Abstract
The structure of auxin-binding protein 1 (ABP1) from maize has been determined at 1.9 Å resolution, revealing its auxin-binding site. The structure confirms that ABP1 belongs to the ancient and functionally diverse germin/seed storage 7S protein superfamily. The binding pocket of ABP1 is predominantly hydrophobic with a metal ion deep inside the pocket coordinated by three histidines and a glutamate. Auxin binds within this pocket, with its carboxylate binding the zinc and its aromatic ring binding hydrophobic residues including Trp151. There is a single disulfide between Cys2 and Cys155. No conformational rearrangement of ABP1 was observed when auxin bound to the protein in the crystal, but examination of the structure reveals a possible mechanism of signal transduction.
Keywords: ABP1/auxin-binding pocket/auxin receptor/crystal structure/signal transduction
Introduction
The plant hormone auxin regulates cell elongation, division, differentiation and morphogenesis. Synthetic auxin analogues are also important selective herbicides. The best candidate for an auxin receptor is auxin-binding protein 1 (ABP1), first studied in membrane fractions 30 years ago and subsequently isolated and purified (Hertel et al., 1972; Venis, 1977; Jones, 1994). ABP1 is a soluble 22 kDa glycoprotein, ubiquitous amongst green plants, found predominantly within the endoplasmic reticulum (ER), and in smaller quantities at the cell surface associated with the plasma membrane (Diekmann et al., 1995). ABP1 is dimeric in solution (Shimomura et al., 1986) and has submicromolar affinity for auxins, consistent with its ability to bind free auxin at physiological concentrations (Löbler and Klämbt, 1985). The most widely used assays for ABP1 receptor activity measure auxin-induced plasma membrane hyperpolarizations and ion fluxes in protoplasts (Barbier-Brygoo et al., 1991; LeBlanc et al., 1999; David et al., 2001). Auxin-induced cell swelling of proteoplasts and intact guard cells can also be attributed to ABP1 (Gehring et al., 1998; Steffens et al., 2001). Ectopic and inducible expression of ABP1 confers auxin-dependent cell expansion in tobacco cells normally lacking auxin responsiveness (Jones et al., 1998); antisense suppression of ABP1 eliminates auxin-induced cell elongation and reduces cell division; and a homozygous null mutation in ABP1 confers embryo lethality in Arabidopsis (Chen et al., 2001). ABP1 is therefore firmly implicated in mediating cell elongation and, directly or indirectly, cell division.
Attempts have been made to deduce the nature and location of the auxin-binding site of ABP1 based on group-reactive reagents (Jones and Venis, 1989), the structure of its ligands (Edgerton et al., 1994), and by using photolabels (Brown and Jones, 1994). Circular dichroism (CD) spectroscopy of ABP1 showed it to be a β-protein (Shimomura et al., 1986), and a change in the CD spectrum on auxin binding has been cited as indicating conformational change. The failure of antibodies to recognize the C-terminus when auxin is bound has also been cited as evidence for conformational change (Napier and Venis, 1990).
ABP1 has two sequence motifs, HXH(X)11G and P(X)4H(X)3N (where X is any amino acid residue), that place it within a family of proteins (cupins) that includes germin and the vicilins (Dunwell and Gane, 1998; Dunwell et al., 2000, 2001). A partial model of ABP1 was proposed by comparative modelling using the vicilin domains as a template (Warwicker, 2001). This model suggested a metal-binding site within a β-barrel structure similar to that of germin (Woo et al., 2000a). Additionally, the modelling suggested that the C-terminus competes for the auxin-binding site, thereby providing a model of conformational change (Warwicker, 2001).
Results
Overall protein fold, disulfide arrangement and glycosylation
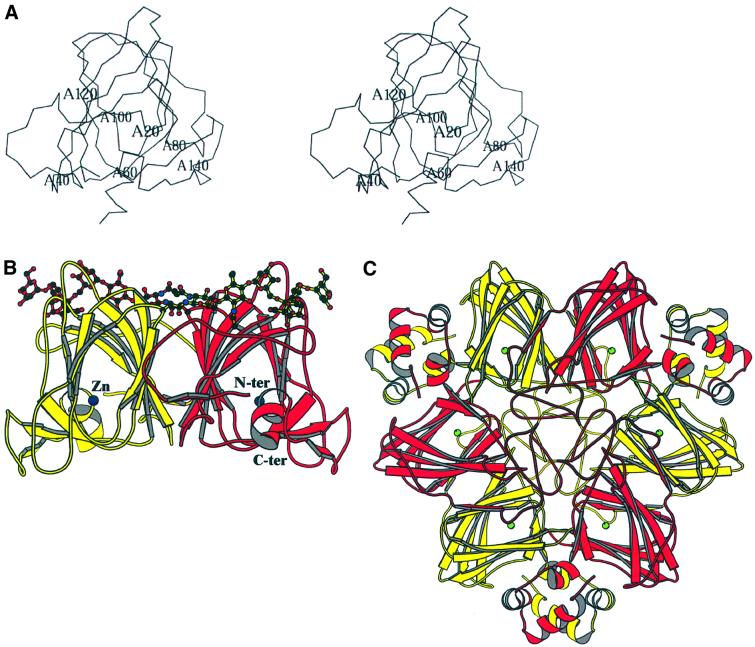
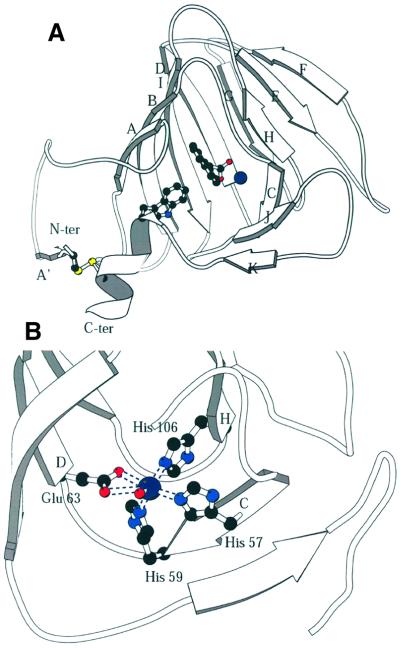
