Abstract
Studies in model organisms have contributed to elucidate multiple levels at which regulation of eukaryotic DNA replication occurs. Cdc7, an evolutionarily conserved serine–threonine kinase, plays a pivotal role in linking cell cycle regulation to genome duplication, being essential for the firing of DNA replication origins. Binding of the cell cycle-regulated subunit Dbf4 to Cdc7 is necessary for in vitro kinase activity. This binding is also thought to be the key regulatory event that controls Cdc7 activity in cells. Here, we describe a novel human protein, Drf1, related to both human and yeast Dbf4. Drf1 is a nuclear cell cycle-regulated protein, it binds to Cdc7 and activates the kinase. Therefore, human Cdc7, like cyclin-dependent kinases, can be activated by alternative regulatory subunits. Since the Drf1 gene is either absent or not yet identified in the genome of model organisms such as yeast and Drosophila, these findings introduce a new level of complexity in the regulation of DNA replication of the human genome.
Keywords: Cdc7/cell cycle/cyclin/Dbf4/DNA replication
Introduction
In eukaryotic organisms, the duplication of the genome requires the action of at least two kinases: a cyclin-dependent kinase (CDK), Cdk2 in metazoans, and the Cdc7 kinase. Both of these kinases need to associate with a regulatory subunit in order to become fully activated. Two related proteins, cyclin E and cyclin A, bind and activate Cdk2 to regulate entry and progression through S phase, respectively. While the levels of Cdk2 do not change during the cell cycle, cyclin E and A have different timing of expression. Cell cycle fluctuation of these cyclins plays a major role in determining the timing of activation and substrate specificity of Cdk2 (reviewed in Sherr, 1993; Harper and Adams, 2001).
The human Dbf4 gene was identified by a ‘two-hybrid’ screening using Cdc7 as bait. Physical interaction between Cdc7 and Dbf4 was confirmed further both in human cells and in insect cells infected with recombinant baculoviruses. Binding of Dbf4 to Cdc7 was shown to activate the kinase (Jiang et al., 1999; Kumagai et al., 1999). Cdc7 and Dbf4 proteins are well conserved during evolution (Johnston et al., 2000; Sclafani, 2000). In budding yeast, Dbf4 protein, like cyclins, oscillates during the cell cycle, it accumulates in S phase and is targeted for ubiquitin-mediated degradation by the anaphase-promoting complex (APC) at the end of mitosis (Oshiro et al., 1999; Weinreich and Stillman, 1999; Ferreira et al., 2000). In human cells, Dbf4 levels also oscillate during the cell cycle: in particular, Dbf4 accumulation, formation of a Cdc7– Dbf4 complex and Cdc7 kinase activity appear to fluctuate with approximately the same kinetics, suggesting that Cdc7 binding to Dbf4 is the main level of regulation of the kinase (Jiang et al., 1999; Kumagai et al., 1999). As in the case of CDKs, phosphorylation events may play an important regulatory role (Masai et al., 2000).
Basic mechanisms that regulate origin activation appear to be highly conserved and have been characterized thoroughly in model organisms such as yeast and Xenopus. At the end of mitosis and in G1, a pre-replicative complex is formed around the DNA origin. This contains the origin recognition complex (ORC), Cdc6, Cdt1 and minichromosome maintenance (MCM) proteins (reviewed in Diffley and Labib, 2002). In budding yeast, Cdc7 kinase is required for the firing of DNA replication origins (Bousset and Diffley, 1998; Donaldson et al., 1998). It is thought that Dbf4 targets the kinase to pre-replicative complexes bound at origins and that, once in S phase, Cdc7 phosphorylates one or more subunits of the MCM complex, thus converting dormant pre-replicative complexes into two active replication forks (Dowell et al., 1994; Lei et al., 1997). Genetic evidence supports this notion, since the bob1-1 mutation in the MCM5 gene allows bypass of the requirement for both Cdc7 and Dbf4 for yeast DNA replication (Hardy et al., 1997). Importantly, once the activation step has occurred, neither Cdc7 nor Dbf4 appears to be necessary for ongoing replication (Bousset et al., 1998; Jares and Blow, 2000). The essential role of human Cdc7 and Dbf4 in the replication of the genome was established through microinjection of neutralizing antibodies (Jiang et al., 1999; Kumagai et al., 1999).
Sequence analysis together with mutagenesis studies have defined a common architecture in the Dbf4-related protein family from different species. Three conserved amino acid boxes that, from the N- to the C-terminus of the protein, are called N, M and C motifs, respectively, can be identified (Masai and Arai, 2000). The M and C motifs define a bipartite module important for binding and activating the catalytic subunit (Masai et al., 2000; Ogino et al., 2001). The function of the N motif is not clear but, because of its similarity to a BRCT domain, it appears to mediate protein–protein interactions, possibly targeting Cdc7–Dbf4 complexes to origins of replication.
The full sequencing of Saccharomyces cerevisiae and Drosophila melanogaster genomes has now revealed that only a single Dbf4-related gene exists in these organisms while, intriguingly, two genes showing significant sequence similarity, Dfp1/Him1 and Spo6, are present in Schizosaccharomyces pombe. Genetic and biochemical studies have now shown that while Dfp1/Him1 binds to the S.pombe Cdc7 homolog Hsk1 and is involved in DNA replication (Masai et al., 1995; Brown and Kelly, 1998), Spo6 associates with a different kinase, Spo4. This kinase is related to Hsk1 and is required for progression through meiosis II and sporulation (Nakamura et al., 2000, 2002).
In order to understand fully the roles of the human Cdc7 kinase in the regulation of the cell cycle, we set out to look for new Cdc7 partners. Using a bioinformatic approach, we have identified a new gene coding for a protein, named Drf1, that has sequence similarity to Dbf4. This work provides experimental evidences that Drf1 is a novel, cell cycle-regulated protein that activates human Cdc7 kinase.
Results
Identification of the Drf1 gene
To identify new partners for the Cdc7 kinase, we screened public and the LGtemplatesFEB2000 Incyte database using the whole human Dbf4 sequence as query. While at the time no matches were found in the public databases, two templates with low similarity to two different regions of Dbf4 were identified in the Incyte database (Incyte unique 150210.1 and Incyte unique 143458.1). Two clones belonging to the first template (3051528 and 477042) were fully sequenced. They were found to contain partially overlapping DNA fragments that defined a single full-length cDNA of 3006 bp. These sequence data have been submitted to the DDBJ/EMBL/GenBank database under accession No. AF448801. Analysis of the assembled sequence revealed the presence of an open reading frame (ORF) coding for a protein of 615 amino acids with a predicted mol. wt of 67 239 Da. Because of its similarity to Dbf4, we decided to give this new gene and the corresponding protein the name Drf1, for Dbf4-related factor 1. The similarity of Drf1 to Dbf4 is significant in the N-terminal regions of the proteins and within the three amino acid boxes that characterize the Dbf4 protein family. The similarity decreases in the C-terminal region (Figure 1A). Prosite scanning of the Drf1 sequence identifies a putative bipartite nuclear localization signal (PS50079) located just before the M box (amino acids 207–224). No other motifs within confidence levels were found, although a putative BRCT domain overlapping with the N box was detected at a low score (amino acids 43–96).
