Abstract
In Ustilago maydis, bidirectional transport of early endosomes is microtubule dependent and supports growth and cell separation. During early budding, endosomes accumulate at putative microtubule organizers within the bud, whereas in medium-budded cells, endosome clusters appear at the growing ends of microtubules at the distal cell pole. This suggests that motors of opposing transport direction organize endosomes in budding cells. Here we set out to identify these motors and elucidate the molecular mechanism of endosome reorganization. By PCR we isolated kin3, which encodes an UNC-104/KIF1-like kinesin from U.maydis. Recombinant Kin3 binds microtubules and has ATPase activity. Kin3–green fluorescent protein moves along microtubules in vivo, accumulates at sites of growth and localizes to endosomes. Deletion of kin3 reduces endosome motility to ∼33%, and abolishes endosome clustering at the distal cell pole and at septa. This results in a transition from bipolar to monopolar budding and cell separation defects. Double mutant analysis indicates that the remaining motility in Δkin3-mutants depends on dynein, and that dynein and Kin3 counteract on the endosomes to arrange them at opposing cell poles.
Keywords: budding pattern/endosomes/kinesin/motility/pathogenic fungus
Introduction
The cytoskeleton is of crucial importance for the eukaryotic cell and is required for many vital processes, including spatial cellular organization and directed growth. Microtubules (MTs) participate in the delivery of components towards the expanding cell region and kinesins and dyneins support MT-dependent traffic (Hirokawa, 1998). The transport direction depends on the polarity of MTs, with most kinesins moving towards the plus-ends, where slow polymerization and rapid depolymerization occur. By contrast, minus-end-directed transport is mainly driven by the dynein complex (Bloom, 1992), although it has recently emerged that specialized kinesins can also participate in membrane traffic towards the minus-ends of MTs (Hanlon et al., 1997; Saito et al., 1997; Noda et al., 2001). Simultaneous activity of kinesins and dyneins along a single MT results in bidirectional motility, which is commonly observed for many types of organelles, including endosomes (Apodaca, 2001).
Endocytosis involves membrane traffic from the plasma membrane towards the lysosome/vacuole. Intermediate endocytic compartments like early endosomes (EEs) are known to be mobile and undergo rapid fission and fusion (Murphy et al., 1996; Nielsen et al., 1999). In vertebrates, this EE motility is based on MTs, while the initial internalization step depends on F-actin (Apodaca, 2001), and a growing body of evidence indicates that cytoplasmic dynein and unspecified kinesins are responsible for bidirectional MT-based transport of EEs (Bananis et al., 2000; Murray et al., 2000).
Recently, it was shown that bidirectional transport of EEs in the corn smut fungus Ustilago maydis is also a MT-dependent process (Wedlich-Söldner et al., 2000). EE motility towards the assumed MT minus-ends at the newly formed bud results in polar EE accumulations (brightly stained dots), suggesting that EE-based recycling participates in polar growth of U.maydis. Interestingly, later in bud formation, EEs are rapidly displaced from the medium-sized bud to the opposite end of the mother cell (Wedlich-Söldner et al., 2000), while MTs still have a unipolar organization (Steinberg et al., 2001). This cell cycle stage-dependent reorganization suggests that opposing motor activities rearrange EEs to support a defined cellular role at the distal end of the cell. However, neither this specific function, nor the underlying transport machinery, is known.
Here we show that cytoplasmic dynein and an UNC-104/KIF1-like kinesin are responsible for EE transport in U.maydis. Our results indicate that, during early bud formation, dynein supports EE traffic towards the growth region, while transport towards the distal cell pole depends on the kinesin motor. Mutant analysis indicates that this rearrangement is required for bipolar budding as well as cell separation.
Results
An UNC-104/KIF1-like kinesin from U.maydis moves rapidly along MTs in vivo
We have recently shown that EEs of U.maydis are transported along MT (Wedlich-Söldner et al., 2000). In a PCR approach with primers designed to amplify members of the UNC-104/KIF1 family of kinesin-like motor molecules, we isolated a U.maydis gene, kin3, which encodes for a putative protein of 1676 amino acids with an estimated mol. wt of 184.5 kDa. The predicted sequence of Kin3 reveals a domain structure typical for members of the UNC-104/KIF1 family (Figure 1A). An N-terminal kinesin motor domain (red; amino acids 4–369, P = 1.18e–171, SMART) is followed by a forkhead-associated domain (FHA domain, yellow; amino acids 510–562, P = 5.9e–5, PFAM) that could be important for protein–protein interactions (Li et al., 1999). The C-terminal part of Kin3 is related to PH domains (blue; amino acids 1570–1670, P = 1.69e–11, SMART) that are thought to be involved in cargo binding (Okada et al., 1995). Kin3 is predicted to form short coiled-coil regions (green; amino acids 423–470 and 755–782). Within the motor domain, Kin3 shares 68% sequence identity with TlKif1 from Thermomyces lanuginosus (Sakowicz et al., 1999) and 63% identity with Kif1A from mouse (Okada et al., 1995). This sequence similarity is not restricted to the head region, as demonstrated by a dot-blot comparison of Kin3 and mouse Kif1A (Figure 1B), and the FHA domain of Kin3 and TlKIF1 still share 43% amino acid identity. Neighbor-joining, parsimony and maximum-likelihood analysis of members of the four families of putative kinesin organelle transporters consistently showed that the Kin3 motor domain groups with representatives of the UNC-104/KIF1 family, although the exact position within this family is not well defined, as indicated by low statistical support values based on the comparison of three different methods (Figure 1C). In summary, we consider it most likely that Kin3 from U.maydis belongs to the UNC-104/KIF1-kinesin family.
Fig. 1. Sequence analysis of Kin3. (A) Domain organization of Kin3 and close relatives. Kin3 contains all domains typical for members of the UNC-104/KIF1 family. This includes the N-terminal motor domain (red), an FHA domain (yellow), a PH domain (blue) and short coiled-coil regions (green). (B) Dot-plot comparison of the predicted amino acid sequence of Kin3 versus MmKIF1A. Note that both motors share sequence similarity along the entire length of the molecule. (C) Nearest-neighbor dendrogram of potential kinesin organelle transporters. Kin3 groups with members of the UNC-104/KIF1 family. The dendogram is based on a distance matrix-based neighbor-joining analysis of the kinesin motor domain. Statistical support for this tree was obtained by bootstrapping (neighbor joining and parsimony, 1000 resamplings each) or Quartet Puzzling. Solid circles indicate a bootstrap and QP support of >80%, while open circles indicate 60–80% support. Accession numbers are given. The bar reflects 0.1 changes per amino acid.