The structure of maize (Zea mays L.) ABP1 was determined by multiple isomorphous replacement using protein expressed in insect cells, and refined at 1.9 Å resolution (see Materials and methods and Table I). With the exception of the three C-terminal residues, which were assumed to be flexible or disordered in the crystal, there was clear electron density for all residues 1–160 (Figure 1A). The protein is a dimer in the crystal (Figure 1B) as it is in solution (Shimomura et al., 1986), and resembles the germin dimer (Figure 1C). Residues 26–148 fold into a β-jellyroll barrel formed by two antiparallel β-sheets ABIDG and KJCHEF (strands labelled in Figure 2A); the N-terminal residues 1–25 are irregular in conformation except for a short β-strand (A′, Figure 2A), and the C-terminal residues are also irregular apart from a short α-helix (residues 152–160). The structural classification according to SCOP (Murzin et al., 1995) is therefore a double-stranded β-helix fold and germin/seed storage 7S protein family. The double-stranded β-helix fold is an ancient protein fold (Anantharaman et al., 2001) and is found throughout the Archaea, Eubacteria and Eukaroyota. The N- and C-terminal extensions of the ABP1 β-jellyroll barrel are linked together by a disulfide bond between Cys2 and Cys155 (Figure 2A). There is no disulfide between Cys2 and Cys61 as recently suggested (Feckler et al., 2001), and ABP1 has two cis-prolines, 127 and 148. There was also evidence in the electron density map for glycosylation of Asn95, with two N-acetyl glucosamine (GlcNAc) and three mannose (Manp) residues clearly visible [Manp(α1,6)–(Manp(α1,3))–Manp(β1,4)–GlcpNAc(β1,4) –GlcpNAc(β1,N)-Asn]; there was weaker density for additional mannose residues.
Table I. Crystallographic data statistics.
| Data collection and phasing | ||||||
|---|---|---|---|---|---|---|
| Crystal | Native | CH3HgCl | UO2Ac2 | PbAc2 | LuCl3 | 1-NAA |
| Concentration | 1 mM | 10 mM | 25 mM | 50 mM | 10 mM | |
| Time | 7 days | 2 h | 3 days | 2 h | 8 weeksa | |
| X-ray source | PX9.6 Daresbury | PX9.5 Daresbury | MacSci. London | MacSci. London | MacSci. London | PX9.6 Daresbury |
| Wavelength (Å) | 0.87 | 1.00851 | 1.5418 | 1.5418 | 1.5418 | 0.87 |
| Dectector | CCD | MAR300 | DIP1030 | DIP1030 | DIP1030 | CCD |
| Resolution (Å) | 1.9 | 2.5 | 2.8 | 3.0 | 2.7 | 1.9 |
| Completeness (%)b | 99.0 | 99.1 | 97.8 | 85.3 | 98.2 | 94.2 |
| (91.1) | (98.3) | (78.9) | (85.5) | (97.4) | (90.0) | |
| Rsym(I) (%)b,c | 4.0 (11.6) | 6.4 (11.1) | 4.2 (7.6) | 5.7 (7.9) | 5.3 (9.1) | 3.8 (9.6) |
| Riso (%)d | 24.9 | 28.2 | 23.1 | 33.8 | 18.2 | |
| Phasing powere,f | 1.5 (1.2) | 1.2 (1.0) | 1.2 (0.9) | 1.1 (0.9) | ||
| Mean figure of merit |
0.56 |
|
|
|
|
|
| Refinement and model stereochemistry |
|
ABP1 |
|
ABP1–1-NAA complex |
|
|
| X-ray data used in refinement | ||||||
| Resolution limits (Å) | 15.0–1.9 | 15.0–1.9 | ||||
| No. of reflections used in refinement | 51 598 | 51 458 | ||||
| No. of reflections used for Rfree calculation | 2761 | 2714 | ||||
| R-factor (%) (Rfree)g | 18.4 (24.1) | 20.0 (24.0) | ||||
| R.m.s.d. (target σ) | ||||||
| 1–1 distances (Å) | 0.014 (0.020) | 0.014 (0.020) | ||||
| 1–2 distances (Å) | 0.033 (0.040) | 0.035 (0.040) | ||||
| 1–4 distances (Å) | 0.033 (0.050) | 0.037 (0.050) | ||||
| Chiral volumes (Å3) | 0.142 (0.150) | 0.150 (0.150) | ||||
| Peptide planes (Å) | 0.025 (0.030) | 0.026 (0.030) | ||||
| Aromatic planes (Å) | 0.011 (0.020) | 0.012 (0.020) | ||||
aThe ABP1–1-NAA complex was co-crystallized.
bValues in parentheses are for the highest resolution shell.
cRsym(I) = ΣhΣi|Ii(h) – <I(h)>|/ΣhΣi Ii(h)
dRiso = Σh|FPH = Fp|/Σh|FP|
ePhasing power = <FH>/<E>, where E is the root mean square lack of closure.
fValues for centrics in parentheses.
gR-factor = Σ|Fobs| – |Fcalc|/Σ|Fobs|. Rfree is obtained from a randomly selected 5% of reflections not included in the refinement.