Fig. 1. Drf1 sequence analysis. (A) Sequence alignment of human Drf1 (top) and Dbf4 (bottom) proteins. Identical amino acids are on a black background; conserved substitutions are in gray. The N, M and C motifs conserved in the Dbf4 protein family are indicated by the boxes. The Drf1 putative bipartite nuclear localization signal is underlined. (B) Phylogenetic tree of Dbf4-related proteins: Hs: Homo sapiens, Mm: Mus musculus, Dm: Drosophila melanogaster, Sc: Saccharomyces cerevisiae, An: Aspergillus nidulans, Sp: Schizosaccharomyces pombe.
Phylogenetic analysis shows a close evolutionary relationship between Drf1 and Dbf4 proteins, indicating that Drf1 belongs to the same protein family (Figure 1B). To date, however, we have not been able to identify other genes related to Drf1 in other species in the available databases. When the Drf1 cDNA sequence was used as bait to search the draft of the human genome, we found that Drf1 identifies the contig _010765 corresponding to a region of chromosome 17q21.
We analyzed Drf1 expression in different tissues and cell lines by northern blotting. Drf1 generally is highly expressed in testis and in all tumor cell lines we analyzed, with the exception of the A549 (small lung cell carcinoma), while it is poorly expressed in most other tissues (data not shown). A similar expression pattern was observed previously for Dbf4 (Kumagai et al., 1999), suggesting that the two molecules might be co-regulated in cells.
Reconstitution of recombinant Cdc7–Drf1 kinase
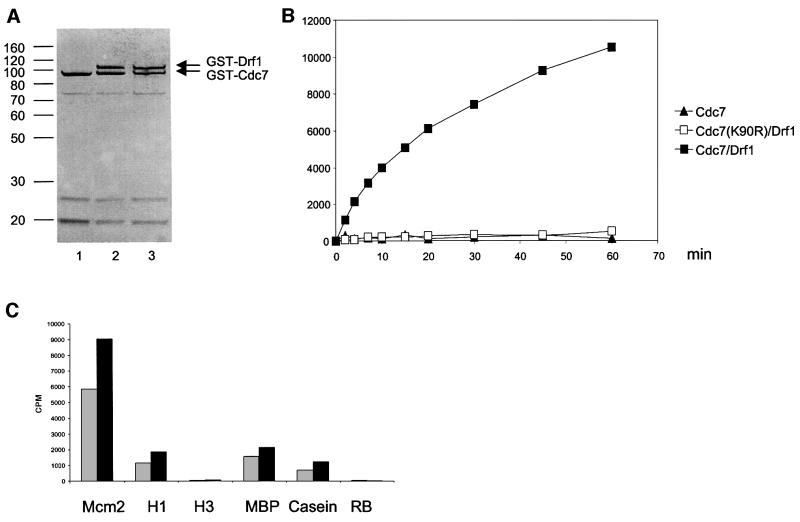
The presence of amino acid motifs in the Drf1 protein similar to those that in Dbf4 are required for binding to Cdc7 (Masai and Arai, 2000) prompted us to test whether Drf1 could also directly bind to Cdc7 kinase. We therefore cloned Drf1, wild-type and mutant Cdc7(K90R) sequences into a baculovirus expression vector, thus obtaining a recombinant virus expressing these proteins as a fusion with GST. Lys90 is a critical residue for the ATP-binding domain, and mutation at this residue to arginine was shown to abolish the enzymatic activity of the Cdc7–Dbf4 complex (Jiang et al., 1999; Masai et al., 2000). Hi5 insect cells were infected with the Cdc7 virus alone or with the Drf1 virus in combination with either wild-type Cdc7 or mutant Cdc7(K90R) virus. Recombinant GST–Cdc7, GST–Cdc7/GST–Drf1 and GST–Cdc7(K90R)/GST–Drf1 proteins were purified in a two-step procedure involving glutathione–Sepharose affinity chromatography followed by size exclusion chromatography. This procedure allowed a purification of >90% and the separation of the Cdc7–Drf1 complex from free Cdc7. Purified proteins were separated on a polyacrylamide gel and visualized by silver staining. In these purified samples, we found that Drf1 and Cdc7 polypeptides were present in approximately the same amount and that mutation of Lys90 does not detectably affect the relative abundance of these proteins (Figure 2A).
Fig. 2. Purification and characterization of the Drf1–Cdc7 kinase complex. (A) Cdc7 (lane 1), Cdc7–Drf1 (lane 2) and Cdc7(K90R)/Drf1 (lane 3) recombinant proteins purified from insect cells as a fusion with GST were separated on a 10% polyacrylamide gel and stained with silver. (B) For each time point, 50 ng of recombinant Cdc7 (closed triangles), Cdc7–Drf1 complex (closed squares) or Cdc7(K90R)/Drf1 (open squares) were incubated in the presence of the Mcm2 N-terminal fragment (amino acids 1–285) and labeled ATP. Protein kinase activity of these proteins was measured using multiscreen plates as described in Materials and methods. (C) The Cdc7–Drf1 complex was incubated in the presence of 6 µM of the indicated proteins as substrate. After 30 (gray bars) or 60 (black bars) min, the reaction was stopped and proteins separated on a polyacrylamide gel. The amount of radioactivity in the different substrates was measured with a Storm 840 Phosphoimager. Mcm2 corresponds to the N-terminal 285 amino acid fragment of the Mcm2 protein, H1 and H3 to histone H1 and H3, respectively, MBP to myelin basic protein; and RB to the retinoblastoma protein.
Next, we tested these samples for the presence of kinase activity using a fragment of the Mcm2 protein (amino acids 1–285) as substrate. Mcm2 (amino acids 1–285) was chosen for these experiments because it is known to be an excellent substrate for the Cdc7–Dbf4 complex in vitro (Sato et al., 1997). As shown in Figure 2, kinase activity was only detected in samples containing wild-type Cdc7 and Drf1 but not in samples containing either Cdc7 alone or a kinase-dead Cdc7–Drf1 complex. The specific activity of the Cdc7–Drf1 complex in this preparation was ∼6.5 nmol of phosphate transferred to the Mcm2 fragment/min/mg. ATP titration indicated that the apparent Km for ATP is ∼0.5 µM, similar to what was observed previously for the Cdc7–Dbf4 complex (Masai et al., 2000). When we used different proteins as substrate for Cdc7–Drf1 kinase at saturating concentration, we observed 4–5 times more activity with the Mcm2 fragment compared with histone H1 and myelin basic protein (MBP), and ∼8 times more when compared with casein; we failed to detect any activity using the retinoblastoma protein and histone H3 (Figure 2). This suggests that in vitro, Cdc7–Drf1, as previously observed for Cdc7–Dbf4, has substrate preference for Mcm2 (Masai et al., 2000; Sclafani, 2000).