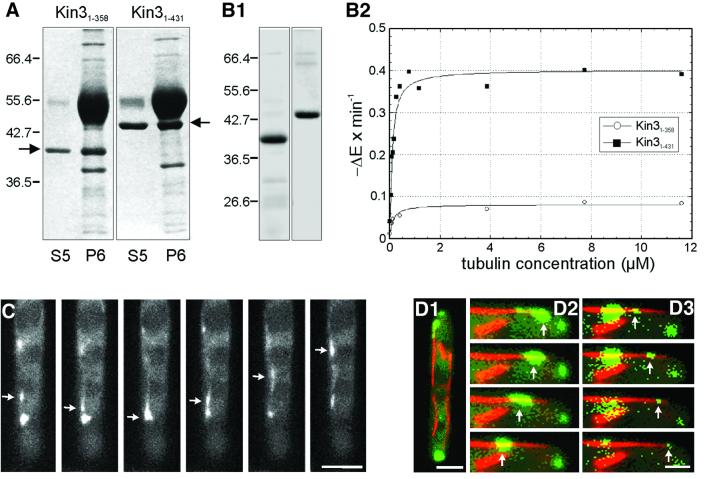
To analyze its motor activity, we expressed recombinant Kin3 protein. Unfortunately, the full-length Kin3 was not soluble (not shown). A truncated polypeptide containing the first 674 amino acids was slightly soluble, but it did not bind MTs or show any ATPase activity in vitro (not shown). A shorter construct containing only the motor domain (amino acids 1–358; Kin31–358) bound to MTs and was released by MgATP (Figure 2A; S5, ATP release; P6, MT pellet after release, right arrow marks Kin31–358), but showed only little ATPase activity (Figure 2B1 and B2). This activity was significantly increased when the neck region was included (amino acids 1–431; Kin31–431). Kin31–431 bound to MTs in an ATP-dependent manner (Figure 2A, left arrow marks Kin31–431), showed a kcat of 4.87 s–1 and a 56.46-fold stimulation by MTs (Figure 2B1 and B2). This behavior is a characteristic feature of MT-associated motors. However, our recombinant proteins showed no MT-gliding or bead-moving activity, indicating that the motor domain alone is not sufficient for in vitro motility.
Fig. 2. MT affinity, ATPase activity and in vivo motility of Kin3. (A) MT affinity of recombinant Kin3 heads. After ATP depletion, bacterial expressed Kin3 motor domains bound to taxol-stabilized MTs, and were partially released by MgATP (S5), while some protein was sedimented with MTs (P6). Kin31–358, amino acids 1–358; Kin31–431, amino acids 1–431 of Kin3 (arrows). (B) His-tagged Kin3 was affinity purified (B1) and ATPase activity, shown as NADH reduction per time in the presence of increasing tubulin concentrations, was measured (B2). Note that the neck region (amino acids 358–431) is required for MT stimulation of truncated motor domain. (C) Movement of Kin3–GFP. The Kin3–GFP fusion protein (arrow) moves rapidly in a bidirectional fashion. Time between frames is 500 ms. Bar: 3 µm. (D) Co-localization of Kin3–YFP- and CFP–Tub1-marked microtubules. MTs are running through the length of an unbudded cell (D1). A series of images of Kin3–YFP (green, arrows) was taken and merged with a single image of CFP–Tub1 (red). Movement of a large accumulation of Kin3–YFP (D2), as well as of small Kin3–YFP-labeled dots (D3) along MTs. Time between frames is 700 ms. Bar: 2 µm (D1) and 1 µm (D2 and D3). A movie for this figure is available as Supplementary data at The EMBO Journal Online.
Finally, we monitored the in vivo motility behavior of Kin3 fused to green fluorescent protein (GFP) or its yellow-shifted derivative (YFP). Either under its native promoter or under the control of the otef promoter, Kin3–GFP rescued the characteristic Δkin3 phenotype (see below), suggesting that the fusion proteins were biologically active. Expressed under its native promoter in RWS6 (Table I), the fusion protein localized to small dots that showed rapid, bidirectional and saltatoric motion (arrows in Figure 2C). However, due to a low expression level, the signal was too faint to analyze the dynamics of single Kin3–GFP dots. Therefore, we expressed the fusion protein under the control of the stronger otef promoter in RWS7. Motility of Kin3–GFP dots in RWS7 was indistinguishable from that of RWS6. Single dots moved at 2.87 ± 0.19 µm/s (n = 20 cells from three experiments), which was not significantly different (P = 0.5667) from movement rates of EEs that carried GFP fused to the endosomal t-SNARE Yup1 (Wedlich-Söldner et al., 2000). This was a first indication that Kin3 participates in EE motility.
Table I. Strains and plasmids used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| FB1 | a1b1 | Banuett and Herskowitz (1989) |
| FB2 | a2b2 | Banuett and Herskowitz (1989 |
| RWS4 | a1b1 /pYup1SG2 | Wedlich-Söldner et al. (2000) |
| FB1otefGFPtub1 | a1b1 /potefGFPTub1 | Steinberg et al. (2001) |
| RWS6 | a2b2 /pKin3GFP1 | This study |
| RWS7 | a2b2 /pKin3GFP2 | This study |
| RWS8 | a1b1 /pKin3YFP /pCFPTub1 | This study |
| RWS9 | a1b1 /pKin3YFP /pPX10-146CFP | This study |
| RWS10 | a2b2 Δkin3::hphR /pYup1SG3 | This study |
| RWS11 | a2b2 Δkin3::hphR /pRU4-Kin3 /pYup1SG3 | This study |
| RWS12 | a2b2 Δkin3::hphR /potefGFPTub1 | This study |
| RWS13 | a2b2 Δkin3::hphR /potefGFPTub1 /pYup1SG3 | This study |
| FB1Dyn2ts | a1b1 Δdyn2::[dyn2ts, natR] | Wedlich-Söldner et al. (2002) |
| RWS14 | a1b1 Δdyn2::[dyn2ts, natR] Δkin3::hphR /pYup1SG2 | This study |
| RWS15 | a1b1 Δdyn2::[dyn2ts, natR] /pYup1SG2 | This study |
| RWS16 | a2b2 Pcrg-kin3, natR /pYup1SG2 | This study |
| RWS17 | a2b2 Pcrg-kin3, natR /potefGFPTub1 | This study |
| RWS18 | a2b2 Δkin3::hphR | This study |
| RWS19 | a2b2 Δkin3::hphR /pYup1SG3 /pcrgKin3G105E | This study |
| RWS20 | a2b2 Δkin3::hphR /pYup1SG3 /pcrgKin3 | This study |
| pKin3GFP1 | Pkin3-kin3-egfp, cbxR | This study |
| pKin3GFP2 | Potef-kin3-egfp, cbxR | This study |
| pKin3YFP | Potef-kin3-yfp, cbxR | This study |
| pCFPTub1 | Potef-cfp-tub1, hygR | This study |
| pPX10-146CFP | Potef-yup1PX1-146-cfp, hygR | This study |
| PYup1SG3 | Potef-yup1-sgfp, bleR | This study |
| pRU4-Kin3 | Pkin3-kin3, cbxR | This study |
| pcrgKin3 | Pcrg-kin3, cbxr | This study |
| pcrgKin3G105E | Pcrg-kin3G105E, cbxr | This study |
| pET-Kin3U1 | PT7-Kin31–358 | This study |
| pET-Kin3U2 | PT7-Kin31–431 | This study |
| potefGFPTub1 | Potef-egfp-tub1, cbxR | Steinberg et al. (2001) |
a, b, mating type loci; Δ, deletion; P, promoter; ::, homologous replacement; -, fusion; ts, temperature-sensitive allele; hphR, hygromycin resistance; bleR, phleomycin resistance; natR, nourseothricin resistance; cbxR, carboxin resistance; /, ectopically integrated; yup1PX1-146, localization domain of the endosomal t-SNARE Yup1 (amino acids 10–146); gfp, green fluorescent protein; yfp, yellow-shifted derivative of green fluorescent protein; cfp, cyan-shifted derivative of green fluorescent protein; Kin31–358 and Kin31–431, motor domain constructs of Kin3; tub1, α tubulin; dyn2, gene encoding the C-terminal third of the dynein heavy chain.
Kin3 is most likely a kinesin motor, suggesting that it utilizes MTs to transport its cargo within the cell. To confirm this assertion, we generated strain RWS8, in which Kin3 is fused to YFP, while Tub1, the α-tubulin of U.maydis (Steinberg et al., 2001), is fused to the cyan-shifted fluorescent protein (CFP). Co-expression of both fusion proteins revealed that Kin3–YFP exclusively moved along CFP-marked MTs (59 signals in 10 cells; Figure 2D1–D3; Kin3–YFP in green, marked by arrows; MTs in red). This MT dependency of the Kin3 motion was confirmed by experiments with the MT-destabilizing drug benomyl, which efficiently disrupts MTs in U.maydis (Steinberg et al., 2001). Applying this drug to RWS7 stopped all Kin3–GFP motility within 30 min (not shown), again suggesting that Kin3 is a MT-dependent motor.
