Abstract
The CDKN2A tumour suppressor locus encodes two distinct proteins, p16INK4a and p14ARF, both of which have been implicated in replicative senescence, the state of permanent growth arrest provoked in somatic cells by aberrant proliferative signals or by cumulative population doublings in culture. Here we describe primary fibroblasts from a member of a melanoma-prone family who is homozygous for an intragenic deletion in CDKN2A. Analyses of the resultant gene products imply that the cells are p16INK4a deficient but express physiologically relevant levels of a frameshift protein that retains the known functions of p14ARF. Although they have a finite lifespan, the cells are resistant to arrest by oncogenic RAS. Indeed, ectopic expression of RAS and telomerase (hTERT) results in outgrowth of anchorage-independent colonies that have essentially diploid karyotypes and functional p53. We find that in human fibroblasts, ARF is not induced demonstrably by RAS, pointing to significant differences between the proliferative barriers implemented by the CDKN2A locus in different cell types or species.
Keywords: anchorage independence/p14ARF/p16INK4a/Ras/senescence
Introduction
Somatic cells have innate defence mechanisms that guard against unrestrained proliferation. Depending on circumstances, a cell in primary culture will commonly undergo either apoptosis or growth arrest when challenged with a single oncogenic agent (Evan and Vousden, 2001). In fibroblasts, at least, the growth arrest is phenotypically similar to that elicited by telomere loss when cells reach the end of their proliferative lifespan, and has therefore been termed ‘premature senescence’ (Serrano et al., 1997; Lundberg et al., 2000; Campisi, 2001). Although the relationship between senescence of cultured cells and ageing in vivo remains uncertain, both oncogene-mediated and lifespan-based senescence are potentially important for tumour suppression, and several accredited tumour suppressors have critical roles in the underlying mechanisms. Prominent examples include p53, the retinoblastoma protein (pRb) and both of the products encoded by the CDKN2A locus.
The CDKN2A locus has the unusual capacity to specify two structurally distinct proteins, designated p16INK4a and p14ARF (p19ARF in the mouse), by exploiting different first exons (1α and 1β) spliced to a common second exon that is translated in alternative reading frames (Ruas and Peters, 1998; Sharpless and DePinho, 1999). The product of the α transcript, p16INK4a, binds directly to Cdk4 and Cdk6, the cyclin-dependent kinases that initiate the phosphorylation and functional inactivation of pRb. Ectopic expression of p16INK4a therefore causes cells to arrest in the G1 phase of the cell cycle, in a pRb-dependent manner (reviewed in Ruas and Peters, 1998). In contrast, the product of the β transcript, p14ARF, interacts directly with MDM2, a multifaceted protein that opposes the function of p53 by blocking its transcriptional activation domain, facilitating its nuclear export and catalysing its ubiquitylation and proteasome-mediated destruction (Ashcroft and Vousden, 1999; Sharpless and DePinho, 1999). Ectopic expression of p14ARF (or p19ARF) therefore stabilizes p53 and causes cells to arrest in G1 and G2, accompanied by increased expression of p53-regulated genes, such as p21CIP1 and MDM2 itself (Sharpless and DePinho, 1999). The expression of the β transcript is negatively regulated both by p53, through an as yet unknown mechanism, and by pRb, through its ability to repress E2F-dependent transcription (Sharpless and DePinho, 1999). Thus, ectopic expression of E2F-1 or the ablation of pRb, for example by DNA tumour virus oncoproteins, can activate a p53 response via the up-regulation of ARF.
As p16INK4a and p14ARF both have the potential to act in tumour surveillance and replicative senescence, it has become important to distinguish their relative significance in different settings. The evidence linking p16INK4a to senescence is compelling. For example, p16INK4a accumulates when human diploid fibroblasts (HDFs) reach the limit of their finite lifespan in culture (Alcorta et al., 1996; Hara et al., 1996; Wong and Riabowol, 1996), and the senescence-like growth arrest provoked by oncogenic signalling through the RAS–RAF–MEK pathway is also accompanied by up-regulation of p16INK4a (Serrano et al., 1997; Lin et al., 1998; Zhu et al., 1998; Ohtani et al., 2001). Indeed, a number of studies have documented the down-regulation of p16INK4a concomitant with the outgrowth of fibroblasts with an extended or indefinite lifespan (Rogan et al., 1995; Vogt et al., 1998). In contrast, immortalization of mouse embryo fibroblasts (MEFs) is invariably accompanied by disruption of either p53 or ARF, with little evidence for selection against p16INK4a (Kamijo et al., 1997). Although MEFs undergo a senescence-like growth arrest accompanied by accumulation of both p19ARF and p16INK4a (Palmero et al., 1997; Zindy et al., 1998), this arrest is not related to telomere erosion, and MEFs immortalize at a much higher frequency than their human counterparts (Wright and Shay, 2000).
In mice, the relative significance of the Cdkn2a gene products has been addressed by elegant genetic experiments in which individual exons of the locus have been ablated by homologous recombination. Thus, mice with disruptions in Cdkn2a exons 2 and 3 (Δ2,3) or exon 1β alone are tumour prone and the resulant MEFs are both immortal and sensitive to transformation by RAS (Serrano et al., 1996; Kamijo et al., 1997). In contrast, while specific ablation of exon 1α may contribute to tumour susceptibility, INK4a-deficient MEFs undergo senescence and are arrested by oncogenic RAS (Krimpenfort et al., 2001; Sharpless et al., 2001). Although it is technically feasible to engineer primary human cells that are defective in specific gene products (Brown et al., 1997; Bunz et al., 1998), such approaches are not applicable to all genes and are unsuited to ablation of the first exon. An alternative is to examine cells derived from individuals with inherited genetic defects. Here, we address the relative contributions of p16INK4a and p14ARF by exploiting dermal HDFs from a rare individual who is homozygous for an intragenic 19 bp deletion in the second exon of CDKN2A (Gruis et al., 1995). From detailed analyses of the resultant gene products, we conclude that these cells lack p16INK4a but retain p14ARF functions. Significantly, the cultured fibroblasts are resistant to RAS-mediated arrest. Upon co-expression of RAS and telomerase (hTERT), they become capable of growth as anchorage-independent colonies that retain an apparently normal p53 response and essentially diploid karyotypes, but do not form tumours in nude mice. Our data point to significant differences in the regulation of the INK4a/ARF locus in human and mouse fibroblasts.
