Abstract
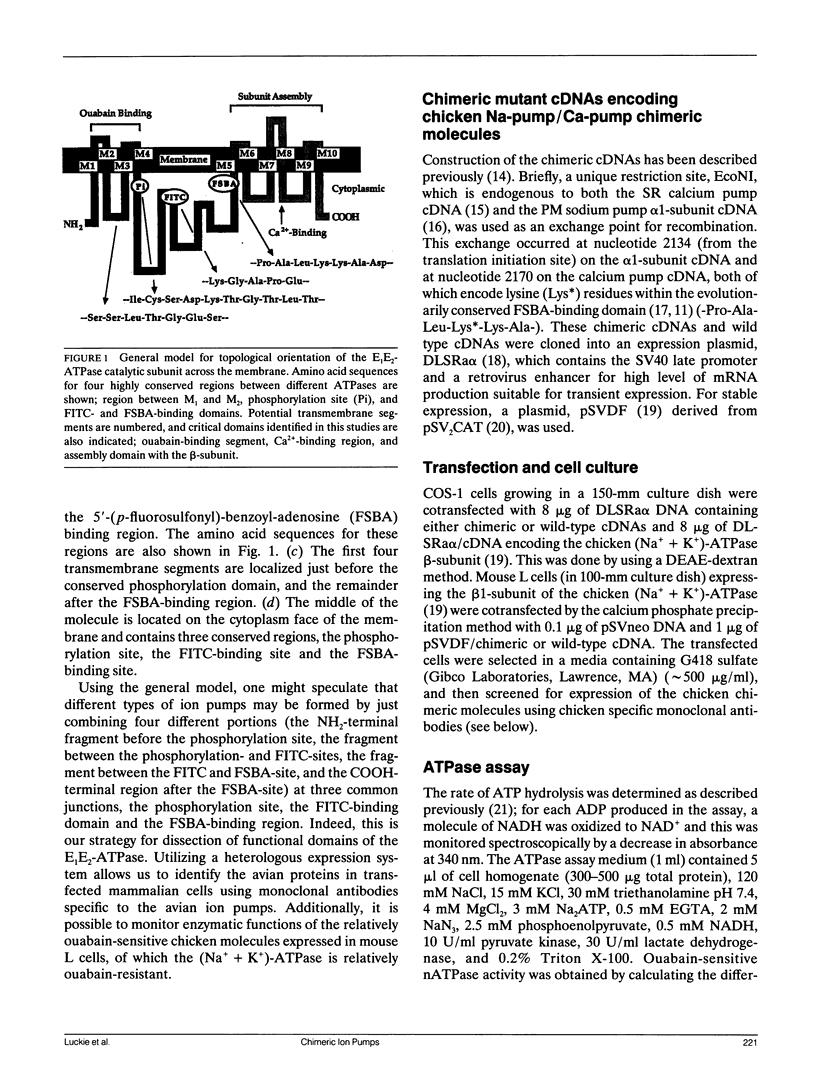
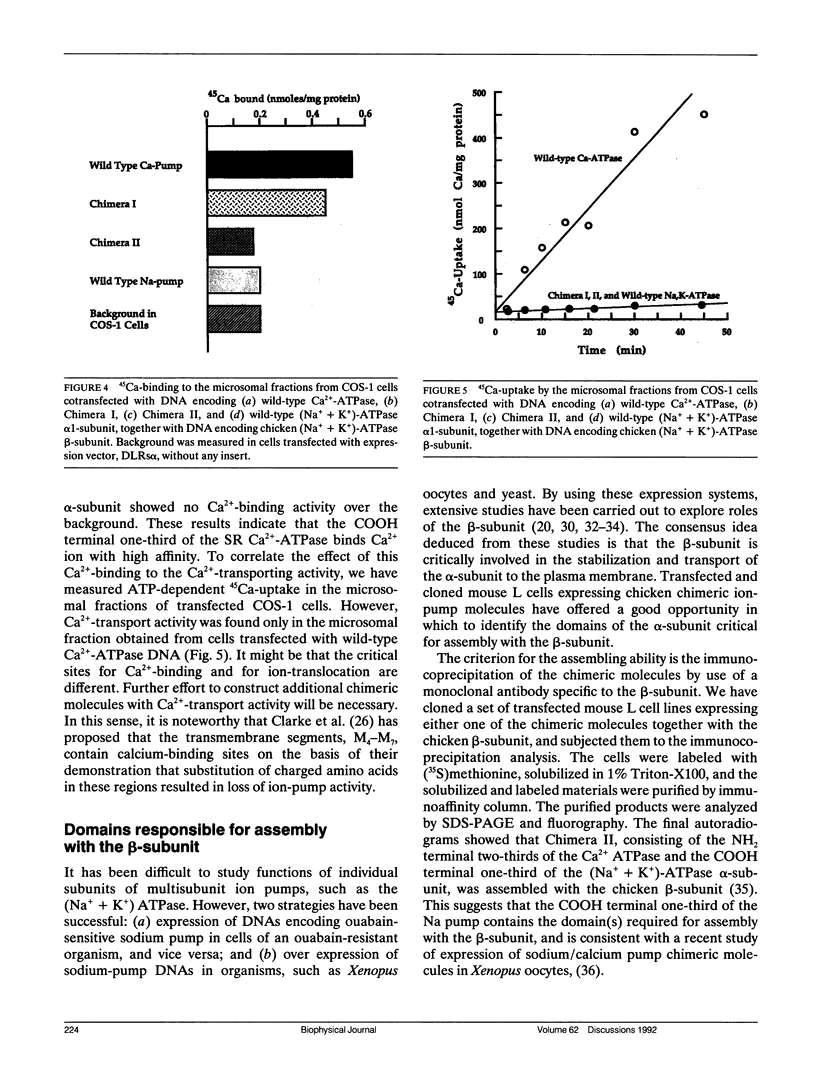
Proposed models for the catalytic subunit of the E1E2-ATPases (ion pumps) predict that the first four transmembrane domains (M1 - M4) reside in the NH2 terminal one-third of the molecule, and the remainder (M5 - M10) in the COOH terminal one-third. The amino-acid sequences for the 5'-(p-fluorosulfonyl)-benzoyl-adenosine (FSBA) binding region residing just before M5 segment are very well conserved among distinct ion pumps. Taking advantage of these models, we have constructed a set of chicken chimeric ion pumps between the (Na++ K+)-ATPase alpha-subunit and the Ca(2+)-ATPase using the FSBA-binding site as an exchange junction, thereby preserving overall topological structure as E1E2 ATPases. From various functional assays on these chimeric ion pumps, including ouabain-inhibitable ATPase activity, Ca2+ binding, Ca2+ uptake, and subunit assembly based on immuno-coprecipitation, the following conclusions were obtained: (a) A (Na++ K+)-ATPase inhibitor, ouabain, binds to the regions before M4 in the alpha-subunit and exerts its inhibitory effect. (b) The regions after M5 of the (Na++ K+)-ATPase alpha-subunit bind the beta-subunit, even when these regions are incorporated into the corresponding domains in the Ca(2+)-ATPase. (c) The corresponding domains of the Ca(2+)-ATPase, the regions after M5, bind 45Ca even when it is incorporated into the corresponding position of the (Na++ K+)-ATPase alpha-subunit.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clarke D. M., Loo T. W., Inesi G., MacLennan D. H. Location of high affinity Ca2+-binding sites within the predicted transmembrane domain of the sarcoplasmic reticulum Ca2+-ATPase. Nature. 1989 Jun 8;339(6224):476–478. doi: 10.1038/339476a0. [DOI] [PubMed] [Google Scholar]
- Clarke D. M., Maruyama K., Loo T. W., Leberer E., Inesi G., MacLennan D. H. Functional consequences of glutamate, aspartate, glutamine, and asparagine mutations in the stalk sector of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem. 1989 Jul 5;264(19):11246–11251. [PubMed] [Google Scholar]