Kin3 supports endosome motility in U.maydis
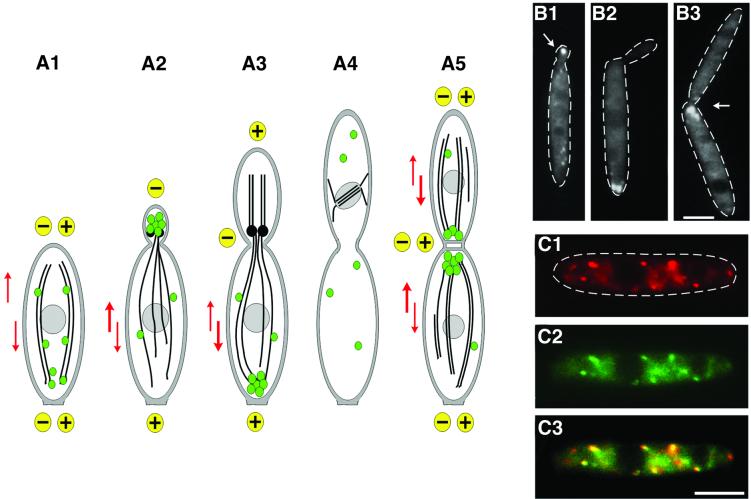
Analysis of the in vivo dynamics of MTs in U.maydis suggested that the tubulin cytoskeleton undergoes extensive rearrangement during the cell cycle (Steinberg et al., 2001; summarized in Figure 3A1–A5). Whereas unbudded and large-budded cells contain antipolar MT bundles (Figure 3A1 and A5, MTs indicated by black lines; growing ends of MTs indicated by ‘+’ and non-growing ends by ‘–’), MTs are unipolar in cells with small buds (Figure 3A2) and medium-sized buds (Figure 3A3), with their growing ends extending to the distal cell poles. Previous studies have shown that EEs (Figure 3, green spheres) move in a bidirectional fashion along these MTs, and it was suggested that this transport leads to a cell cycle-dependent accumulation at the poles (Wedlich-Söldner et al., 2000). To support this model, we carried out a quantitative analysis of endosome motility. This revealed that in all stages depicted in Figure 3A2, A3 and A5, significantly more EEs entered the EE cluster than escaped from it (Figure 3A2: 51.7 ± 3.0 versus 48.3 ± 3.0, P = 0.0081; Figure 3A3: 52.5 ± 2.1 versus 47.5 ± 2.1, P < 0.0001; Figure 3A5: 52.4 ± 3.3 versus 47.6 ± 3.3, P = 0.0004; each value is based on >200 EEs from >11 cells). This supports the notion that slight changes in the balance of bidirectional transport account for the polar accumulation of EEs.
Fig. 3. Early endosomes and Kin3. (A) Overview of the organization of EEs and MTs during the cell cycle of U.maydis. Unbudded cells show bidirectional traffic of EEs (green spheres) along antipolar MTs (A1; orientation of MTs indicated by ‘+’ and ‘–’; motility indicated by red arrows). Polar budding is accompanied by the formation of an EE cluster at the minus-end of the MTs (A2), which is due to a significant net movement towards the bud (bold red arrow). A similar situation is found in medium-budded cells that contain EE clusters at their rear cell pole (A3). No motility occurs during mitosis (A4). Finally, septum formation is accompanied by one and, in later stages, two endosomal clusters at the septa (A5). During this stage, antipolar MT bundles are found. MT polarity is based on in vivo observation of dynamics of GFP-MTs (Steinberg et al., 2001), and EE behavior is described in Wedlich-Söldner et al. (2000). (B) Cell cycle-dependent accumulation of Kin3–GFP fusion protein under the control of its native promoter (strain RWS16). Kin3–GFP localizes to small buds during early budding (B1). In medium-budded cells, a strong signal of Kin3–GFP appears at the distal cell pole (B2), which finally locates to the region of septum formation (B3). Note that Kin3–GFP localization corresponds well with endosome rearrangement (A). Bar: 3 µm. (C) Co-localization of EEs and Kin3–GFP. In pulse–chase experiments, FM4-64 appears in EEs (C1) that also carry Kin3–GFP (C2; overlay in C3). Note that, occasionally, EEs do not co-localize with Kin3–GFP fusion protein. Bar: 3 µm.
To investigate whether Kin3–GFP follows the cell cycle-specific rearrangements of EEs, we analyzed the Kin3–GFP distribution in RWS6 cells. Comparable to EEs, the fusion protein was dispersed in most unbudded cells (not shown) and located to small buds (Figure 3B1, arrow), whereas it accumulated at the distal cell pole in medium-budded cells (Figure 3B2) and appeared at the septum during cell division (Figure 3B3, cleavage plane indicated by an arrow). To obtain more direct evidence for Kin3–GFP association with EEs, we performed pulse– chase experiments using strain RWS6 and the endocytic marker dye FM4-64, which allows the endocytic pathway in U.maydis to be followed (Wedlich-Söldner et al., 2000). Immediately after internalization, the marker appears in rapidly moving EEs (Figure 3C1). Of these EEs, 89.5% co-localized with Kin3–GFP (Figure 3C2, overlay in Figure 3C3; n = 362 EEs, 56 cells), whereas only 10.5 and 4% were stained either with FM4-64 or Kin3–GFP, respectively. However, in our pulse–chase experiment, FM4-64 also stained some stationary dots in the plasma membrane as well as later structures at the vacuole (Wedlich-Söldner et al., 2000), suggesting that some FM4-64 signals are not early EEs. Finally, Kin3–YFP co-localized with Yup1–CFP in strain RWS9 (not shown).
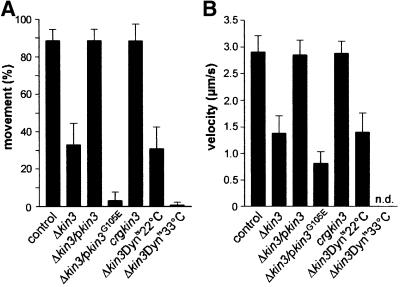
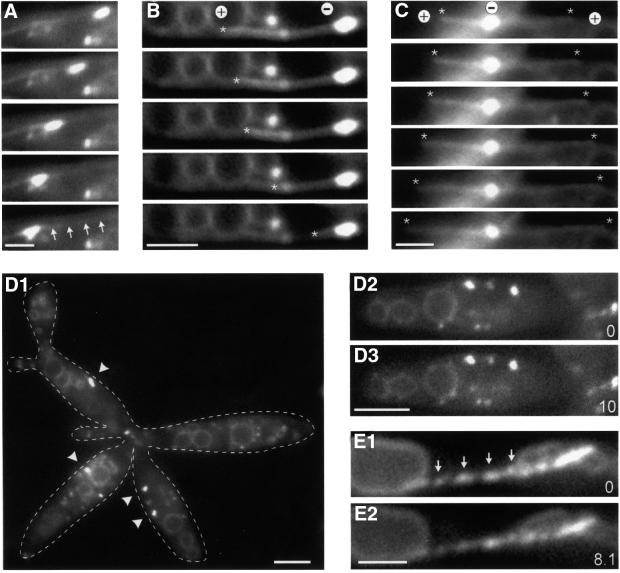
Further evidence for an active role of Kin3 in EE motility came from quantitative analysis of a Δkin3 mutant (RWS10). In control strain RWS4, almost all EEs were moving within 30 s observation time (Figure 4A, control; 88.6 ± 6.1%, n = 15 cells). By contrast, only one-third of the remaining EEs were moving in Δkin3 cells (Figure 4A; 32.9 ± 11.5%, n = 15 cells). Most of the residual motion of EEs in Δkin3 cells was unidirectional, slower than that of the control (Figure 4B) and occurred exclusively along MTs in a strain expressing a GFP–α-tubulin fusion protein (RWS13; Figure 5A, arrows mark MT). Only rarely was bidirectional motility observed (3.95%, n = 127 cells) that always occurred along MT bundles and was restricted to older cells within the Δkin3 cell aggregate (see below), which most likely show the antipolar MT organization depicted in Figure 3A5 (not shown; see also Figure 6D). All defects of the Δkin3 strain were completely rescued after integration of a single copy of kin3 under the control of its native promoter (strain RWS11; Figure 4, Δkin3/pkin3), confirming that the observed phenotype was due to the absence of Kin3.
