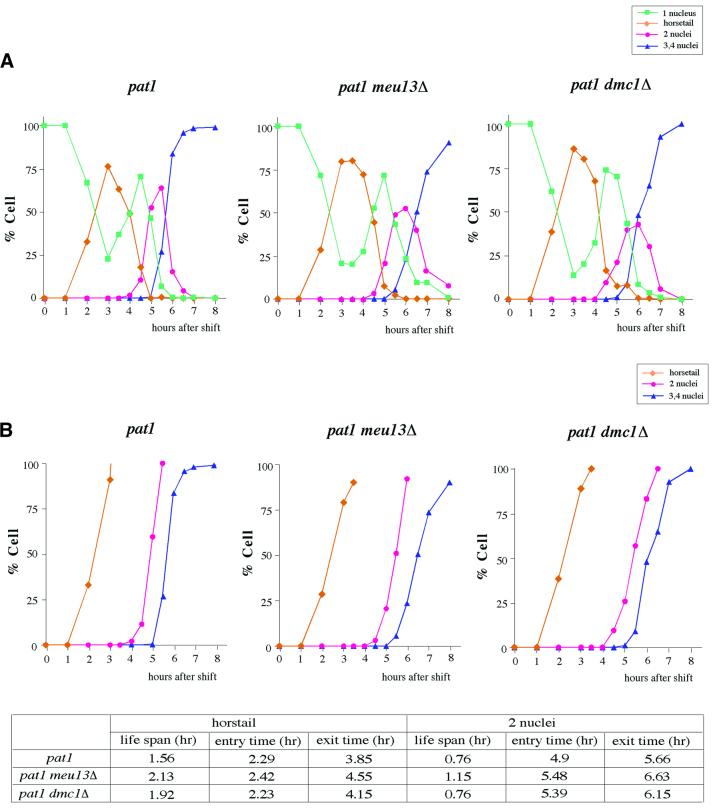
In budding yeast, nematodes (Caenorhabditis elegans), flies (Drosophila melanogaster) and mice, some of the mutant cells that are defective in recombination and/or synapsis will arrest at the pachytene stage (Roeder and Bailis, 2000). In fission yeast, however, so far it has not been determined whether a failure to complete meiotic recombination will affect the meiotic progression. To investigate whether meiotic recombination-deficient mutants show any defect in meiotic progression, we checked the meiotic progression of two meiotic recombination-deficient mutants, meu13Δ and dmc1Δ. meu13+ and dmc1+ have both been identified as meiosis-specific genes (Watanabe et al., 2001). meu13+ is a budding yeast HOP2 homolog and is required for proper homologous pairing and recombination (Nabeshima et al., 2001). dmc1+, a meiotic RecA homolog conserved among a variety of organisms, also participates in fission yeast meiotic recombination (Fukushima et al., 2000). To synchronize meiosis effectively, we used the pat1-114 temperature-sensitive strain, which enters meiosis in a highly synchronous manner when it is shifted to its restrictive temperature (Iino and Yamamoto, 1985). Homozygous diploid pat1, pat1 meu13Δ and pat1 dmc1Δ cells were arrested at the G1 stage by nitrogen starvation and then shifted to the restrictive temperature to induce synchronous meiosis (Figure 1A). Meiotic recombination occurs at the horsetail stage (orange line in Figure 1A), where the nucleus repeatedly moves backwards and forwards several times. The time at which the horsetail stage developed was similar for all three strains (∼2 h after temperature shift, peaking at ∼3 h). Thus, neither meu13Δ nor dmc1Δ appear to affect the entry into meiotic prophase in pat1 cells. However, in the pat1 meu13Δ and pat1 dmc1Δ double mutants, meiosis I peaked 6 h after the temperature shift, 30 min later than for pat1. Cumulative curves (Hunter and Kleckner, 2001) clearly showed that in pat1 meu13Δ and pat1 dmc1Δ mutants, entry into the horsetail stage was similar to that in pat1, but both exit from the horsetail stage and entry into meiotic division were delayed (Figure 1B). This delay in meiosis I initiation was observed in more than four independent experiments performed identically.
Fig. 1. Meiotic recombination-deficient mutants, meu13Δ and dmc1Δ, are delayed in entering meiosis I. (A) Homozygous diploid pat1 cells were cultured to mid log phase, transferred to EMM-N medium for 16 h at 25°C and then shifted to 34°C to inactivate Pat1 and synchronize meiosis. Progression of meiosis was monitored by DAPI staining of samples that were collected every 30 min or 1 h after temperature shift. At least 150 cells were scored by fluorescence microscopy for each time point. (B) Cumulative curves and the time indicating the life span, entry into and exit from each DNA stage of pat1, pat1 meu13Δ and pat1 dmc1Δ. Cumulative curves are expressed as a percentage of maximum values against time after temperature shift (Hunter and Kleckner, 2001). (C) Meiotic delay of meu13Δ is dependent on the rec12+ gene and checkpoint rad+ genes but this is not the case for dmc1Δ. Cells were induced to enter meiosis and the progression of meiosis was monitored by DAPI staining. Shown are the average ratios of cells in meiosis I at 5 and 6 h in three or four independent experiments. (D) γ-ray-irradiated cells are delayed in entering meiosis I and this delay is also dependent on rad17+. Cells were induced to enter meiosis and irradiated with γ-rays 3 h after temperature shift, when the cells were in the horsetail period. Shown are the average ratios of cells in meiosis I at 5.5 h in three independent experiments. Strains: pat1 (JZ670), pat1 meu13Δ (KN8), pat1 dmc1Δ (MS276-1), pat1 rec12Δ (MS123-5), pat1 rec12Δ meu13Δ (MS110-2), pat1 rec12Δ dmc1Δ (MS189-1), pat1 rad17Δ (MS101-4), pat1 rad17Δ meu13Δ (MS107-1), pat1 rad17Δ dmc1Δ (MS190-7), pat1 cds1Δ meu13Δ (MS231-1) and pat1 mek1Δ meu13Δ (MS217-1).
The delay in meiotic progression of meu13Δ but not dmc1Δ is dependent on Rec12/Spo11
The observations above suggest that the deficiency in meiotic homologous recombination in the meu13Δ and dmc1Δ mutants is responsible for the delay in meiotic progression. If so, this delay should be dependent on the initiation of recombination. It is known that the formation of DSBs is required for the initiation of meiotic recombination and that rec12+ of S.pombe, which encodes a protein homologous to Spo11 of S.cerevisiae, is essential for the generation of DSBs (Cervantes et al., 2000). Thus, we examined whether the delay in the meiotic progression of the meu13Δ and dmc1Δ mutants is dependent on rec12+. The average percentages of cells in meiosis I at 5 and 6 h are shown in Figure 1C. Deletion of rec12+ eliminated the delay that was observed in pat1 meu13Δ cells and allowed these cells to enter into meiosis I with the same kinetics as seen in pat1. Thus, the delay in meiotic progression observed in meu13Δ is dependent on rec12+, namely on the initiation of recombination. This suggests that recombination intermediates in meu13Δ trigger a meiotic delay.