- Emanuel J. R., Schulz J., Zhou X. M., Kent R. B., Housman D., Cantley L., Levenson R. Expression of an ouabain-resistant Na,K-ATPase in CV-1 cells after transfection with a cDNA encoding the rat Na,K-ATPase alpha 1 subunit. J Biol Chem. 1988 Jun 5;263(16):7726–7733. [PubMed] [Google Scholar]
- Fambrough D. M., Bayne E. K. Multiple forms of (Na+ + K+)-ATPase in the chicken. Selective detection of the major nerve, skeletal muscle, and kidney form by a monoclonal antibody. J Biol Chem. 1983 Mar 25;258(6):3926–3935. [PubMed] [Google Scholar]
- Fukuda K., Kubo T., Maeda A., Akiba I., Bujo H., Nakai J., Mishina M., Higashida H., Neher E., Marty A. Selective effector coupling of muscarinic acetylcholine receptor subtypes. Trends Pharmacol Sci. 1989 Dec;Suppl:4–10. [PubMed] [Google Scholar]
- Geering K. The functional role of the beta-subunit in the maturation and intracellular transport of Na,K-ATPase. FEBS Lett. 1991 Jul 22;285(2):189–193. doi: 10.1016/0014-5793(91)80801-9. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen P. L., Andersen J. P. Structural basis for E1-E2 conformational transitions in Na,K-pump and Ca-pump proteins. J Membr Biol. 1988 Jul;103(2):95–120. doi: 10.1007/BF01870942. [DOI] [PubMed] [Google Scholar]
- Kaprielian Z., Fambrough D. M. Expression of fast and slow isoforms of the Ca2+-ATPase in developing chick skeletal muscle. Dev Biol. 1987 Dec;124(2):490–503. doi: 10.1016/0012-1606(87)90502-1. [DOI] [PubMed] [Google Scholar]
- Karin N. J., Kaprielian Z., Fambrough D. M. Expression of avian Ca2+-ATPase in cultured mouse myogenic cells. Mol Cell Biol. 1989 May;9(5):1978–1986. doi: 10.1128/mcb.9.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent R. B., Emanuel J. R., Ben Neriah Y., Levenson R., Housman D. E. Ouabain resistance conferred by expression of the cDNA for a murine Na+, K+-ATPase alpha subunit. Science. 1987 Aug 21;237(4817):901–903. doi: 10.1126/science.3039660. [DOI] [PubMed] [Google Scholar]
- Kobilka B. K., Kobilka T. S., Daniel K., Regan J. W., Caron M. G., Lefkowitz R. J. Chimeric alpha 2-,beta 2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science. 1988 Jun 3;240(4857):1310–1316. doi: 10.1126/science.2836950. [DOI] [PubMed] [Google Scholar]