Drf1 interacts with Cdc7 kinase in mammalian cells
To examine the ability of Drf1 to bind to Cdc7 in mammalian cells, the Drf1-coding sequence was cloned into the expression vector pCDNA-HA. With this construct, expression of Drf1 protein tagged with a single hemagglutinin (HA) epitope is driven by the cytomegalovirus (CMV) promoter. Flag-tagged wild-type or mutant Cdc7(K90R) expression plasmids were also prepared. These constructs were transfected in 293 cells in various combinations and cells were harvested 3 days post-transfection. Transfected HA-Drf1 protein normally migrates in polyacrylamide gels with an apparent mol. wt of ∼75 kDa. However, when it is co-transfected with Cdc7, the mobility of Drf1 is greatly reduced and it can be detected as a smear of bands between 75 and 105 kDa. The extent of this mobility shift appears to be variable, depending on the ratio of Drf1 and Cdc7 plasmids and on the time of cell collection after transfection (data not shown). Since Drf1 protein mobility shift is detected in the presence of a functional Cdc7 but not with kinase-dead Cdc7 (Figure 3A, lanes 1 and 2), we reasoned that the HA-Drf1 mobility shift may be due to Cdc7-dependent phosphorylation. This appears to be the case, since Drf1 mobility shift is reversed by treatment with intestinal calf phosphatase in a reaction that is blocked by addition of phosphatase inhibitors (Figure 3A,lanes 3–5).
Fig. 3. Characterization of Cdc7–Drf1 complexes in human cells. (A) Drf1 is phosphorylated when co-transfected with functional Cdc7. 293 cells were transiently transfected with HA-Drf1 in combination with either wild-type or kinase-deficient (K90R)Cdc7 constructs as indicated. Protein samples were either kept on ice (lanes 1 and 2) or incubated with or without calf intestinal phosphatase in the absence or presence of phosphatase inhibitors as indicated (lanes 3–5) before western blot analysis with anti-HA antibodies. (B and C) Formation of active Cdc7–Drf1 complexes in human cells. Protein extracts from 293 cells transfected with the indicated constructs were immunoprecipitated with either anti-HA (lanes 1–4) or anti-Flag (lanes 5–8) antibodies and subsequently incubated with radioactive ATP and a fragment of Mcm2 protein for 15 min before SDS–PAGE. In (B), Drf1 and Cdc7 were visualized with anti-HA or anti-Cdc7 antibodies, respectively. In (C), labeled proteins were visualized by autoradiography. Images correspond to different exposure times: a longer exposure was required to detect phosphorylated Drf1 and Dbf4 compared with phospho-MCM2. (D) Dbf4 does not immunoprecipitate with Drf1. Protein extracts from HeLa cells transfected with the indicated constructs were probed with anti-HA and anti-Cdc7 antibodies in western blot experiments in lanes 1 and 2. Drf1 and Dbf4, both tagged with the HA epitope, are indicated and migrate differentially in these gels. In lanes 3–5, extracts were immunoprecipitated with either anti-Drf1 mAb 5G4 or unrelated murine IgGs before western blot analysis. (E) Anti-Drf1 mAbs pull-down Cdc7 from extracts prepared from exponentially growing HeLa cells. In each lane, 4 mg of whole-cell extract were used in immunoprecipitation experiments with the anti-Drf1 mAbs 5G4 and 5H4 or control IgG. Western blot was performed with affinity-purified anti-Cdc7 polyclonal antibodies.
Protein extracts prepared from transfected 293 cells as above were used in immunoprecipitation experiments with either anti-HA or anti-Flag antibodies. Before separating the proteins on acrylamide gel and western blot analysis, immunocomplexes were also incubated for 15 min in the presence of radioactive ATP and the Mcm2 fragment in kinase reaction buffer. The presence of HA-Drf1 and Cdc7 in the immunocomplexes was detected using anti-HA antibodies or anti-Cdc7 polyclonal antibodies, respectively. As shown in Figure 3B, anti-HA antibodies were able to immunoprecipitate Drf1 together with Flag-Cdc7. We also observed a strong immunoreactive band that migrates slightly faster than Flag-Cdc7 (lanes 1–3). This band corresponds to the endogenous Cdc7, since it cross-reacts with anti-Cdc7 antibodies and shows the same electrophoretic mobility as Cdc7 (data not shown). Anti-HA antibodies did not precipitate Cdc7 when HA-Drf1-expressing plasmid was not used in the transfection (lane 4). Reciprocally, anti-Flag antibodies efficiently precipitated transfected Flag-Cdc7 together with HA-Drf1 (lanes 6 and 7). As a negative control, immunoprecipitation was also performed from extract overexpressing only Cdc7 in the absence of Ha-Drf1 (lane 8). Both wild-type and mutant Cdc7 were able to co-precipitate with Drf1 with the same efficiency (Figure 3B, compare lanes 1 and 2, and 6 and 7), indicating that the interaction between these two proteins is not affected by the mutation in the catalytic site as previously observed with baculovirus-expressed proteins. We also observed that both modified and unmodified Drf1 were found in the anti-HA immunoprecipitate (lane 2) while only modified Drf1 is detected in the anti-Flag immunoprecipitate (lane 7). This result suggests that only when Drf1 is bound to Cdc7 can it be phosphorylated efficiently on those residues responsible for the change in the mobility. Finally, since we did not find endogenous Cdc7 in the anti-Flag immunoprecipitates (Figure 3B, lanes 5–8), dimerization of Cdc7 subunits does not occur in either the presence or absence of Drf1.
Labeled phosphoproteins on the same filter were visualized subsequently by autoradiography. When both HA-Drf1 and wild-type Cdc7 were present in the immunocomplex, we observed efficient phosphorylation of Mcm2, irrespective of whether anti-HA antibody or anti-Flag antibody had been used for the immunoprecipitation (Figure 3C). As previously shown, phosphorylation of Mcm2 protein by Cdc7 kinase results in a reduced electrophoretic mobility of the substrate (compare lane 2 with lane 1, and lanes 5 and 7 with lane 6) (Masai et al., 2000). Since Cdc7 is not active in the absence of a regulatory subunit (Jiang et al., 1999; Kumagai et al., 1999) and overexpression of mutant Cdc7(K90R) strongly reduces the activity (compare lane 1 with lane 2, and lane 6 with lane 7), we reason that the kinase activity observed in the immunoprecipitates containing both Drf1 and wild-type Cdc7 is due to the formation of Cdc7–Drf1 active complexes as observed for Cdc7–Dbf4 (Figure 3C, lane 5; Jiang et al., 1999; Kumagai et al., 1999). Furthermore, a labeled phosphoprotein migrating at the same molecular weight as Drf1 could be detected only when Drf1 was transfected in combination with wild-type but not with the kinase-dead Cdc7.
Altogether, these results indicate that Drf1 associates with Cdc7, that it activates the kinase and that Drf1 itself is a substrate of Cdc7 kinase.