Interestingly, the deletion of rec12+ did not eliminate the dmc1Δ delay but caused more delay than pat1 dmc1Δ (Figure 1C), suggesting that the meiotic delay in these cells is independent of the initiation of recombination and that it may be triggered by a signal other than recombination intermediates (see Discussion).
rad17+, rad3+, rad1+ and rad9+ genes participate in delaying meiotic progression in meu13Δ but not in dmc1Δ
Checkpoint rad+ genes (rad1+, rad3+, rad9+, rad17+, rad26+ and hus1+), crb2+/rhp9+, cds1+ and chk1+ act in the DNA damage and/or DNA replication checkpoint controls (Caspari and Carr, 1999). Unrepaired DNA or a block in replication are sensed by these genes, which then act to stop or delay the cell cycle progression by transmitting the checkpoint signals to Cdc2 kinase, a major regulator of the G2/M transition. In S.cerevisiae, it has been reported that some of the homologs of fission yeast checkpoint genes also control meiotic progression (Lydall et al., 1996). Thus, we investigated if these fission yeast checkpoint genes could be participating in the delay of meiotic progression observed in the meu13+ and dmc1+ mutants. As shown in Figure 1C, the deletion of rad17+ abolished the delay observed in pat1 meu13Δ cells and allowed these cells to enter meiosis I with a timing similar to that observed in pat1. Deletion of rad1+, rad3+ and rad9+ also eliminated the delay of pat1 meu13Δ (data not shown). Thus, rad17+, rad3+, rad1+ and rad9+ genes participate in the delay of meiotic progression in meu13Δ. In the damage checkpoint, checkpoint rad+ genes are thought to monitor DNA damage such as DSBs and respond to this damage by delaying cell cycle progression (Caspari and Carr, 1999). Thus, these data, together with the observation in the previous section that lack of the ability to produce DSBs normalizes meu13Δ meiosis, suggest that meu13Δ cells may accumulate DSBs that are monitored by these checkpoint genes.
Saccharomyces cerevisiae dmc1Δ cells are known to accumulate DSBs during recombination intermediate stages (Bishop et al., 1992). Given that this mutant, like the dmc1Δ mutant of S.pombe (Fukushima et al., 2000), also exhibits a decreased frequency of meiotic recombination, we expected that S.pombe dmc1Δ cells would also accumulate DSBs and that these might trigger the meiotic delay in dmc1Δ cells. However, the deletion of rad17+, rad1+, rad3+ and rad9+ did not eliminate the delay in meiotic progression of the pat1 dmc1Δ mutant but caused a much greater delay than pat1 dmc1Δ (Figure 1C and data not shown). This again indicates that the regulation of the meiotic delay observed in dmc1Δ is different from that in meu13Δ cells.
rad3+ and rad17+ increase the viability of the meiotic product and the recombination frequency in recombination mutants
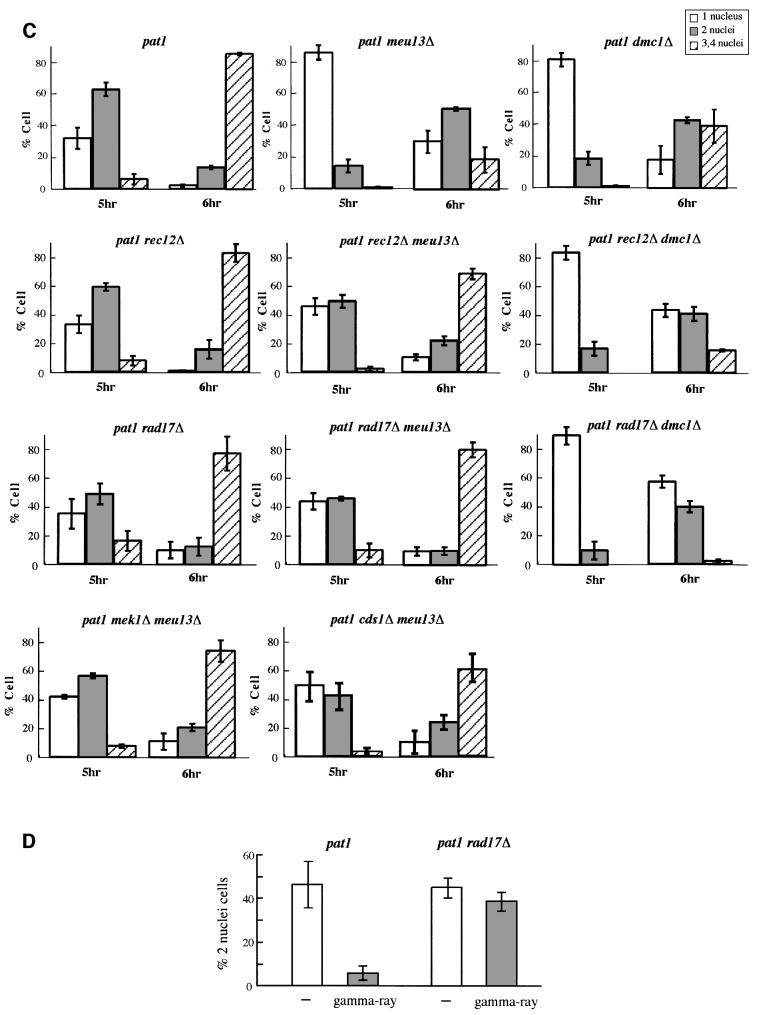
Because checkpoint rad+ genes participate in controlling meiotic progression in meu13Δ and dmc1Δ, checkpoint rad+ deletion in meu13Δ and dmc1Δ would affect their production of viable meiotic products. As shown in Figure 2A, the spore viability of rad3Δ and rad17Δ was similar to that of wild-type cells, whereas rad3Δ meu13Δ, rad17Δ meu13Δ and rad17Δ dmc1Δ double mutants showed a reduced spore viability when compared with their corresponding single mutant. The number of spores with more than four 4′,6-diamidino-2-phenylindole (DAPI)-stained masses was greater in the rad17Δ meu13Δ and rad17Δ dmc1Δ double mutants than in the single mutants (Figure 2C), suggesting more frequent fragmentation in double mutants. Thus, spore viabilities were reduced by the disruption of rad3+ and rad17+ in meiotic recombination mutants, although the mechanism operating in meu13Δ is different from that in dmc1Δ.