- Lax I., Bellot F., Howk R., Ullrich A., Givol D., Schlessinger J. Functional analysis of the ligand binding site of EGF-receptor utilizing chimeric chicken/human receptor molecules. EMBO J. 1989 Feb;8(2):421–427. doi: 10.1002/j.1460-2075.1989.tb03393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzon P., Jackson R., Wallmark B., Sachs G. Inhibition of (H+ + K+)-ATPase by omeprazole in isolated gastric vesicles requires proton transport. Biochim Biophys Acta. 1987 Feb 12;897(1):41–51. doi: 10.1016/0005-2736(87)90313-0. [DOI] [PubMed] [Google Scholar]
- MacLennan D. H., Brandl C. J., Korczak B., Green N. M. Amino-acid sequence of a Ca2+ + Mg2+-dependent ATPase from rabbit muscle sarcoplasmic reticulum, deduced from its complementary DNA sequence. Nature. 1985 Aug 22;316(6030):696–700. doi: 10.1038/316696a0. [DOI] [PubMed] [Google Scholar]
- McDonough A. A., Geering K., Farley R. A. The sodium pump needs its beta subunit. FASEB J. 1990 Apr 1;4(6):1598–1605. doi: 10.1096/fasebj.4.6.2156741. [DOI] [PubMed] [Google Scholar]
- Noguchi S., Mishina M., Kawamura M., Numa S. Expression of functional (Na+ + K+)-ATPase from cloned cDNAs. FEBS Lett. 1987 Dec 10;225(1-2):27–32. doi: 10.1016/0014-5793(87)81125-0. [DOI] [PubMed] [Google Scholar]
- Nørby J. G. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- Ohta T., Nagano K., Yoshida M. The active site structure of Na+/K+-transporting ATPase: location of the 5'-(p-fluorosulfonyl)benzoyladenosine binding site and soluble peptides released by trypsin. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2071–2075. doi: 10.1073/pnas.83.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedemonte C. H., Kaplan J. H. Chemical modification as an approach to elucidation of sodium pump structure-function relations. Am J Physiol. 1990 Jan;258(1 Pt 1):C1–23. doi: 10.1152/ajpcell.1990.258.1.C1. [DOI] [PubMed] [Google Scholar]
- Price E. M., Lingrel J. B. Structure-function relationships in the Na,K-ATPase alpha subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme. Biochemistry. 1988 Nov 1;27(22):8400–8408. doi: 10.1021/bi00422a016. [DOI] [PubMed] [Google Scholar]
- Reuben M. A., Lasater L. S., Sachs G. Characterization of a beta subunit of the gastric H+/K(+)-transporting ATPase. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6767–6771. doi: 10.1073/pnas.87.17.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs G., Munson K., Hall K., Hersey S. J. Gastric H+,K(+)-ATPase as a therapeutic target in peptic ulcer disease. Dig Dis Sci. 1990 Dec;35(12):1537–1544. doi: 10.1007/BF01540572. [DOI] [PubMed] [Google Scholar]