In order to understand whether Cdc7 can bind to both Drf1 and Dbf4 simultaneously, HeLa cells were transiently transfected with plasmids overexpressing Flag-Cdc7, HA-Drf1 and HA-Dbf4. HA-Drf1 and HA-Dbf4 proteins in these extracts were detected simultaneously using anti-HA antibodies and can be identified by their different molecular weights (Figure 3D, lanes 1 and 2). Immunoprecipitations were performed with either anti-Drf1 monoclonal antibody (mAb) 5G4 or control IgG. Using mAb 5G4, we were able to pull-down Drf1 together with Cdc7, independent of the presence of Dbf4 (Figure 3D, lanes 4 and 5). However, we failed to detect any Dbf4 protein in the immunoprecipitate when it was present in the extract (lane 5). This observation suggests that Drf1, Dbf4 and Cdc7 do not form ternary complexes efficiently and the binding of Drf1 and Dbf4 to Cdc7 is mutually exclusive under these conditions. When unrelated mouse IgGs were used instead, neither Drf1, Cdc7 nor Dbf4 were found in the beads (Figure 3D, lane 3). Further experiments with purified proteins will be required to understand fully the binding mode of Drf1 and Dbf4 to Cdc7.
Finally, we asked if Drf1–Cdc7 interaction could be observed with endogenous proteins. To this end, extract prepared from exponentially growing HeLa cells was immunoprecipitated with either two different mAbs generated against Drf1 protein (mAb 5G5 and mAb 5H4) or unrelated mouse IgG. These immunocomplexes were then tested for the presence of Cdc7 protein using anti-Cdc7 polyclonal antibodies. Figure 3E shows that endogenous Cdc7 can be detected in both samples immunoprecipitated with the anti-Drf1 mAbs (lanes 1 and 2) but not in the control sample (lane 3), indicating that binding of endogenous Drf1 to endogenous Cdc7 occurs in HeLa cells.
Cell cycle regulation of Drf1 expression
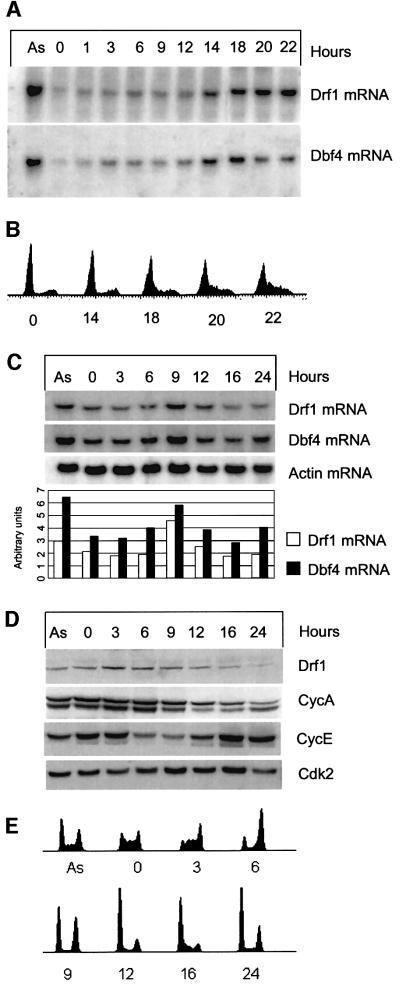
It was observed previously that human Cdc7 kinase activity fluctuates during the cell cycle, peaking in S phase. It is believed that this cell cycle regulation occurs mainly at the level of binding with the Dbf4 regulatory subunit that is per se cell cycle regulated at both the mRNA and protein level (Jiang et al., 1999; Kumagai et al., 1999). We therefore investigated the possibility that Drf1 expression might also be cell cycle regulated. Levels of Drf1 mRNA during the cell cycle were studied by northern blot analysis in normal human dermal fibroblasts (NHDFs). Cells were arrested by serum starvation and stimulated to enter into the cell cycle by the addition of 10% serum. RNA samples were taken at different times after stimulation. Expression of both Dbf4 and Drf1 was low in G0-arrested cells and throughout G1; however, when cells reach the G1/S border, ∼14 h post-stimulation, both Dbf4 and Drf1 mRNAs begin to accumulate. Interestingly, we observed that while Drf1 mRNA keeps accumulating in S phase, Dbf4 levels begin to decrease at 20 h post-stimulation. Therefore, induction of Drf1 and Dbf4 upon re-entry into the cell cycle is a late G1 event that precedes initiation of S phase (Figure 4A and B). To expand the timing studies, we performed thymidine block and release experiment with the same fibroblasts. Samples for RNA, protein and fluorescence-activated cell sorting (FACS) analysis were taken. FACS analysis shows that thymidine-treated NHDFs are arrested in S phase, with a broad distribution of early, middle and late S phase cells. After release, cells recover DNA synthesis and synchronously go through the cell cycle. At 6 h post-release, most of the cells have a 4N DNA content; by 9 h, half of the population has completed mitosis; and by 12 h the majority are in G1. By 24 h post-release, cell synchrony appears to be lost. Under these conditions, Drf1 and Dbf4 mRNAs exhibit a moderate fluctuation during the cell cycle. Their levels peak at 9 h post-release, showing a 2- to 2.5-fold change between the lowest and highest levels (Figure 4C). Western blot analysis of protein extracts of the same experiment with the anti-Drf1 mAb 5H4 showed that Drf1 protein levels, in contrast to mRNA levels, first increase after the release from the thymidine block and then decrease when most of the cells appear to leave mitosis, with kinetics similar to the cyclin A decrease. In the same experiment, cyclin E protein fluctuates during the cell cycle as previously described (Ohtsubo et al., 1995; Ekholm and Reed, 2000).
Fig. 4. Cell cycle analysis of Drf1 and Dbf4 expression. (A and B) Drf1 and Dbf4 mRNAs are induced upon re-entry into the cell cycle. NHDFs were arrested in G0 by serum starvation and then stimulated by addition of 10% serum. At the indicated times, RNA was prepared and Drf1 and Dbf4 mRNA levels analyzed by northern blotting. (B) The DNA content at the indicated times measured by FACS. (C, D and E) Fluctuation of S-phase cyclins and Cdc7 regulatory subunits during the cell cycle in normal human fibroblasts. NHDFs were blocked in S phase with thymidine and released into fresh medium. (C) Drf1, Dbf4 and β-actin mRNA levels were analyzed by northern blotting. The bar chart represents the quantification of Drf1 and Dbf4 levels at different times after normalization with respect to β-actin. Arbitrary units are given on the y-axis. Data were obtained by analyzing radioactivity present in each lane for each hybridization using the Molecular imager FX phosphoimager and Quantity One software (Bio-Rad). (D) Western blot analysis of Drf1, cyclin E, cyclin A and cdk2 levels. (E) The DNA content at the indicated times measured by FACS.
This experiment indicates that Drf1 protein is regulated during the cell cycle and suggests that, since Drf1 protein levels do not fully mirror mRNA levels, post-transcriptional mechanisms regulating Drf1 might exist.