Results
Novel fusion proteins created by intragenic deletion in CDKN2A
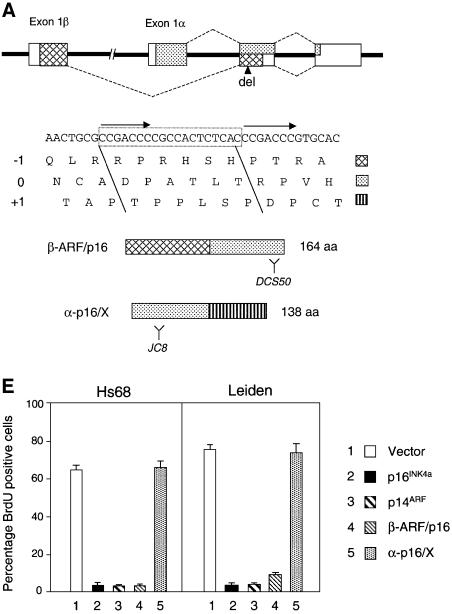
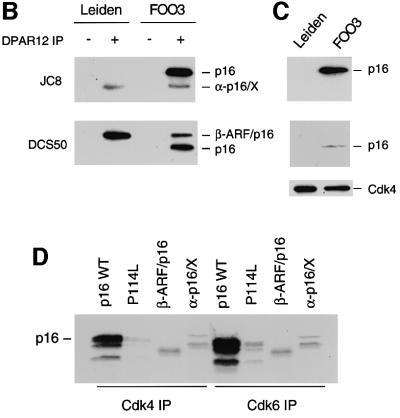
Dermal fibroblasts were isolated from a 24-year-old male who is homozygous for a 19 bp deletion in exon 2 of the CDKN2A gene (Gruis et al., 1995), and from a first degree relative who is heterozygous for the mutation. These cell strains are designated Leiden and FOO3 HDFs, respectively. In addition to removing six amino acids, the lesion creates a frameshift that affects both of the proteins encoded by the locus (Figure 1A). As a result, the transcript initiating in exon 1α encodes a 138 amino acid protein (α-p16/X) comprising the first 74 residues of p16INK4a followed by 64 residues specified by the +1 reading frame of exon 2, whereas the exon 1β transcript encodes a fusion protein (β-ARF/p16) containing the first 88 residues of p14ARF followed by the last 76 residues of p16INK4a. The two frameshift proteins were detected in Leiden and FOO3 cell lysates by immunoprecipitating with a polyclonal antiserum against recombinant p16INK4a and immunoblotting with monoclonal antibodies (mAbs) directed against different regions of p16INK4a (Figure 1B). The α-p16/X fusion protein, visualized with the mAb JC8, appeared slightly smaller than authentic p16INK4a, whereas the β-ARF/p16 product, recognized by the DCS50 mAb, was significantly larger than p16INK4a. The α-p16/X product was consistently more difficult to detect than wild-type p16INK4a, suggesting that it may be less stable.
Fig. 1. Leiden deletion and resultant CDKN2A fusion proteins. (A) Schematic representation of the CDKN2A locus with the exons shown as boxes. The sequences encoding p16INK4a are identified by stippling, and those encoding p14ARF by cross-hatches. The nucleotide sequence of the relevant region of exon 2 is shown, with direct repeats identified by arrows and the 19 bp Leiden deletion boxed. The encoded amino acids in the three different reading frames are shown in single letter code. The –1 (p14ARF), 0 (p16INK4a) and +1 frames are identified by the distinctive shading, and their contributions to the β-ARF/p16 and α-p16/X fusion proteins are indicated. Y defines the regions of p16INK4a recognized by the DCS50 and JC8 mAbs. (B) Lysates from the Leiden and FOO3 strains of HDFs were immunoprecipitated with either non-immune serum (– lanes) or a polyclonal antibody (DPAR12) against p16INK4a (+ lanes). After fractionation by SDS–PAGE, the samples were immunoblotted with JC8 (upper panel) and DCS50 (lower panel). The positions of wild-type p16INK4a and the β-ARF/p16 and α-p16/X fusion proteins are shown. (C) Immunoprecipitation of Cdk4 from the same Leiden and FOO3 lysates followed by immunoblotting with JC8, DCS50 and a mAb against Cdk4. (D) In vitro binding assay using unlabelled Cdk4 (lanes 1–4) and Cdk6 (lanes 5–8) mixed with [35S]methionine-labelled wild-type p16INK4a (lanes 1 and 5), the P114L variant (lanes 2 and 6), β-ARF/p16 (lanes 3 and 7) or α-p16/X (lanes 4 and 8). The mixtures were immunoprecipitated with polyclonal antisera against Cdk4 or Cdk6 and the labelled proteins were fractionated by SDS–PAGE and visualized by autoradiography. The multiple bands reflect initiation at internal methionines. (E) BrdU incorporation assays in Hs68 and Leiden cells infected with retroviruses encoding p16INK4a, p14ARF, β-ARF/p16 or α-p16/X as indicated.
Effects of the Leiden deletion on p16INK4a function
To determine whether either of the fusion proteins retained p16INK4a function, the Leiden and FOO3 cell lysates were immunoprecipitated with a polyclonal antiserum against Cdk4. Whereas the wild-type p16INK4a co-precipitated with Cdk4 in FOO3 cells, neither of the fusion proteins was detected in the immunoprecipitates from Leiden or FOO3 HDFs (Figure 1C). Similar negative findings were obtained with Cdk6 antiserum (not shown). For a more comprehensive analysis, the two frameshift proteins, tagged at their N-terminus with a His6 epitope, were expressed by coupled transcription and translation of the respective cDNAs. The products were then mixed with Cdk4 and Cdk6, and immunoprecipitated with polyclonal antisera against the respective kinases. Whereas wild-type p16INK4a bound efficiently to Cdk4 and Cdk6, only background levels of the α-p16/X and β-ARF/p16 fusion proteins were detected in the immune complexes (Figure 1D). In this respect, the two fusion proteins resembled P114L, a well-documented loss-of-function mutant of p16INK4a (Parry and Peters, 1996). Analogous results were obtained in reciprocal immunoprecipitations with a polyclonal antiserum against p16INK4a (data not shown).
We also asked whether either of the fusion proteins was capable of causing growth arrest in primary diploid fibroblasts. The relevant cDNAs were transferred into a retroviral vector that provided an N-terminal 2× haemagglutinin (HA) tag, and the packaged virus particles were used to infect either Hs68 or Leiden cells that had been engineered to express the receptor for ecotropic viruses (Ruas et al., 1999). Wild-type p16INK4a and p14ARF were used as controls and the expression of the various HA-tagged proteins was confirmed by immunoblotting (data not shown). As illustrated in Figure 1E, the β-ARF/p16 retrovirus had a profound effect on cell proliferation, as measured by the reduced incorporation of bromodeoxyuridine (BrdU) in the pools of drug-resistant cells. Similar effects were elicited by wild-type p16INK4a and p14ARF, as expected. In contrast, the retrovirus encoding α-p16/X had no discernible effect on BrdU incorporation.