Fig. 2. Spore viability and gene conversion frequencies of meiosis-defective mutants. The spore viabilities (A) and allelic gene conversion frequencies (B) between ade6-469 and ade6-M26 of double mutants (rad3Δ meu13Δ, rad17Δ meu13Δ, cds1Δ meu13Δ, mek1Δ meu13Δ and rad17Δ dmc1Δ) and single mutants were measured. The various strains were generated by crossing wild-type (MS111-W1 × MS105-1B), rad3Δ (MS126-4 × MS162-1), rad17Δ (MS111-1 × MS105-22D), cds1Δ (MS233-4 × MS245-3), mek1Δ (MS203-2 × MS202-11), meu13Δ (MS111-m13 × MS105–22C), dmc1Δ (MS133-1 × MS114-3), rad3Δ meu13Δ (MS128-15 × MS163-10), rad17Δ meu13Δ (MS111-6 × MS105-25C), cds1Δ meu13Δ (MS233-3 × MS245-7), mek1Δ meu13Δ (MS203-2 × MS202-10) and rad17Δ dmc1Δ (MS133-1 × MS114-3). All data presented above are the average of three independent experiments with standard errors. (C) The phenotype of wild-type, meu13Δ, rad17Δ, rad17Δ meu13Δ, dmc1Δ and rad17Δ dmc1Δ cells. Typical microscopic images of the asci stained by DAPI are shown in the right panels. The scale bar represents 5 µm.
Next we examined whether the disruption of rad3+ and rad17+ decreases the frequency of recombination in the meu13Δ and dmc1Δ mutants by measuring the frequency of allelic gene conversion between two different mutant alleles of ade6, namely ade6-M26 and ade6-469, in these double mutants (Figure 2B). We found that the rad3Δ and rad17Δ single mutants already had a reduced recombination frequency, as shown by a decrease in the percentage of Ade+ recombinant spores relative to the wild-type strain. Thus, the rad3+ and rad17+ genes are required to maintain a normal level of meiotic recombination even in normal cells. The double mutants also exhibited a decrease in recombination. For example, recombination in rad3Δ meu13Δ and rad17Δ meu13Δ was decreased to 44 and 34% of that of meu13Δ. The rad17Δ dmc1Δ mutant also showed a reduced recombination frequency as compared with the single mutants. Thus, Rad3 and Rad17 are needed in the meu13Δ and dmc1Δ mutants to allow sufficient levels of recombination to occur so that viable spores can be produced.
cds1+ and mek1+ participate in maintaining spore viability and recombination frequency and delaying meiotic progression in the meu13Δ mutant
The MEK1 gene of S.cerevisiae encodes a meiosis-specific protein kinase that participates in the pachytene checkpoint (Rockmill and Roeder, 1991). We isolated a fission yeast homolog (mek1+) of MEK1 by computer-assisted homologous searching in GenBank. The mek1+ gene was then isolated by PCR using appropriate primers and the genomic DNA of S.pombe as a substrate (T.Tougan, M.Shimada and H.Nojima, unpublished data). We found that the Ade+ frequencies of cds1Δ and mek1Δ were only 48 and 33% that of the wild-type strain, respectively, suggesting that Cds1 and the fission yeast Mek1 homolog are also required to maintain normal levels of meiotic recombination in wild-type cells (Figure 2B). However, the viabilities of cds1Δ and mek1Δ spores were 91 and 94% that of the wild-type strain, respectively. Thus, spore viability was not affected by these mutations as by the rad3Δ and rad17Δ mutations (Figure 2A). Both cds1Δ meu13Δ and mek1Δ meu13Δ double mutants, however, exhibited reduced levels of spore viability and meiotic recombination. Thus, when there is already a partial defect in meiotic recombination, the cds1+ and mek1+ genes are also required to generate enough meiotic recombination to ensure spore viability (Figure 2A and B). Furthermore, deletion of cds1+ and mek1+genes also eliminated the delay of pat1 meu13Δ cells, indicating that both cds1+ and mek1+ genes have a function in controlling meiotic progression in pat1 meu13Δ cells like checkpoint rad+ genes.
Meiotic DSB repair is delayed in meu13Δ
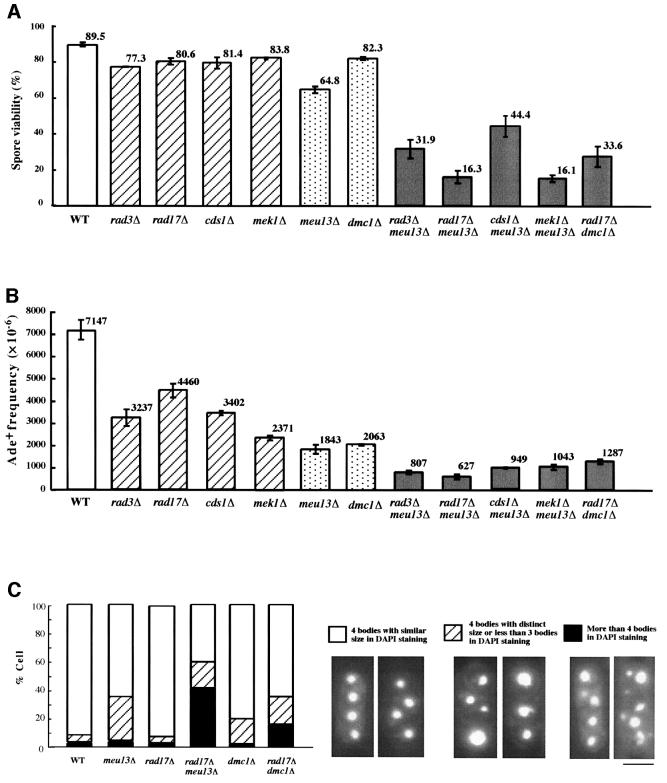
Considering that the meiotic delay of meu13Δ is dependent on rec12+ as well as on checkpoint rad+ genes, which monitor such damage, we surmised that the meiotic delay is triggered by the accumulation of recombination intermediates such as DSBs. To test this possibility directly, we examined the formation of DSBs during meiosis in the meu13Δ strain by pulsed-field gel electrophoresis (PFGE). In fission yeast, meiotic DSBs can be detected as smeared bands by PFGE because they produce broken chromosomes with variable length (Cervantes et al., 2000). In the pat1 meu13Δ and pat1 dmc1Δ double mutants, as well as in the pat1 single mutant strain, smeared bands appeared 3 h after the temperature shift (Figure 3). This result is consistent with that shown in Figure 1A, which suggests that the double mutant cells enter the meiotic prophase at approximately the same time after shifting the temperature as pat1 cells. Smeared bands were no longer observed in pat1 cells 4.5 h after the temperature shift but they continued to be observed in pat1 meu13Δ cells. Thus, meiotic DSB repair is delayed in pat1 meu13Δ.