Drf1 is a protein with a short half-life
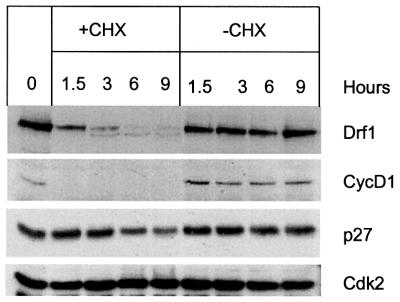
Previous experiments show that Drf1 protein acts as a ‘cyclin’ for Cdc7 kinase: it binds to Cdc7, activates Cdc7 kinase and fluctuates during the cell cycle, although with different kinetics compared with its mRNA. Cyclin regulation is achieved both by cell cycle-dependent transcription and by controlled proteolysis (Ekholm and Reed, 2000; Tyers and Jorgensen, 2000). Dbf4, at least in budding yeast, has also been shown to be an unstable protein specifically degraded at the end of mitosis in an APC-dependent manner (Oshiro et al., 1999; Weinreich and Stillman, 1999; Ferreira et al., 2000). We therefore measured the half-life of Drf1 protein in human cells. HeLa cells were transiently transfected with HA-Drf1 plasmid, and 48 h later the protein synthesis inhibitor cycloheximide was added. Cells were collected at different times, and protein extracts were prepared and analyzed by western blot with anti-Drf1 antibodies. Figure 5 shows that Drf1 levels sharply decrease after addition of cycloheximide, while constant levels are seen in mock-treated samples. This result indicates that Drf1, at least when overexpressed, is a highly unstable protein. Densitometric analysis of the same blot suggests that Drf1 has a half life ≤1.5 h. In the same experiment, the half-lives of cyclin D1 and the CDK inhibitor p27 were <90 min and ∼6 h, respectively, as previously reported (Matsushime et al., 1992; Rodier et al., 2001). Cdk2 levels were found to be constant throughout the experiment. Intriguingly, we observed that a very small fraction of HA-Drf1 appears to be stable since it is detected with the same intensity at 6 and 9 h after cycloheximide treatment. We cannot exclude at this stage the possibility that the binding to a limiting cellular component might influence Drf1 protein stability.
Fig. 5. Drf1 is a protein with a short half-life. HeLa cells overexpressing HA-Drf1 were either treated with cycloheximide or mock treated. At the indicated times, protein extracts were prepared and levels of the indicated protein analyzed by western blot.
Drf1 is a nuclear protein
Consistent with their role in regulation of DNA synthesis, both Cdc7 and Dbf4 are nuclear proteins (Jiang and Hunter, 1997; Sato et al., 1997; Kumagai et al., 1999). We therefore tested whether Drf1 was also localized in the nuclear compartment. HeLa cells were transiently transfected with plasmid expressing HA-Drf1 and, after 3 days, they were stained with anti-HA antibodies. Figure 6A shows that overexpressed Drf1 can be clearly detected in the nucleus of transfected cells and it is generally excluded from the nucleolus. In a fraction of cells, Drf1 protein accumulates in particular subnuclear compartments as it appears as a punctuate staining. This pattern is reminiscent of both Cdc7 and Dbf4 protein staining (Sato et al., 1997; Kumagai et al., 1999). In cells overexpressing Drf1 together with Cdc7, we observed that Drf1 and Cdc7 staining was significantly overlapping, as expected from the fact that the two proteins can form stable complexes (Figure 6B). Finally, in order to understand the nature of Drf1 punctuate staining, we performed co-localization experiments with sites of DNA synthesis. HeLa cells were transfected with HA-Drf1 plasmid and, 24 h post-transfection, bromodeoxyuridine (BrdU) was added to the medium for 1 h. Cells were fixed and processed for the simultaneous detection of Drf1 protein and BrdU foci by confocal microscopy. Again we found that all Drf1-positive cells showed nuclear staining. Nuclear localization is not linked to the S phase of the cell cycle, since it was observed in both BrdU-positive and BrdU-negative cells (data not shown). Analysis of the images of transfected cells that are actively duplicating their DNA indicates that Drf1 does not co-localize with replication foci but, intriguingly, it appears to be excluded (Figure 6C).
Fig. 6. Drf1 is a nuclear protein. (A) Staining of HeLa cells expressing Ha-Drf1. Staining with anti-HA antibodies is shown in red, while DAPI staining is shown in blue. (B) Confocal microscopy analysis of Drf1 and Cdc7 localization in HeLa cells overexpressing HA-Drf1 and V5-Cdc7. Staining with anti-V5 antibodies is shown in red, while staining with anti-HA antibodies is shown in green. The last picture represents a merged image of the previous two. (C) Confocal microscopy analysis of Drf1 staining and replication foci. HA-Drf1 is shown in red and the BrdU staining in green. The last picture represents a merged image of the previous two.
Discussion
Cell cycle progression is driven by orderly activation of CDKs. The first step of CDK activation is the binding of the cyclin to the catalytic subunit. This binding causes several changes in the conformation of the catalityc subunit, particularly in the ATP-binding pocket (Jeffrey et al., 1995). G1/S transition and S phase progression also require the Cdc7 kinase activity. Although structural information is not available to date for Cdc7 kinase, many biochemical studies indicate that binding of a regulatory subunit to Cdc7 strongly affects its enzymatic activity. Different cyclins that can bind to a given CDK have been identified. Some of the redundancy in the number of cyclins is due to the fact that similar cyclin subtypes are expressed at different phases of development and in different cellular lines, but, most importantly for regulation of cell cycle events, the binding of alternative cyclins allows determination of substrate specificity and timing of activation of the kinase (reviewed in Harper and Adams, 2001). In contrast to cyclins and CDKs, studies in model organisms seem to indicate that only one regulatory subunit binds and activates Cdc7, and this is required for entry into S phase (reviewed in Johnston et al., 2000). Since the basic mechanisms that regulate DNA replication appear to be highly conserved during evolution, it was conceivable that human Cdc7 also had a single cell cycle-regulated activating subunit. Our findings now challenge this assumption and indicate that Cdc7, just like some CDKs, makes use of at least two alternative subunits, Dbf4 and Drf1.
Characterization of recombinant Cdc7–Drf1 complexes unambiguously demonstrates that Drf1 activates Cdc7 kinase. Also, when overexpressed in human cells, we have observed that Drf1 binds to Cdc7 and activates the enzyme. Drf1 itself becomes a substrate for the kinase, probably only when bound to Cdc7, and this phosphorylation produces a significant electrophoretic mobility shift, just like previously observed for Dbf4 (Jiang et al., 1999; Kumagai et al., 1999). Whether Drf1 and Dbf4 phosphorylation by Cdc7 has functional implications is not known to date.