Effects of the Leiden deletion on p14ARF function
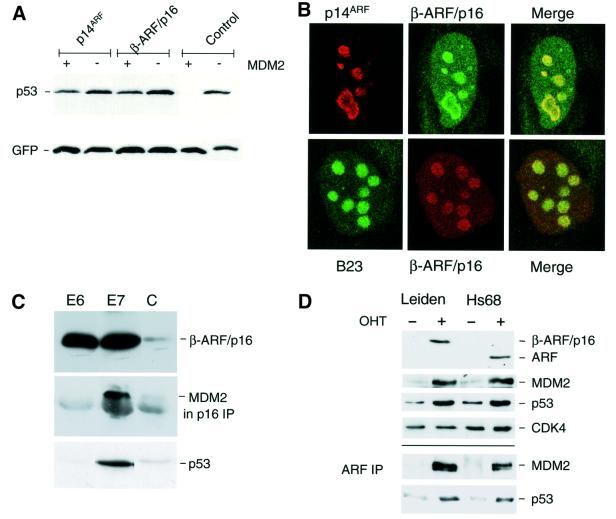
As neither of the fusion proteins was capable of interacting with Cdk4 and Cdk6, we reasoned that the growth-inhibitory properties of β-ARF/p16 were more likely to reflect the ARF sequences present in the fusion protein. The known functions of human p14ARF are attributable to the N-terminal 64 residues or even smaller peptides (Stott et al., 1998; Zhang et al., 1998; Lohrum et al., 2000; Midgley et al., 2000; Rizos et al., 2000; Llanos et al., 2001), all of which are retained in β-ARF/p16. In line with these predictions, we found that ectopic β-ARF/p16 was able to protect p53 from MDM2-mediated degradation in a transient transfection assay (Figure 2A). Moreover, when transfected into NARF2 cells, a clone of U20S cells that express p14ARF from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter (Stott et al., 1998), β-ARF/p16 was found to co-localize with the wild-type protein and with the nucleolar marker B23 (Figure 2B). In this experiment, the β-ARF/p16 fusion protein was detected with p16INK4a-specific antibodies and the wild-type protein was observed with a mAb against p14ARF (Llanos et al., 2001). We also infected Leiden HDFs with retroviruses encoding either human papilloma virus (HPV)-16 E6 or E7, which target p53 and pRb, respectively. As anticipated, both viruses markedly increased the expression of the β-ARF/p16 fusion protein (Figure 2C). In the E7-infected cells, it was possible to immunoprecipitate a complex of β-ARF/p16 and MDM2 using an antibody against p16INK4a, confirming that the fusion protein is capable of binding to and stabilizing endogenous MDM2. Moreover, the E7-infected cells showed increased levels of p53 (Figure 2C, lower panel), the predicted consequence of ARF induction. No p53 activation or MDM2 complex was detected in the E6-infected cells because the E6-mediated degradation of p53 would have negated the effects of ARF. Finally, to confirm that the levels of the β-ARF/p16 fusion protein in Leiden cells are comparable with those of authentic p14ARF in control fibroblasts, we made use of a polyclonal antiserum against residues 54–75 of human ARF that can recognize both proteins. In the experiment shown in Figure 2D, we induced ARF by using a retrovirus encoding E2F-1 fused to a modified hormone-binding domain from the oestrogen receptor (Müller et al., 2001). Upon addition of 4-hydroxy tamoxifen, to release functional E2F-1, both the β-ARF/p16 fusion protein and authentic ARF were up-regulated (Figure 2D). Significantly, the β-ARF/p16 signal was comparable with if not higher than that of authentic p14ARF. Induction of ARF was accompanied by up-regulation of both MDM2 and p53. Furthermore, the amounts of MDM2 and p53 that co-precipitated with β-ARF/p16, using the 54–75 antibody, were comparable with those observed with authentic p14ARF in Hs68 cells. Thus, not only is β-ARF/p16 expressed at physiologically relevant levels, but it is functionally equivalent to wild-type ARF in these assays.
Fig. 2. Functional appraisal of the β-ARF/p16 fusion protein. (A) U20S cells were transiently transfected with plasmids encoding p53 and green fluorescent protein, with and without human MDM2. As shown in the control lanes, MDM2 causes the marked destruction of p53 in this assay. Introduction of either wild-type p14ARF or β-ARF/p16 protected p53 from MDM2-mediated degradation. (B) NARF2 cells (U20S cells expressing p14ARF from an IPTG-inducible promoter) were transfected with a plasmid encoding β-ARF/p16 and analysed by indirect immunofluorescence. In the upper panels, p14ARF was detected with the mouse mAb 4C6/4 (red) whereas the β-ARF/p16 fusion protein was detected with a rabbit polyclonal antibody against p16INK4a (green). In the lower panels, β-ARF/p16 fusion protein was detected with the mouse mAb DCS50 (red) and the nucleolar protein B23 was visualized using a goat polyclonal antibody (green). (C) Samples (20 µg) of total protein from Leiden cells infected with retroviruses encoding the E6 or E7 proteins of HPV-16 (as indicated) or control virus (C) were fractionated by SDS–PAGE and immunoblotted with the mAbs DCS50 to detect the β-ARF/p16 fusion protein (upper panel) and DO-1 to detect p53 (lower panel). A sample (500 µg) of each lysate was immunoprecipitated with polyclonal antiserum against p16INK4a (DPAR12) and immunoblotted with mAb IF2 against human MDM2 (middle panel). (D) Control (Hs68) and Leiden HDFs expressing E2F-1 fused to the hormone-binding domain of the oestrogen receptor were treated (+) or not (–) with 4-hydroxy tamoxifen (OHT) for 24 h. Samples (20 µg) of cell lysate were fractionated by SDS–PAGE and immunoblotted with an antiserum raised against amino acids 54–75 of human p14ARF, and with mAbs against MDM2 and p53. Cdk4 served as a loading control. Samples (500 µg) of each lysate were immunoprecipitated with the same p14ARF antiserum and immunoblotted for MDM2 and p53.
Resistance of Leiden cells to RAS-mediated growth arrest
Although we cannot be certain that ARF function is completely normal in the homozygous individual, the weight of evidence suggested that the Leiden HDFs are specifically deficient for p16INK4a. In terms of morphology, growth rates and plating efficiency, Leiden HDFs appeared no different from FOO3 and other strains of dermal fibroblasts obtained from normal age-matched donors. They also underwent M1-like senescence (data not shown) after ∼60 population doublings (PDs), which is at the upper extreme of the normal distribution for HDFs from healthy adults (Cristofalo et al., 1998). In comparison, the heterozygous FOO3 cells senesced after ∼40 PDs. Thus, while p16INK4a may contribute to the timing or mechanics of telomere-driven senescence in HDFs, it does not appear to be essential. It was therefore of interest to ask whether p16INK4a deficiency compromises the senescence-like growth arrest engendered by oncogenic RAS.
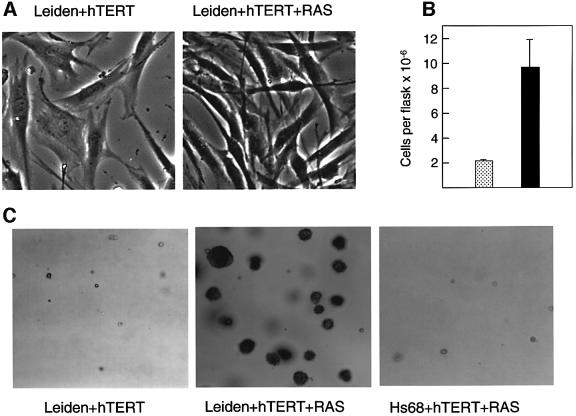
To separate RAS-induced from lifespan-based events, Leiden and control HDFs were first infected with a retrovirus encoding hTERT, the catalytic component of telomerase. Expression of hTERT enables HDFs to bypass M1 replicative senescence (Bodnar et al., 1998; Kiyono et al., 1998; Vaziri and Benchimol, 1998) but does not prevent RAS-mediated arrest (Morales et al., 1999; Wei et al., 1999). Pools of hTERT-expressing HDFs were then infected with recombinant retroviruses encoding the G12V mutant of H-RAS and different drug resistance markers. After a period of selection and expansion (generally ∼4 weeks), there was a striking difference in the behaviour of the Leiden cells compared with other strains of HDF. Whereas control HDFs underwent a senescence-like growth arrest, as judged by their flattened morphology and markedly reduced BrdU incorporation (Table I and data not shown), the Leiden fibroblasts continued to proliferate vigorously, adopted characteristics reminiscent of morphological transformation and had a saturation density ∼4.5-fold higher than that of the corresponding cells infected with the empty vector (Figure 3A and B). Moreover, when pools of RAS + hTERT-expressing Leiden cells were seeded in semi-solid medium, a significant proportion appeared capable of growing as anchorage-independent colonies (Figure 3C; Table I). Leiden cells expressing only hTERT did not grow appreciably in such conditions, and multicellular colonies were rarely observed in the control fibroblasts expressing RAS and hTERT (Table I). Note, however, that the RAS + hTERT-expressing Leiden cells did not form tumours in nude mice.