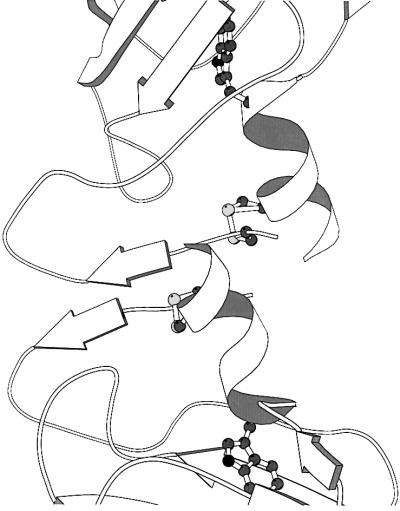
Fig. 1. The overall structure of maize ABP1. (A) Stereoview of an ABP1 subunit with the N- and C-terminal extensions closest to the viewer and with labels every 20 residues. (B) Schematic representation of the overall fold of the ABP1 dimer with the β-strands drawn as arrows, the α-helices as helices, and with the bound zinc ions represented as dark blue spheres, one at the centre of each subunit. The orientation of the red subunit is similar to that shown in (A). The subunits comprising the dimer are related by a vertical 2-fold rotation axis. Asn95 and the observed N-linked sugar residues [Manp(α1,6)–(Manp(α1,3))–Manp(β1,4)–GlcpNAc(β1,4)–GlcpNAc(β1,N)-Asn] are also shown. (C) Schematic representation of the germin hexamer to illustrate the relationship between the ABP1 dimer shown in (B) and the trimer of dimers in germin. The germin subunit has additional α-helical clasps that lock the three dimers together into the trimer of dimers (Woo et al., 2000a; Protein Data Bank accession code 1FI2). The subunit sequences of ABP1 and germin are 24% identical, and the r.m.s.d. for 101 equivalent α-carbon atoms is 1.85 Å. The green spheres are manganese ions, one at the active centre of each of the germin subunits. Note that (B) and (C) are not on the same scale. Figures 1, 2 and 5 were drawn using MOLSCRIPT (Kraulis, 1991).
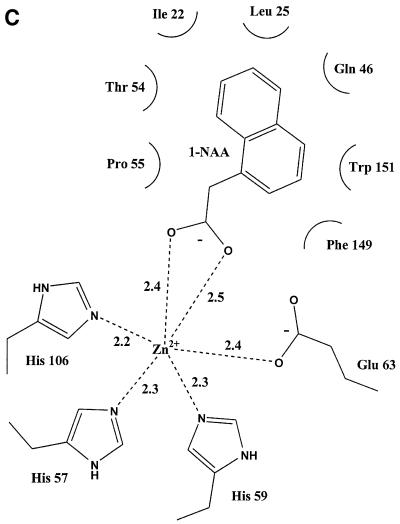
Fig. 2. The subunit structure and binding site of ABP1. (A) The β-strands of the ABP1 subunit are labelled A′–K; the dimer interface is formed by strands ABIDG and the outer sheet by strands KJCHEF. This figure shows the disulfide between residues 2 and 155 that links the N- and C-terminal extensions; the sulfur atoms are the yellow spheres. Also shown is the relationship between the disulfide, the C-terminal α-helix, Trp151 (drawn as ball-and-stick model), 1-NAA (also ball and stick) and the zinc ion (dark blue sphere). The subunit is viewed from the direction of the opening to the auxin-binding pocket. (B) The zinc-binding site in detail. The protein ligands are His57 (strand C), His59 (at the end of strand C), Glu63 (at the beginning of strand D) and His106 (at the beginning of strand H). A single water molecule completes the octahedral coordination sphere. The zinc–nitrogen (His NE2) distances are between 2.2 and 2.3 Å and the zinc–oxygen distances are 2.4 Å to Glu63 OE1, 3.1 Å to Glu63 OE2 and 2.2 Å to the water molecule (shown as a small red sphere).
ABP1 is a dimer in solution and appears as an intimate dimer in the crystals where the subunits are related by non-crystallographic symmetry. The dimer interface is formed principally by sheet ABIDG (Figure 2A), and the total area buried at the interface is 2390 Å2 per subunit, corresponding to 24% of the subunit solvent-accessible surface area. The high degree of burial of hydrophobic residues (isoleucines, leucines and valines) at the interface, the interaction of carbohydrate from one subunit with the other subunit (Figure 1B), the exclusion of water from the interface, the presence of a salt bridge (Asp40 with 2-fold-related Arg125) and the similarity to the germin dimer (see below) all suggest that the dimer is biologically authentic.
Evolutionary relationship to other proteins
The β-jellyroll barrel subunit structure of ABP1 is similar to that of germin with which it shares 24% sequence identity (the r.m.s.d. for 101 equivalent α-carbons is 1.85 Å). ABP1 forms a dimer similar to that of germin but lacks the additional C-terminal helices of germin (Figure 1B and C). Comparing the ABP1 and germin subunits, the strands furthest from the N- and C-termini are structurally the most similar (strands IDG and HEF) and those nearest to the N- and C-termini (A and J) the least similar (Figure 2A). The role of the extensions to the β-barrel in germin is to stabilize the trimer of dimers; the additional C-terminal helices locking the dimers together and the N-terminal extensions filling the centre of the hexamer (Figure 1C). Whereas ABP1 has a zinc-binding histidine cluster (see below), the corresponding cluster in germin binds manganese. The ABP1 dimer, like germin, is also structurally similar to the vicillin monomer that has internal pseudo-dimer structure (Lawerence et al., 1994) but lacks the histidine cluster and therefore the metal-binding site. The cupins have a wide range of functions, from storage protein to enzyme to essential protein in auxin signalling.
