Abstract
Yeast Rad51 promotes homologous pairing and strand exchange in vitro, but this activity is inefficient in the absence of the accessory proteins, RPA, Rad52, Rad54 and the Rad55–Rad57 heterodimer. A class of rad51 alleles was isolated that suppresses the requirement for RAD55 and RAD57 in DNA repair, but not the other accessory factors. Five of the six mutations isolated map to the region of Rad51 that by modeling with RecA corresponds to one of the DNA-binding sites. The other mutation is in the N-terminus of Rad51 in a domain implicated in protein–protein interactions and DNA binding. The Rad51-I345T mutant protein shows increased binding to single- and double-stranded DNA, and is proficient in displacement of replication protein A (RPA) from single-stranded DNA, suggesting that the normal function of Rad55–Rad57 is promotion and stabilization of Rad51–ssDNA complexes.
Keywords: DNA repair/Rad51/Rad55/Rad57/recombination
Introduction
The repair of DNA double-strand breaks (DSBs) is essential to maintain genome integrity and for the accurate segregation of homologous chromosomes during meiosis. Recombinases of the RecA family play a central role in homology-dependent DSB repair (Gasior et al., 2001). These proteins assemble into filaments on single-stranded DNA (ssDNA) that is formed at stalled replication forks or by nucleolytic processing of DSBs (Flory et al., 1984; Egelman and Stasiak, 1986; Ogawa et al., 1993; Sung and Robberson, 1995). The nucleoprotein filament is the active form of the recombinase and is capable of searching for homology within intact duplex DNA to initiate synapsis and strand exchange (Kowalczykowski et al., 1994; Sung and Robberson, 1995).
Yeast and human encode two RecA homologs, Rad51 and Dmc1, as well as Rad51-related proteins, referred to as Rad51 paralogs (Gasior et al., 2001; Thompson and Schild, 2001). Yeast RAD51 is expressed in mitotic and meiotic cells and is required for resistance to ionizing radiation, spontaneous and induced mitotic recombination and for meiotic recombination (Aboussekhra et al., 1992; Basile et al., 1992; Shinohara et al., 1992). Deletion of RAD51 in vertebrates results in cell inviability and early embryonic lethality in mice (Lim and Hasty, 1996; Tsuzuki et al., 1996). Depletion of Rad51 using a conditional chicken DT40 cell line results in a G2/M phase arrest, the accumulation of cytologically visible chromosomal breaks and eventual cell death (Sonoda et al., 1998). These data suggest that the essential role of RAD51 in vertebrates is to repair breaks generated during DNA replication. DMC1 is expressed only during meiosis and, although it has some redundant functions with RAD51, it also has unique functions and acts specifically in the inter-homolog recombination pathway (Bishop et al., 1992; Schwacha and Kleckner, 1997; Shinohara et al., 1997). The Rad51 paralogs of Saccharomyces cerevisiae are encoded by the RAD55 and RAD57 genes and are determined by genetic studies to function in the same pathway as RAD51 (Kans and Mortimer, 1991; Lovett, 1994; Rattray and Symington, 1995). The vertebrate Rad51 paralogs are encoded by the RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3 genes (Thompson and Schild, 2001). Mutation of any of these genes in the chicken DT40 cell lines does not cause inviability, but the cells show high sensitivity to DNA cross-linking agents and increased frequencies of spontaneous chromosome aberrations (Takata et al., 2001).
Purified yeast Rad51 forms right-handed helical filaments on ssDNA and double-stranded DNA (dsDNA) with structural similarity to those formed by RecA (Ogawa et al., 1993; Sung and Robberson, 1995). Formation of filaments on ssDNA is stimulated in the presence of the heterotrimeric DNA-binding protein, replication protein A (RPA) (Sung and Robberson, 1995; Sugiyama et al., 1997). The addition of RPA to the Rad51 reaction is thought to allow the formation of continuous filaments by removal of secondary structures from ssDNA (Sugiyama et al., 1997). Once assembled, the Rad51 nucleoprotein filament is capable of interacting with a second DNA molecule to initiate strand exchange. The in vitro strand exchange assay is used frequently as a metric for pre-synapsis, synapsis and strand exchange steps of the reaction. Rad51 by itself has very weak strand exchange activity and can be stimulated by the addition of RPA, but only if Rad51 is allowed to nucleate on the ssDNA prior to the addition of RPA (Sung, 1994). If RPA is added before, or simultaneously with Rad51, then RPA inhibits the reaction (Sung, 1997b). The inhibition by RPA is presumed to be a consequence of the higher affinity of RPA for ssDNA and faster binding kinetics compared with Rad51.
The inhibitory effect of RPA can be overcome by the inclusion in the reaction of accessory proteins, also known as mediators. Rad52 is the best characterized of the mediators of Rad51 filament assembly (Sung, 1997a; Benson et al., 1998; New et al., 1998; Shinohara and Ogawa, 1998). Rad52 interacts with both RPA and Rad51, and the interaction with Rad51 is necessary for overcoming the RPA inhibitory effect (Shinohara and Ogawa, 1998). Rad52 is thought to replace RPA bound to ssDNA with Rad51, or provide a seeding site within the RPA-bound ssDNA for subsequent cooperative binding by Rad51 (Song and Sung, 2000). The Rad55 and Rad57 proteins, which form a stable heterodimer, can also overcome the inhibition to Rad51-promoted strand exchange imposed by RPA (Sung, 1997b), but the mechanism of mediation is unknown. Consistent with their in vitro roles in Rad51-promoted strand exchange, rad52, rad55 and rad57 mutants fail to assemble Rad51 foci during meiotic recombination (Gasior et al., 1998). However, in mitotic cells, there appears to be some redundancy between Rad52 and Rad55–Rad57 for the assembly of radiation-induced Rad51 foci (Gasior et al., 2001). The role of Rad55 and Rad57 as accessory proteins for Rad51 is also supported by the observation that RAD51 expressed from a high copy number plasmid partially suppresses the radiation sensitivity of rad55, rad57 and rad55 rad57 mutants (Hays et al., 1995; Johnson and Symington, 1995). The cold sensitivity for DNA repair conferred by rad55 and rad57 deletion mutants is suggestive of a role for these proteins in stabilizing a protein complex (Game and Mortimer, 1974; Lovett and Mortimer, 1987; Hays et al., 1995; Johnson and Symington, 1995).
Mediators for recombinase functions are also found in bacteriophage T4 and bacteria. The strand exchange activity of T4 UvsX is stimulated by UvsY, which interacts directly with UvsX and gene 32 protein (ssDNA-binding protein) to replace gene 32 protein bound to ssDNA with UvsX (Bleuit et al., 2001). In Escherichia coli, the RecBCD nuclease–helicase complex facilitates loading of RecA onto 3′ ssDNA generated at Chi sites (Anderson and Kowalczykowski, 1997), and the RecF, RecO and RecR proteins overcome the inhibition to RecA-mediated strand exchange by ssDNA-binding protein (SSB) (Umezu et al., 1993). The UV sensitivity of recF, recO and recR mutants can be suppressed by high copy expression of recA, consistent with their roles as accessory proteins. Furthermore, several extragenic suppressors of the UV sensitivity of recF mutants (srf mutants) map to recA (Volkert and Hartke, 1984). The recA srf alleles also suppress the UV sensitivity of recO and recR mutants, suggesting that RecFOR functions at the same step in recombination (Wang et al., 1993). Alleles of recA that are constitutive for the SOS response (tif alleles) also confer the srf phenotype (Wang et al., 1993). Most of the srf and tif alleles map to regions of RecA involved in monomer–monomer or polymer–polymer interactions, and the mutant proteins are better at displacing SSB from ssDNA (Kowalczykowski et al., 1994).