Table I. BrdU incorporation and agar colony formation of HDFs expressing RAS and hTERT.
| Cell strain/virus | % BrdU-positive cells | % of cells forming agar colonies |
|---|---|---|
| Leiden + hTERT | 86 | 1 |
| Leiden + hTERT + RASBleo | 64 | 50 |
| Leiden + hTERT | 47 | ND |
| Leiden + hTERT + RASNeo | 47 | 65 |
| Hs68 + hTERT | 82 | 0 |
| Hs68 + hTERT + RASBleo | 11 | 0.5 |
| Hs68 + hTERT | 68 | 0 |
| Hs68 + hTERT + RASBleo | 4 | 0 |
| TIG3 + hTERT | 78 | 0 |
| TIG3 + hTERT + RASBleo | 0 | 0 |
| 293 | ND | 57 |
Fig. 3. Phenotypic effects of RAS + hTERT in Leiden HDFs. (A) Pools of Leiden HDFs expressing RAS + hTERT or the respective empty vector controls were examined by phase microscopy ∼4–5 weeks post-selection. (B) The saturation densities of Leiden cells expressing hTERT alone (stippled bar) or RAS + hTERT (filled bar) were compared by counting the numbers of cells per 75 cm2 flask at 4–7 days post-confluence. The figure presents the results of four independent comparisons, and the error bars indicate the standard deviation. (C) Leiden cells expressing RAS + hTERT formed macroscopically visible colonies in soft agar (centre) whereas Leiden cells expressing hTERT alone did not (left). No significant agar growth was observed with control fibroblasts expressing RAS + hTERT (right).
Cytogenetic analyses of anchorage-independent Leiden HDFs
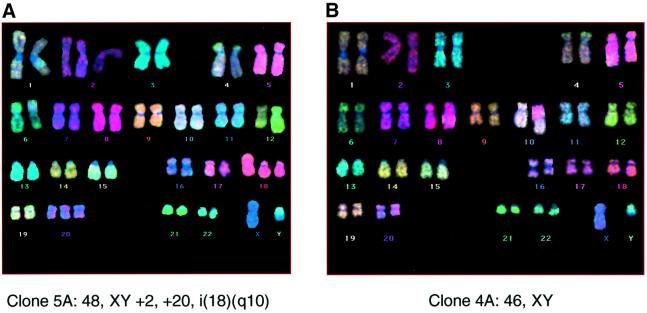
At face value, our results imply that Leiden cells achieve anchorage independence with lesions in three cellular genes: INK4a, RAS and TERT. The high frequency with which the agar colonies arose would be incompatible with the occurrence of random de novo mutations but did not exclude the possibility that during prolonged culture, a subpopulation had emerged carrying a mutation that rendered them ultrasensitive to the effects of RAS. Alternatively, loss of p16INK4a might have made the Leiden cells genetically unstable. To try to address these possibilities, pools of Leiden cells, before and after the introduction of hTERT and RAS, as well as anchorage-independent colonies recovered from the agarose, were analysed by standard G-banding of metaphase spreads and by fluorescence in situ hybridization using multicolour chromosome paints (M-FISH). The cell pools generally had normal diploid karyotypes, although a number of the anchorage-independent colonies contained additional or aberrant chromosomes (Figure 4A; Table II). However, there were no consistent genetic abnormalities, and some of the colonies had no visible defects (Figure 4B; Table II). We therefore conclude that the response of Leiden cells to RAS + hTERT does not reflect gross genetic instability or a pre-existing aberration detectable at this level.
Fig. 4. Cytogenetic analyses of anchorage-independent colonies. Representative metaphases from RAS + hTERT-expressing Leiden HDF colonies recovered from semi-solid medium and analysed by M-FISH. Clone 5A contained additional and rearranged chromosomes (A), whereas clone 4A showed a normal diploid karyotype (B).
Table II. Karyotypes of anchorage-independent colonies of Leiden cells expressing RAS + hTERT.
| Clone No. | Karyotype | Method |
|---|---|---|
| 1 | 46,XY,+3,–2 | G-banding |
| 2 | 46,XY,t(6:?)(qter:?) | G-banding |
| 4A | 46,XY | M-FISH |
| 4B | 47,XY,+20[47]/46,XY[3] | M-FISH |
| 5A | 48,XY,+2,+20,i(18)(q10) | M-FISH |
| 6C | 47,XY,+del(3)(?) | M-FISH |
Integrity of the p53 pathway in Leiden HDFs
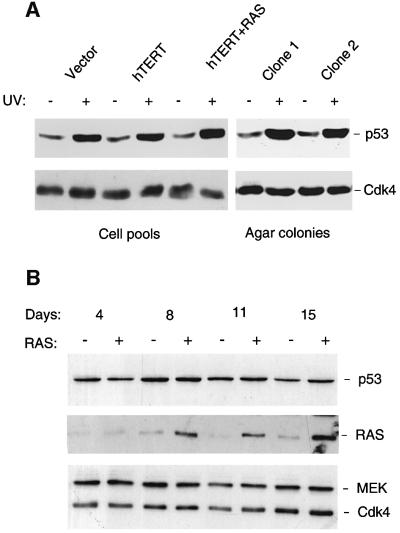
Such analyses did not rule out single gene defects and, in particular, it was surprising to find that Leiden cells could resist RAS-induced senescence without intentional disruption of the p53 pathway (Serrano et al., 1997). Studies on MEFs have established that inactivation of either p53 or p19ARF renders the cells sensitive to transformation by RAS alone (Kamijo et al., 1997). As the data in Figure 2 imply that ARF remains functional in Leiden HDFs, logic would dictate that some other component of the p53 pathway must be defective. To test whether the p53 response remained operational, pools of Leiden HDFs expressing empty vector, hTERT or RAS + hTERT, as well as cells recovered from anchorage-independent colonies, were subjected to UV irradiation and subsequently immunoblotted for p53. The cell pools and colonies showed a similar induction of p53 in response to DNA damage (Figure 5A) as well as up-regulation of the p53 target gene p21CIP1 (not shown). At this level, therefore, we have no reason to suspect a defect in p53 function.
Fig. 5. Induction of p53 in Leiden and FOO3 cells by UV irradiation but not RAS. (A) Anchorage-independent colonies of Leiden cells recovered from agarose (clones 1 and 2), and pools of Leiden cells infected with empty vector controls (39 PDs), expressing hTERT alone (41 PDs) or RAS + hTERT (46 PDs), were exposed to UV radiation (15 J/m2). Samples (20 µg) of total protein were then subjected to SDS–PAGE in a 10% gel and immunoblotted with a mAb against p53 (DO-1). Cdk4 served as a loading control. (B) FOO3 cells were infected with either pBABE-RAS-puro retrovirus (+ lanes) or empty vector (– lanes) and harvested at various times post-infection, as indicated, without drug selection. Samples (30 µg) were fraction ated by SDS–PAGE on a 10% gel and immunoblotted for the indicated proteins.
