Abstract
E2F transcription factors play a major role in controlling mammalian cell cycle progression. We recently reported that a potential tumor suppressor, prohibitin, which interacts with retinoblastoma protein (Rb), regulates E2F function and this activity correlates with its growth-suppressive activity. We show here that prohibitin recruits Brg-1/Brm to E2F-responsive promoters, and that this recruitment is required for the repression of E2F-mediated transcription by prohibitin. Expression of a dominant-negative Brg-1 or Brm releases prohibitin-mediated repression of E2F and relieves prohibitin-mediated growth suppression. Although prohibitin associates with, and recruits, Brg-1 and Brm independently of Rb, prohibitin/Brg-1/Brm-mediated transcriptional repression requires Rb. A viral oncoprotein, SV40 large T antigen, can reverse prohibitin-mediated suppression of E2F-mediated gene transcription, and targets prohibitin through interruption of the association between prohibitin and Brg-1/Brm without affecting the prohibitin–E2F interaction.
Keywords: Brg-1/Brm/E2F/prohibitin/SV40 T antigen
Introduction
Prohibitin is a potential tumor suppressor gene that has been linked to human cancers. The prohibitin gene encodes a protein of 275 amino acids that is highly evolutionarily conserved. The prohibitin gene is located at 17q21, close to the BRCA1 locus, and four mutations have been reported in a screen of 23 sporadic breast cancers, suggesting a role for prohibitin in breast cancer (Sato et al., 1992, 1993). The prohibitin gene was originally cloned based on its anti-proliferative activity, causing cell cycle arrest at G1/S (McClung et al., 1992, 1995; Asamoto and Cohen, 1994; Ikonen et al., 1995; Dell’Orco et al., 1996; Coates et al., 1997). We recently reported that prohibitin physically interacts with E2F and regulates E2F function, and that this interaction is necessary for the growth-suppressive activity of prohibitin (Wang et al., 1999a,b).
The E2F family of transcription factors plays a major role in regulating mammalian cell cycle progression and is involved in differentiation, transformation and apoptosis (Chellappan et al., 1991; Nevins, 1992, 1998; Adams and Kaelin, 1995, 1996; Brehm et al., 1999; Harbour and Dean, 2000; Muller and Helin, 2000). Many cellular genes required for progression through the S phase contain E2F-binding sites in their promoters, and E2F activity is essential for their expression (Adams and Kaelin, 1995). It has been established that the retinoblastoma protein (Rb) family of tumor suppressors interacts with E2F and regulates their function (Chellappan et al., 1991). Recent studies have shown that Rb represses E2F-mediated transcriptional activation through recruitment of chromatin-remodeling complexes, such as histone deacetylase (HDAC) and Brg-1/Brm (Dunaief et al., 1994; Brehm et al., 1998; Brehm and Kouzarides, 1999).
We have found that prohibitin interacts with all members of the Rb family (Wang et al., 1999a) and demonstrated important functional and mechanistic differences between Rb- and prohibitin-mediated repression of E2F (Wang et al., 1999b). Rb and prohibitin respond to different signaling pathways (Wang et al., 1999b). Regulators such as adenovirus E1A and cyclin-dependent kinases affect Rb and not prohibitin. Stimulation of B cells through IgM releases prohibitin-mediated repression of E2F specifically (Wang et al., 1999b). Furthermore, we have found that prohibitin targets the conserved marked-box region of E2Fs 1–5, unlike Rb, which targets the transcriptional activation domain of E2Fs 1–3 (Wang et al., 1998, 1999a,b,c; Harbour and Dean, 2000; Wang and Chellappan, 2000).
The molecular mechanism of prohibitin-mediated repression of E2F is not yet fully defined. We recently established that prohibitin requires recruitment of HDAC for transcriptional repression and that co-repressors such as NcoR mediate this process (Wang and Chellappan, 2000). However, some E2F-responsive promoters appear to be unaffected by the recruitment of HDAC (Luo et al., 1998). These observations suggest that alternative mechanisms may be involved in E2F repression. Indeed, in addition to HDAC, general mechanisms of transcriptional repression involving recruitment of other repressors, such as Brg-1/Brm or CtIP/CtBP, have been elucidated recently (Dunaief et al., 1994; Trouche et al., 1997; Meloni et al., 1999; Strobeck et al., 2000). We have found previously that prohibitin was unable to repress E2F in certain cancer cell lines, including SW13 and C33A, which are Brg-1 and Brm negative (Strobeck et al., 2000). In this report, we demonstrate that prohibitin requires Brg-1/Brm for transcription repression of E2F-1. The recruitment of Brg-1 and Brm by prohibitin is independent of Rb, while the transcriptional repression mediated by prohibitin/Brg-1/Brm requires Rb, and is reconstituted when Rb is co-transfected in Rb-deficient Saos2 cells. A viral oncoprotein, SV40 T antigen (SV40T), inactivates prohibitin by blocking the interactions of prohibitin with Brg-1 and Brm.