The metal-binding site of ABP1
The experimental electron density map clearly indicated the presence of a metal ion coordinated by three histidines and a glutamate in each subumit (Figure 2B). The coordinating histidines are those of the HXH(X)11G and P(X)4H(X)3N fingerprints, the first two of which are on β-strand C (His57 and His59) and the third on β-strand H (His106); the glutamate (Glu63) is on strand D (Figure 2B). The metal-binding site is buried deep within the β-barrel. Proton-induced X-ray emission analysis (PIXE; Garman, 1999) of recombinant ABP1 dialysed against EDTA revealed that the tightly bound metal was zinc. The coordination number of the zinc is six; the three histidines coordinate via their NE2 atoms, the glutamate via OE1 and more distantly OE2, and a single water molecule completes the coordination sphere (Figure 2B). With the exception of OE2 of Glu63 (3.1 Å from the zinc), these zinc–ligand distances are between 2.2 and 2.4 Å, with the metal–nitrogen bonds being shorter than the metal–oxygen bonds. PIXE measurements on maize ABP1 purified from plant material indicated the presence of both zinc and copper in roughly equal quantities. Zn(II) and Cu(II) have closely similar atomic radii, so Cu(II) may be able to substitute for Zn(II) at the metal-binding site; there is no evidence, however, that ABP1 catalyses a redox reaction.
Auxin binding to ABP1
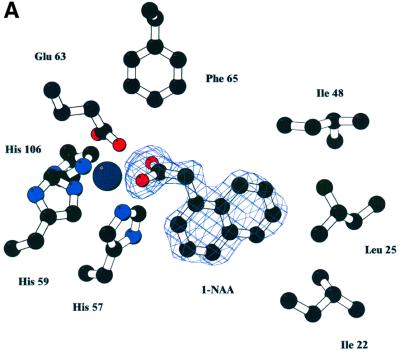
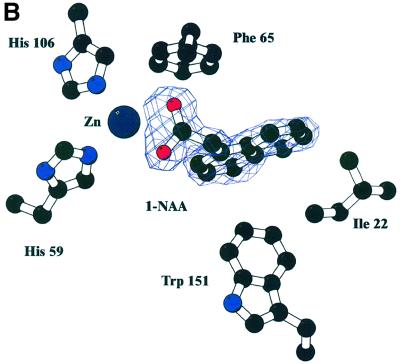
Purified ABP1 had a KD of 1.5 × 10–7 M for the synthetic auxin 1-naphthalene acetic acid (1-NAA) at pH 5.5, and Scatchard analysis suggested one auxin molecule bound per monomer (results not shown; Bauly et al., 2000), in agreement with previous results (Hesse et al., 1989). The highest binding affinity was observed at pH 5.5 and this was therefore the pH used for co-crystallization of the ABP1–auxin complex. Crystals of the ABP1–1-NAA complex formed at pH 5.5 grew from conditions similar to the uncomplexed protein and were isomorphous with the native protein. Difference Fourier maps calculated using protein phases calculated from the ABP1 structure with the waters in the internal pocket omitted clearly revealed one 1-NAA molecule per monomer (Figure 3). The structure of 1-NAA (Rajan, 1978) was obtained from the Chemical Database (Allen and Kennard, 1993). The structure of the complex was refined at 1.9 Å to an R-factor of 20% and Rfree of 24% (Table I). The carboxylate group of 1-NAA made symmetric bidentate contacts with the zinc deep inside the binding pocket (zinc–oxygen distances of 2.4 and 2.5 Å) and replaced the zinc–water interaction (2.2 Å). The OE2 of Glu63 moved away from a distance of 3.1 to 3.3 Å in the complex. The naphthalene ring of 1-NAA packed against a number of hydrophobic residues inside the β-barrel including a face to end interaction with Trp151, with Thr54 and Pro55 forming the other side of the binding pocket and with Ile22 and Leu25 forming the side distal from the metal (Figure 3). Gln46 and Phe149 also line the pocket. The bound auxin is virtually inaccessible to solvent (99% of the 350 Å2 solvent-accessible surface of 1-NAA is buried when bound). There is, however, a clear route by which auxin enters the binding pocket, between the N- and C-terminal extensions, as seen in Figure 2A, which also shows the relationship between bound 1-NAA, the zinc ion and Trp151. Assuming natural auxin, indole acetic acid (IAA), binds similarly to 1-NAA, its affinity would be expected to be lower because the indole ring has fewer carbons to bind the hydrophobic pocket and the indole amide does not have an obvious hydrogen-binding partner; both these factors would attenuate binding affinity for IAA as reported previously (Löbler and Klämbt, 1985).