We noted, however, that in the various cell pools analysed in Figure 5A and in several similar experiments (not shown), the basal levels of p53 were relatively constant, whether or not the cells expressed hTERT or RAS. This did not concur with reports that oncogenic RAS causes up-regulation of p19ARF and p53 in primary mouse cells (Serrano et al., 1997; Palmero et al., 1998; Groth et al., 2000; Lin and Lowe, 2001). Until recently, it has been tacitly assumed that the same situation prevails in HDFs, because several studies have noted activation of p53 in response to sustained RAS or MEK signalling (Serrano et al., 1997; Lin et al., 1998; Ferbeyre et al., 2000; Pearson et al., 2000; Wei et al., 2001). However, this is not a universal finding (Zhu et al., 1998; Ries et al., 2000), and we therefore sought to determine whether our inability to see an effect of RAS on p53 levels had a technical explanation or reflected a peculiarity of Leiden HDFs. To address these concerns, we infected various HDF strains with retroviruses encoding RAS and harvested the cells at different intervals. No selection was applied in order to exclude potential effects of the drug (either puromycin or bleomycin) on the p53 pathway. In parallel experiments, we confirmed that >50% of cells were infected and that RAS was functional (see below). Under these conditions, oncogenic RAS had little impact on p53 levels as detected by immunoblotting (Figure 5B). Representative data for FOO3 cells are shown but equivalent results were obtained with Leiden and Hs68 cells, with different p53 antibodies, and over time courses of between 3 and 30 days.
Lack of ARF induction in RAS-expressing HDFs
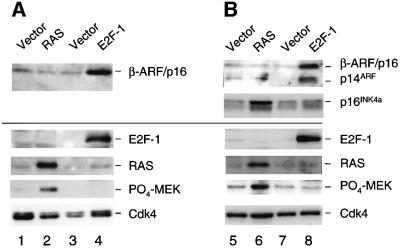
While it is well established that ARF provides a link between RAS signalling and p53 in mouse cells (Palmero et al., 1998; Groth et al., 2000; Lin and Lowe, 2001), there is no evidence that such a link operates in human cells (Ferbeyre et al., 2000; Wei et al., 2001). The issue is clouded by the difficulty in detecting the low levels of p14ARF in normal cells, but it has recently been reported that RAS does not have a measurable effect on ARF RNA levels in normal HDFs (Wei et al., 2001). Our ability to visualize the β-ARF/p16 fusion protein with antibodies against p16INK4a (Figure 1) and ARF (Figure 2D) enabled us to address this question. Leiden and FOO3 HDFs were infected with a retrovirus encoding RAS, without drug selection, and cell lysates prepared at different times post-infection were immunoblotted for ARF and p16INK4a. Parallel cultures were infected with a retrovirus encoding E2F-1, to provide a positive control for ARF activation. As expected, E2F-1 caused up-regulation of β-ARF/p16 in Leiden cells (Figure 6A, lane 4) and of both β-ARF/p16 and wild-type p14ARF in FOO3 cells (Figure 6B, lane 8). In Figure 6A, the fusion protein was detected with the DCS50 mAb against the C-terminal domain of p16INK4a whereas, in Figure 6B, both ARF products were visualized with the polyclonal antiserum against residues 54–75 of p14ARF. Note that β-ARF/p16 is more abundant than wild-type p14ARF in FOO3 cells, suggesting that it may be more stable (also suggested in Figure 2D).
Fig. 6. Induction of CDKN2A gene products by RAS and E2F-1. Leiden (A) and FOO3 HDFs (B) were infected with retroviruses encoding RAS (lanes 2 and 6) or E2F-1 (lanes 4 and 8), or with appropriate vector controls. Because of the extensive apoptosis induced by E2F-1, the cells were analysed at 3 days post-infection, whereas the RAS-expressing cells were harvested at 11 days to maximize the effects (see Figure 6B). Samples (30 µg) were fractionated by SDS–PAGE and immunoblotted for the indicated proteins, using a 15% gel in the case of RAS, phospho-MEK and p14ARF- or p16INK4a-related proteins, and a 10% gel for E2F-1, p53 and Cdk4.
Significantly, the levels of β-ARF/p16 and p14ARF were not altered detectably by RAS (Figure 6, lanes 2 and 6). Here we show the consequences at 11 days post-infection, a time at which exogenous RAS is maximally expressed. This was confirmed both by the increased phosphorylation of MEK (Figure 6, lanes 2 and 6), without concomitant changes in total MEK levels (see Figure 5B), and by the up-regulation of the normal p16INK4a allele in FOO3 cells (Figure 6, lane 6). In the experiment shown, the α-p16/X fusion protein also appeared to be up-regulated by RAS, but the two p16INK4a-related products were not always resolved on gels and the inherent instability of this fusion protein made it difficult to draw definitive conclusions. In summary, under conditions in which levels of ectopic RAS are sufficient to cause p16INK4-mediated arrest of normal HDFs, and over extensive time courses, we saw no changes in ARF levels in either Leiden or FOO3 cells.
Discussion
The identification of a homozygous carrier of the 19 bp deletion in CDKN2A (Gruis et al., 1995) and access to the Leiden and FOO3 HDFs provided a unique opportunity to study the biological properties of p16INK4a and p14ARF and their relative contributions to senescence. The two fusion proteins expressed in Leiden cells can be viewed as generic examples of other frameshift events that affect the CDKN2A locus. Although the consequences will depend on where the lesion occurs and which reading frames are involved, loss of any of the ankyrin repeats that underlie its structure appears sufficient to abrogate p16INK4a function (Ruas and Peters, 1998). In contrast, the functions of ARF can be mimicked by relatively short N-terminal peptides (Lohrum et al., 2000; Midgley et al., 2000; Rizos et al., 2000; Llanos et al., 2001), making it more likely that frameshift products will retain some activity. Here we provide evidence that the β-ARF/p16 fusion protein is present at physiologically relevant levels in Leiden HDFs and show that, by currently available criteria, it is functionally indistinguishable from wild-type ARF (Figure 2). Nevertheless, our rudimentary understanding of the ARF pathway and the lack of a strictly normal baseline in the human population makes it impossible for us to prove that the β-ARF/p16 fusion protein is fully functional; nor can we exclude the possibility that the fusion protein has gained some function not shared by wild-type ARF.
With these caveats in mind, our data imply that the Leiden cells are specifically deficient for INK4a and provide the first genetic evidence that, in HDFs at least, the senescence-like arrest engendered by oncogenic RAS operates via p16INK4a rather than ARF. In practice, lingering doubts about the functionality of β-ARF/p16 are immaterial, because in these cells ARF levels are not demonstrably affected by RAS. Similar conclusions have been reported in other fibroblast strains (Ferbeyre et al., 2000; Wei et al., 2001), and it therefore seems unlikely that Leiden and FOO3 cells have specific defects in the signalling pathways upstream of ARF. Both the wild-type ARF and β-ARF/p16 proteins responded predictably to HPV E6 and E7, E2F1 and MYC (Figure 2 and S.Drayton, personal communication), and the RAS retroviruses used in these studies were clearly able to activate p16INK4a in FOO3 and control HDFs (Figure 6B; Ohtani et al., 2001), with an efficiency that did not require the selection of drug-resistant cell pools.