Our studies indicate that Drf1 is regulated at multiple levels, in a similar manner to S phase cyclins. Drf1 expression responds to growth signals, and its mRNA is induced after serum stimulation of quiescent cells. Under these conditions, induction of Drf1 and Dbf4 mRNAs occurs prior to entry into S phase, as has also been observed for several genes involved in the initiation of DNA replication (Ohtani et al., 1996; Leone et al., 1998). In a thymidine block and release experiment, we observed a similar fluctuation for both Drf1 and Dbf4 mRNAs. Under these conditions, the peak of expression is visible at the time at which half of the population is preparing to enter mitosis while half of it is already in G1. We speculate that the peak of Dbf4 and Drf1 expression occurs before cell division, since previous studies have already shown that Dbf4 mRNA is present during most of the cell cycle, with the exception of early G1 phase (Kumagai et al., 1999), and we observed high levels of the two mRNAs in nocodazole-treated HeLa cells (data not shown). Timing of Drf1 and Dbf4 expression is very similar, although it appears that after serum stimulation, Dbf4 mRNA declines before Drf1 mRNA. Nevertheless, Drf1 protein accumulates when cells are in S phase and decreases when cells exit mitosis. The discrepancy observed between Drf1 mRNA and protein levels suggests additional levels of regulation. Drf1 protein decreases with similar kinetics to cyclin A, whose levels are known to be regulated through ubiquitin-mediated proteolysis by the APC (Peters, 1999; Geley et al., 2001). Drf1 itself, when overexpressed, is a protein with a short half-life. These observations suggest that a similar regulated degradation might be the main mechanism controlling Drf1 protein levels. This idea is supported further by the notion that the stability of the only budding yeast homolog, Dbf4 protein, is also regulated (Oshiro et al., 1999; Weinreich and Stillman, 1999; Ferreira et al., 2000). Further work will be necessary to elucidate mechanisms of Drf1 degradation and to understand whether the binding of Drf1 to other cellular factors might contribute to regulate its stability.
Drf1 function is not yet understood, although, on the grounds of its interaction with Cdc7, some speculations can be made. A great deal of evidence indicates that Cdc7 kinase is important for the firing of replication origins in eukaryotic cells. Origin firing occurs in a predictable temporal pattern, with some origins firing early and some late in S phase (Fangman and Brewer, 1992). The finding that Cdc7 kinase in human cells has two regulatory subunits, Dbf4 and Drf1, that can bind and activate the enzyme, together with the fact that both subunits are nuclear and expressed in S phase, could suggest that different Cdc7 complexes might be responsible for activation of specific subsets of replication origins, e.g. Cdc7–Dbf4 is important for firing of early origins while Cdc7–Drf1 is important for the firing of late origins. The lack of co-localization of Drf1 with the sites of BrdU incorporation might therefore imply that Drf1, as was demonstrated for Cdc7 in budding yeast (Bousset et al., 1998; Donaldson et al., 1998), is involved only in the initiation and not in the elongation reaction. A corollary for this hypothesis is that either activation of Cdc7–Drf1 and Cdc7–Dbf4 kinases occurs at different times in S phase or these proteins localize in different nuclear territories, correlating with early and late replicating domains (Dimitrova and Gilbert, 1999). Localization studies with endogenous proteins should address this interesting aspect of Cdc7 regulation. We have observed, using overexpressed proteins, that Drf1 and Dbf4 do not bind to the same Cdc7 molecule in human cells. In the near future, detailed biochemical studies will determine whether endogenous Cdc7–Dbf4 and Cdc7–Drf1 complexes are assembled at the same time in the cell cycle or if the presence of these two complexes at any given time is mutually exclusive. It is also plausible at this stage that Cdc7 kinase activity might be differentially regulated by other events apart from the binding to the activating subunits. Finally, at this time, we cannot exclude that Drf1 and Dbf4 proteins might have different roles in cellular metabolism. This second hypothesis is supported by the observation that Cdc7 kinase participates either directly or indirectly in other processes such as regulation of induced mutagenesis and cell cycle checkpoint control in budding and fission yeast, respectively (Ostroff and Sclafani, 1995; Jares et al., 2000; Takeda et al., 2001). Further experimental work will be required to define the functions and regulation of Drf1, Dbf4 and Cdc7 and to verify whether other molecules that bind and regulate Cdc7, either by activating or functioning as inhibitors, exist in human cells.
Materials and methods
Plasmids and antibodies
Drf1- and Cdc7-coding sequences were amplified and introduced into plasmid pDONOR201 according to Gateway Cloning Technology (Invitrogen) and subsequently moved into a derivative of pVL1393 (PharMingen) for production of recombinant baculoviruses. For expression of Drf1 in mammalian cells, the Drf1-coding sequence was subcloned into the BglII site of the vector pCDNA1-HA (modified from the original Invitrogen pCDNA1). The same vector was used for expression of HA-tagged Dbf4. The pcDNA3-mycDbf4 construct was from K.Helin. For Cdc7 expression, the Cdc7-coding region was cloned into the vector pCMV-FLAG Tag 2-5 vector (Stratagene) and, for immunofluorescence studies, in pcDNA3.1/V5/His-TOPO vector (Invitrogen). In order to generate the K90R Cdc7 mutant, a site-directed mutagenesis kit was used (Amersham Biosciences). To produce Mcm2 N-terminal fragment used as substrate in kinase assays, a DNA fragment corresponding to residues 1–285 of Mcm2 protein was cloned into the bacterial expression vector pGEX6P and expressed as a GST fusion protein. GST was then removed by preScission protease cleavage according to the manufacturer’s instructions (Amersham Biosciences).
Rat monoclonal antibody (4F10) against influenza HA was from Roche. Anti-Flag monoclonal (M2) was from Sigma. Anti-cyclin E (HE12), anti-cyclin A and anti-Cdk2 antibodies were from Santa Cruz. Anti-p27 and anti-cyclinD1 (AM29) antibodies were from Transduction Laboratories and Zymed, respectively. Anti-Cdc7 polyclonal antibodies were generated in collaboration with Primm (http://www.primm.it) by immunizing rabbits with a synthetic peptide (RITAEEALLHPFFKD MSL) coupled to keyhole limpet hemocyanin. Anti-Drf1 5G4 and 5H4 mAbs were developed in collaboration with Areta international (http://www.aretaint.com) using as antigen a Drf1-His-tagged protein produced in bacteria and purified using an Ni-NTA affinity columns (Qiagen) according to the manufacturer’s instructions. For bacterial expression, the Drf1-coding region was subcloned into the BglII site of the pRSET plasmid (Invitrogen). In western blot experiments, mAbs 5G4 and 5H4 recognize a single band of the predicted molecular weight for Drf1 protein in extracts prepared from several cell lines.
Purification of Cdc7–Drf1 complexes from insect cells
Hi5 cells, grown in flasks at 27°C in Ultimate Serum Free Medium (Invitrogen), were infected with recombinant virus pVLCdc7-pVLDbf4, pVLCdc7-pVLDrf1, pVLCdc7(K90R)-pVLDrf1 or pVLCdc7 alone at an m.o.i. of 2.