Fig. 3. Auxin binding to ABP1. (A) Omit electron density map at 1.9 Å resolution and contoured at 3σ revealing 1-NAA bound to ABP1. The view in (B) is rotated ∼60° around the x-axis compared with the view in (A), and His57, Glu63, Leu25 and Ile48 are omitted from this view for clarity. The bidentate binding of the 1-NAA carboxylate to the zinc (dark blue sphere) can be clearly seen, as can the hydrophobic environment of the naphthalene ring. (C) Simplified representation of the contacts between 1-NAA and ABP1 and between the zinc ion and ABP1 in the complex. Distances shown are in angstroms. (A) and (B) were prepared using BOBSCRIPT (Esnouf, 1997).
Superimposing the structures with and without bound auxin indicates that there was no substantial conformational change as a result of binding (r.m.s.d. of 0.186 Å for 158 equivalent Cα atoms). Four water molecules were expelled from the binding pocket, including the water molecule binding to the zinc, and there were no other appreciable changes observed on auxin binding. The disulfide bridge between the N- and C-terminal polypeptides will restrict the movement of the C-terminal helix. There is no evidence to suggest that the disulfide is reduced in either the ER or on the surface of the plasma membrane.
Sequence variation
Sequence alignment (Altschul et al., 1997) reveals considerable conservation in the sequence of mature ABP1 across a broad range of plant species (Figure 4). ABP1 has no homologues outside the Streptophyta (green algae) and Embryophyta (land plants). The selective herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) is lethal towards many dicots, but not monocots. A molecular model of strawberry ABP1 built using the maize ABP1 structure as a template using MODELLER (Šali and Blundell, 1993) reveals a striking substitution in the auxin-binding site of strawberry ABP1, arginine for Ile22, a substitution that occurs in all dicots with the exception of Raphanus (Figure 4). Additionally, tyrosine is substituted for Phe149 with the same exception. Ile22 forms a hydrophobic contact with the region of the naphthalene ring distal from the metal in the monocot ABP1 (Figure 3). Phe149 is close to C2 of the naphthalene ring. These substitutions may have implications for herbicide selectivity.
Fig. 4. Multiple alignment of ABP1 sequences. The first column gives the species from which the ABP1 originates: Zea ABP1 (Zea Mays; accession No. P13689); Avena (Avena sativa, T07797); Hordeum*, a compilation of EST entries (Hordeum vulgare, BE456042, BF259480, AL508794 and AV834303); Triticum*, a compilation of EST entries (Triticum aestivum, BE426626, BE446060, BE425494, BG274987 and BE516927); Arabidopsis (Arabidopsis thaliana, P33487); Raphanus (Raphanus sativus, AB000706); Nicotiana (Nicotiana tobacum, P33490); Lycopersicon (Lycopersicon esculentum, CAA09882); Capsicum (Capsicum annuum, CAA88361); Glycine*, an EST entry (Glycine max, BG045594); Malus (Malus x domestica, AAB47752); Fragaria (Fragaria x ananassa, CAA62956); Ceratopteris*, an EST entry (Ceratopteris richardii, BE641066); Ceratodon (Ceratodon purpureus, AAF37576); and Chlamydomonas*, a compilation of EST entries (Chlamydomonas reinhardtii, BG847560, BG860232, AV635532 and AV633592). Note that the sequence for Ceratodon extends past the KDEL terminus of other ABP1s, but the extension is not shown. The first four sequences are from monocot, the next eight from dicot plants and the last three from a fern, a moss and a green alga. The approximate positions of the β-strands identified in the text and Figure 2 are illustrated as arrows. Motif indicates the signal peptide, cupin motif and KDEL sequence. The residues for which there are clear differences between monocots and dicots are highlighted in blue. Red and cyan indicate strict and less strict conservation, respectively. The Clamydomonas ABP1 sequence may not be aligned correctly as it differs considerably from the other sequences; it is not known whether this ABP1 binds a metal. Both monocots and dicots have a glycosylation site at Asn95; dicots have an additional glycosylation site at Asn11.
Orientation of ABP1 on membranes
The C-terminal helices of the ABP1 dimer and the succeeding KDEL ER retention sequences (the last three residues are not seen in the electron density map, see above, but their location can be readily inferred) are positioned on the same side of the molecule such that they could both interact simultaneously with one or possibly two membrane-bound ER retention proteins (Figure 1B). The ER membrane would be horizontal with respect to the orientation of ABP1 shown in Figure 1B, with the flat lower surface of the dimer sitting on the membrane surface and with the KDEL sequence interacting with the retention protein. The location of the glycosylation on the surface of ABP1 protruding into the lumen of the ER is consistent with this suggestion. ABP1 may be located on the plasma membrane of the cell surface in a similar orientation.
Crystal packing
When the non-crystallographic symmetry restraints are removed during maximum-likelihood refinement of the structure, differences in the conformation of the four molecules in the asymmetric unit of the crystal may be detected. These differences are found to be rather small and reflect differences in the crystal contacts. The C-terminal residues beyond Cys155 do differ in mobility as indicated by the crystallographic temperature factors, but do not differ significantly in position. The observed C-terminal residues (156–160 and 156–159 in molecule B) are more ordered in molecules A and D due to the presence of crystal contacts. In molecules B and C, there are no crystal contacts close to the C-terminus and, as a result, the C-terminal residues are more mobile, as indicated by the temperature factors for the α-carbon atom of residue 159 which are 26, 53, 42 and 21 Å2 for molecules A, B, C and D, respectively. There is, however, no evidence of differential mobility in the C-terminal segment between the auxin-bound and the auxin-free structures in any of the four ABP1 molecules, showing that the change in mobility is a characteristic of the environment in the crystal rather than the presence or absence of auxin.