The similarity between Rad55, Rad57 and RecFOR in their roles as mediators of the strand exchange activity of their cognate recombinases suggested the possibility that alleles of RAD51 with altered function could be isolated as suppressors of the DNA repair defect conferred by rad55 and rad57 mutations. Here, we describe six alleles of RAD51 that partially suppress the radiation sensitivity of rad55 and rad57 mutants, but not rad52 or rad54 mutants. Biochemical characterization of one of the mutant Rad51 proteins revealed higher affinity and more stable binding to DNA, suggesting that one function of Rad55 and Rad57 is promotion and stabilization of the Rad51 nucleoprotein filament.
Results
Identification of alleles of RAD51 that partially suppress the ionizing radiation sensitivity of a rad57 strain
Based on the observation that RAD51 present on a high copy number plasmid partially suppresses the γ-ray sensitivity of a rad55 rad57 strain, we sought to identify alleles of RAD51 that when expressed at low copy (from a CEN plasmid) could also suppress the rad55 rad57 mutant phenotype. Because Rad55 and Rad57 act as an obligate heterodimer and the phenotype of the double mutant is no more severe than that of the single mutants, the screen was conducted in a rad51 rad57 yeast strain. The strain utilized contained a deletion of the RAD51 gene in case the mutations were recessive. A CEN/ARS/HIS3 plasmid carrying the RAD51 open reading frame (ORF) and promoter elements was mutagenized randomly and the pool of mutant plasmids transformed into the double mutant strain. Transformants were selected and individual isolates were assayed at 23°C for their resistance to γ-irradiation. Suppression by mutant rad51 plasmids was expected to be more obvious at 23°C due to the cold sensitivity for DNA repair of rad57 mutants. Those transformants that survived irradiation at frequencies equal to or higher than that seen with the double mutant strain harboring wild-type RAD51 expressed from a high copy number (2µ) plasmid were analyzed further.
To establish that the phenotype was the result of the plasmid and not some other change in the strain, the plasmid containing the putative rad51 mutation was isolated from yeast, amplified in E.coli and then retested in the rad51 rad57 strain. Rad51 protein levels from each suppressing CEN plasmid were assayed by western blot analysis and shown to be equal to wild-type RAD51 expressed from the same CEN plasmid (data not shown).
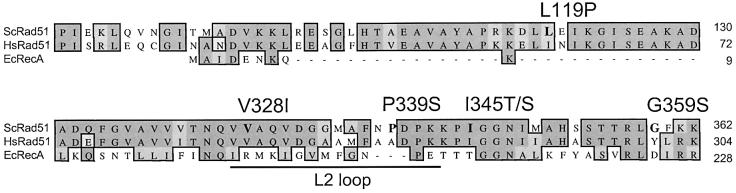
Of ∼3000 transformants screened, six plasmids were obtained capable of conferring increased γ-ray resistance without overexpressing Rad51. From the DNA sequence, it was determined that each contained a single point mutation within the RAD51 ORF. The residues altered and the putative function of these residues based on alignment with RecA are listed in Table I. Two alleles were identified that contained different point mutations in codon 345; one resulted in alteration of Ile345 to a threonine while the other change was to serine. Figure 1 is a partial alignment of the amino acid sequence of S.cerevisiae Rad51 compared with E.coli RecA and human Rad51. Each of the amino acid changes is indicated. Val328 and Ile345 are both conserved in human Rad51, and the corresponding region of RecA is implicated in DNA binding and/or ATP-induced conformational changes (Story et al., 1992, 1993). Leu119 corresponds to Ile61 in the human protein. The N-terminal domain of human Rad51, which is highly conserved among Rad51 proteins, has DNA-binding activity (Aihara et al., 1999). The N-terminal region of yeast Rad51 is implicated in monomer–monomer interactions and interaction with Rad52 and Rad54 (Krejci et al., 2001). Although none of the altered residues is conserved with E.coli RecA, Ile345 immediately precedes two invariant glycine residues that are thought to be involved in DNA binding or ATP-induced conformational changes in RecA (Story and Steitz, 1992). These alleles of rad51 will be referred to as srp (suppressor of rad51 paralogs) alleles.
Table I. rad51 alleles recovered from the screen.
| Mutated residue | Putative functiona |
|---|---|
| Rad51-L119P | Rad51 self-interaction, Rad52–Rad54 interaction, DNA interactionb |
| Rad51-V328I | DNA interaction (L2 loop) |
| Rad51-P339S | DNA interaction (L2 loop) |
| Rad51-I345T | DNA interaction and/or ATP-induced conformational change |
| Rad51-I345S | DNA interaction and/or ATP-induced conformational change |
| Rad51-G359S | β-strand 6 |
aPutative function based on alignment with RecA (Kowalczykowski et al., 1994).
bPutative function for Leu119 based on studies of human and yeast Rad51 (Aihara et al., 1999; Krejci et al., 2001).
Fig. 1. Partial alignment of ScRad51 with HsRad51 and RecA. The srp mutations are indicated above each mutated amino acid (in bold). The L2 loop of RecA, corresponding to one of the DNA-binding sites, is indicated.
The rad51 srp alleles suppress the γ-ray sensitivity of rad55 and rad57 mutants, but not rad52 or rad54 mutants
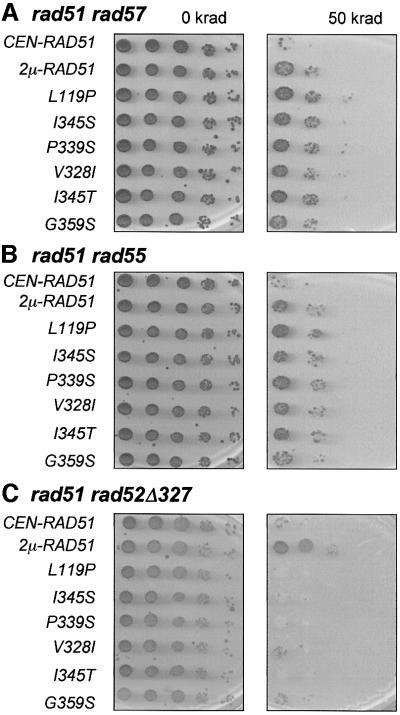
Each of the plasmids containing the srp alleles was able to complement the radiation sensitivity of a rad51 strain (data not shown), indicating that they have retained RAD51 function. Rad57 acts as an obligate heterodimer with Rad55; therefore, the srp alleles were expected to suppress the radiation sensitivity conferred by a rad55 mutation as well as a rad57 mutation. The CEN plasmids expressing the srp alleles, wild-type RAD51 expressed from the same CEN plasmid or RAD51 expressed from a 2µ plasmid were used to transform rad51 rad55 and rad51 rad57 strains. Serial dilutions of the strains containing each plasmid were grown at 23°C after 50 krad γ-irradiation and compared with an unirradiated control (Figure 2). When RAD51 was expressed at low copy number from a CEN plasmid, only a few colonies were formed from undiluted rad51 rad55 or rad51 rad57 cells after irradiation. When RAD51 was expressed from a 2µ plasmid, a 10- to 50-fold suppression of the γ-ray sensitivity was observed in both strain backgrounds. Each of the rad51 srp plasmids conferred partial suppression of both the rad55 and the rad57 mutant phenotypes when expressed from a CEN plasmid, with ∼10- to 50-fold increased plating efficiency after γ-irradiation when compared with wild-type RAD51.