However, the involvement of INK4a in RAS-mediated senescence in HDFs has to be set against the compelling evidence that ARF performs this role in mouse cells. Thus, MEFs from Arf nullizygous mice are immortal and sensitive to transformation by RAS, whereas MEFs from Ink4a nullizygous mice are mortal and undergo arrest in response to RAS (Kamijo et al., 1997; Krimpenfort et al., 2001; Sharpless et al., 2001). Significantly, oncogenic RAS has been shown to up-regulate p19Arf expression (Palmero et al., 1998; Groth et al., 2000), in striking contrast to the situation we and others have observed in HDFs (Ferbeyre et al., 2000; Wei et al., 2001). At this point, it remains unclear whether these differences reflect diversity in the regulation of the INK4a/ARF locus between different species or different cell types. For example, whereas MEFs spontaneously immortalize by disabling the Arf/p53 pathway rather than Ink4a, the immortalization of Arf nullizygous mouse macrophages seems to require concomitant disruption of Ink4a (Randle et al., 2001).
The absence of a link between RAS and ARF in HDFs could of course explain why we failed to see any consistent effects of RAS on p53, ARF-independent pathways notwithstanding, and why dominant-negative p53 fails to prevent RAS-mediated arrest of normal HDFs (Serrano et al., 1997). However, we remain puzzled by the conflicting reports regarding p53 and the RAS–RAF– MEK pathway from laboratories doing ostensibly similar experiments (Figure 6B; Serrano et al., 1997; Lin et al., 1998; Zhu et al., 1998; Ferbeyre et al., 2000). In our hands, different HDF strains, different RAS retroviruses and different p53 antibodies have given the same neutral outcome, but we have not formally excluded RAS-specific modifications of p53 that render it insensitive to particular antibodies (Webley et al., 2000). However, the antibodies used in our analyses did detect accumulation of p53 in response to E7 (Figure 2C) and UV irradiation (Figure 6A). Another potential confounding factor is that p53 levels are inextricably linked to those of MDM2, which promotes the ubiquitin-mediated degradation of p53 but is itself transcriptionally activated by p53 (Ashcroft and Vousden, 1999) and by RAS (Ries et al., 2000). It would therefore be reasonable to expect that in the absence of any protection by ARF, RAS could cause a reduction in p53 levels by activating MDM2. Such an outcome was indeed reported for human cell lines that lack ARF (Ries et al., 2000).
It was recently demonstrated that hTERT and RAS in combination with either SV40 T-antigen or HPV-16 E6 + E7 will not enable HDFs to form anchorage-independent colonies without some additional function provided by SV40 small tumour antigen (Morales et al., 1999; Hahn et al., 2002). By binding to pRb, T-antigen or E7 should render the cells insensitive to p16INK4a and hence resistant to RAS-mediated arrest, but inactivation of pRb would also be pro-apoptotic without the concomitant inactivation of p53 by T-antigen or E6. We would argue that p16INK4a deficiency will have fewer pleiotropic effects on cell physiology than these DNA tumour virus oncoproteins. In our hands, p16INK4a-deficient cells escaped RAS-mediated arrest and formed anchorage-independent colonies without requiring small t or ablation of the ARF/p53 pathway, but they did not grow as tumours in nude mice. With relatively young Leiden cells, it was also possible to observe anchorage-independent colonies without ectopic expression of hTERT, but these colonies did not have sufficient proliferative capacity to permit a thorough biochemical analysis.
In summary, our results reinforce the notion that the CDKN2A locus operates a front-line defence mechanism against chronic mitogenic or oncogenic stresses, and provide insights into its role in tumour suppression. Whereas these defences seem to be reliant on the ARF product in MEFs, a different scenario prevails in HDFs where p16INK4a may be the more significant player. In view of these striking differences, it will be interesting to investigate these issues in relation to the immortalization and transformation of cells from other lineages and organisms.
Materials and methods
Cell culture
The male patient from whom the Leiden HDFs were isolated was born in 1972 and from the age of 6 developed multiple prominent naevi. By puberty, these had become clinically atypical, several reaching a diameter of 10–12 mm, and seven superficial spreading melanomas have been removed from the back, chest and forehead. Dermal fibroblasts were isolated from a region of normal epidermis adjacent to one of the dysplastic naevi when the patient was 24 years of age. A similar fibroblast culture was isolated from the patient’s brother, at 30 years of age. The TIG-3 strain of embryonic lung fibroblasts and the Hs68 strain of neonatal foreskin fibroblasts (ATCC: CRL 1635) have been described previously (Ohtani et al., 2001). Cell stocks were maintained at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS). The HDF cultures were passaged routinely at a 1:4 split ratio as soon as they reached confluence, and were therefore assumed to have undergone two PDs at each passage.
Retroviral infections
HDF strains were rendered sensitive to mouse ecotropic retroviruses by infecting them with amphotropic retroviruses encoding the mouse basic amino acid transporter, and maintained in medium containing either 150 µg/ml G418 or 1.25 µg/ml puromycin as previously described (McConnell et al., 1998). Ecotropic retroviral stocks were prepared by transient transfection of BOSC-23 cells. For retroviral infections, HDFs expressing the ecotropic receptor were plated at 25–50% confluence and incubated overnight. The culture medium was replaced with 5 ml of filtered viral supernatant together with 3 ml of fresh medium and the equivalent of 4 µg/ml polybrene. After 24 h, the medium was replaced and selection in medium containing either 1.25 µg/ml puromycin (Calbiochem), 50–100 µg/ml hygromycin (Sigma) or 150 µg/ml bleomycin/zeocin (Invitrogen) was initiated on day 2 post-infection.
The pBABE retroviral vectors with different drug selection markers were generously provided by Harmut Land who also supplied pBABE-neo-encoding G12V H-RAS. pBABE puro vectors containing HPV-16 E6 or E7 were constructed by subcloning the relevant EcoRI–SalI restriction fragments. pBABE-hygro-hTERT was provided by Parmjit Jat, and was based on an hTERT cDNA (EcoRI–SalI) originally obtained from Matthew Meyerson. pBABE-bleo-RAS was constructed by subcloning a BamHI–XhoI fragment encoding RAS into the BamHI–SalI sites in pBABE-bleo. Retroviruses encoding E2F-1 and E2F-1-ER were generously provided by Kristian Helin.
Agar colony assays
Between 1 and 2 × 104 cells were suspended in 1 ml of 2× DMEM supplemented with 20% FCS and mixed with an equal volume of 0.4% molten agarose, held at 60°C. The mixture was then layered on top of 2 ml of solidified 1% agarose in DMEM + 10% FCS in a 35 mm dish. The cells were incubated at 37°C and fed with fresh 0.2% agarose/DMEM/FCS every 7 days. Colonies were counted after 21 days. Multiple fields of >100 cells were observed microscopically, and the number of multicellular colonies was estimated as a percentage of the total number of cells visualized.
BrdU incorporation
Cells were seeded in 35 mm dishes and labelled for 18 or 20 h with 5 mM BrdU. Cells were then fixed and stained for BrdU incorporation using a histochemical assay (Boehringer). The percentage of BrdU-positive cells was calculated after viewing 100–500 cells in multiple fields.