The N-terminal extension of the ABP1 subunit is, however, involved in quite extensive interactions with the N-terminal extension of a symmetry-related subunit. The interaction is between the short β-strand (labelled A′ in Figure 2A) and its symmetry-related equivalent which come together to form a two-stranded parallel β-sheet with low twist stabilized by four main chain hydrogen bonds and five side chain stacking interactions (Figure 5). The side chain stacking interactions are cysteine, valine, arginine, aspartate and asparagine of subunit A against serine, cysteine, valine, arginine and aspartate, respectively, of the symmetry equivalent to subunit D, where AD is a germin-like dimer. The interactions are not so extensive as to bring into question the identity of the biologically relevant dimer, but may be sufficient to imply that crystallization is selecting a conformation of the N-terminal extension. Because of the disulfide between the N- and C-terminal extensions, a change in the conformation of the N-terminal extension could affect the conformation of the C-terminal extension. This packing interaction is found in both crystals grown in the absence of auxin and those grown in its presence, and could lead to the selection of a single conformation of the N- and C-terminal extensions in both. It is possible, therefore, that the two structures reported here represent the bound form of the ABP1 receptor in the presence and absence of auxin, the bound conformation being selected by the packing in the crystal. The crystal contacts involving the N-terminal extensions cannot occur if ABP1 is located on the membrane’s surface as shown in Figure 1B, as adjacent dimers would be inverted and could not therefore interact with proteins in the same membrane through their C-terminal residues. Alternatively, the interaction of the C-terminal extension with another protein, possibly membrane bound, might affect the conformation of the extensions, holding ABP1 in an unbound conformation not seen in the crystal structures.
Fig. 5. The crystal contact involving the N-terminal extensions of symmetry-related molecules in type II crystals of ABP1. The disulfide bridge and Trp151 are drawn to illustrate how this crystal contact might influence the position of the C-terminal α-helix in the protein.
A putative model of signal transduction
Assuming that the conformation of the terminal extensions of ABP1 is unaffected by crystal packing, or interactions with other proteins, then the principal effect of auxin binding will be to stabilize the folded structure of the protein and directly, via interaction with Trp151, the stability of the terminal extensions.
On the other hand, assuming that the conformation of the N- and C-terminal extensions is that of the bound form of ABP1 in both the presence and absence of auxin, due either to crystal contacts involving the N-terminal extension or to the interaction of ABP1 with another protein, we can then suggest how the structure might change in the absence of auxin and additional interactions. Trp151 might move away slightly from the binding pocket, causing the C-terminal α-helix also to move away from the binding pocket in the direction of the helix axis. The α-helix would act as a rigid rod conveying movement to the protein’s surface, and the disulfide would limit the extent of the movement and possibly cause rearrangement of the N-terminal extension (Figure 2A). Auxin binding would then cause Trp151 to move fully into the binding pocket, as seen in the structures, pulling the C-terminal α-helix with it, and thereby signalling the presence of auxin to the membrane or other proteins.
Discussion
The crystal structure reveals the architecture, disulfide arrangement and glycosylation of ABP1, and the structure of the ABP1 auxin-binding site. The overall similarity of the ABP1 subunit to that of germin is confirmed; however, substantial differences are also revealed. Previous work has suggested a disulfide bridge between Cys2 and Cys61 of maize ABP1 (Feckler et al., 2001) and, for tobacco ABP1 expressed in Escherichia coli, a disulfide involving the residue equivalent to 155 (David et al., 2001). The crystal structure reveals the single disulfide is between Cys2 and Cys155, with the third cysteine, Cys61, buried close to the zinc-binding site. The selectivity of ABP1 for auxins is explained by the characteristics of the binding pocket. The binding pocket is deep and hydrophobic except for the zinc ion at the bottom of the pocket.
No change in the conformation of ABP1 was observed when auxin binds in the crystal. The obvious conclusion is that there is no change in the structure of ABP1 when auxin binds in solution. Certainly the evidence for conformational change from far UV CD is not compelling, and the near UV CD spectral changes probably reflect the changing environment of aromatics when auxin binds (Shimomura et al., 1986). Conformational change is the most obvious explanation for the antibody-binding data. The antibody binds the auxin-free form but not the auxin-bound form of ABP1 (Napier and Venis, 1990; David et al., 2001). There is an alternative explanation for this observation; the antibody might induce a change in the structure of ABP1, one that is resisted by the more stable complex. However, if antibody binding does induce a change, this itself might suggest that conformational rearrangement of ABP1 is biologically important.
The contacts between the N-terminal extensions in the crystal lattice imply that conformational change involving the N- and C-terminal regions of ABP1 cannot be ruled out. The crystal contacts may be selecting a ‘crystallizable conformation’ of the protein in which the auxin-binding pocket is fully formed whether auxin is bound or not. It is unlikely that these crystal contacts could exist on the surface of a membrane. Alternatively, the interaction of ABP1 with membrane proteins may favour an alternative conformation of the extensions. We can therefore suggest a mechanism by which ABP1 may signal the presence of auxin to membrane-bound proteins. The auxin-free conformation of the extensions in ABP1 would be different, possibly because of the interaction of ABP1 with other components, possibly simply because of the lack of crystal contacts. The binding of auxin would then alter the position of Trp151 and cause the C-terminal helix to move. Because of the disulfide bridge, the N-terminal extension may also undergo some rearrangement (Figure 2A). The rearrangement of the N- and C-terminal extensions would then signal the presence of auxin to membrane proteins, resulting in the observed changes in ion fluxes across the plasma membrane (Theil et al., 1993).