Fig. 3. DSB repair is retarded in meu13Δ. Cells were induced to enter meiosis as shown in Figure 2. Samples were taken after the temperature shift at the indicated time points, analyzed by PFGE and stained with ethidium bromide to detect DNA. Chr I, Chr II and Chr III indicate the position of chromosomes 1, 2 and 3, respectively. The smear bands represent the DSBs that appear during meiosis.
Unexpectedly, the smeared bands in pat1 dmc1Δ cells were diminished 4.5 h after the temperature shift, as in the pat1 strain (Figure 3). This suggests that defects other than a delay in DSB repair may be triggering the delay in meiosis in dmc1Δ, which is consistent with the fact that the checkpoint rad+ genes also do not appear to be involved.
pat1 rad17Δ meu13Δ cells had smeared bands at the time point when they would be entering meiosis I (Figures 3 and 1), suggesting that pat1 rad17Δ meu13Δ cells probably enter meiosis I without completing DSB repair, which would cause chromosome fragmentation. In line with this is that this strain produced abnormal spores with an irregular number of DAPI-stained bodies, as shown in Figure 2C. To assess whether the smeared bands observed here are specific to meiotic recombination, we examined the effect of disrupting rec12+ on the presence of smeared bands, as rec12+ is responsible for introducing DSBs during meiosis. The rec12Δ meu13Δ double mutant never shows smeared bands at any time (Figure 3). These results strongly support the idea that the broken chromosomes, which were observed in rad17Δ meu13Δ meiosis I, were caused by meiotic DSBs, and that rad17+ acts to delay meu13Δ from entering meiosis I to allow the DSBs to be repaired.
γ-ray irradiation of wild-type cells during the horsetail period delays their entry into meiosis I in a rad17+-dependent manner
If the delay in entering meiosis I in meu13Δ cells is caused by the persistent presence of DSBs, one would expect that an increased number of DSBs formed during the meiotic prophase would cause even wild-type cells to experience a delay in entering meiosis I. To examine this possibility, we irradiated meiotic pat1 cells during the horsetail period with γ-rays (3 h after temperature shift) and determined when these cells entered meiosis I. As shown in Figure 1D, the onset of meiosis I was delayed in irradiated pat1 cells relative to non-irradiated cells. However, when rad17+ was deleted from pat1 cells, γ-ray irradiation no longer affected the timing with which meiosis I was initiated. Thus, DSBs appear to be monitored by a checkpoint pathway involving rad17+, and their presence delays the cell’s entry into meiosis I.
Phosphorylation of Tyr15 in Cdc2 is maintained for a longer period in pat1 meu13Δ than in pat1
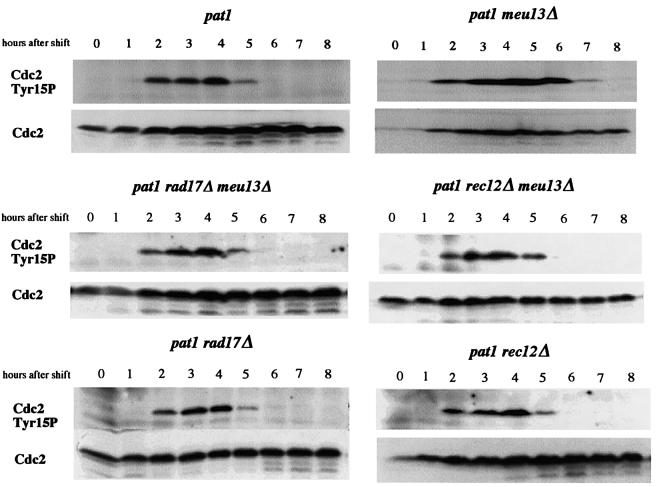
Phosphorylation of Tyr15 in Cdc2 plays a key role in blocking the onset of mitosis when DNA replication is inhibited or DNA is damaged (Rhind et al., 1997; Rhind and Russell, 1998). To investigate whether this phosphorylation participates in delaying meiosis in meu13Δ cells, we monitored Cdc2 Tyr15 phosphorylation in pat1 and pat1 meu13Δ cells during meiosis and sporulation (Figure 4). The level of the phosphorylated form of Cdc2 increased during the pre-meiotic S phase and decreased during meiotic division in pat1 cells. In pat1 meu13Δ cells, however, Cdc2 was phosphorylated for 1 h longer than in pat1 cells, consistent with the length of the delay in meiosis I onset in pat1 meu13Δ cells. Furthermore, we found that this extended period of phosphorylation was not present in pat1 rad17Δ meu13Δ and pat1 rec12Δ meu13Δ cells, which do not experience a delay in meiosis, as shown in Figure 1C. Thus, the regulatory pathway delaying the onset of meiosis I in meu13Δ employs the phosphorylation of Tyr15 in Cdc2.
Fig. 4. Phosphorylation of the Tyr15 in Cdc2 is extended in pat1 meu13Δ. pat1 (JZ670), pat1 meu13Δ (KN8), pat1 rec12Δ meu13Δ (MS110–2), pat1 rad17Δ meu13Δ (MS107-1), pat1 rec12Δ (MS123-5) and pat1 rad17Δ (MS101-4) cells were induced to enter meiosis as shown in Figure 1. Samples were taken after the temperature shift at the indicated time points and western blot analysis was performed to detect the amount of Cdc2 and phosphorylated Cdc2.