Fig. 2. Suppression of the γ-ray sensitivity of rad55 and rad57 mutants by the plasmid-borne rad51 srp alleles. (A) Serial dilutions of rad51 rad57 strains containing each of the rad51 mutant plasmids, wild-type RAD51 expressed from the same CEN/HIS/ARS plasmid (CEN-RAD51) and wild-type RAD51 expressed from a 2µ plasmid (2µ-RAD51) were spotted onto YPD plates and left unirradiated or irradiated at 50 krad. (B) Serial dilutions of rad51 rad55 strains containing each of the plasmids described above were spotted onto YPD plates and treated with 0 or 50 krad. (C) Serial dilutions of rad51 rad52Δ327 plasmid-containing strains irradiated with 0 or 50 krad. For strains tested in (A–C), all survival was assessed following incubation at 23°C.
According to the in vitro strand exchange data, both the Rad55–Rad57 heterodimer and Rad52 play some role in enabling Rad51 to overcome the inhibition imposed by RPA (Sung, 1997a,b; Benson et al., 1998; New et al., 1998; Shinohara and Ogawa, 1998). To determine whether the Rad52 and Rad55–Rad57 mediators play redundant roles in the Rad51-promoted reaction, we tested the ability of the rad51 srp alleles to suppress a rad52 phenotype. In S.cerevisiae, rad52 mutants show the most severe recombination and repair phenotypes of all the rad52 group mutants. Consequently, Rad52 protein is thought to have other roles in recombination in addition to its mediator function. rad52Δ327 is a C-terminal truncated allele of RAD52 encoding amino acids 1–327. The truncated protein was shown to have partial DSB repair activity in vivo, but to be defective in its interaction with Rad51 and unable to function as a mediator in vitro (Boundy-Mills and Livingston, 1993; Milne and Weaver, 1993). As in the case of rad55 and rad57 mutants, overexpression of RAD51 partially suppresses the methyl methanesulfonate (MMS) sensitivity of a rad52Δ327 strain (Boundy-Mills and Livingston, 1993; Milne and Weaver, 1993). To assess whether any of the rad51 srp alleles also suppressed the Rad52 mediator defect, each was transformed into a rad51 rad52Δ327 strain. As expected, RAD51 expressed from a 2µ plasmid suppressed the radiation sensitivity of the rad51 rad52Δ327 strain, but none of the rad51 srp alleles did (Figure 2).
Rad54 protein stimulates the Rad51-promoted pairing reaction primarily during the synapsis and strand exchange phases, thus the rad51 srp alleles were not expected to suppress the requirement for RAD54 in DNA repair (Petukhova et al., 1998; Mazin et al., 2000; Van Komen et al., 2000; Solinger et al., 2001). As anticipated, no suppression of the DNA repair defect was observed when each of the rad51 srp alleles was expressed in a rad51 rad54 strain (data not shown).
To determine whether there is synergism between the rad51-L119P mutation and alleles in the C-terminal region of RAD51, a rad51-L119P, I345T double mutant was constructed. However, the level of suppression of the rad57 DNA repair defect was the same as observed for the two single mutations (data not shown). The rad51 srp alleles showed the same level of suppression of the rad55 and rad57 DNA repair defects whether expressed from a high copy number plasmid or from a CEN plasmid (data not shown), suggesting that RAD55 and RAD57 have additional roles in DNA repair, or that other factors are limiting in their absence.
Suppression of the rad57 DNA repair defect by rad51-I345T expressed in single copy
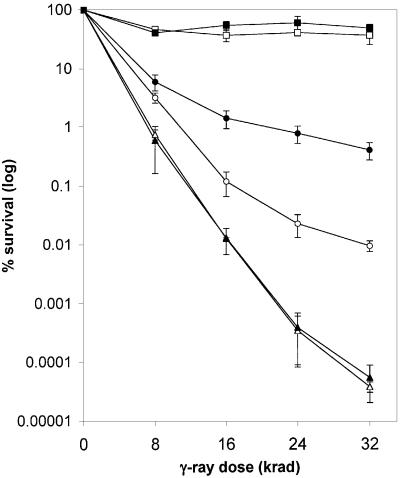
Although CEN plasmids are maintained at low copy number, some mutations confer a slightly different phenotype when present on a CEN plasmid compared with the native chromosomal locus (U.Mortensen and R.Rothstein, personal communication). To test whether suppression of the rad57 phenotype could occur when the srp alleles were expressed from the chromosomal RAD51 locus, a representative srp allele was used to replace the native RAD51 locus. As the mutations in most of the srp alleles map to the region of Rad51 implicated in DNA binding, the mechanism of suppression is probably the same. Therefore, the rad51-I345T allele was chosen as representative because two independent mutations were identified affecting this residue, which is also conserved in the human Rad51 protein. The strain containing the rad51-I345T allele is as resistant as the wild-type strain at both 30 (Figure 3) and 23°C (data not shown), suggesting that this allele retains RAD51 function. At 30°C, strains containing rad51::LEU2 and rad51::LEU2 rad57::LEU2 are both equally sensitive, while the rad57:: LEU2 strain is moderately more resistant. The rad51- I345T rad57::LEU2 strain, when compared with rad57:: LEU2, shows an increase in survival of 1–2 orders after 32 krad γ-irradiation, similar to the level of suppression seen when this allele is expressed from a CEN plasmid (Figure 3). At 23°C, the rad57::LEU2 strain is as sensitive as the rad51::LEU2 and rad51::LEU2 rad57::LEU2 strains due to the cold sensitivity conferred by the rad57 mutation. The rad51-I345T rad57::LEU2 strain, when compared with the rad57::LEU2 strain, shows a 50-fold increase in resistance after 32 krad of γ-irradiation at 23°C (data not shown). Thus the rad51-I345T mutation shows similar suppression of the DNA repair defect of the rad57::LEU2 strain at both temperatures. However, it should be noted that the rad51-I345T rad57::LEU2 strain, like the rad57::LEU2 strain, is more sensitive to γ-irradiation at 23°C than at 30°C.
Fig. 3. Partial suppression of the γ-ray sensitivity conferred by the rad57 mutation by rad51-I345T expressed from the chromosome. Survival was assessed following incubation at 30°C. The log percentage survival was plotted as a function of dose for each strain (filled squares, rad51-I345T; open squares, wild-type; filled circles, rad51-I345T rad57::LEU2; open circles, rad57::LEU2; filled triangles, rad51:: LEU2; open triangles, rad51::LEU2 rad57::LEU2).
The rad51-I345T rad57 strain was crossed to a RAD51 rad57 strain to test for dominance. The resulting diploid was ∼2-fold less resistant to γ-irradiation than the rad51-I345T rad57 homozygous diploid, but >20-fold more resistant than the RAD51 rad57 diploid (data not shown). Thus, the rad51-I345T mutation is dominant.