Studying the conformation of the extensions of ABP1 in other crystal forms and in the antibody (Fab)–ABP1 complex will illuminate the effect of crystal packing and antibody binding on the conformation of the extensions of ABP1 and lead to a fuller understanding of the role of ABP1 in auxin signal transduction. Ultimately, the goal is to identify the proteins with which ABP1 interacts in vivo and study their interactions in detail.
Materials and methods
Expression, crystallization and purification
ABP1 was expressed, purified and crystallized as described previously (Macdonald et al., 1994; Woo et al., 2000b). A mutant of ABP1 in which the C-terminal ER retention sequence (KDEL) was substituted by KEQL was used to promote secretion into the culture medium and thereby facilitate purification (Macdonald et al., 1994). Crystallization trials gave three types of crystals of which type II grew from 23% polyethylene glycol (PEG) 4000 in the pH range 5.5–7.5 and belonged to the monoclinic space group P21 with unit cell parameters a = 62.4 Å, b = 82.3 Å, c = 69.8 Å, β = 94.2°, with two homodimers per asymmetric unit. The type II crystals diffracted to 1.9 Å resolution, somewhat better than the other crystal types. Before data collection, the crystals were cryo-cooled at 100 K using a cryoprotectant consisting of mother liquor supplemented with 12% 2-propanol and 8% 2-methyl-2-4-pentane diol (MPD). The crystals used to solve the native structure were grown at pH 7.5.
Structure solution
The structure was solved by multiple isomorphous replacement using protein phases calculated from four heavy atom derivatives. Data collected at synchrotrons and in the laboratory were processed and merged using DENZO and SCALEPACK (Otwinowski and Minor, 1997). The positions of the four major mercury sites were determined from the difference Patterson map, and the other heavy atom positions were found using cross-phased difference Fourier maps. The CCP4 program suite (CCP4, 1994) was used for these calculations. The non-crystallographic symmetry was exploited to improve the clarity of the maps using DM (Cowtan and Main, 1998). A partial model was constructed using wARP (Perrakis et al., 1997). REFMAC (Murshudov et al., 1997) and O (Jones et al., 1991) were used for further refinement and manual rebuilding.
Complex formation
The ABP1–1-NAA complex was prepared by mixing 10 mM 1-NAA with the protein before crystallization with the pH maintained at 5.5, the optimum for auxin binding. 1-NAA was used in preference to IAA as it is more stable in solution and has the same biological effect. The crystals grew in the same conditions as ligand-free ABP1 and had similar cell parameters.
Coordinates
Coordinates and structure factor amplitudes for the ligand-free ABP1 and the 1-NAA complex have been deposited in the Protein Data Bank with accession codes 1LR5 and 1LRH, respectively.
Acknowledgments
Acknowledgements
We are grateful to Dr H.Macdonald (University of West of England) for use of the baculovirus expression vector, and Dr G.Grime (Oxford University) for assistance with PIXE. We acknowledge use of beamlines PX 9.6 at the Synchrotron Radiation Source Daresbury, and EMBL BW7A at the DORIS storage ring, DESY, Hamburg. We also acknowledge use of the EPSRC’s Chemical Database Service at Daresbury. This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC).
References
- Allen F.H. and Kennard,O. (1993) 3D search and research using the Cambridge Structural Database. Chemical Design Automation News, 8, 1 and 31–37. [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J.H., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V., Koonin,E.V. and Aravind,L. (2001) Regulotory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol., 307, 1271–1292. [DOI] [PubMed] [Google Scholar]
- Barbier-Brygoo H. Ephritikhine,G., Klambt,D., Maurel,C., Palme,K., Schell,J. and Guern,J. (1991) Perception of the auxin signal at the plasma membrane of tobacco mesophyll protoplasts. Plant J., 1, 83–93. [DOI] [PubMed] [Google Scholar]
- Bauly J.M. et al. (2000) Overexpression of ABP1 heightens the sensitivity of guard cells to auxin. Plant Physiol., 124, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.C. and Jones,A.M. (1994) Mapping the auxin-binding site of auxin-binding protein-1. J. Biol. Chem., 269, 21136–21140. [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 program suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chen J.-G., Ullah,H., Young,J.C., Sussman,M.R. and Jones,A.M. ( 2001) ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev., 15, 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. and Main,P. (1998) Miscellaneous algorithms for density modification. Acta Crystallogr. D, 54, 487–493. [DOI] [PubMed] [Google Scholar]
- David K., Carnero-Diaz,E., Leblanc,N., Monestiez,M., Grosclaude,J. and Perrot-Rechenmann,C. (2001) Conformational dynamics underlie the activity of the auxin-binding protein, Nt-abp1. J. Biol. Chem., 276, 34517–34523. [DOI] [PubMed] [Google Scholar]
- Diekmann W., Venis,M.A. and Robinson,D.G. (1995) Auxins induce clustering of auxin-binding protein at the surface of maize coleoptile protoplasts. Proc. Natl Acad. Sci. USA, 92, 3425–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell J.M. and Gane,P.J. (1998) Microbial ancestors of seed storage proteins: conservation of motifs in a functionally diverse superfamily of enzymes. J. Mol. Evol., 46, 147–154. [DOI] [PubMed] [Google Scholar]