Three protein kinases, Cds1, Chk1 and Mek1, are expressed and phosphorylated during meiosis
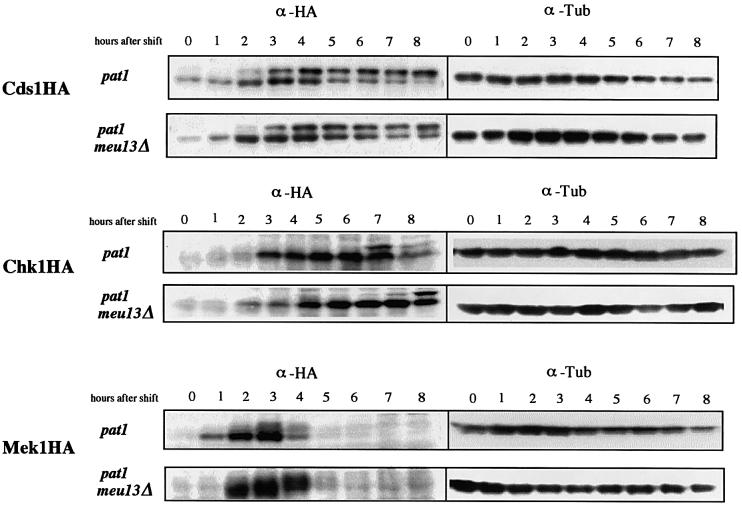
Cds1 and Chk1 play important roles in checkpoint controls and are phosphorylated when the checkpoints are activated. Thus, we examined the expression of Cds1, Chk1 and Mek1 (the meiotic paralog of Cds1) during meiosis and sporulation in pat1 and pat1 meu13Δ cells (Figure 5). In both pat1 and pat1 meu13Δ cells, Cds1 was phosphorylated from 2 h after the temperature shift, which continued to the end of sporulation. In both cell types, Chk1 was not phosphorylated during early meiosis but was phosphorylated slightly in the later stage. However, Mek1 was expressed only during pre-meiotic replication and meiotic recombination and showed a mobility shift during these periods, suggesting that phosphorylation occurred. Furthermore, the extent of Mek1 mobility shift in pat1 meu13Δ is greater than that in pat1, whereas the expression patterns of Cds1 and Chk1 are similar in these cells at the time point just before the onset of meiosis I (Figure 5; 4 h). These results suggest that Cds1, Chk1 and Mek1 have different functions during meiosis.
Fig. 5. Three protein kinases, Cds1, Chk1 and Mek1, are expressed and phosphorylated during meiosis and sporulation in fission yeast. pat1 Cds1HA (MS197-6), pat1 meu13Δ Cds1HA (MS198-1), pat1 Chk1HA (MS108-1), pat1 meu13Δ Chk1HA (MS196-2), pat1 Mek1HA (MS199-4) and pat1 meu13Δ Mek1HA (MS200-5) cells were induced to enter meiosis as shown in Figure 1. Samples were taken after the temperature shift at the indicated time points and proteins were prepared and detected with anti-HA and anti α-tubulin antibodies.
Discussion
The meiotic recombination checkpoint exists in fission yeast
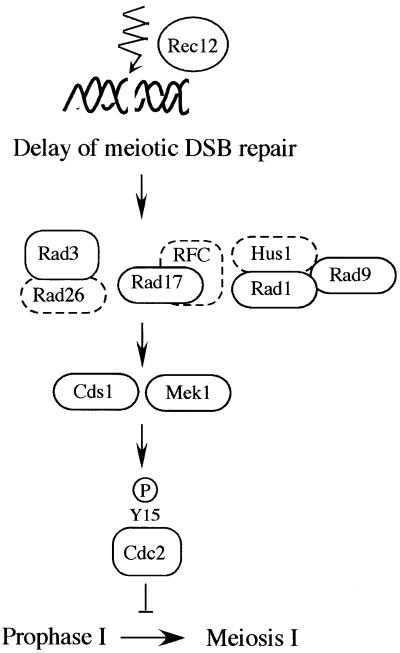
In this study, we found that meu13Δ cells harboring a defect in meiotic recombination had a delay in the entry into meiosis I. The delay was dependent on rec12+/SPO11, which is required for initiation of meiotic recombination, rad17+, rad1+, rad3+ and rad9+, which are required for damage and replication checkpoints in mitosis, cds1+, and mek1+, a meiosis-specific cds1+ paralog. Furthermore, we found that there is a delay in meiotic DSB repair in meu13Δ. From these results, we surmised that a delay, elicited by DNA damage and replication checkpoint genes, was required in meu13Δ cells to repair meiotic DSBs before their entry into meiosis I. Consistent with this hypothesis, deletion of rad17+ allowed meu13Δ cells to progress into meiosis I without completing DSB repair. These results strongly suggest that, when meiotic DSB repair is delayed in S.pombe, DSBs are detected by DNA damage and replication checkpoint genes, which retards meiotic progression. To test this hypothesis directly, we artificially introduced an unusually large amount of DSBs into meiotic cells. This triggered a delay in meiotic progression, which was dependent on rad17+ (Figure 1D). Taken together, we propose that a meiotic recombination checkpoint exists in S.pombe to monitor meiotic DSB repair and regulate entry into meiosis I (Figure 6). This regulation seems to be responsible for the maintenance of the optimal level of spore viability and meiotic recombination frequency in cells that experience a delay in DSB repair (Figure 2).
Fig. 6. A model of the meiotic recombination checkpoint in fission yeast. When repair of meiotic DSBs is retarded, checkpoint rad+, cds1+ and mek1+ genes delay cells to enter into meiosis I through phosphorylation of Cdc2 Tyr15 to provide enough time to repair DSBs.
In budding yeast, S.cerevisiae, it has been reported that the pachytene checkpoint operates to arrest cells at the pachytene stage in response to a defect in recombination and/or synapsis. It has not been shown clearly, however, whether meiotic delay is triggered by a defect in DSB repair or synaptonemal complex formation, or by defects in both of them together (Roeder and Bailis, 2000). Of note is that in striking contrast to S.cerevisiae, S.pombe does not form any SC (Bahler et al., 1993). Considering that many genes required for the meiotic recombination checkpoint in S.pombe have homologs that are required for the pachytene checkpoint in S.cerevisiae, and the strong analogy between the observations in these two yeasts, our results strongly suggest that the checkpoint actually monitors defects in meiotic recombination.
The role of the checkpoint rad+ genes, cds1+ and mek1+ in meiotic recombination
We report here that S.pombe rad17+, rad3+, cds1+ and mek1+ genes are required for maintaining a normal level of meiotic recombination, although they are not required for the maintenance of viable spores (Figure 2 and Murakami and Nurse, 1999). This is consistent with the notion that many DNA damage checkpoint genes may play a direct role in meiotic recombination in other species. In S.cerevisiae, genes homologous to S.pombe rad17+, rad3+ and mek1+ are required for meiotic recombination (Kato and Ogawa, 1994; Lydall et al., 1996; Xu et al., 1997). Mammalian Rad1, ATR, ATM and Chk1 (S.pombe rad1+, rad3+, tel1+ and chk1+ homologs, respectively) have been found to localize to synapsed and/or unsynapsed meiotic chromosomes (Roeder and Bailis, 2000) and are suggested to play a direct role in meiotic recombination.