Partial suppression of the rad57 mating-type switching defect by the rad51-I345T allele
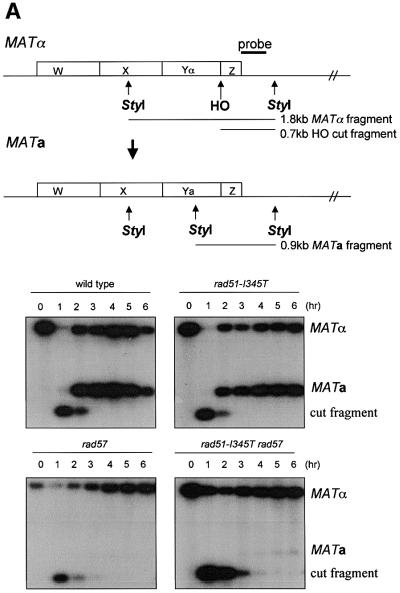
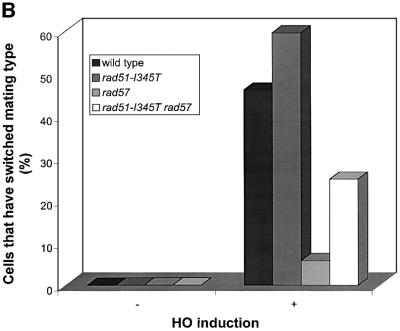
Ionizing radiation produces a variety of DNA lesions in addition to DSBs. To determine the effect of the rad51- I345T allele on the repair of a single DSB, a mating-type switching assay was performed. The repair of an HO endonuclease-induced DSB was monitored at the DNA level after induction of HO endonuclease for 1 h. To measure the formation of switched products, the DNA samples were digested with StyI, which cuts within MAT Ya but not Yα sequences. The appearance of a 0.9 kb StyI fragment is indicative of repair of the DSB from the HMRa locus (Figure 4A). In the wild-type and rad51-I345T strains, switching was efficient and completed 3 h after induction of HO. In the rad57 mutant, the 0.7 kb cut fragment persisted for several hours, but disappearance of the cut DNA was not concomitant with conversion to the MATa product. In the rad51-I345T rad57 double mutant, a low level of switched product was detected 3 h after induction of HO, indicating partial suppression of the rad57 mating-type switching defect.
Fig. 4. (A) The rad51-I345T mutation partially suppresses the mating-type switching defect of a rad57 strain. Schematic representation of the MATα and MATa loci indicating the locations of StyI sites and the hybridization probe is shown at the top of the figure. HO endonuclease produces a 0.7 kb fragment from the 1.8 kb MATα StyI fragment. A 0.9 kb StyI fragment is produced when the mating-type switches from MATα to MATa. DNA was isolated from cultures grown at 30°C prior to galactose induction (0 h time point) and at 1 h intervals after HO induction. The strains tested are indicated above each autoradiogram. (B) Suppression of the rad57 mating-type switching defect quantified genetically. HO endonuclease was induced in each strain for 3 h. Cells plated on YPD and incubated at 30°C were assessed for their mating type. Shown is a representative experiment displaying the reproducible 4-fold suppression of rad57 by rad51-I345T. Black bars, wild-type; dark gray bars, rad51-I345T; light gray bars, rad57; white bars, rad51-I345T rad57.
To quantitate mating-type switching, HO endonuclease was induced in cultures of each strain for 3 h. Cells were then plated on YPD to repress expression of HO and survivors were tested for their mating type. Approximately 50% of wild-type and 60% of rad51-I345T cells switched from MATα to MATa following HO induction whether grown at 23 (data not shown) or 30°C (Figure 4B). The strain containing the rad51-I345T mutation consistently showed slightly increased switching compared with the wild-type strain. At 30°C, the rad51-I345T mutation suppressed the mating-type switching defect of the rad57 strain by ∼4-fold (Figure 4B). At 23°C, suppression of the rad57 defect by rad51-I345T was weaker, with a 2-fold increase in the number of switched products compared with the rad57 strain (data not shown). Low levels of switched products (2–6%) were detected in the rad57 mutant, consistent with a previous study suggesting that RAD55 and RAD57 are not essential for mating-type switching (Johnson and Symington, 1995).
Rad51-I345T overcomes the inhibition imposed by RPA
The mediator role of the Rad55–Rad57 heterodimer was discovered using an in vitro strand exchange assay (Sung, 1997b). The addition of the Rad55–Rad57 heterodimer enables Rad51 to overcome the inhibition imposed by RPA, presumably by mediating Rad51 filament formation. Consistent with this model, the formation of meiosis-specific Rad51 foci requires Rad55 and Rad57 (Gasior et al., 1998). Because the rad51 srp alleles partially suppress the γ-ray sensitivity of rad57 mutants, one possibility is that the Rad51 srp mutant proteins are capable of overcoming the inhibition imposed by RPA in the absence of Rad55 and Rad57. This notion was tested biochemically using the purified Rad51-I345T protein (Figure 5A).
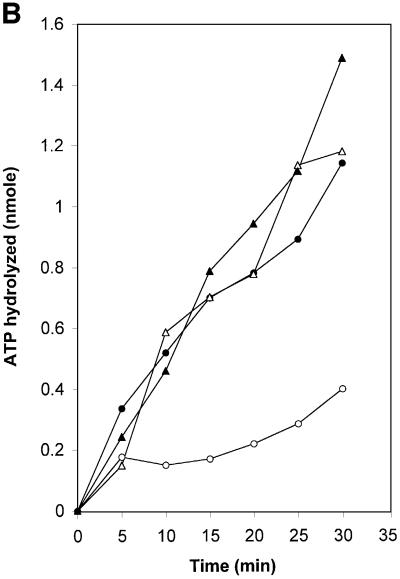
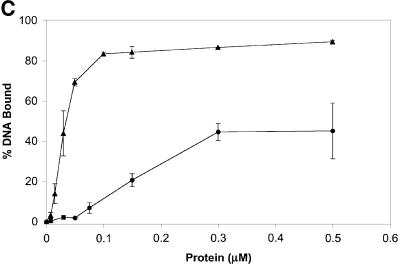
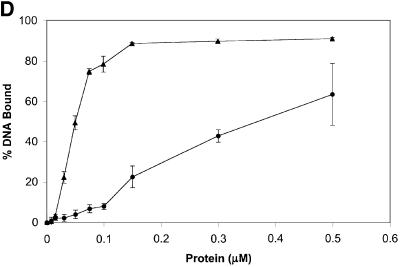
Fig. 5. Rad51-I345T overcomes the inhibition to ssDNA binding imposed by RPA and exhibits higher affinity for single- and double-stranded DNA. (A) Purified proteins used in the in vitro assays. Lane 1, RPA (3.75 µg); lane 2, molecular weight size standards; lane 3, Rad51 (7.5 µg); lane 4, Rad51-I345T (9 µg). (B) ssDNA-dependent ATPase was compared for wild-type Rad51 versus Rad51-I345T using poly(dT) as the ssDNA substrate. Rad51 (filled circles), Rad51-I345T (filled triangles), Rad51 with RPA-coated DNA (open circles), Rad51-I345T with RPA-coated DNA (open triangles). (C) Increasing concentrations of either wild-type Rad51 (filled circles) or Rad51-I345T (filled triangles) were incubated with a constant amount of ssDNA and passed through alkali-treated nitrocellulose and DEAE filters. The percentage bound represents the amount of DNA retained on nitrocellulose compared with the total DNA. (D) As (C) except using dsDNA as the substrate.
The ability of wild-type and mutant proteins to overcome the RPA inhibition of pre-synaptic filament formation was examined by measuring the ssDNA-dependent ATPase activity of Rad51. If ssDNA is pre-coated with RPA, the ATPase activity of Rad51 is inhibited because Rad51 is unable to compete with RPA for binding to the ssDNA. ATP hydrolysis with RPA-coated ssDNA as a cofactor is then a measure of the ability of Rad51 to overcome the inhibition imposed by RPA. The wild-type and Rad51-I345T proteins showed similar rates of ATP hydrolysis using as a DNA cofactor poly(dT), which has little or no secondary structure (Figure 5B). As expected, ATP hydrolysis by Rad51 was inhibited when the DNA was coated with RPA. However, in the case of Rad51-I345T, ATP hydrolysis was the same with either naked or RPA-coated ssDNA (Figure 5B). Thus, RPA does not prevent Rad51-I345T from gaining access to DNA.