Fig. 4. Quantitative analysis of endosome motility. (A) Percentage of moving EEs. In strain RWS4 (control), almost all Yup1–GFP-stained EEs showed movement. After deletion of kin3 (Δkin3; RWS10) endosome motility decreased to ∼33%. This phenotype could be restored by expression of kin3 under the control of its native promoter (Δkin3/pkin3; RWS11). Expression of a mutant allele of kin3 with a defect in the P-loop of the motor domain in Δkin3 cells (Δkin3/pkin3G105E; RWS19) resulted in a drastic decrease in the residual motility. High levels of Kin3 did not increase movement rates (crgkin3; RWS16). In a conditional dynein strain that was deleted for kin3 (Δkin3Dynts; RWS14), movement of EEs at permissive temperature was similar to that of Δkin3. However, shift to 33°C decreased EE motility to <1%. Bars represent mean percentage ± SD (n = 15 cells). (B) Velocity of EE motion. In the reference strain RWS4, EEs moved at ∼3 µm/s (control). This velocity decreased by >50% after deletion of kin3 (Δkin3). Both the expression of kin3 under the control of its native promoter (Δkin3/pkin3), as well as overexpression (crgkin3), restored this defect. Overexpression of the point-mutated kin3 allele (Δkin3/pkin3G105E) resulted in a clear decrease in velocity. In the temperature-sensitive dynein–Δkin3 double mutant (Δkin3Dynts) at 22°C, EEs move at the expected velocity (compare with Δkin3). Almost all motility was abolished at 33°C, and the velocity was therefore not determined (n.d.). Bars represent mean velocities ± SD (n = 20 EEs, and 7 EEs for Δkin3/pkin3G105E).
Fig. 5. Motion of EEs in the kin3 deletion strain RWS13. (A–C) Co-expression of a GFP–Tub1 fusion protein and Yup1–GFP in a Δkin3 strain allowed us to analyze the role of MTs in the remaining motility of EEs. Brightly stained EE clusters move along MTs (A; arrows mark the MT). In the absence of Kin3, they were located at the ends of MTs. In all cases (n = 31), MTs rapidly shortened towards these cluster (B; end of MT marked by asterisk), suggesting that EEs are located at the minus-ends (labeled ‘+’ and ‘–’). Consistent with this, MT elongation was found to be directed away from the BSDs (C; ends of MTs marked by asterisks, orientation indicated by ‘+’ and ‘–’). Time between frames is 1.4 s. Bar: 2 µm. (D) Endosome organization in the temperature-sensitive dynein–Δkin3 double mutant (RWS14). After 2 h at 33°C, endosome clusters (arrowheads) are smaller or even absent from the cell (D1; right cell). Note that cells have a separation defect due to the deletion of kin3. In these structures, almost no EE motility was found (D2, D3; time in seconds is given in the bottom right corner). Bars: 3 µm. (E) Expression of a dominant-negative kin3 mutant allele that is described to result in a rigid binding of the motor to the MT leads to ‘pearl-string’-like arrangement of EEs (arrows) and abolished almost all motion (compare E1 and E2). Bar: 2 µm; time in seconds is given in the bottom right corner. Movies are available as Supplementary data.
Fig. 6. Kin3 and dynein activity and endosome accumulation in small-budded cells. (A) In small-budded cells of control strain RWS4, accumulations of EEs are located within the bud (A1; arrow indicates bud; arrowhead marks BSD), and only ∼4% of the cells contain the BSD at the distal pole. Previous studies on the MT dynamics suggested that the minus-ends of MTs are located in the bud (Steinberg et al., 2001), indicating that cytoplasmic dynein moves EEs towards the growth region. EE motility was found to be bidirectional, indicating that Kin3 is also active (A2; MTs indicated by red lines, MT orientation marked by ‘+’ and ‘–’, and motor activity and direction indicated by arrows). (B) Heat inactivation of Dyn2ts in the conditional mutant strain RWS15 for 30–60 min shifted EEs to the distal cell pole (B1; arrow indicates bud; arrowhead marks BSD). This localization is in contrast to control cells and is most likely due to an alteration in the balance of Kin3 and dynein activity (B2). (C) High levels of Kin3 also result in strong accumulations of EEs at the distal cell pole of small-budded cells (C1; arrow indicates bud; arrowhead marks BSD). This reorganization is most likely the result of a disturbed balance between Kin3 and dynein (C2). (D) Deletion of kin3 led to randomly positioned EE accumulations (D1; arrowhead marks BSD). Δkin3 cells fail to separate and have a MT organization that is typical for unbudded cells (compare with Figure 3A5). Endosome clusters are located at the minus-ends of these MTs, which is most likely due to the activity of dynein. Occasionally, bidirectional motility was observed, which occurred exclusively along bundles, suggesting that this motility is also dynein based (D2). Bar: 2 µm. Movies are available as Supplementary data.
To further prove that Kin3 is an active motor for EEs, we generated a dominant-negative mutant allele, Kin3G105E, which carried a point mutation in the putative ATP-binding site of the motor (GX4GKT to GX4EKT) and expressed this under the strong crg promoter in a Δkin3 background. This mutation was shown to result in a tight binding of kinesins to MTs (Meluh and Rose, 1990). Assuming that Kin3 is an EE motor, we predicted that overexpression of Kin3G105E in a Δkin3 background should anchor the EEs to MTs, thereby inhibiting all residual motion. Indeed, high levels of Kin3G105E (not shown) abolished almost all motility (Figure 4A, Δkin3/pkin3G105E) and led to a ‘pearl-string’-like arrangement of the immobile EEs (Figure 5E1 and E2, arrows mark EEs). Expression of a wild-type Kin3 allele under the same conditions fully rescued motility (strain RWS20, not shown). In addition, Kin3G105E was not able to complement the morphological phenotype of this mutant (see below), indicating that the motor activity of Kin3 is essential for its cellular function. These data strongly support an active role of Kin3 in transport of EEs in U.maydis.
Deletion of kin3 led to large and randomly positioned EE clusters (Figure 6D1, arrowhead) that no longer appeared at the cell poles or septa. In these cells, the number of individual EEs per area was reduced to ∼43% of the control (15 cells), indicating that EEs gather to form larger aggregates in the absence of Kin3. To support this conclusion, we estimated the average volume of the early endosomal compartment relative to the volume of both wild-type and Δkin3 cells. On average, EEs comprised ∼0.5% of the cell, and no differences was found after deletion of Kin3 (n = 10 cells; P = 0.4031), indicating that Kin3 participates in endosome distribution rather than in their formation.