Results
Brg-1/Brm enhances prohibitin-mediated transcriptional repression
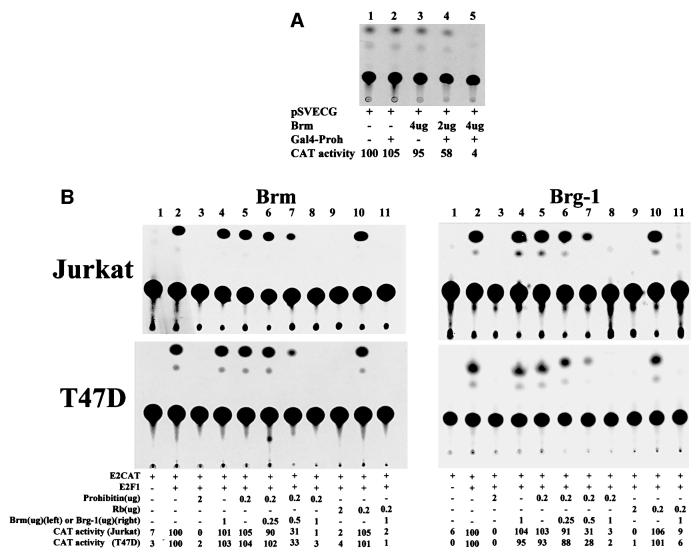
Our earlier studies have shown that prohibitin fails to repress E2F-mediated transcription in SW13 and C33A cells, both of which lack Brg-1 and Brm (Strobeck et al., 2000) (Figure 1). Brg-1 and Brm are members of the mammalian SWI/SNF complex, which has ATP-dependent chromatin remodeling activities (Luo and Dean, 1999; Jaskelioff et al., 2000). Brg-1 and Brm were reported recently to be required for Rb-mediated transcriptional repression (Strober et al., 1996; Strobeck et al., 2000). We tested whether transcriptional repression by prohibitin in SW13 cells might be reconstituted by expression of Brm, acting as a co-repressor. The pSVECG CAT reporter, which carries a GAL4-binding site and is driven by an SV40 promoter, was transfected into SW13 cells either alone, or together with pCR3.1GAL4DBD-prohibitin or pCGhBrm, or with both. As we found before, the expression of prohibitin alone did not affect the basal activity of the reporter, nor did Brm alone, but co-expression of prohibitin with hBrm caused strong repression (Figure 1A). We next tested whether Brg-1 or Brm participate in prohibitin-mediated repression of E2F-induced transcription using Jurkat and T47D cell lines. Cells were transfected with the E2CAT reporter, the activity of which is induced by E2F1 (Figure 1B). Co-transfection of 2 µg of a prohibitin expression vector completely repressed E2F-induced CAT activity, while transfection of a 10-fold lower amount of the prohibitin expression vector (0.2 µg) failed to suppress E2F-driven CAT activity. Co-transfection of either Brg-1 or Brm restored prohibitin-related repression of E2F-mediated CAT activity, in a dose-dependent fashion. Collectively, these results indicate that prohibitin-mediated transcription repression requires, or is enhanced by, Brg-1 or Brm.
Fig. 1. Prohibitin-mediated transcription repression involves Brg-1 and Brm. (A) SW13 cells were transfected with the pSVECG CAT reporter, which is driven by the SV40 promoter and carries a GAL4-binding site along with pCR3.1GAL4DBD-prohibitin, pCGhBrm or both. The relative CAT activities were estimated by directly measuring the radioactivity recovered from the TLC plate and was calculated based on ‘pSVECG’ as ‘100’. The basal activity of the reporter was reduced an estimated 42 and 96% by co-transfection of prohibitin with 2 and 4 µg of Brm expression vector (lanes 4 and 5), while prohibitin alone (lane 2) or Brm alone (lane 3) did not cause strong repression. (B) Jurkat and T47D cells were transfected with the E2CAT reporter, the activity of which is induced by co-transfected E2F1 (lanes 1 and 2). Co-transfection of 2 µg of prohibitin completely repressed the CAT activity, while transfection of a 10-fold lower amount (0.2 µg) of prohibitin expression vector failed to repress the CAT activity. Co- transfection of increasing amounts of Brg-1 or Brm with 0.2 µg of prohibitin expression vector restored the repression of E2F-mediated CAT activity. The radioactivity of TLC was measured and the relative CAT activity was calculated based on ‘E2CAT + E2F1’ as ‘100’.