- Sagara Y., Inesi G. Inhibition of the sarcoplasmic reticulum Ca2+ transport ATPase by thapsigargin at subnanomolar concentrations. J Biol Chem. 1991 Jul 25;266(21):13503–13506. [PubMed] [Google Scholar]
- Shull G. E., Schwartz A., Lingrel J. B. Amino-acid sequence of the catalytic subunit of the (Na+ + K+)ATPase deduced from a complementary DNA. Nature. 1985 Aug 22;316(6030):691–695. doi: 10.1038/316691a0. [DOI] [PubMed] [Google Scholar]
- Shull G. E. cDNA cloning of the beta-subunit of the rat gastric H,K-ATPase. J Biol Chem. 1990 Jul 25;265(21):12123–12126. [PubMed] [Google Scholar]
- Takebe Y., Seiki M., Fujisawa J., Hoy P., Yokota K., Arai K., Yoshida M., Arai N. SR alpha promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988 Jan;8(1):466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeyasu K., Tamkun M. M., Renaud K. J., Fambrough D. M. Ouabain-sensitive (Na+ + K+)-ATPase activity expressed in mouse L cells by transfection with DNA encoding the alpha-subunit of an avian sodium pump. J Biol Chem. 1988 Mar 25;263(9):4347–4354. [PubMed] [Google Scholar]
- Takeyasu K., Tamkun M. M., Siegel N. R., Fambrough D. M. Expression of hybrid (Na+ + K+)-ATPase molecules after transfection of mouse Ltk-cells with DNA encoding the beta-subunit of an avian brain sodium pump. J Biol Chem. 1987 Aug 5;262(22):10733–10740. [PubMed] [Google Scholar]
- Tamkun M. M., Fambrough D. M. The (Na+ + K+)-ATPase of chick sensory neurons. Studies on biosynthesis and intracellular transport. J Biol Chem. 1986 Jan 25;261(3):1009–1019. [PubMed] [Google Scholar]
- Tanabe T., Adams B. A., Numa S., Beam K. G. Repeat I of the dihydropyridine receptor is critical in determining calcium channel activation kinetics. Nature. 1991 Aug 29;352(6338):800–803. doi: 10.1038/352800a0. [DOI] [PubMed] [Google Scholar]
- Wolitzky B. A., Fambrough D. M. Regulation of the (Na+ + K+)-ATPase in cultured chick skeletal muscle. Modulation of expression by the demand for ion transport. J Biol Chem. 1986 Jul 25;261(21):9990–9999. [PubMed] [Google Scholar]