Cytoplasmic dynein drives minus-end-directed endosome traffic
Deletion of kin3 resulted in a reduction of EE motility to ∼33% of wild-type levels and this motility occurred along MTs. We confirmed this result by disrupting MTs in Δkin3 mutants using the inhibitor benomyl. After 30 min incubation of the cells in 10 µM benomyl, all EE transport was abolished, and could be restored by washing with fresh medium (not shown), confirming that the residual motility is a MT-dependent process. Interestingly, EE clusters were often located at the end of GFP-stained MTs in the kin3 deletion strain RWS13, suggesting that they preferentially accumulate at either the plus- or the minus-ends of MTs. These polymers are dynamic structures that slowly elongate at their plus-ends and rapidly depolymerize towards their minus-ends. Based on this difference, we determined the putative orientation of MTs in RWS13. In all cases analyzed (n = 31), MTs rapidly shortened towards the EE clusters (Figure 5B, the asterisk marks the end of the MT) or slowly grew away from the endosome accumulations (Figure 5C, asterisks mark the ends of two growing MTs). This indicates that, in the absence of Kin3, EEs accumulate at the minus-ends of MTs (orientation marked by ‘+’ and ‘–’ in Figure 5B and C).
The position of EE clusters at the minus-ends of MTs suggested that a minus-end-directed motor molecule was responsible for the residual motion in the absence of Kin3. A good candidate for such a minus-directed motor is cytoplasmic dynein, which was recently described for U.maydis (Straube et al., 2001). The heavy chain of Ustilago dynein consists of two polypeptides, Dyn1 and Dyn2, which are both essential for dynein function in vivo. To investigate the role of dynein in endosome motility, we used a temperature-sensitive allele of dyn2 (Wedlich-Söldner et al., 2002), which we introduced into a Δkin3 strain carrying Yup1–GFP (strain RWS14). At 22°C, RWS14 cells showed EE motility that was indistinguishable from that of the kin3 deletion strain RWS10 (Figure 4, Δkin3Dynts22°C; not significantly different, P = 0.5133). However, after 2–2.5 h incubation of RWS14 at restrictive temperature (33°C), virtually all movement was abolished (Figures 4A and 5D1–D3, Δkin3Dynts33°C; 0.7 ± 1.5% moving EEs, n = 15 cells), and clusters at MT minus-ends became smaller or disappeared (Figure 5D1, arrowheads). This suggests that dynein is responsible for the residual minus-end-directed endosome motility (Figure 6D2, MTs in red, EE clusters in green, orientation of MTs indicated by ‘+’ and ‘–’).
A balance between Kin3 and dynein activity organizes polar endosomes during budding
Our data indicated that bidirectional EE motility in U.maydis is driven by Kin3 and cytoplasmic dynein. During early budding, the assumed minus-ends of MTs as well as EE clusters are localized at the growing end of the cell (see Figure 3A2), although EEs still show bidirectional motion, suggesting that Kin3 and dynein are simultaneously active in these cells. This led us to test the hypothesis that an increased dynein activity results in a net movement of EEs to the small bud and EE cluster formation in this cell cycle stage (Figure 6A2, EE cluster in green, MTs in red, orientation of MTs is indicated by ‘+’ and ‘–’).
In control strain RWS4, almost all EE clusters were located at the small bud (Figure 6A1, the bud is marked by a black arrow and the EE cluster by an arrowhead), while only in 4.2 ± 8.6% of all cells (n = 3 experiments, 100 cells each) was an accumulation of EEs found at the distal cell pole. To test whether this localization is dynein dependent we constructed strain RWS15, which contained a temperature-sensitive dyn2 allele and expressed Yup1–GFP. At permissive temperature, EE distribution was found to be similar to wild type with only 10.7 ± 2.5% of the EE clusters at the distal cell end (n = 3 experiments, 50 cells each; not significantly different, P = 0.3356). However, after 20–40 min at restrictive temperature, 82.0 ± 4.2% (n = 3 experiments, 50 cells each) of all small-budded cells showed a strong EE accumulation at the distal cell pole (Figure 6B1, arrowhead marks EE cluster). In addition, fewer individual EEs were found and only 8.3% of these showed motility. This motion was always directed towards the distal end of the cell and was found to be slower than in control cells (2.05 ± 0.42, n = 8; P = 0.0010). These results are consistent with the view that, in the absence of functional dynein, EEs were moved by Kin3, which trapped EEs at the growing ends of MTs at the distal cell pole of small-budded cells (Figure 6B2).
To strengthen this conclusion, we generated the kin3 overexpression strain RWS16. In this strain, kin3 is under the control of the strong crg promoter, leading to ∼200-fold overexpression of Kin3 (A.Straube and G.Steinberg, unpublished). This high level of Kin3 slightly affected cell shape (Figure 6C1, small bud marked by a black arrow), but did not alter the dynamics of EEs (Figure 4). However, there was a strong effect on the organization of EEs. In small-budded cells, high levels of Kin3 shifted the EE clusters away from the small buds towards the distal cell pole (96.9 ± 2.5% of all cells, n = 3 experiments with >40 cells each; Figure 6C1 and C2), where the growing ends of MTs are located. EE clusters were also found to be significantly larger than in the control strain (compare Figure 6A1 and C1; P < 0.0001). Correspondingly, the number of free EEs was reduced (P = 0.0025). This suggests that an increased amount of Kin3 disturbed the balance between dynein and Kin3, and led to net EE transport towards the distal end of the cell, where they were trapped in large accumulations (Figure 6C2).
Next, we asked for the detailed mechanism by which Kin3 and dynein generate the polar accumulations of EEs. As described above, significantly more EEs reached the EE cluster within the bud than were moving towards the distal cell pole. Interestingly, the velocity of EE motility did not depend on its direction (towards bud: 2.76 ± 0.28 µm/s, n = 20; towards distal cell pole: 2.81 ± 0.27 µm/s, n = 20; P < 0.0001), but we found that the processivity of EE transport towards the clusters was increased. In small-budded cells, the average running distance towards the EE cluster was found to be 6.80 ± 2.46 µm (n = 23), whereas EEs moved significantly shorter distances towards the distal cell pole (4.50 ± 1.71 µm, n = 22; P = 0.0008). A similar situation was found in medium-budded cells, with EEs moving 5.27 ± 1.43 µm (n = 22) towards the bud, whereas the running distance towards the cluster at the distal cell pole was significantly increased (7.81 ± 3.30 µm, n = 20; P = 0.0003). This suggests that alterations in the transport distance are responsible for the rearrangement of EEs during budding. High levels of Kin3 led to a net motility of EEs towards the distal cell pole of small-budded cells. We therefore checked whether overexpression of Kin3 affected the processivity of EE motions. As expected, high levels of Kin3 reduced the minus-end-directed running length to 3.73 ± 1.48 µm (n = 20), whereas the distance of plus-end-directed motility was clearly increased (7.81 ± 3.3 µm, n = 20; P < 0.0001). This suggests that differences in activity of Kin3 or dynein influence the duration of EE transport, thereby leading to a cell cycle-specific net movement towards either end of the cell.
Finally, we checked whether inactivation of Dyn2 or high levels of Kin3 had an influence on MT orientation, which might account for the observed differences in EE cluster formation in these strains. For this purpose, we analyzed the orientation of GFP-labeled MTs based on their dynamic behavior in strain FB1Dyn2tsGT (I.Schulz and G.Steinberg, unpublished) and RWS17. Inactivation of Dyn2ts, as well as high levels of Kin3, had an influence on the frequency with which MTs switched between elongation and shrinkage, which led to longer MTs (I.Schulz and G.Steinberg, unpublished), but based on their dynamic behavior the orientation of these MTs was not different from that of the control strain (not shown).