- Dunwell J.M., Khuri,S. and Gane,P.J. (2000) Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev., 64, 153–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell J.M., Culham,A., Carter,C.E., Sosa-Aguirre,C.R. and Goodenough,P.W. (2001) Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci., 26, 740–746. [DOI] [PubMed] [Google Scholar]
- Edgerton M.D., Tropsha,A. and Jones,A.M. (1994) Modeling the auxin-binding site of auxin-binding protein-1 of maize. Phytochemistry, 35, 1111–1123. [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph., 15, 132–134. [DOI] [PubMed] [Google Scholar]
- Feckler C., Muster,G., Feser,W., Römer,A. and Palme,K. (2001) Mass spectrometric analysis reveals a cysteine bridge between residues 2 and 61 of the auxin-binding protein 1 from Zea mays L. FEBS Lett., 509, 446–450. [DOI] [PubMed] [Google Scholar]
- Garman E. (1999) Leaving no element of doubt: analysis of proteins using microPIXE. Structure, 7, 291–299. [DOI] [PubMed] [Google Scholar]
- Gehring C.A., McConchie,R.M., Venis,M.A. and Parish,R.W. (1998) Auxin-binding-protein antibodies and peptides influence stomatal opening and alter cytoplasmic pH. Planta, 205, 585–586. [DOI] [PubMed] [Google Scholar]
- Hertel R., Thompson,K.-S. and Russo,V.E.A. (1972) In vitro auxin binding to particulate cell fractions from corn coleoptiles. Planta, 107, 325–340. [DOI] [PubMed] [Google Scholar]
- Hesse T. et al. (1989) Molecular cloning and structural analysis of a gene from Zea mays (L.) coding for a putative receptor for the plant hormone auxin. EMBO J., 8, 2453–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M. (1994) Auxin-binding proteins. Annu. Rev. Plant Physiol., 45, 393–420. [Google Scholar]
- Jones A.M. and Venis,M.A. (1989) Photoaffinity labelling of auxin-binding proteins in maize. Proc. Natl Acad. Sci. USA, 86, 6153–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Im,K.H., Savka,M.A., Wu,M.J., DeWitt,N.G., Shillito,R. and Binns,A.N. (1998) Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science, 282, 1114–1117. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-T., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT—a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lawerence M.C., Izard,T., Beuchat,M., Blagrove,R.J. and Colman,P.M. (1994) Structure of phaseolin at 2.2 Å resolution. Implications for a common vicilin/legumin structure and the genetic engineering of seed storage proteins. J. Mol. Biol., 238, 748–776. [DOI] [PubMed] [Google Scholar]
- LeBlanc N., David,K., Grosclaude,J., Pradier,J.M., Barbier-Brygoo,H., Labiau,S. and Perrot-Rechenmann,C. (1999) A novel immunological approach establishes that the auxin-binding protein, Nt-abp1, is an element involved in auxin signalling at the plasma membrane. J. Biol. Chem., 274, 28314–28320. [DOI] [PubMed] [Google Scholar]
- Löbler M. and Klämbt,D. (1985) Auxin-binding protein from coleoptile membranes of corn (Zea mays L.) I. Purification by immunological methods and characterisation. J. Biol. Chem., 260, 9848–9853. [PubMed] [Google Scholar]
- Macdonald H., Henderson,J., Napier,R.M., Venis,M.A., Hawes,C. and Lazarus,C.M. (1994) Authentic processing and targeting of active maize ABP1 in the baculovirus expression system. Plant Physiol., 105, 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Napier R.M. and Venis,M.A. (1990) Monoclonal antibodies detect an auxin-induced conformational change in the auxin-binding protein. Planta, 182, 313–318. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Sixma,T.K., Wilson,K.S. and Lamzin,V.S. (1997) wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr. D, 53, 448–455. [DOI] [PubMed] [Google Scholar]
- Rajan S.S. (1978) Napthalene acetic acid. Acta Crystallogr. B, 34, 998–1000. [Google Scholar]
- Šali A. and Blundell,T.L. (1993) Comparative modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Shimomura S., Sotobayashi,T., Futai,M. and Fukui,T. (1986) Purification and properties of an auxin-binding protein from maize shoot membranes. J. Biochem., 99, 1513–1524. [DOI] [PubMed] [Google Scholar]
- Steffens B., Feckler,C., Palme,K., Christian,M., Böttger,M. and Lüthen,H. (2001) The auxin signal for protoplast swelling is perceived by extracellular ABP1. Plant J., 27, 591–599. [DOI] [PubMed] [Google Scholar]
- Theil G., Blatt,M.R., Fricker,M.D., White,I.R. and Millner,P. (1993) Modulation of K+ channels in Vicia stomatal guard cells by peptide homologues to the auxin-binding protein C-terminus. Proc. Natl Acad. Sci. USA, 90, 11493–11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venis M.A. (1977) Solubilisation and partial purification of auxin-binding sites from corn membranes. Nature, 66, 268–269. [Google Scholar]
- Warwicker, J. (2001) Modelling of auxin-binding protein 1 suggests that its C-terminus and auxin could compete for a binding site that incorporates a metal ion and tryptophan residue 44. Planta, 212, 343–347. [DOI] [PubMed] [Google Scholar]
- Woo E.-J., Dunwell,J.M., Goodenough,P., Marvier,A.C. and Pickersgill,R.W. (2000a) Germin is a manganese containing homohexamer with germin and superoxide dismutase activities. Nat. Struct. Biol., 7, 1036–1040. [DOI] [PubMed] [Google Scholar]
- Woo E.-J. et al. (2000b) Crystallization and preliminary X-ray analysis of the auxin receptor. Acta Crystallogr. D, 56, 1476–1478. [DOI] [PubMed] [Google Scholar]