Schizosaccharomyces pombe Mek1, which is expressed only during meiosis, is required for efficient meiotic recombination. Mek1 is also required for the meiotic recombination checkpoint (Figure 1C). In S.cerevisiae, Mek1 is considered to be the meiotic counterpart of Rad53 (S.pombe Cds1 homolog). However, the meiotic function of Rad53 is not fully understood. Schizosaccharomyces pombe Cds1 is required for a normal level of meiotic recombination. Cds1 is also required for maintaining spore viability and recombination efficiency in meu13Δ (Figure 2). Therefore, we conclude that both Mek1 and Cds1 are required for the meiotic recombination checkpoint in S.pombe.
Phosphorylation of Cds1 is observed from the initiation of pre-meiotic replication (2 h after temperature shift) through to the end of sporulation. Chk1 expression strongly increases after the first meiotic division, but Chk1 only becomes phosphorylated at the sporulation stage (Figure 5). Mek1 expression begins at the start of pre-meiotic replication and continues until about the time of meiosis I. Mek1 displays a mobility shift during these periods, which is probably due to its phosphorylation, as was shown previously in S.cerevisiae (Rockmill and Roeder, 1991). Thus, the meiotic regulation of each of these three protein kinases, Cds1, Chk1 and Mek1, is possibly different in terms of expression and phosphorylation, suggesting that each has a distinct role to play in meiosis. From these results, we speculate that Rad17, Rad3, Cds1 and Mek1 may be involved in some part of the DNA recombination process as well as in the meiotic recombination monitoring system.
In addition to the homology to the above-mentioned genes, the targets of these checkpoints seem to be the same. In S.cerevisiae, entry into meiosis I is inhibited by maintaining the Tyr19-phosphorylated state of Cdc28 (S.pombe Cdc2 homolog) in response to activation of the pachytene checkpoint because the Tyr19-non-phosphorylated form mutants cannot arrest properly in such a situation (Leu and Roeder, 1999; Tung et al., 2000). We confirmed that similar regulation of Cdc2 operates in S.pombe; the timing of Tyr15 phosphorylation of Cdc2 is retarded when entry into meiosis I is delayed in the meu13Δ mutant (Figure 4).
What is the defect that triggers a meiotic delay in dmc1Δ cells?
We showed here that S.pombe dmc1Δ cells display the following unexpected phenotypes as compared with those of S.cerevisiae. First, dmc1Δ cells show a delay in meiotic progression but do not arrest at prophase as seen in S.cerevisiae (Figure 1). Second, they do not exhibit accumulation of DSBs (Figure 3). Third, the level of reduction in recombination or spore viability is much less than those of the dmc1 mutant of S.cerevisiae (Figure 2).
Meiotic delay in dmc1Δ cells is not alleviated by elimination either of DSBs by deletion of rec12+ or of the checkpoint genes involved in the delay in meu13Δ cells (Figure 1C). These results imply that Dmc1 plays a role in S.pombe meiosis that is independent of DSB formation. They also mean that the absence of this DSB-independent role triggers a delay in meiotic progression which is distinct from that mediated by the functions of the DNA damage checkpoint.
We surmise the following possibilities to explain why dmc1Δ shows meiotic delay. First, S.pombe dmc1+ may play a role in recombination independent of DSB formation, which might be single-strand break-induced recombination. Second, the defect of dmc1+ function may alter chromatin structure to inhibit proper division of the nucleus, which requires time to be repaired. Third, like S.cerevisiae SPO11 and REC8, mutations of which affect the duration of pre-meiotic replication (Cha et al., 2000), dmc1+ could have a function in unexpected cell cycle progression which is unrelated to recombination.
The meiotic delay observed in dmc1Δ cells is not dependent on checkpoint rad genes (rad17+, rad3+, rad1+ and rad9+); however, rad17+ is required for maintenance of viable spores and meiotic recombination frequency in dmc1Δ cells (Figure 2). One possible explanation of this phenotype is that dmc1+ and rad17+ might have a function in recombination between homologous chromosomes in an independent way. Thus, when both genes are defective, meiotic recombination between homologous chromosomes would strongly decrease, resulting in reduced spore viability. Further examination of Dmc1 function should provide new insights into the regulatory mechanism of meiotic progression.
Materials and methods
Yeast strains, media and genetic methods
The S.pombe strains used in this study are listed in Table I. Standard S.pombe genetic procedures were followed (Moreno et al., 1991). Complete media YPD or YE containing adenine sulfate (75 µg/ml), synthetic minimal medium EMM2, sporulation medium ME or EMM2-N and germination media YEAde or EMMG were used. The homozygous diploid strains were constructed by cell fusion.
Table I. Strains used in this study.