Cells were collected 42 h post-infection and resuspended in ice-cold lysis buffer [50 mM Tris–HCl pH 7.6, 20% glycerol, 150 mM NaCl, 0.2% CHAPS, 20 mM dithiothreitol (DTT), 1 mM orthovanadate and 1× Complete Protease Inhibitor Cocktail (Roche)]. After sonication, the protein extract was clarified at 33 000 g for 30 min. The suspension was loaded onto a glutathione–Sepharose 4 fast flow column (Pharmacia Amersham) pre-equilibrated in lysis buffer. The column was washed extensively and proteins were eluted with 50 mM Tris–HCl pH 8.0, 150 mM NaCl and 20% glycerol containing 20 mM glutathione. Cdc7-enriched fractions subsequently were loaded onto a Superdex S200 column (Pharmacia Amersham) pre-equilibrated with buffer containing 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 20% glycerol and 0.02% CHAPS.
Cdc7 kinase activity was measured in a 25 µl reaction mixture consisting of 50 mM HEPES pH 7.9, 15 mM MgCl2, 0.2 mg/ml bovine serum albumin (BSA), 2 mM DTT, 10 µM ATP in the presence of 1 µCi of [γ-32P]ATP at 37°C; substrates were used at a final concentration of 6 µM. For multiscreen assay, reactions were performed in a 40 µl volume and stopped by the addition of EDTA to 25 mM. After addition of 40 µl of 150 mM H3PO4, the mixture was transferred into multiscreen plates (Millipore). The plates were washed three times with 75 mM H3PO4, and 100 µl of Microscint 0 (Packard) was added. The results were analyzed with a TopCount XNT β-counter. MBP, α-casein and histone H1 were from Sigma, histone H3 from Upstate Biotechnology, and retinoblastoma protein from Santa Cruz Biotechnology.
Cell culture, cell synchronization, cell cycle analysis and transfection
The 293 and HeLa cells (ATCC) were cultured in modified Eagle’s medium (MEM) supplemented with 10% fetal calf serum (FCS). NHDFs were maintained in fibroblast basal medium (FBM) supplemented with growth factors (Promocell) with 10% FCS and 1 ng/ml of human dermal fibroblast growth factor (FGF). NHDFs were serum starved for 72 h in FBM plus FGF and 0.1% FCS. NHDFs were blocked in G1/S phase by 2 mM thymidine treatment for 14 h. For FACS analysis, cells were fixed in 70% ethanol and stained with propidium iodide (40 ng/ml) plus RNase A (50 ng/ml) for 30 min at 37°C. Samples were then analyzed to determine the DNA content with a FACScan (Beckton-Dickinson). 293 cells and HeLa cells were transfected using either Lipofectamine plus reagent (Gibco-BRL) or Effectene reagent (Qiagen) following the manufacturer’s instructions.
Preparation of extracts, immunoprecipitation, protein kinase and phosphatase assays.
For western blot analysis, cell extracts were prepared in RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM DTT) containing protease (Roche) and phosphatase inhibitor cocktails (Sigma cocktails I and II). For co-immunoprecipitation experiments, cells were lysed in NP-40 buffer (0.1% NP-40, 50 mM Tris-Cl pH 7.5, 300 mM NaCl, 10 mM MgCl2, 1 mM DTT) containing protease and phosphatase inhibitor cocktails. Lysates were cleared by centrifugation and incubated at 4°C with specific antibodies. Immunocomplexes were recovered using protein G– Sepharose beads and were washed three times with lysis buffer before SDS–PAGE.
For kinase assays after immunoprecipitation, the beads were equilibrated in kinase buffer (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM DTT) and incubated in 25 µl of the same buffer containing 0.5 µg of MCM2 N-terminal fragment, 100 µM ATP and 10 µCi of [γ-32P]ATP. The reaction was performed at 30°C for 15 min and it was terminated by addition of 25 µl of 2× sample buffer and incubation at 95°C for 5 min.
For the phosphatase experiment, 15 µg of cell extract was incubated with 1 U of calf intestinal phosphatase (Roche) in the absence or presence of phosphatase inhibitors (Sigma phosphatase cocktails I and II) for 40 min at 30°C.
To determine the half-life of Drf1, 60% confluent HeLa cells were transfected with HA-Drf1 plasmid and 48 h later treated with cycloheximide at 50 µg/ml.
Northern blot analysis
The RNA for northern blotting was prepared using the RNeasy mini extraction kit (Qiagen). A 20 µg aliquot of total RNA was separated on 1% agarose/formaldeyde gels, transferred onto nitrocellulose (Hybond N, Amersham) and hybridized with random primed radiolabeled DNA probes using ExpressHyb solution from Clontech. Drf1 and Dbf4 probes were generated by amplifying the first 630 nucleotides of the corresponding ORFs. The β-actin probe was from Invitrogen. Radioactivity was quantified using the Molecular imager FX phosphoimager and Quantity One software (Bio-Rad).
Immunofluorescence
HeLa cells were grown to medium density on glass coverslips and transfected with HA-Drf1 plasmid. At 24 h post-transfection, the cells were treated with BrdU for 1 h and fixed with cold methanol. After permeabilization in 0.5% Triton X-100, the cells were washed and incubated with anti-HA rat antibody diluted 1:25 in medium containing 10% serum for 1 h. After three washes with phosphate-buffered saline (PBS), the samples were incubated for 1 h with the anti-rat Texas Red-conjugated antibody (PharMingen) diluted 1:200 in medium with 10% serum. After three washes in PBS, coverslips were fixed with methanol again, treated with 2 M HCl for 20 min, permeabilized with 0.5% Triton X-100, blocked with 0.3% BSA for 30 min and incubated for 30 min with fluorescein-linked anti-BrdU mAb (Becton Dickinson). After three more washes in PBS, coverslips were mounted with Mowiol (adapted from Pagano et al., 1994; Montecucco et al., 1998). For Drf1:Cdc7 co-localization studies, HeLa cells were co-transfected with HA-Drf1 and V5-Cdc7 plasmids. At 48 h post-transfection, cells were fixed in methanol, permeabilized as before and incubated in medium containing 10% serum and 0.1% BSA for 90 min. The antibodies were used as follows: anti-V5 (Invitrogen) at 1:200 dilution; Texas Red-conjugated anti-mouse antibody (Vector Laboratories) at 1:200 dilution; and Alexa 488-conjugated anti-HA (Molecular Probes) at 5 µg/ml. Antibody dilutions were in medium containing 10% serum and incubation time was 1 h for each step. Images were taken using a Confocal laser mounted on a Zeiss microscope (Radiance 2000, Bio-Rad) with an objective 40×.
Acknowledgments
Acknowledgements
We thank Clara Albanese and Paolo Cappella for FACS analysis, Jan Malyszko for DNA sequencing, and Francesco Sola for technical help with confocal microscopy experiments. We also thank Rodrigo Bravo, Antonella Isacchi and Giulio Draetta for helpful discussions, and Sandra Healy for critical reading of the manuscript. M.R. was supported by an EU-funded Marie Curie Host Industry Fellowship under the ‘Improving the Human Potential Research & Socio-Economic Knowledge Base’ program.