Prohibitin physically associates with Brg-1 and Brm in vivo
To test whether Brg-1, Brm and prohibitin physically interact, co-immunoprecipitation studies were carried out. Extracts from Jurkat cells, which express prohibitin endogenously, were immunoprecipitated with anti-c-Myc (control), anti-Brg-1 or anti-Brm antibodies, and the precipitated proteins were then immunoblotted for prohibitin (Figure 2B). Endogenous cellular prohibitin was found in complexes with Brg-1 or Brm, but not with c-Myc. These immunoprecipitation/immunoblotting assays were repeated in a reciprocal fashion (Figure 2A), with prohibitin-associated complexes being immunoprecipitated first. Association of Brg-1 and Brm with prohibitin was again detected by immunoblotting of these complexes.
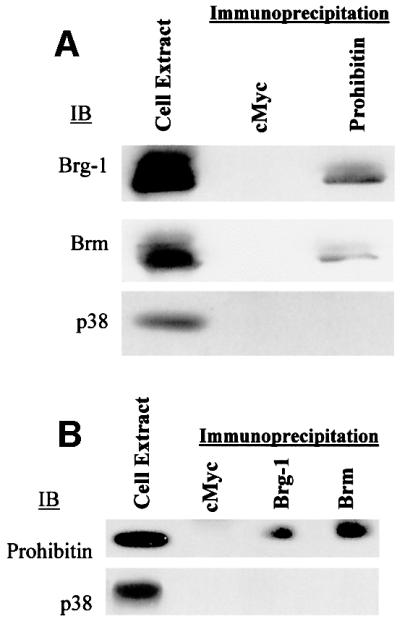
Fig. 2. Prohibitin interacts with Brg-1 and Brm in vivo. (A) Extracts from Jurkat cells were immunoprecipitated by c-Myc (control) or prohibitin antibodies followed by immunoblot analysis (IB) using Brg-1 or Brm antibodies. (B) The reverse immunoprecipitation (IP)/IB was carried out by using cMyc, Brg-1 or Brm antibodies for the IP and prohibitin antibody for the IB. p38 antibody failed to detect any band in the immunoprecipitates.
Prohibitin recruits Brg-1 and Brm to an E2F-responsive promoter
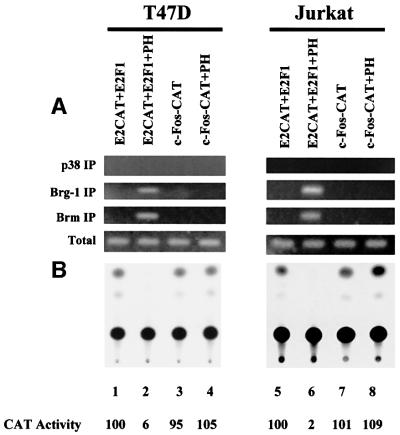
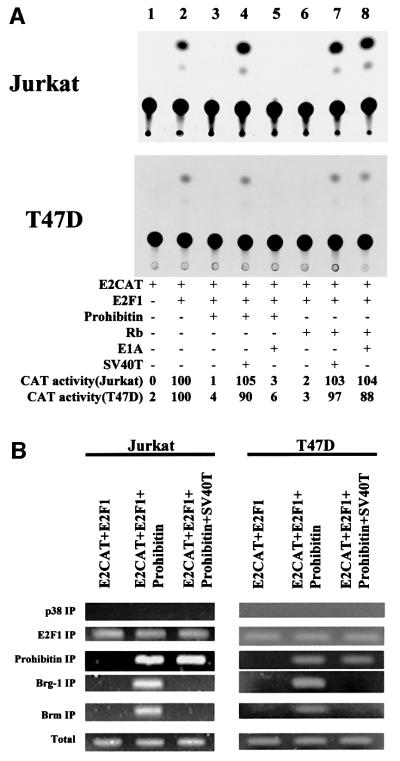
To assess whether prohibitin can recruit Brg-1 or Brm to E2F-responsive promoters, chromatin immunoprecipitation (CHIP) assays, using anti-Brg-1 and anti-Brm anti bodies, were performed. The analyses were first carried out on transiently expressed E2F-responsive promoters using HSF, T47D and Jurkat cells. Representative results from T47D and Jurkat cell transfections are shown. When an E2CAT vector and an E2F1 expression vector were co-transfected into the cells, an association of Brg-1 or Brm with the transfected E2CAT promoter was not detected by CHIP/PCR (Figure 3A). Upon co-transfection of prohibitin, however, E2F-driven CAT activity was repressed (Figure 3B) and associations of both Brg-1 and Brm with the E2CAT reporter gene were detected, demonstrating the prohibitin-dependent recruitment of Brg-1 and Brm to this E2F-responsive promoter. A control experiment was performed in parallel, using a c-Fos promoter–CAT gene, which carries no E2F-binding site. Co-transfection of prohibitin did not repress the c-Fos promoter activity (Figure 3B), as we have demonstrated previously (Wang et al., 1999a), and there was no CAT DNA detectable in the Brg-1 or Brm immunoprecipitates (Figure 3A), indicating that the recruitment of Brg-1 and Brm in this system has some specificity to prohibitin-regulated promoters.