Kin3-dependent transport is required for cell separation and bipolar budding
In cells with medium-sized buds, EEs accumulate at the distal cell pole, while bud growth continues (Wedlich-Söldner et al., 2000). Although this distribution is a characteristic feature of this cell cycle stage, nothing is known about the cellular role of EEs at this end of the cell. Therefore, we carried out a detailed analysis of the morphological phenotype of the kin3 deletion strain (RWS18), in which EE accumulations were absent from the septa in dividing cells and from the distal cell pole during bud growth (not shown).
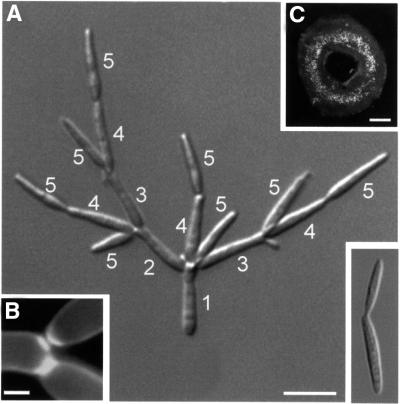
The most prominent defect of RWS18 cells was the formation of large tree-like cell aggregates (Figure 7A, the small image on the lower right refers to wild type). Calcofluor staining demonstrated that two septa were formed (Figure 7B), and cells could be separated by mild sonication, suggesting that late steps of cell separation are impaired in Δkin3 mutants. In addition, new buds were almost exclusively formed at one cell pole (Figure 7A, numbers refer to order of bud appearance), indicating that Δkin3 cells follow a mono-polar budding pattern, which results in a ring-shaped colony appearance (Figure 7C). This budding pattern is in contrast to wild-type strain FB2. Based on the appearance of bud scars visualized with rhodamine-conjugated wheatgerm agglutinin (WGA), most cells formed buds alternating at both poles of the cell (95.7 ± 2.1%, n = 3 experiments with >100 cells each; Figure 8A, bud scars marked by arrows, growth region marked by arrowhead). By contrast, only 16.6 ± 0.8% (n = 3 experiments with >100 cells each) of the Δkin3 cells followed a bipolar pattern, and cells showed many bud scars or septa at one cell pole but only one at the opposite end (Figure 8B, bud scars and septa marked by arrows). Together with the described cell separation defect, this altered budding pattern explains the tree-like phenotype of the kin3 deletion mutants. As the phenotype of kin3 null mutants indicates that this kinesin is required for EE cluster formation at distal cell poles and at the cleavage region, we conclude that Kin3 supports bipolar budding and cell separation.
Fig. 7. Morphology and colony phenotype of a Δkin3 mutant strain. (A) In contrast to wild-type cells (small image at bottom right), the Δkin3 strain forms tree-like aggregates in liquid culture. This phenotype is most likely due to a cell separation defect in combination with a mono-polar budding pattern. The order of bud formation is indicated by white numbers. Bar: 10 µm. (B) Calcofluor staining showed that septa are formed, indicating that deletion of kin3 affects late steps of cell separation. Bar: 2 µm. (C) Morphology of RWS18 leads to ring-like colonies on agar plates. Bar: 0.4 mm.
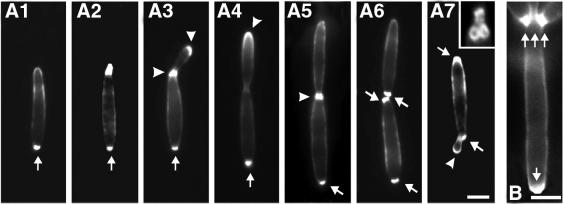
Fig. 8. Distribution of chitin and bud-site selection during the cell cycle of U.maydis. (A) Chitin was stained with WGA in strain FB2. Chitin accumulates strongly at the bud scars in all stages of the cell cycle (arrow in A1–A7). The new bud emerges at the cell pole opposite the bud scar (A2, A3), and chitin localizes to the tip and to the neck region (arrowheads in A3). In cells with larger buds, the chitin ring at the neck becomes more faint, while the tip still carries chitin (arrowhead in A4). Subsequently, chitin appears at the septa (arrowhead in A5), and two bud scars remain after cell separation (pair of arrows in A6). The next appears next to the old bud scar (arrowhead in A7), finally leading clusters of ring-like bud scars (small image in A7) at both ends of the cell. Bar: 3 µm. (B) In Δkin3 cells, many bud scars accumulate at one cell pole (arrows). Bar: 3 µm.
Discussion
In this study we have investigated the molecular basis for rapid motility of EEs in the corn smut fungus U.maydis. Bidirectional transport of EEs was found during all stages of interphase, while MTs switch from an antipolar orientation in unbudded cells to a unipolar organization during budding (Steinberg et al., 2001; Figure 3A). In addition, cell growth is accompanied by the rearrangement of endosome clusters from the small bud towards the distal cell pole (Wedlich-Söldner et al., 2000), suggesting that MT-dependent motors of opposing activity drive endosome motility in U.maydis.
An UNC-104/KIF1-like kinesin is the major motor for endosome motility in U.maydis
Previous studies have shown that EEs move along MTs in U.maydis (Wedlich-Söldner et al., 2000). Such a role of MTs in endosome motility is unknown for lower eukaryotes but established for vertebrate cells, although the underlying transport machinery is not well understood (Apodaca, 2001). In U.maydis, EEs move rapidly along MTs at ∼3 µm/s, and several lines of evidence indicate that a kinesin-like protein, Kin3, drives plus-end-directed endosome motility in this fungus. First, biologically active Kin3–GFP fusion protein localizes to EEs, suggesting that Kin3 is an EE transporter. Secondly, EE motility rates and velocity are significantly reduced in kin3 deletion strains. Thirdly, Kin3G105E, which carries a point mutation in the motor domain that confers a rigid binding to MTs, inhibited all remaining EE motility in Δkin3 cells. Finally, overexpression of Kin3 drives EEs towards the growing ends of MTs at the distal end of budding cells. Because these ends are most likely the MT plus-ends, Kin3-based EE transport to this cell end is consistent with the transport direction of kinesins.
Based on its sequence organization, Kin3 is a fungal representative of UNC-104/KIF1-like kinesins. Motors of this family of kinesins have been identified in a broad range of organisms (see Bloom, 2001 and references therein) and are known to move organelles, such as synaptic vesicles (Hall and Hedgecock, 1991; Okada et al., 1995), mitochondria (Nangaku et al., 1994), or intermediates between Golgi and the endoplasmic reticulum (Dorner et al., 1998). We demonstrate here that Kin3 most likely participates in endosome motility in haploid cells of U.maydis. This adds a new cellular role for UNC-104/KIF1-like motors and is the first molecular identification of a kinesin involved in early endosome traffic. It remains to be determined whether this class of kinesins is of general importance for endosome motility in eukaryotes.