Immunofluoresence
NARF2 cells (Stott et al., 1998) were transfected with a pcDNA3-based plasmid encoding β-ARF/p16 and, the following day, the cells were washed and placed in medium containing 1 mM IPTG. After a further 24 h, the cells were fixed and analysed by indirect immunofluorescence as described (Llanos et al., 2001).
Cytogenetic analyses
Pools of Leiden cells infected with retroviruses encoding hTERT, or RAS + hTERT and a number of anchorage-independent colonies recovered from the agarose, were analysed by either G-banding or M-FISH using 24 combinatorially labelled probes with five fluorophores to identify individual chromosomes (Eils et al., 1998) as recommended by the supplier (Vysis). Image processing was performed using Vysis Quips Spectravysion software.
Immunoprecipitation and immunoblotting
Cell monolayers were lysed by suspension in NP-40 lysis buffer (Stott et al., 1998), clarified by centrifugation and the protein concentrations were determined using the BCA assay (Pierce). Immunoprecipitations were performed with 0.5–1.0 mg of lysate plus 1 µg of antibody and 20 µl of a 50% slurry of protein A– or protein G–Sepharose beads (Pierce) by rotating overnight at 4°C. Immunoprecipitates were resuspended in Laemmli sample buffer and boiled. For direct immunoblotting, cells were lysed in 62.5 mM Tris–HCl pH 6.8 containing 2% (w/v) SDS, and the protein concentration determined as above. Mercaptoethanol and Bromophenol Blue were added to make the final composition equivalent to Laemmli sample buffer. Samples were fractionated by SDS–PAGE, blotted onto Immobilon-P membrane (Millipore) and processed as previously described (Stott et al., 1998). Depending on the experiment, sheep horseradish peroxidase (HRP)-coupled anti-mouse (1:1000 dilution) and donkey HRP-coupled anti-rabbit (1:2000 dilution) antibodies were used as secondary antibodies (Amersham). Antibody binding was visualized using Amersham enhanced chemiluminescence (ECL) reagents.
The mAb DO-1 (sc-126) against p53, the mouse mAb against E2F-1 (sc-251), the goat polyclonal antibody against B23 (sc-6013) and rabbit polyclonal antibodies against Cdk4 (sc-601) and Cdk6 (sc-177) were obtained from Santa Cruz. Rabbit polyclonal antisera against MEK1/2 (9122) and phospho-MEK (9121S) were from Cell Signalling, and mAbs against Ras (OP41) and MDM2 (OP46) were from Oncogene Research Products. The mAbs against p14ARF (4C6/4) and p16INK4a (DCS50), and the polyclonal serum (DPAR12) against bacterially expressed p16INK4a have been described previously (Parry and Peters, 1996; Llanos et al., 2001).
In vitro binding assay
Protein–protein interaction assays were performed as previously described using components synthesized in vitro by coupled transcription and translation of plasmid DNAs (Parry and Peters, 1996).
UV treatment of cells
Cells at 60–80% confluence in 90 mm dishes were washed twice in phosphate-buffered saline (PBS) and then overlaid with 2 ml of PBS, at 37°C. The lids were removed and the cells were exposed to 254 nm UV light (15 J/m2). The medium was then replaced and the cells incubated for a further 18 h before harvesting in SDS sample buffer as described above.
p53 stabilization assays
MDM2-mediated degradation of p53 was analysed in transiently transfected cells as previously described (Stott et al., 1998).
Acknowledgments
Acknowledgements
We are grateful to Manuel Serrano, Hartmut Land, Parmjit Jat and Kristian Helin for retroviral constructs, to Jim Koh and Ed Harlow for supplying the JC8 monoclonal antibody, to Denise Sheer and Jill Williamson for help with cytogenetics, and to John Sedivy for communicating results prior to publication. Thanks also to Julian Downward, Eiji Hara, Mike Fried and Alison Sinclair for critical reading of the manuscript. Cancer Research UK London Research Institute comprises the Lincoln’s Inn Fields and Clare Hall Laboratories of the former Imperial Cancer Research Fund (ICRF) following the merger of the ICRF with the Cancer Research Campaign (CRC) in February 2002.
References
- Alcorta D.A., Xiong,Y., Phelps,D., Hannon,G., Beach,D. and Barrett,J.C. (1996) Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence. Proc. Natl Acad. Sci. USA, 93, 13742–13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashcroft M. and Vousden,K.H. (1999) Regulation of p53 stability. Oncogene, 18, 7637–7643. [DOI] [PubMed] [Google Scholar]
- Bodnar A.G. et al. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- Brown J.P., Wei,W. and Sedivy,J.M. (1997) Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science, 277, 831–834. [DOI] [PubMed] [Google Scholar]
- Bunz F., Dutriaux,A., Lengauer,C., Waldman,T., Zhou,S., Brown,J.P., Sedivy,J.M., Kinzler,K.W. and Vogelstein,B. (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science, 282, 1497–1501. [DOI] [PubMed] [Google Scholar]
- Campisi J. (2001) From cells to organisms: can we learn about aging from cells in culture. Exp. Gerontol., 36, 607–618. [DOI] [PubMed] [Google Scholar]
- Cristofalo V.J., Allen,R.G., Pignolo,R.J., Martin,B.G. and Beck,J.C. (1998) Relationship between donor age and the replicative lifespan of human cells in culture: a reevaluation. Proc. Natl Acad. Sci. USA, 95, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eils R., Uhrig,S., Saracoglu,K., Satzler,K., Petersen,I., Chassery,J.M., Ganser,M. and Speicher,M.R. (1998) An optimized, fully automated system for fast and accurate identification of chromosomal rearrangements by multiplex-FISH (M-FISH). Cytogenet. Cell Genet., 82, 160–171. [DOI] [PubMed] [Google Scholar]
- Evan G. and Vousden,K.H. (2001) Proliferation, cell cycle and apoptosis in cancer. Nature, 411, 342–348. [DOI] [PubMed] [Google Scholar]
- Ferbeyre G., de Stanchina,E., Querido,E., Baptiste,N., Prives,C. and Lowe,S.W. (2000) PML is induced by oncogenic ras and promotes premature senescence. Genes Dev., 14, 2015–2027. [PMC free article] [PubMed] [Google Scholar]
- Groth A., Weber,J.D., Willumsen,B.M., Sherr,C.J. and Roussel,M.F. (2000) Oncogenic Ras induces p19ARF and growth arrest in mouse embryo fibroblasts lacking p21Cip1 and p27Kip1 without activating cyclin D-dependent kinases. J. Biol. Chem., 275, 27473–27480. [DOI] [PubMed] [Google Scholar]
- Gruis N.A., van der Velden,P.A., Sandkuiji,L.A., Prins,D.E., Weaver-Feldhaus,J., Kamb,A., Bergman,W. and Frants,R.R. (1995) Homozygotes for CDKN2 (p16) germline mutations in Dutch familial melanoma kindreds. Nature Genet., 10, 351–353. [DOI] [PubMed] [Google Scholar]
- Hahn W.C., Dessain,S.K., Brooks,M.W., King,J.E., Elenbaas,B., Sabatini,D.M., DeCaprio,J.A. and Weinberg,R.A. (2002) Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol., 22, 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E., Smith,R., Parry,D., Tahara,H., Stone,S. and Peters,G. (1996) Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol. Cell. Biol., 16, 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T., Zindy,F., Roussel,M.F., Quelle,D.E., Downing,J.R., Ashmun,R.A., Grosveld,G. and Sherr,C.J. (1997) Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell, 91, 649–659. [DOI] [PubMed] [Google Scholar]