| Strain | Genotype |
|---|---|
| MS111w1 | h+ ura4-D18 leu1-32 ade6-469 his2 |
| MS105-1B | h– ura4-D18 ade6-M26 |
| MS126-4 | h+ ura4-D18 leu1-32 ade6-469 his2 rad3::ura4+ |
| MS162-1 | h– ura4-D18 ade6-M26 rad3::ura4+ |
| MS111-1 | h+ ura4-D18 leu1-32 ade6-469 his2 rad17::ura4+ |
| MS105-22D | h– ura4-D18 ade6-M26 rad17::ura4+ |
| MS203-2 | h+ ura4-D18 leu1-32 ade6-469 his2 mek1::ura4+ |
| MS202-11 | h– ura4-D18 ade6-M26 mek1::ura4+ |
| MS233-4 | h+ ura4-D18 leu1-32 ade6-469 his2 cds1::ura4+ |
| MS245-3 | h– ura4-D18 ade6-M26 cds1::ura4+ |
| MS111m13 | h+ ura4-D18 leu1-32 ade6-469 his2 meu13::ura4+ |
| MS105-22C | h– ura4-D18 ade6-M26 meu13::ura4+ |
| MS133-1 | h+ ura4-D18 leu1-32 ade6-469 his2 dmc1::ura4+ |
| MS114-3 | h– ura4-D18 ade6-M26 dmc1::ura4+ |
| MS128-15 | h+ ura4-D18 leu1-32 ade6-469 his2 rad3::ura4+ meu13::ura4+ |
| MS163-10 | h– ura4-D18 ade6-M26 rad3::ura4+ meu13::ura4+ |
| MS111-6 | h+ ura4-D18 leu1-32 ade6-469 his2 rad17::ura4+ meu13::ura4+ |
| MS105-25C | h– ura4-D18 ade6-M26 rad17::ura4+ meu13::ura4+ |
| MS203-2 | h+ ura4-D18 leu1-32 ade6-469 his2 mek1::ura4+ meu13::ura4+ |
| MS202-10 | h– ura4-D18 ade6-M26 mek1::ura4+ meu13::ura4+ |
| MS233-3 | h+ ura4-D18 leu1-32 ade6-469 his2 cds1::ura4+ meu13::ura4+ |
| MS245-7 | h– ura4-D18 ade6-M26 cds1::ura4+ meu13::ura4+ |
| MS133-1 | h+ ura4-D18 leu1-32 ade6-469 his2 rad17::ura4+ dmc1::ura4+ |
| MS114-3 | h– ura4-D18 ade6-M26 rad17::ura4+ dmc1::ura4+ |
| JZ670a | h–/h– pat1-114/pat1-114 leu1-32/ leu1-32 ade6-M210/ade6-M216 |
| KN8 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 meu13::ura4+/meu13::ura4+ |
| MS276-1 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 dmc1::ura4+/dmc1::ura4+ |
| MS123-5 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 rec12::LEU2/ rec12::LEU2 |
| MS110-2 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 rec12::LEU2/rec12::LEU2 meu13::ura4+/meu13::ura4+ |
| MS189-1 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 rec12::LEU2/rec12::LEU2 dmc1::ura4+/dmc1::ura4+ |
| MS101-4 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 rad17::ura4+/rad17::ura4+ |
| MS107-1 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 rad17::ura4+/rad17::ura4+ meu13::ura4+/meu13::ura4+ |
| MS190-7 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 rad17::ura4+/rad17::ura4+ dmc1::ura4+/dmc1::ura4+ |
| MS231-1 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 cds1::ura4+/cds1::ura4+ meu13::ura4+/meu13::ura4+ |
| MS217-1 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 mek1::ura4+/mek1::ura4+ meu13::ura4+/meu13::ura4+ |
| MS197-6 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 Cds1:2HA: ura4+/Cds1 |
| MS198-1 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 meu13::ura4+/meu13::ura4+ Cds1:2HA: ura4+/Cds1 |
| MS108-1 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 Chk1:3HA/Chk1 |
| MS196-2 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 meu13::ura4+/meu13::ura4+ Chk1:3HA/Chk1 |
| MS199-4 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 Mek1:3HA:LEU2/Mek1 |
| MS200-5 | h–/h– pat1-114/pat1-114 ura4-D18/ ura4-D18 leu1-32/ leu1-32 ade6-M210/ade6-M216 meu13::ura4+/meu13::ura4+ Mek1:3HA:LEU2/Mek1 |
| TP4-1D | h+ ura4-D18 leu1-32 ade6-M216 his2 |
| TP4-5A | h– ura4-D18 leu1-32 ade6-M210 |
aProvided by M.Yamamoto.
Spore viability and recombination assays
Haploid parental strains were grown on YPD plates at 33°C, cultured in YE-Ade and harvested at the stationary phase. Cells were mated and sporulated on ME plates at 28°C (zygotic meiosis). After 3–4 days of incubation, spores were treated with 0.5% glusulase (NEN Life Science Products, Inc.) for 16 h at 36°C and checked microscopically for the complete digestion of contaminating vegetative cells. The glusulase-treated spores were washed with water and then used for the spore viability assay and the intragenic recombination assay.
Spore viability assays. A total of 160 spores were spotted with a micro-manipulator (MS series 200; Singer Instrument, UK) on EMMG containing supplements and germinated at 30°C for 4 days. Colonies were counted and the spore viability was expressed as a percentage relative to the 160 spores. We repeated this assay three times for each strain.
Intragenic recombination assays. We determined recombination frequencies as described previously (Nabeshima et al., 2001). To examine the frequency of intragenic recombination, we used two ade6 alleles (ade6-M26 and ade6-469) as the intragenic recombination between these alleles produces the ade6+ allele.
Synchronous meiosis
Fresh homozygous pat1-114 diploid strains were grown in EMM with supplements at 25°C for >24 h. Cells at the mid-log phase were collected, washed and then transferred at a density of 3–4 × 106 cells/ml to EMM2 containing supplements Leu (60 µg/ml) and Ura (40 µg/ml) but lacking NH4Cl. After 16 h of incubation at 25°C, NH4Cl (5 mg/ml) and supplements Leu (250 µg/ml) and Ura (75 µg/ml) were added to the culture medium and then the temperature was raised to 34°C to induce meiosis. The progression of meiosis was monitored by staining the methanol-fixed cells with DAPI (Wako). For γ-ray analysis, horsetail cells (3 h after temperature shift) were irradiated from a 60Co source at a dose rate of 468 Gy/h for ∼20 min (170 Gy).
Meiotic DSB assays
The procedures for PFGE have been described previously (Cervantes et al., 2000). Meiosis was induced in pat1 diploid cells by shifting the temperature, and 14 ml of cultured cells were collected at the indicated times. PFGE was conducted in a 0.8% chromosomal grade agarose gel (Bio-Rad) in a Bio-Rad CHEF-Mapper system at 14°C for 48 h with 2 V/cm, 100°C-induced angle in 1× TAE buffer (40 mM Tris-acetate pH 8.0, 1 mM EDTA), with a switch time of 30 min.
Protein extraction and western blotting
Protein extracts were prepared as described previously (Caspari et al., 2000) and western blots were performed as published (Shimada et al., 1999). For the detection of Cds1HA, Chk1HA and Mek1HA, the blots were probed with mouse monoclonal antibodies 12CA5 (Boehringer Mannheim for Chk1HA and Mek1HA) and 16B12 (COVANCE Inc. for Cds1HA), then stripped and reprobed with anti-tubulin antibody (Sigma T5168) as a loading control. For the detection of Cdc2 Tyr15 phosphorylation, the blots were probed with an anti-phospho-Cdc2 (Tyr15) rabbit polyclonal antibody (NEB 9111), then stripped and reprobed with the anti-Cdc2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Mek1 disruption
The mek1+ gene was disrupted by replacing it with the ura4+ gene. To do this, we performed PCR and obtained a DNA fragment carrying the 5′ upstream region and 3′ downstream region of the mek1+ gene. For this purpose, we synthesized the following four oligonucleotides and used them as primers: mek1-5F, 5′-CGGGGTACCTGCAGAATTGAAAATACGTCAAACCGAAC-3; mek1-5R, 5′-CCGCTCGAGCACTTTGCAAAACGGTGATGCGCGTAAGC-3′; mek1-3F, 5′-AAAACTG CAGCTCGAGCCACGTAAGAAAATATTCCGATAAACTTG-3′; and mek1-3R, 5′-CCCGAGCTCTAATTTAATATATCTTTGCTTGAATTATCG-3′. The underlined sequences denote the artificially introduced restriction enzyme sites for KpnI, XhoI, PstI–XhoI and SacI, respectively. These PCR products and the 1.8 kb HindIII fragment containing the ura4+ gene (Grimm et al., 1988) were inserted into the pBluescriptII KS(+) vector via the KpnI–XhoI, PstI–SacI and HindIII sites, respectively. This plasmid construct was digested with KpnI and SacI and the resulting construct was introduced into the diploid strain TP4-5A/TP4-1D. The Ura+ transformants were then screened by Southern blot analysis to identify the disrupted strain.