A balance between Kin3 and cytoplasmic dynein determines polar endosome clustering
EEs in U.maydis show bidirectional motility along MTs. Such a behavior is described for many types of organelles in vertebrate cells, including endocytic organelles (Bananis et al., 2000; Murray et al., 2000). Bipolar transport along unipolar oriented MTs is probably based on counteracting dynein- and kinesin-related motors that appear to localize to the same organelle (Pol et al., 1997; Rogers et al., 1997; Martin et al., 1999). Here we show that bidirectional transport of EEs along unipolar MTs in U.maydis is powered by two opposing motors: an UNC-104/KIF1-related kinesin and cytoplasmic dynein. Quantitative analysis of this motility revealed that significantly more EEs move towards the growing cell pole during early budding. Consequently, >95% of all small-budded cells contain an endosome cluster (BSD) at their growth region. Analysis of GFP-marked MTs indicated that minus-ends are focused at the bud (Steinberg et al., 2001), suggesting that minus-end-directed dynein supports endosome traffic towards the bud. However, Kin3 was found on almost all EEs and we observed oscillatory endosome motility during all stages of growth, indicating that the opposing Kin3 is always active. Our model implies that simultaneous action of Kin3 and dynein underlies bidirectionality of EE motility, suggesting that both motors are located on a single endosome. A close relationship of Kin3 and dynein is also suggested by the fact that both motors appear to require their counterpart to reach their maximum transport velocity. Independently of direction, EE motility occurred at ∼2.9 µm/s, whereas the absence of Kin3 or heat inactivation of Dyn2ts resulted in 30–50% reduced velocity, which argues for a physical or functional interaction between both motors. In addition, our data demonstrate that increased activity or levels of either motor affect the average running length of EEs, which results in polar accumulation of these organelles. In this model, reorganization from the bud towards the distal cell pole occurs at a critical threshold, where Kin3 dominates over its counterpart. Consistent with this idea, we found that inactivation of dynein moves EEs from the bud towards the MT plus-ends at the distal cell pole, and the same effect was achieved by overexpression of kin3, indicating that EE reorganization is due to the motor activity of Kin3. This adds further support to the conclusion that alterations in a dynamic balance of dynein and Kin3 activity result in longer transport distance and, thereby, in net movement of EEs to either end of the cell.
Reorganization of endosomes between cell poles is required for bipolar budding
Ustilago maydis wild-type strain FB2 follows a bipolar budding pattern, and this appears to depend on Kin3-based transport of endosomes towards the distal end of the cell, where they might participate in bud-site selection. Evidence exists that the actin cytoskeleton and endosome-based recycling and subsequent redirection of bud-site tag components participate in determination and focusing of the bud site in Saccharomyces cerevisiae (Yang et al., 1997; Ni and Snyder, 2001). These results correspond well with an assumed role of Kin3-driven transport of EEs in bud-site selection in U.maydis. This argues for a general cytoskeleton-based mechanism of placing spatial cues at the cell cortex that subsequently orients the secretory machinery towards the new bud site. Another alternative is that Kin3 moves as yet unidentified vesicles of the secretory pathway that are responsible for the defects in budding and cell separation. The Golgi apparatus also localizes to septa and the growing bud (Wedlich-Söldner et al., 2002), and it remains to be seen whether EE-based recycling or Golgi-derived secretion is responsible for the observed defects in bipolar budding and cell separation.
Materials and methods
Identification of Kin3 and cloning procedures
Primers against the motor domain of UNC-104/KIF1-like kinesins (UNC2, MGIGTIMGIGARCAYCC; UNC6, TTRTTRATRTTNGCD CCYTC) were used in a PCR with genomic DNA of strain FB1 (35 cycles, 94°C for 45 s, 58°C for 45 s, 72°C for 45 s). This was followed by a second PCR using 1 µl of this reaction, 5% glycerol and 10% DMSO at an annealing temperature of 55°C. The amplified DNA fragments were used to obtain the complete genomic DNA from a cosmid library (Bölker et al., 1995). Cloning and double-stranded sequencing of kin3 followed standard procedures.
Strains, plasmids and growth conditions
All strains had the genetic background of FB1 (a1b1) or FB2 (a2b2; Table I). Deletion of kin3 was achieved by homologous replacement of amino acids 12–885 with a hygromycin resistance cassette. To generate the kin3-overexpressing strains (RWS16, RWS17) a nourseothricin resistance cassette followed by the strong 3.6 kb crg promoter (Bottin et al., 1996) was integrated in front of the start codon of kin3. All homologous integrations were confirmed by Southern blot analysis. For complementation or visualization of subcellular structures, strains were transformed with plasmids carrying derivatives of kin3 or GFP fusion proteins (Table I). In brief, pKin3GFP1 contains a kin3–egfp fusion construct behind the kin3 promoter, pKin3GFP2 contains a kin3–egfp fusion construct behind the otef promoter, pKin3YFP contains a kin3–yfp fusion construct behind the otef promoter, pCFPTub1 contains a cfp–tub1 fusion construct behind the otef promoter and a hygromycin resistance cassette, pPX10-146CFP contains a PX10-146–cfp fusion construct (PX domain of yup1) behind the otef promoter, pYup1SG2 contains a yup1–sgfp fusion construct behind the otef promoter, pYup1SG3 contains a yup1–sgfp fusion construct behind the otef promoter and a phleomycin resistance cassette, pRU4-Kin3 contains the kin3 gene behind the native promoter, pcrgKin3G105E contains the carboxin resistance cassette and the 3.5 kb crg promoter fused to the kin3 open reading frame which carried two point mutations in the region encoding the putative P-loop (G314A and C315G), followed by 0.5 kb of the 3′ untranslated region of kin3, potefGFPTub1 contains a gfp–tub1 fusion construct behind the otef promoter. Ectopic integration of plasmids was analyzed for morphology, growth and fluorescence in at least 10 transformants to minimize possible defects due to integration, and those without defects were used for further analysis. If not stated otherwise, strains were grown at 28°C in 2.5% potato dextrose medium or complete medium supplemented with either 1% arabinose (CM-A) or 1% glucose (CM-G). Solid media contained 2% (w/v) bacto agar. Temperature-sensitive strains (RWS14, RWS15) were grown at 22°C and shifted to 33°C for up to 4 h. Medium shift experiments were carried out as described (Straube et al., 2001).
Sequence analysis
The motor domains of Kin3 and TlKIF1 (Sakowicz et al., 1999) were fit into an existing alignment of 140 kinesin motor-head domain sequences available from the kinesin website (http://www.proweb.org/kinesin//KinesinAlign.html). Trees of a subset of these sequences were reconstructed and statistical support was estimated by using neighbor-joining (PHYLIP), parsimony (PHYLIP) and maximum-likelihood (PUZZLE) methods (for further details see Friedrich, 2002). Further analysis was carried out using BLAST (http://www.ncbi.nlm.nih.gov/blast/), PFAM (http://www.sanger.ac.uk/Software/Pfam/index.shtml), SMART (http://smart.ambl-heidelberg.de), PROSITE (http://www. isrec.isb-sib.ch/software/PFSCAN_form.html) and COILS (http://www. ch.embnet.org/software/COILS_form.html).
Expression of recombinant Kin3 protein and ATPase activity measurements
For expression of Kin3 head regions in Escherichia coli, pET15-derived plasmids were transformed into strain BL21-CodonPlusTM-RIL (Stratagene). Expression from the T7 promoter was induced by the addition of 0.5 mM IPTG. His6-tagged proteins were bound to Ni2+–Sepharose, washed with 0.5 and 5 mM imidazole, and eluted with 1 M imidazole. MT-dependent ATPase was measured with a coupled enzymatic assay according to Huang and Hackney (1994) using 1 µg of recombinant Kin3 protein and taxol-stabilized MTs. The decrease in extinction was determined immediately after addition of the motor. MT binding assays were performed essentially as described (Straube et al., 2001).
Light microscopy and image processing
Microscopic analysis was performed using a Zeiss Axiophot microscope. Frames were taken with a cooled CCD camera (Hamamatsu, C4742-95). Epifluorescence was observed using standard FITC, DAPI and rhodamine filter sets. For co-localization studies, eGFP fluorescence was observed with a specific filter set (BP 470/20, FT493, BP505-530). YFP and CFP were also analyzed with specific filter sets (YFP: BP500/20, FT515, BP535/30; CFP: BP436, FT455, BP480-500). Quantification and image processing were performed with Image Pro Plus (Media Cybernetics), Photoshop (Adobe) and Canvas (Deneba).