- Kiyono T., Foster,S.A., Koop,J.I., McDougall,J.K., Galloway,D.A. and Klingelhutz,A.J. (1998) Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature, 396, 84–88. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P., Quon,K.C., Mool,W.J., Loonstra,A. and Berns,A. (2001) Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature, 413, 83–86. [DOI] [PubMed] [Google Scholar]
- Lin A.W. and Lowe,S.W. (2001) Oncogenic ras activates the ARF–p53 pathway to suppress epthelial cell transformation. Proc. Natl Acad. Sci. USA, 98, 5025–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A.W., Barradas,M., Stone,J.C., van Aelst,L., Serrano,M. and Lowe,S.W. (1998) Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev., 12, 3008–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos S., Clark,P.A., Rowe,J. and Peters,G. (2001) Stabilisation of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nature Cell Biol., 3, 445–452. [DOI] [PubMed] [Google Scholar]
- Lohrum M.A.E., Ashcroft,M., Kubbutat,M.H.G. and Vousden,K.H. (2000) Contribution of two independent MDM2-binding domains in p14ARF to p53 stabilization. Curr. Biol., 10, 539–542. [DOI] [PubMed] [Google Scholar]
- Lundberg A.S., Hahn,W.C., Gupta,P. and Weinberg,R.A. (2000) Genes involved in senescence and immortalization. Curr. Opin. Cell Biol., 12, 705–709. [DOI] [PubMed] [Google Scholar]
- McConnell B.B., Starborg,M., Brookes,S. and Peters,G. (1998) Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr. Biol., 8, 351–354. [DOI] [PubMed] [Google Scholar]
- Midgley C.A., Desterro,J.M.P., Saville,M.K., Howard,S., Sparks,A., Hay,R.T. and Lane,D.P. (2000) An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene, 19, 2312–2323. [DOI] [PubMed] [Google Scholar]
- Morales C.P., Holt,S.E., Ouellette,M., Kaur,K.J., Yan,Y., Wilson,K.S., White,M.A., Wright,W.E. and Shay,J.W. (1999) Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nature Genet., 21, 115–118. [DOI] [PubMed] [Google Scholar]
- Müller H. et al. (2001) E2Fs regulate the expression of genes involved in differentiation, development, proliferation and apoptosis. Genes Dev., 15, 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N., Zebedee,Z., Huot,T.J.G., Stinson,J.A., Sugimoto,M., Ohashi,Y., Sharrocks,A.D., Peters,G. and Hara,E. (2001) Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature, 409, 1067–1070. [DOI] [PubMed] [Google Scholar]
- Palmero I., McConnell,B., Parry,D., Brookes,S., Hara,E., Bates,S., Jat,P. and Peters,G. (1997) Accumulation of p16INK4a in mouse fibroblasts as a function of replicative senescence and not of retinoblastoma gene status. Oncogene, 15, 495–503. [DOI] [PubMed] [Google Scholar]
- Palmero I., Pantoja,C. and Serrano,M. (1998) p19ARF links the tumour suppressor p53 to Ras. Nature, 395, 125–126. [DOI] [PubMed] [Google Scholar]
- Parry D. and Peters,G. (1996) Temperature-sensitive mutants of p16CDKN2 associated with familial melonoma. Mol. Cell. Biol., 16, 3844–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M. et al. (2000) PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature, 406, 207–210. [DOI] [PubMed] [Google Scholar]
- Randle D.H., Zindy,F., Sherr,C.J. and Roussel,M.F. (2001) Differential effects of p19Arf and p16Ink4a loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc. Natl Acad. Sci. USA, 98, 9654–9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries S., Biederer,C., Woods,D., Shifman,O., Shirasawa,S., Sasazuki,T., McMahon,M., Oren,M. and McCormick,F. (2000) Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell, 103, 321–330. [DOI] [PubMed] [Google Scholar]
- Rizos H., Darmanian,A.P., Mann,G.J. and Kefford,R.F. (2000) Two arginine rich domains in the p14ARF tumour suppressor mediate nucleolar localization. Oncogene, 19, 2978–2985. [DOI] [PubMed] [Google Scholar]
- Rogan E.M. et al. (1995) Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li–Fraumeni syndrome fibroblasts. Mol. Cell. Biol., 15, 4745–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas M. and Peters,G. (1998) The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta, 1378, 115–177. [DOI] [PubMed] [Google Scholar]
- Ruas M., Brookes,S., McDonald,N.Q. and Peters,G. (1999) Functional evaluation of tumour-specific variants of p16INK4a/CDKN2A: correlation with protein structure information. Oncogene, 18, 5423–5434. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lee,H.-W., Chin,L., Cordon-Cardo,C., Beach,D. and DePinho,R.A. (1996) Role of the INK4a locus in tumor suppression and cell mortality. Cell, 85, 27–37. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin,A.W., McCurrach,M.E., Beach,D. and Lowe,S.W. (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell, 88, 593–602. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E. and DePinho,R.A. (1999) The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev., 9, 22–30. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Bardeesy,N., Lee,K.-H., Carrasco,D., Castrillon,D.H., Aguirre,A.J., Wu,E.A., Horner,J.W. and DePinho,R.A. (2001) Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature, 413, 86–91. [DOI] [PubMed] [Google Scholar]
- Stott F.J. et al. (1998) The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J., 17, 5001–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H. and Benchimol,S. (1998) Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative lifespan. Curr. Biol., 8, 279–282. [DOI] [PubMed] [Google Scholar]
- Vogt M., Haggblom,C., Yeargin,J., Christiansen-Weber,T. and Haas,M. (1998) Independent induction of senescence by p16INK4a and p21CIP1 in spontaneously immortalized human fibroblasts. Cell Growth Differ., 9, 139–146. [PubMed] [Google Scholar]
- Webley K., Bond,J.A., Jones,C.J., Blaydes,J.P., Craig,A., Hupp,T. and Wynford-Thomas,D. (2000) Posttranslational modifications of p53 in replicative senescence overlapping but distinct from those induced by DNA damage. Mol. Cell. Biol., 20, 2803–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Wei,W. and Sedivy,J.M. (1999) Expression of catalytically active telomerase does not prevent premature senescence caused by overexpression of oncogenic Ha-Ras in normal human fibroblasts. Cancer Res., 59, 1539–1543. [PubMed] [Google Scholar]
- Wei W., Hemmer,R.M. and Sedivy,J.M. (2001) The role of p14ARF in replicative and induced senescence of human fibroblasts. Mol. Cell. Biol., 21, 6748–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. and Riabowol,K. (1996) Differential CDK-inhibitor gene expression in aging human diploid fibroblasts. Exp. Gerontol., 31, 311–325. [DOI] [PubMed] [Google Scholar]
- Wright W. and Shay,J.W. (2000) Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nature Med., 6, 849–851. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xiong,Y. and Yarbrough,W.G. (1998) ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell, 92, 725–734. [DOI] [PubMed] [Google Scholar]
- Zhu J., Woods,D., McMahon,M. and Bishop,J.M. (1998) Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev., 12, 2997–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F., Eischen,C.M., Randle,D.H., Kamijo,T., Cleveland,J.L., Sherr,C.J. and Roussel,M.F. (1998) Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev., 12, 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]