Construction of Mek1-3HA strain
To prepare the Mek1-3HA construct, we performed PCR and obtained a DNA fragment carrying the open reading frame (ORF) region and the 3′ downstream region of the mek1+ gene. For this purpose, we synthe sized the following four oligonucleotides and used them as primers: mek1-ORF-F, 5′-ATAGGCGCGCCGTCGACTATGGACTTTTTATCACATGCCATGC-3′; and mek1-ORF-R, 5′-TATTCTTAGCGGCCGCCGTAGCCGGGAATGTTTAAGAGG-3′. The underlined sequences denote the artificially introduced restriction enzyme sites for AscI–SalI and NotI, respectively. To obtain the 3′ downstream region, we used the same primers as described above. These PCR products were inserted into the pIL(II) vector (T.Nakamura, unpublished), which is designed to allow one-step integration via SalI–NotI and XhoI–SacI sites, respectively. This plasmid construct was digested with MluI. The resulting construct was introduced into the haploid strains TP4-5A and TP4-1D. We then screened the Leu+ transformants by Southern blot analysis to identify the Mek1-3HA tagged strain.
Acknowledgments
Acknowledgements
We are grateful to Dr Tony Carr for strains and critical discussions, and Dr Gerry Smith for strains and technical suggestions. We also thank Dr T.J.Kim for general support and encouragement, Drs T.Nakamura, Y.Watanabe and E.Hartsuiker for critical discussions, Drs P.Russell, F.Ishikawa, M.Yamamoto and C.Shimoda for strains, Dr P.Nurse for the Cdc2 antibody, Dr V.Wood for cosmid clones of the S.pombe genome, and Dr P.Hughes for critically reading the manuscript. This work was supported by a Grant-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan and grants from The Uehara Memorial Foundation to H.N. M.S. is a Research Fellow of the Japan Society for the Promotion of Science.
References
- Bahler J., Wyler,T., Loidl,J. and Kohli,J. (1993) Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol., 121, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) DMC1: a meiosis-specific yeast homolog of recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- Caspari T. and Carr,A.M. (1999) DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie, 81, 173–181. [DOI] [PubMed] [Google Scholar]
- Caspari T., Dahlen,M., Kanter,S.G., Lindsay, HD., Hofmann,K., Papadimitriou,K., Sunnerhagen,P. and Carr,A.M. (2000) Characteriz ation of Schizosaccharomyces pombe Hus1; a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol., 20, 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes M.D., Farah,J.A. and Smith,G.R. (2000) Meiotic DNA breaks associated with recombination in S.pombe. Mol. Cell, 5, 883–888. [DOI] [PubMed] [Google Scholar]
- Cha R.S., Weiner,B.M., Keeney,S., Dekker,J. and Kleckner,N. (2000) Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by SPO11p and positively by Rec8p. Genes Dev., 14, 493–503. [PMC free article] [PubMed] [Google Scholar]
- Fukushima K., Tanaka,Y., Nabeshima,K., Yoneki,T., Tougan,T., Tanaka,S. and Nojima,H. (2000) Dmc1 of Schizosaccharomyces pombe plays a role in meiotic recombination. Nucleic Acids Res., 28, 2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Kohli,J., Murray,J. and Maundrell,K. (1988) Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol. Gen. Genet., 215, 81–86. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. and Weinert,T.A. (1989) Checkpoints: controls that ensure the order of cell cycle events. Science, 246, 629–634. [DOI] [PubMed] [Google Scholar]
- Hunter N. and Kleckner,N. (2001) The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell, 106, 59–70. [DOI] [PubMed] [Google Scholar]
- Iino Y. and Yamamoto,M. (1985) Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol. Gen. Genet., 198, 416–421. [DOI] [PubMed] [Google Scholar]
- Kato R. and Ogawa,H. (1994) An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res., 22, 3104–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. (1996) Meiosis: how could it work? Proc. Natl Acad. Sci. USA, 93, 8167–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu J.Y. and Roeder,G.S. (1999) The pachytene checkpoint in S.cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell, 4, 805–814. [DOI] [PubMed] [Google Scholar]
- Lydall D., Nikolsky,Y., Bishop,D.K. and Weinert,T. (1996) A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature, 383, 840–843. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Murakami H. and Nurse,P. (1999) Meiotic DNA replication checkpoint control in fission yeast. Genes Dev., 13, 2581–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Kakihara,Y., Hiraoka,Y. and Nojima,H. (2001) A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J., 20, 3871–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N. and Russell,P. (1998) Tyrosine phosphorylation of cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol. Cell. Biol., 18, 3782–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N., Furnari,B. and Russell,P. (1997) Cdc2 tyrosine phosphoryl ation is required for the DNA damage checkpoint in fission yeast. Genes Dev., 11, 504–511. [DOI] [PubMed] [Google Scholar]
- Rockmill B. and Roeder,G.S. (1991) A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev., 5, 2392–2404. [DOI] [PubMed] [Google Scholar]
- Roeder G.S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev., 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Roeder G.S. and Bailis,J.M. (2000) The pachytene checkpoint. Trends Genet., 16, 395–403. [DOI] [PubMed] [Google Scholar]
- Shimada M., Okuzaki,D., Tanaka,S., Tougan,T., Tamai,K.K., Shimoda,C. and Nojima,H. (1999) Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol. Biol. Cell, 10, 3991–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D. and Wittenberg,C. (1998) CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev., 12, 2698–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung K.S., Hong,E.J. and Roeder,G.S. (2000) The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl Acad. Sci. USA, 97, 12187–12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Miyashita,K., Saito,T.T., Yoneki,T., Kakihara,Y., Nabeshima,K., Kishi,Y.A., Shimoda,C. and Nojima,H. (2001) Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res., 29, 2327–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Weiner,B.M. and Kleckner,N. (1997) Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev., 11, 106–118. [DOI] [PubMed] [Google Scholar]