Quantification and statistical analysis
For microscope observation, cells from logarithmic cultures were embedded in 1% low melt agarose and digital sequences of 30–60 frames were taken with an exposure time of 300–800 ms. During observation, fluorescence intensity was set to 20–40% (AttoArc, 100W HBO; Zeiss) to minimize radiation damage and photobleaching. Observations were made for no longer than 15 min to prevent defects due to oxygen depletion. For speed measurements, only EEs that showed continuous motion over at least 2 µm were analyzed. To determine the percentage of mobile EEs, only those that showed directed motion over >1 µm in one direction were considered. The relative volume of the early endosmal compartment was estimated from three images at different focus planes of individual cells, assuming that EEs and EE clusters are spherical, whereas the cell volume was assumed to be that of an ellipsoid. All measurements were made using Image Pro Plus (Media Cybernetics). All statistical analysis was carried out using PRISM (GraphPad).
Staining procedures and inhibitor studies
For cytological studies, logarithmically growing cultures were used. Benomyl was stored at –20°C at 1 mg/ml in DMSO and added to the cells at 10 µM. FM4-64, WGA and calcofluor staining were performed as described (Wedlich-Söldner et al., 2000). To visualize GFP after fixation, cells were treated with 1% formaldehyde (EM grade; Polyscience) for 30 min at room temperature.
Accession number
The DDBJ/EMBL/GenBank accession No. for kin3 is AF480446.
Supplementary data
Supplementary movies are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank Michael Artmeier for technical support and Dr G.Wöhlke for help with ATPase assays. We are grateful to Dr R.Kahmann for helpful comments on the manuscript and for generous support. This work was supported by a grant to the DFG through SFB 413.
References
- Apodaca G. (2001) Endocytic traffic in polarized epithelial cells: role of the actin and microtubule cytoskeleton. Traffic, 2, 149–159. [DOI] [PubMed] [Google Scholar]
- Bananis E., Murray,J.W., Stockert,R.J., Satir,P. and Wolkoff,A.W. (2000) Microtubule and motor-dependent endocytic vesicle sorting in vitro. J. Cell Biol., 151, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannuett F. and Herskowitz,I. (1989) Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl Acad. Sci. USA, 86, 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom G.S. (1992) Motor proteins for cytoplasmic microtubules. Curr. Opin. Cell Biol., 4, 66–73. [DOI] [PubMed] [Google Scholar]
- Bloom G.S. (2001) The UNC-104/KIF1 family of kinesins. Curr. Opin. Cell Biol., 13, 36–40. [DOI] [PubMed] [Google Scholar]
- Bölker M., Bohnert,H.U., Braun,K.H., Gorl,J. and Kahmann,R. (1995) Tagging pathogenicity genes in Ustilago maydis by restriction enzyme-mediated integration (REMI). Curr. Opin. Cell Biol., 248, 547–552. [DOI] [PubMed] [Google Scholar]
- Bottin A., Kamper,J. and Kahmann,R. (1996) Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet., 253, 342–352. [DOI] [PubMed] [Google Scholar]
- Dorner C., Ciossek,T., Muller,S., Moller,P.H., Ullrich,A. and Lammers,R. (1998) Characterization of KIF1C, a new kinesin-like protein involved in vesicle transport from the Golgi apparatus to the endoplasmic reticulum. J. Biol. Chem., 273, 20267–20275. [DOI] [PubMed] [Google Scholar]
- Friedrich M.W. (2002) Phylogenetic analysis reveals multiple lateral transfers of adenosine-5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J. Bacteriol., 184, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D.H. and Hedgecock,E.M. (1991) Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell, 65, 837–847. [DOI] [PubMed] [Google Scholar]
- Hanlon D.W., Yang,Z. and Goldstein,L.S. (1997) Characterization of KIFC2, a neuronal kinesin superfamily member in mouse. Neuron, 18, 439–451. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. (1998) Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science, 279, 519–526. [DOI] [PubMed] [Google Scholar]
- Huang T.G. and Hackney,D.D. (1994) Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J. Biol. Chem., 269, 16493–16501. [PubMed] [Google Scholar]
- Li J., Smith,G.P. and Walker,J.C. (1999) Kinase interaction domain of kinase-associated protein phosphatase, a phosphoprotein-binding domain. Proc. Natl Acad. Sci. USA, 96, 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Iyadurai,S.J., Gassman,A., Gindhart,J.G.,Jr, Hays,T.S. and Saxton,W.M. (1999) Cytoplasmic dynein, the dynactin complex and kinesin are interdependent and essential for fast axonal transport. Mol. Biol. Cell, 10, 3717–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B. and Rose,M.D. (1990) KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell, 60, 1029–1041. [DOI] [PubMed] [Google Scholar]
- Murphy C. et al. (1996) Endosome dynamics regulated by a Rho protein. Nature, 384, 427–432. [DOI] [PubMed] [Google Scholar]
- Murray J.W., Bananis,E. and Wolkoff,A.W. (2000) Reconstitution of ATP-dependent movement of endocytic vesicles along microtubules in vitro: an oscillatory bidirectional process. Mol. Biol. Cell, 11, 419–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M., Sato-Yoshitake,R., Okada,Y., Noda,Y., Takemura,R., Yamazaki,H. and Hirokawa,N. (1994) KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell, 79, 1209–1220. [DOI] [PubMed] [Google Scholar]
- Ni L. and Snyder,M. (2001) A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell, 12, 2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Severin,F., Backer,J.M., Hyman,A.A. and Zerial,M. (1999) Rab5 regulates motility of early endosomes on microtubules. Nat. Cell Biol., 1, 376–382. [DOI] [PubMed] [Google Scholar]
- Noda Y., Okada,Y., Saito,N., Setou,M., Xu,Y., Zhang,Z. and Hirokawa,N. (2001) KIFC3, a microtubule minus end-directed motor for the apical transport of annexin XIIIb-associated Triton-insoluble membranes. J. Cell Biol., 155, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Yamazaki,H., Sekine-Aizawa,Y. and Hirokawa,N. (1995) The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell, 81, 769–780. [DOI] [PubMed] [Google Scholar]
- Pol A., Ortega,D. and Enrich,C. (1997) Identification of cytoskeleton-associated proteins in isolated rat liver endosomes. Biochem. J., 327, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.L., Tint,I.S., Fanapour,P.C. and Gelfand,V.I. (1997) Regulated bidirectional motility of melanophore pigment granules along microtubules in vitro. Proc. Natl Acad. Sci. USA, 94, 3720–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito N., Okada,Y., Noda,Y., Kinoshita,Y., Kondo,S. and Hirokawa,N. (1997) KIFC2 is a novel neuron-specific C-terminal type kinesin superfamily motor for dendritic transport of multivesicular body-like organelles. Neuron, 18, 425–438. [DOI] [PubMed] [Google Scholar]
- Sakowicz R., Farlow,S. and Goldstein,L.S. (1999) Cloning and expression of kinesins from the thermophilic fungus Thermomyces lanuginosus. Protein Sci., 8, 2705–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G., Wedlich-Söldner,R., Brill,M. and Schulz,I. (2001) Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci., 114, 609–622. [DOI] [PubMed] [Google Scholar]
- Straube A., Enard,W., Berner,A., Wedlich-Söldner,R., Kahmann,R. and Steinberg,G. (2001) A split motor domain in a cytoplasmic dynein. EMBO J., 20, 5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Bölker,M., Kahmann,R. and Steinberg,G. (2000) A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J., 19, 1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R., Schulz,I., Straube,A. and Steinberg,G. (2002) Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell, 13, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Ayscough,K.R. and Drubin,D.G. (1997) A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J. Cell Biol., 136, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]