Rad51-I345T shows increased affinity for DNA
Most of the rad51 srp mutations map to the region of Rad51 that by alignment with RecA corresponds to the L2 DNA-binding domain. This observation suggests that the mutant proteins might have an altered affinity for DNA. Consistent with this hypothesis, generation of random mutations within the region corresponding to the L2 loop of the human Rad51 protein resulted in proteins with increased ssDNA-binding activity in vitro (Kurumizaka et al., 1999). Binding to ssDNA and dsDNA by Rad51-I345T was tested using a nitrocellulose filter-binding assay. When increasing concentrations of protein were incubated with a constant amount of either ssDNA or dsDNA, 6- to 8-fold less of the Rad51-I345T protein was required to achieve half-maximal binding compared with wild-type Rad51 (Figure 5C and D). This increased binding to DNA was evident for both ssDNA and dsDNA. The sigmoidal curves for both wild-type Rad51 and Rad51-I345T suggest cooperative binding to both substrates. The shift of the curve for the Rad51-I345T protein indicates a higher affinity for both ssDNA and dsDNA. The increased level of complex formation may reflect a more stable interaction between the Rad51-I345T protein and DNA compared with wild-type Rad51.
Discussion
The nucleoprotein filament formed by interaction between Rad51 and ssDNA is essential for homologous pairing and strand exchange. Formation of the nucleoprotein filament is impeded by RPA, which has higher affinity for ssDNA and is more abundant than Rad51 (Sugiyama et al., 1997). Consequently, additional factors, known as mediators, are required to promote assembly of the Rad51 nucleoprotein filament in the presence of RPA. Rad52 and Rad55–Rad57 have both been identified as mediators based on their ability to stimulate Rad51-promoted strand exchange in the presence of RPA (Sung, 1997a,b; New et al., 1998; Shinohara and Ogawa, 1998), but these factors are clearly non-redundant in vivo (Rattray and Symington, 1995). Efforts to understand the mechanism of mediation by Rad55–Rad57 have been hampered by the difficulty in purifying the heterodimer. To shed light on this problem, we isolated gain-of-function alleles of RAD51 that partially suppress the requirement for RAD55 and RAD57 in DNA repair, but not the need for RAD52. Thus, the mechanism by which the two mediators stimulate Rad51 can be separated genetically.
Rad51-I345T protein, a representative of the gain-of-function mutants, was compared with wild-type Rad51 protein for its ability to overcome the inhibition imposed by RPA. As Rad51 is an ssDNA-dependent ATPase, pre-incubating the DNA cofactor with RPA inhibits Rad51 ATPase activity by competing with Rad51 for binding sites on the ssDNA (Sugiyama et al., 1997). Pre-coating ssDNA with RPA fails to inhibit the ATPase activity of the Rad51-I345T protein (Figure 5B). Thus, Rad51-I345T is able to compete with RPA for binding to ssDNA and displace it. The RecA-803 protein, identified genetically as a suppressor of the UV sensitivity conferred by recF mutations, also shows increased ability to compete with SSB for binding to ssDNA (Lavery and Kowalczykowski, 1992; Madiraju et al., 1992). These data suggest that the primary function of the mediator proteins in prokaryotes and eukaryotes is to assist loading, or stabilization, of the recombinase to ssDNA in the presence of SSBs. The purified RecO and RecR proteins facilitate loading of RecA onto ssDNA in the presence of SSB by a direct interaction between RecO and SSB (Umezu and Kolodner, 1994). The RecF, RecO and RecR proteins also play additional roles in stabilization of the RecA filament and preventing RecA polymerization onto dsDNA from adjacent ssDNA (Shan et al., 1997; Webb et al., 1997).
Formation of joint molecules in reactions employing ssDNA circles and linear duplex molecules as substrates was the assay originally used to define the Rad55–Rad57 heterodimer as a mediator in enabling Rad51 to overcome the inhibition imposed by RPA. When Rad51-I345T was used in the strand exchange assay, even under stimulatory conditions, joint molecules were not resolved to nicked circles (data not shown). Instead, large aggregates of DNA were observed. This could result from an inability to complete strand exchange due to unproductive binding of Rad51 to dsDNA, or perhaps the products of reinvasion by the displaced strand (Chow et al., 1988). RecA mutant proteins that show increased binding to DNA also promote formation of aggregates when tested in the in vitro strand exchange assay (Lavery and Kowalczykowski, 1990). DNA aggregates are likely to represent the products of invasion of the displaced strand from the linear duplex into another duplex molecule (Chow et al., 1988). The continuing cycle of re-invasion is thought to result in the large DNA networks seen on agarose gels as smeared species with slower migration than the predicted product bands. As mentioned earlier, based on sequence alignment with RecA, Ile345 maps to a region of Rad51 implicated in DNA binding. Studies with the human Rad51 protein have shown that the I287S mutation, corresponding to Ile345 of yeast Rad51, results in a protein with higher affinity for single-stranded DNA (Kurumizaka et al., 1999). Thus, it seemed plausible that the rad51-I345T allele may act to suppress the defects caused by the rad57 mutation by more stable binding of the mutant protein to DNA.
Rad51 binding to both ssDNA and dsDNA exhibits a sigmoidal dependence on protein concentration indicative of cooperative binding (Shinohara et al., 1992). Rad51- I345T also appears to bind ssDNA and dsDNA cooperatively, but the concentration of protein required to exhibit half-maximal binding to the DNA is 6- to 8-fold less than wild-type protein (Figure 5). This observation suggests that Rad51-I345T has a higher affinity for DNA. Also different between wild-type and mutant proteins was the maximum amount of DNA bound. This property could be explained in terms of the dynamic interaction between Rad51 and DNA. With wild-type protein, the equilibrium between bound and unbound protein would appear to be such that the maximum amount of DNA trapped in complexes was never more than 60%. An intrinsic decrease in the Keq of Rad51-I345T could result in formation of a higher level of trappable complex. The RecA mutant proteins encoded by the suppressors of recF, recO and recR mutants (recA srf alleles) have been determined to have higher affinity for ssDNA (Lavery and Kowalczykowski, 1992). However, none of the mutations in the srf alleles maps to residues implicated in DNA binding by RecA. Instead, the mutations are located in the regions thought to be involved in filament–filament interactions (Story et al., 1992). In this case, the mechanism of suppression appears to operate through decreasing the formation of large RecA protein aggregates and promoting productive association with ssDNA (Kowalczykowski et al., 1994).
The cold sensitivity of rad55 and rad57 mutants has suggested that the Rad55–Rad57 heterodimer is involved in stabilizing a protein or protein–DNA complex (Lovett and Mortimer, 1987; Hays et al., 1995; Johnson and Symington, 1995). A critical early step in recombinational repair is the formation of the Rad51 filament on ssDNA. Therefore, it is quite plausible that the Rad55–Rad57 heterodimer serves to stabilize the interaction of Rad51 with ssDNA. Consistent with this idea, the requirement for Rad55–Rad57 is partially alleviated by the Rad51-I345T protein, which exhibits more stable binding to DNA than wild-type Rad51.