References
- Bousset K. and Diffley,J.F. (1998) The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev., 12, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.W. and Kelly,T.J. (1998) Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J. Biol. Chem., 273, 22083–22090. [DOI] [PubMed] [Google Scholar]
- Diffley J.F. and Labib,K. (2002) The chromosome replication cycle. J. Cell Sci., 115, 869–872. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S. and Gilbert,D.M. (1999) The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell, 4, 983–993. [DOI] [PubMed] [Google Scholar]
- Donaldson A.D., Fangman,W.L. and Brewer,B.J. (1998) Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev., 12, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell S.J., Romanowski,P. and Diffley,J.F. (1994) Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science, 265, 1243–1246. [DOI] [PubMed] [Google Scholar]
- Ekholm S.V. and Reed,S.I. (2000) Regulation of G1 cyclin-dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol., 12, 676–684. [DOI] [PubMed] [Google Scholar]
- Fangman W.L. and Brewer,B.J. (1992) A question of time: replication origins of eukaryotic chromosomes. Cell, 71, 363–366. [DOI] [PubMed] [Google Scholar]
- Ferreira M.F., Santocanale,C., Drury,L.S. and Diffley,J.F. (2000) Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol., 20, 242–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S., Kramer,E., Gieffers,C., Gannon,J., Peters,J.M. and Hunt,T. (2001) Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol., 153, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C.F., Dryga,O., Seematter,S., Pahl,P.M. and Sclafani,R.A. (1997) mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl Acad. Sci. USA, 94, 3151–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W. and Adams PD (2001) Cyclin-dependent kinases. Chem. Rev., 101, 2511–2526. [DOI] [PubMed] [Google Scholar]
- Jares P. and Blow,J.J. (2000) Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev., 14, 1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Jares P., Donaldson,A. and Blow,J.J. (2000) The Cdc7/Dbf4 protein kinase: target of the S phase checkpoint? EMBO rep., 1, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey P.D., Russo,A.A., Polyak,K., Gibbs,E., Hurwitz,J., Massague,J. and Pavletich,N.P. (1995) Mechanism of CDK activation revealed by the structure of a cyclinA–CDK2 complex. Nature, 376, 313–320. [DOI] [PubMed] [Google Scholar]
- Jiang W. and Hunter,T. (1997) Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc. Natl Acad. Sci. USA, 94, 14320–14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., McDonald,D., Hope,T.J. and Hunter,T. (1999) Mammalian Cdc7–Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J., 18, 5703–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.H., Masai,H. and Sugino,A. (2000) A Cdc7p–Dbf4p protein kinase activity is conserved from yeast to humans. Prog. Cell Cycle Res., 4, 61–69. [DOI] [PubMed] [Google Scholar]
- Kumagai H., Sato,N., Yamada,M., Mahony,D., Seghezzi,W., Lees,E., Arai,K. and Masai,H. (1999) A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol. Cell. Biol., 19, 5083–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Kawasaki,Y., Young,M.R., Kihara,M., Sugino,A. and Tye,B.K. (1997) Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis. Genes Dev., 11, 3365–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G., DeGregori,J., Yan,Z., Jakoi,L., Ishida,S., Williams,R.S. and Nevins,J.R. (1998) E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev., 12, 2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H. and Arai,K. (2000) Dbf4 motifs: conserved motifs in activation subunits for Cdc7 kinases essential for S-phase. Biochem. Biophys. Res. Commun., 275, 228–232. [DOI] [PubMed] [Google Scholar]
- Masai H., Miyake,T. and Arai,K. (1995) hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J., 14, 3094–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Matsui,E., You,Z., Ishimi,Y., Tamai,K. and Arai,K. (2000) Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 by Cdks. J. Biol. Chem., 275, 29042–29052. [DOI] [PubMed] [Google Scholar]
- Matsushime H., Ewen,M.E., Strom,D.K., Kato,J.Y., Hanks,S.K., Roussel,M.F. and Sherr,C.J. (1992) Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell, 71, 323–334. [DOI] [PubMed] [Google Scholar]
- Montecucco A. et al. (1998) DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J., 17, 3786–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Kishida,M. and Shimoda,C. (2000) The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells, 5, 463–479. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Nakamura-Kubo,M., Nakamura,T. and Shimoda,C. (2002) Novel fission yeast Cdc7–Dbf4-like kinase complex required for the initiation and progression of meiotic second division. Mol. Cell. Biol., 22, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K., Takeda,T., Matsui,E., Iiyama,H., Taniyama,C., Arai,K. and Masai,H. (2001) Bipartite binding of a kinase activator activates Cdc7-related kinase essential for S phase. J. Biol. Chem., 276, 31376–31387. [DOI] [PubMed] [Google Scholar]
- Ohtani K., DeGregori,J., Leone,G., Herendeen,D.R., Kelly,T.J. and Nevins,J.R. (1996) Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell. Biol., 16, 6977–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M., Theodoras,A.M., Schumacher,J., Roberts,J.M. and Pagano,M. (1995) Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol., 15, 2612–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro G., Owens,J.C., Shellman,Y., Sclafani,R.A. and Li,J.J. (1999) Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Mol. Cell. Biol., 19, 4888–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroff R.M. and Sclafani,R.A. (1995) Cell cycle regulation of induced mutagenesis in yeast. Mutat. Res., 329, 143–152. [DOI] [PubMed] [Google Scholar]
- Pagano M., Theodoras,A.M., Tam,S.W. and Draetta,G.F. (1994) Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev., 8, 1627–1639. [DOI] [PubMed] [Google Scholar]
- Peters J.M. (1999) Subunits and substrates of the anaphase-promoting complex. Exp. Cell Res., 248, 339–349. [DOI] [PubMed] [Google Scholar]
- Rodier G., Montagnoli,A., Di Marcotullio,L., Coulombe,P., Draetta,G.F., Pagano,M. and Meloche,S. (2001) p27 cytoplasmic localization is regulated by phosphorylation on Ser10 and is not a prerequisite for its proteolysis. EMBO J., 20, 6672–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Arai,K. and Masai,H. (1997) Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J., 16, 4340–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani R.A. (2000) Cdc7p–Dbf4p becomes famous in the cell cycle. J. Cell Sci., 113, 2111–2117. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. (1993) Mammalian G1 cyclins. Cell, 73, 1059–1065. [DOI] [PubMed] [Google Scholar]
- Takeda T., Ogino,K., Tatebayashi,K., Ikeda,H., Arai,K. and Masai,H. (2001) Regulation of initiation of S phase, replication checkpoint signaling and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol. Biol. Cell, 12, 1257–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M. and Jorgensen,P. (2000) Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev., 10, 54–64. [DOI] [PubMed] [Google Scholar]
- Weinreich M. and Stillman,B. (1999) Cdc7p–Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J., 18, 5334–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]