The current model for Rad52 in Rad51-mediated strand exchange is that Rad52 is required to target Rad51 to the ssDNA and displace RPA (Song and Sung, 2000). A Rad51 mutant that can compete with RPA for binding to ssDNA should bypass the need for Rad52 in forming the pre-synaptic filament. One possible explanation for why Rad51-I345T still requires Rad52 in vivo is that the mutant protein shows higher affinity and more stable binding to both ssDNA and dsDNA. Therefore, Rad52 is still required to target Rad51-I345T to ssDNA and prevent unproductive association of Rad51-I345T with duplex DNA. However, once targeted to the ssDNA, Rad51- I345T is bound more stably and no longer requires stabilization by the Rad55–Rad57 heterodimer. Based on the properties of the Rad51-I345T mutant protein, we suggest that the Rad52 and Rad55–Rad57 mediators have distinct functions in the formation of the Rad51 nucleoprotein filament. ssDNA formed by resection of double-stranded ends is immediately bound by RPA. Rad52 associates with RPA and Rad51, and targets Rad51 to ssDNA. We propose that Rad55 and Rad57 stabilize the initial Rad51–ssDNA complex to facilitate cooperative binding by additional Rad51 monomers, resulting in filament formation.
The role of the vertebrate Rad51 paralogs in recombinational repair is currently unknown. Like rad55 and rad57 mutants, rad51b, rad51c, rad51d, xrcc2 and xrcc3 cell lines show less severe defects in recombinational repair than rad51 mutants, and overexpression of RAD51 suppresses the sensitivity of the paralog mutant cell lines to DNA-damaging agents (Takata et al., 2001). Given these similarities, it seems likely that mutations in human RAD51 corresponding to the yeast rad51 srp alleles might suppress the requirement for the vertebrate Rad51 paralogs in DNA repair and genome stability.
Materials and methods
Media, growth conditions and genetic methods
Standard genetic methods were followed. Rich medium (YPD) and synthetic complete (SC) medium lacking the appropriate amino acid or nucleic acid base were prepared as described previously (Sherman et al., 1986). Selection for Ura– cells was performed on SC medium containing 5-fluoro-orotic acid (5-FOA) at 1 mg/ml (Boeke et al., 1987). Yeast mating, sporulation and tetrad dissection were performed as previously described (Sherman et al., 1986). The DNA repair and recombination defects of rad55 and rad57 strains are greater at lower temperatures, and most of the assays using these mutants were performed at room temperature (23°), unless otherwise indicated. For most purposes, yeast cells were grown at 30°C. Transformations were performed by the lithium acetate method (Ito et al., 1983).
Yeast strains and plasmids
Saccharomyces cerevisiae strains used in this study are listed in Table II. All strains except RDKY2275 are derivatives of strains W303-1A or W303-1B containing the corrected RAD5 allele (W1588-4C and W1588-4A) except where noted. Double mutants were identified by phenotype among haploid progeny generated by crossing the appropriate haploid parents. To construct strain LSY991, LSY411 was co-transformed with a BamHI fragment from plasmid pRS413::rad51-I345T and a replicating vector containing the HIS3 gene, and selecting for His+ transformants. Recombination of the fragment with the homologous chromosomal locus results in loss of the URA3 marker and 5-FOA resistance. His+ transformants were screened for 5-FOA resistance, and positive clones were verified further by their resistance to 50 krad γ-irradiation (Gammacell-220 60Co irradiator, Atomic Energy of Canada). The rad51 ORF of 5-FOA-resistant, γ-ray-resistant colonies was amplified by PCR and directly sequenced to confirm the presence of the rad51-I345T allele. Segregation of rad51-I345T in crosses could be distinguished from RAD51 by the increased resistance to γ-irradiation in the presence of a rad57 mutation. Alternatively, LSY991 was crossed to strains containing a marked rad51 allele that would segregate from the rad51-I345T allele during genetic crosses. To construct strain LSY1007, HKY595-1C was crossed to J883. Haploid progeny were irradiated with 50 krad, and tetrads showing 2:2 segregation for γ-ray sensitivity were scored as rad51::LEU2 rad52Δ327 double mutants. To construct strain LSY1009, LSY679 (ade2-1) was transformed to Ura+ with BstEII-linearized plasmid YIPade3::HO (Sandell and Zakian, 1993). Ura+ transformants were grown on non-selective medium and then streaked onto medium containing 5-FOA to select for pop-out recombination events. White colonies contain the ade3::HO disruption, whereas red colonies contain the undisrupted ADE3 allele. The presence of the GAL10-regulated HO gene was confirmed by the poor growth of white colonies on medium containing galactose.
Table II. Yeast strains.
| Strain | Genotypea | Reference or source |
|---|---|---|
| W1588-4C | MATa | R.Rothstein |
| W1588-4A | MATα | R.Rothstein |
| HKY595-1C | MATα rad51::LEU2 | H.Klein |
| HKY595-3B | MATa rad51::LEU2 | H.Klein |
| HKY597-2B | MATa rad55::LEU2 | H.Klein |
| HKY597-2C | MATα rad55::LEU2 | H.Klein |
| HKY587-2C | MATa rad57::LEU2 | H.Klein |
| HKY598-8B | MATα rad57::LEU2 | H.Klein |
| J883 | MATa rad52Δ327 | R.Rothstein |
| YAR179-2C | MATa rad51::LEU2 rad54::LEU2 ade2::hisG-URA3-hisG | Rattray and Symington (1995) |
| LSY411 | MATα rad51::URA3 rad5-535 | Rattray and Symington (1994) |
| LSY516-1C | MATα rad51::URA3 rad57::LEU2 | This study |
| LSY516-2D | MATa rad51::URA3 rad57::LEU2 | This study |
| LSY989-1 | MATa rad51::LEU2 rad57::LEU2 | This study |
| LSY989-2 | MATα rad51::LEU2 rad57::LEU2 | This study |
| LSY990 | MATa rad51::LEU2 rad55::LEU2 | This study |
| LSY991 | MATα rad51-I345T rad5-535 | This study |
| LSY992-1 | MATa rad51-I345T rad57::LEU2 | This study |
| LSY992-2 | MATα rad51-I345T rad57::LEU2 | This study |
| LSY1007-1 | MATa rad51::LEU2 rad52Δ327 | This study |
| LSY1007-2 | MATα rad51::LEU2 rad52Δ327 | This study |
| LSY1009 | MATα ade3::GAL10-HO | This study |
| LSY1022-2 | MATα rad57::LEU2 ade3::GAL10-HO | This study |
| LSY1023-1 | MATα rad51-I345T rad57::LEU2 ade3::GAL10-HO | This study |
| LSY1024-2 | MATα rad51-I345T ade3::GAL10-HO | This study |
| RDKY2275 | MATa ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1D1.6R can1 GAL pRDK273, pRDK274, pRDK275 | Nakagawa et al. (2001) |
| MCY14 | MATa suc2-437 lys2-801 | S.Jinks-Robertson |
| SJR13 | MATα lys2-802 | S.Jinks-Robertson |
aAll strains are in the RAD5 corrected W303 background (his3-11, 15 leu2-3,112 trp1-1 ura3-1 ade2-1 can1-100 RAD5) except RDKY2275, MCY14 and SJR13; only differences from this genotype are noted.
Plasmid pRS413:RAD51 was generated by cloning the 3.7 kb BamHI fragment from pRS423::RAD51 into the BamHI site of pRS413. The rad51-I345T point mutation was reconstituted in plasmid pEZ5139 (Zaitseva et al., 1999) using the Gene Editor in vitro Site-Directed Mutagenesis System (Promega, Madison, WI) and mutagenic oligonucleotide 5′-CCAGATCCAAAGCCTACCGGTGGTAATATT-3′.
Genetic screen for rad57 suppressors
Plasmid pRS413::RAD51 was mutagenized randomly by growth for ∼30 generations in E.coli strain XL1-Red [endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT Tn10 (Tetr) Stratagene]. Two different pools were screened. Each pool of mutagenized plasmids was used to transform yeast strain LSY516-2D to His+. Cultures (1 ml) of individual His+ transformants were grown to saturation, harvested and resuspended in 1 ml of water. A 5 µl aliquot of each suspension was spotted onto selective plates, irradiated at 50 krad and incubated at 23°C for 5 days. Plasmids were recovered and re-transformed into LSY516-2D. Rad51 protein levels were measured for each transformant capable of growth after 50 krad. Cell extracts were prepared (Strahl-Bolsinger et al., 1997) and analyzed by western blotting using affinity-purified rabbit anti-Rad51 (kindly provided by P.Sung).
γ-irradiation survival assays
Cells were grown in liquid medium to mid-log phase. The cultures were serially diluted and aliquots of each dilution were plated on solid medium. The plates were irradiated in a Gammacell-220 containing 60Co for the designated dose. The dose rate of the Gammacell-220 was 53 rad/s. The plates were incubated for 3 days at 30°C or 5 days at 23°C, before survivors were counted. Each strain was assayed three separate times and the mean values are presented (Figure 3). For the spot assays, strains were grown as described above, serial dilutions were spotted onto YPD plates, left unirradiated or irradiated at 50 krad, and were incubated at 23°C for 5 days.
Analysis of mating-type switching
The physical analysis of mating-type switching was performed as described previously with strains LSY1009-1, LSY1022-2, LYS1023-1 and LSY1024-2 grown at 30°C (White and Haber, 1990). Mating-type switching was assayed genetically by inducing HO endonuclease in the above strains for 3 h at 23 or 30°C and then plating diluted cultures onto YPD. Following incubation at either 23 or 30°C, survivors were assayed for mating type by the ability to mate with tester strains MCY14 (MATa) or SJR13 (MATα). Approximately 100 colonies were assayed for each strain at each temperature, and the experiment was repeated twice.
Protein purification
RPA was purified from strain RDKY2275 as described previously (Nakagawa et al., 2001). Rad51 was expressed in E.coli strain BL21 (DE3)/pLysS (Novagen Inc., Madison, WI) using plasmid pEZ3951 and purified as described (Zaitseva et al., 1999). The Rad51-I345T protein was purified by the same procedure as Rad51.
DNA binding assay
DNA binding was measured by retention of protein–DNA complexes on alkali-treated nitrocellulose filters as described previously (Wong and Lohman, 1993). Linear DNA labeled at the 3′ ends was obtained by digesting pUC19 with EcoRI and filling-in the recessed 3′ ends with DNA polymerase I (Klenow fragment) in the presence of [α-32P]dATP. Labeled DNA was made single stranded by heat denaturation and then quenched on ice prior to use in ssDNA-binding assays. Increasing amounts of protein (0.0075, 0.015, 0.03, 0.05, 0.075, 0.1, 0.15, 0.3 and 0.5 µM) were incubated with a fixed amount (0.6 µM nucleotides) of either ssDNA or dsDNA in buffer A [20 mM Tris acetate pH 7.9, 10 mM magnesium acetate, 1 mM dithiothreitol (DTT), 0.5 mM EDTA, 7.5 µg/ml bovine serum albumin (BSA), 5 mM adenosine 5′-[γ-thio]triphosphate (ATP[γ-S])]. Reactions were incubated at 37°C for 15 min and then passed through the nitrocellulose and DEAE membranes in a vacuum manifold apparatus. DNA binding was monitored as the signal retained on the alkali-treated nitrocellulose filter over the total amount of DNA retained on both filters. Data were quantified with a Molecular Dynamics Storm 445 SI phosphoimager and IMAGE-QUANT software. Numbers presented are the mean of two trials.
ATPase assay
Reactions contained 25 mM Tris acetate pH 7.9, 10 mM magnesium acetate, 1 mM ATP, 1 mM DTT, 10 µCi/ml [γ-32P]ATP, 0.6 µM poly(dT) DNA and 0.2 µM Rad51 or rad51-I345T. Poly(dT) was either free or pre-incubated with 0.06 µM RPA for 2 min at 37°C. Reactions were incubated at 37°C for the time indicated and then quenched with 10 mM EDTA. A 2 µl aliquot of each reaction was spotted onto polyethyleneimine cellulose thin-layer chromatography plastic sheets. The plates were developed in 0.8 M acetic acid, 0.8 M LiCl, dried and the amounts of 32Pi and [γ-32P]ATP quantitated using a Molecular Dynamics Storm 445 SI phosphoimager and IMAGE-QUANT software.
Acknowledgments
Acknowledgements
We thank members of the Symington laboratory, C.S.H.Young and W.K.Holloman for helpful discussions, and S.Jinks-Robertson, H.Klein, R.Kolodner, S.Kowalczykowski, J.Nickoloff, R.Rothstein, P.Sung and V.Zakian for gifts of antibodies, plasmids and yeast strains. The research described in this article was supported by grants from the National Institutes of Health (GM54099, T32 AI07161 and T32 CA09503).
References
- Aboussekhra A., Chanet,R., Adjiri,A. and Fabre,F. (1992) Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol., 12, 3224–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aihara H., Ito,Y., Kurumizaka,H., Yokoyama,S. and Shibata,T. (1999) The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol., 290, 495–504. [DOI] [PubMed] [Google Scholar]
- Anderson D.G. and Kowalczykowski,S.C. (1997) The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell, 90, 77–86. [DOI] [PubMed] [Google Scholar]
- Basile G., Aker,M. and Mortimer,R.K. (1992) Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol. Cell. Biol., 12, 3235–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson F.E., Baumann,P. and West,S.C. (1998) Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature, 391, 401–404. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) DMC1: a meiosis-specific yeast homolog of E.coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- Bleuit J.S., Xu,H., Ma,Y., Wang,T., Liu,J. and Morrical,S.W. (2001) Mediator proteins orchestrate enzyme–ssDNA assembly during T4 recombination-dependent DNA replication and repair. Proc. Natl Acad. Sci. USA, 98, 8298–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., Trueheart,J., Natsoulis,G. and Fink,G.R. (1987) 5-Fluoro orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol., 154, 164–175. [DOI] [PubMed] [Google Scholar]
- Boundy-Mills K.L. and Livingston,D.M. (1993) A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: activities and intragenic complementation of missense mutations. Genetics, 133, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow S.A., Rao,B.J. and Radding,C.M. (1988) Reversibility of strand invasion promoted by recA protein and its inhibition by Escherichia coli single-stranded DNA-binding protein or phage T4 gene 32 protein. J. Biol. Chem., 263, 200–209. [PubMed] [Google Scholar]
- Egelman E.H. and Stasiak,A. (1986) Structure of helical RecA–DNA complexes. Complexes formed in the presence of ATP-γ-S or ATP. J. Mol. Biol., 191, 677–697. [DOI] [PubMed] [Google Scholar]
- Flory J., Tsang,S.S. and Muniyappa,K. (1984) Isolation and visualization of active presynaptic filaments of recA protein and single-stranded DNA. Proc. Natl Acad. Sci. USA, 81, 7026–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J.C. and Mortimer,R.K. (1974) A genetic study of X-ray sensitive mutants in yeast. Mutat. Res., 24, 281–292. [DOI] [PubMed] [Google Scholar]
- Gasior S.L., Wong,A.K., Kora,Y., Shinohara,A. and Bishop,D.K. (1998) Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev., 12, 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior S.L., Olivares,H., Ear,U., Hari,D.M., Weichselbaum,R. and Bishop,D.K. (2001) Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl Acad. Sci. USA, 98, 8411–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays S.L., Firmenich,A.A. and Berg,P. (1995) Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55 and Rad57 proteins. Proc. Natl Acad. Sci. USA, 92, 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.D. and Symington,L.S. (1995) Functional differences and interactions among the putative RecA homologs Rad51, Rad55 and Rad57. Mol. Cell. Biol., 15, 4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kans J.A. and Mortimer,R.K. (1991) Nucleotide sequence of the RAD57 gene of Saccharomyces cerevisiae. Gene, 105, 139–140. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S.C., Dixon,D.A., Eggleston,A.K., Lauder,S.D. and Rehrauer,W.M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L., Damborsky,J., Thomsen,B., Duno,M. and Bendixen,C. (2001) Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol. Cell. Biol., 21, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kurumizaka H., Aihara,H., Kagawa,W., Shibata,T. and Yokoyama,S. (1999) Human Rad51 amino acid residues required for Rad52 binding. J. Mol. Biol., 291, 537–548. [DOI] [PubMed] [Google Scholar]
- Lavery P.E. and Kowalczykowski,S.C. (1990) Properties of recA441 protein-catalyzed DNA strand exchange can be attributed to an enhanced ability to compete with SSB protein. J. Biol. Chem., 265, 4004–4010. [PubMed] [Google Scholar]
- Lavery P.E. and Kowalczykowski,S.C. (1992) Biochemical basis of the constitutive repressor cleavage activity of recA730 protein. A comparison to recA441 and recA803 proteins. J. Biol. Chem., 267, 20648–20658. [PubMed] [Google Scholar]
- Lim D.S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S.T. (1994) Sequence of the RAD55 gene of Saccharomyces cerevisiae: similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene, 142, 103–106. [DOI] [PubMed] [Google Scholar]
- Lovett S.T. and Mortimer,R.K. (1987) Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics, 116, 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju M.V., Lavery,P.E., Kowalczykowski,S.C. and Clark,A.J. (1992) Enzymatic properties of the RecA803 protein, a partial suppressor of recF mutations. Biochemistry, 31, 10529–10535. [DOI] [PubMed] [Google Scholar]
- Mazin A.V., Bornarth,C.J., Solinger,J.A., Heyer,W.D. and Kowalczykowski,S.C. (2000) Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell, 6, 583–592. [DOI] [PubMed] [Google Scholar]
- Milne G.T. and Weaver,D.T. (1993) Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev., 7, 1755–1765. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Flores-Rozas,H. and Kolodner,R.D. (2001) The MER3 helicase involved in meiotic crossing over is stimulated by single-stranded DNA binding proteins and unwinds DNA in the 3′ to 5′ direction. J. Biol. Chem., 276, 31487–31493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New J.H., Sugiyama,T., Zaitseva,E. and Kowalczykowski,S.C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature, 391, 407–410. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Yu,X., Shinohara,A. and Egelman,E.H. (1993) Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science, 259, 1896–1899. [DOI] [PubMed] [Google Scholar]
- Petukhova G., Stratton,S. and Sung,P. (1998) Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature, 393, 91–94. [DOI] [PubMed] [Google Scholar]
- Rattray A.J. and Symington,L.S. (1994) Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics, 138, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A.J. and Symington,L.S. (1995) Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics, 139, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L.L. and Zakian,V.A. (1993) Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell, 75, 729–739. [DOI] [PubMed] [Google Scholar]
- Schwacha A. and Kleckner,N. (1997) Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell, 90, 1123–1135. [DOI] [PubMed] [Google Scholar]
- Shan Q., Bork,J.M., Webb,B.L., Inman,R.B. and Cox,M.M. (1997) RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol., 265, 519–540. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink,G. and Hicks,J. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shinohara A. and Ogawa,T. (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature, 391, 404–407. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S.cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Gasior,S., Ogawa,T., Kleckner,N. and Bishop,D.K. (1997) Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells, 2, 615–629. [DOI] [PubMed] [Google Scholar]
- Solinger J.A., Lutz,G., Sugiyama,T., Kowalczykowski,S.C. and Heyer,W.D. (2001) Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol., 307, 1207–1221. [DOI] [PubMed] [Google Scholar]
- Song B. and Sung,P. (2000) Functional interactions among yeast Rad51 recombinase, Rad52 mediator and replication protein A in DNA strand exchange. J. Biol. Chem., 275, 15895–15904. [DOI] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Buerstedde,J.M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchi-Iwai,Y. and Takeda,S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story R.M., Bishop,D.K., Kleckner,N. and Steitz,T.A. (1993) Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science, 259, 1892–1896. [DOI] [PubMed] [Google Scholar]
- Story R.M. and Steitz,T.A. (1992) Structure of the recA protein–ADP complex. Nature, 355, 374–376. [DOI] [PubMed] [Google Scholar]
- Story R.M., Weber,I.T. and Steitz,T.A. (1992) The structure of the E.coli recA protein monomer and polymer. Nature, 355, 318–325. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht,A., Luo,K. and Grunstein,M. (1997) SIR2 and SIR4 interactions differ in core and extended telomeric hetero chromatin in yeast. Genes Dev., 11, 83–93. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Zaitseva,E.M. and Kowalczykowski,S.C. (1997) A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem., 272, 7940–7945. [DOI] [PubMed] [Google Scholar]
- Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science, 265, 1241–1243. [DOI] [PubMed] [Google Scholar]
- Sung P. (1997a) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem., 272, 28194–28197. [DOI] [PubMed] [Google Scholar]
- Sung P. (1997b) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- Sung P. and Robberson,D.L. (1995) DNA strand exchange mediated by a RAD51–ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell, 82, 453–461. [DOI] [PubMed] [Google Scholar]
- Takata M., Sasaki,M.S., Tachiiri,S., Fukushima,T., Sonoda,E., Schild,D., Thompson,L.H. and Takeda,S. (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol., 21, 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L.H. and Schild,D. (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res., 477, 131–153. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T., Fujii,Y., Sakumi,K., Tominaga,Y., Nakao,K., Sekiguchi,M., Matsushiro,A., Yoshimura,Y. and Morita,T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA, 93, 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K. and Kolodner,R.D. (1994) Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem., 269, 30005–30013. [PubMed] [Google Scholar]
- Umezu K., Chi,N.W. and Kolodner,R.D. (1993) Biochemical interaction of the Escherichia coli RecF, RecO and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl Acad. Sci. USA, 90, 3875–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen S., Petukhova,G., Sigurdsson,S., Stratton,S. and Sung,P. (2000) Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell, 6, 563–572. [DOI] [PubMed] [Google Scholar]
- Volkert M.R. and Hartke,M.A. (1984) Suppression of Escherichia coli recF mutations by recA-linked srfA mutations. J. Bacteriol., 157, 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.C., Chang,H.Y. and Hung,J.L. (1993) Cosuppression of recF, recR and recO mutations by mutant recA alleles in Escherichia coli cells. Mutat. Res., 294, 157–166. [DOI] [PubMed] [Google Scholar]
- Webb B.L., Cox,M.M. and Inman,R.B. (1997) Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell, 91, 347–356. [DOI] [PubMed] [Google Scholar]
- White C.I. and Haber,J.E. (1990) Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J., 9, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong I. and Lohman,T.M. (1993) A double-filter method for nitrocellulose-filter binding: application to protein–nucleic acid interactions. Proc. Natl Acad. Sci. USA, 90, 5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva E.M., Zaitsev,E.N. and Kowalczykowski,S.C. (1999) The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem., 274, 2907–2915. [DOI] [PubMed] [Google Scholar]