Fig. 3. Prohibitin recruits Brg-1 and Brm to an E2F-responsive promoter. Jurkat and T47D cells were transfected with E2CAT and E2F1 or c-Fos–CAT reporters, with or without a prohibitin expression vector, as indicated. (A) CHIP assay using a pair of PCR primers against the CAT gene detected bands from the whole-cell lysate (‘Total’), indicating equivalent transfection efficiencies. The PCR assay using DNA recovered from the Brg-1 or Brm immunoprecipitates generated products only when prohibitin was co-transfected with E2CAT and E2F1, but not with c-Fos–CAT. PCR using DNA recovered from the p38 immunoprecipitate failed to detect any CAT gene. (B) CAT assay was performed using the same batches of cell lysates. Transfection of 2 µg of E2CAT along with 2 µg of E2F1 vector or 12 µg of c-Fos–CAT resulted in basal CAT activity (lanes 1 and 3). Co-transfection of a prohibitin expression vector (PH) repressed the transcription of E2CAT (lane 2), but not that of c-Fos–CAT (lane 4). Quantitation of CAT activity: the radioactivity recovered from the TLC plate was measured and the relative CAT activity was calculated based on ‘E2CAT + E2F1’ as ‘100’.
Dominant-negative Brg-1 and Brm release prohibitin-mediated repression of E2F-induced transcription and growth arrest
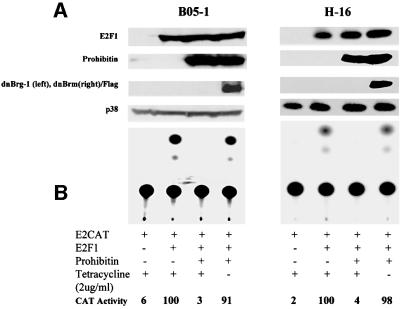
To assess further the necessity for Brg-1 or Brm in prohibitin-mediated repression of E2F1 activity, we adopted a system in which dominant-negative (dn) Brg-1 and Brm expression is conditional. 3T3 cells were stably transfected with dominant-negative mutants of Brg-1 or Brm (deficient in the ATP-binding sites), and controlled by a tetracycline-responsive promoter (de La Serna et al., 2000). Withdrawal of the tetracycline from the medium induces expression of the dominant-negative Brg-1 (B05-1 cells) or Brm (H-16 cells) (Figure 4A). In the presence of tetracycline, when dnBrg-1 or dnBrm were not expressed, ectopic expression of prohibitin inhibited E2F-mediated CAT activity in both B05-1 and H-16 cells (Figure 4B, lanes 3 and 7). In contrast, ectopic expression of prohibitin did not significantly inhibit E2F activity when tetracycline was withdrawn and dnBrg-1 or dnBrm were expressed in B05-1 and H-16 cells, respectively (Figure 4B, lanes 4 and 8). These data demonstrate that the activity of Brg-1 or Brm is required for prohibitin-mediated repression of E2F-mediated transcriptional activation.
Fig. 4. Expression of Flag-tagged dnBrg-1 or dnBrm preverses prohibitin-mediated repression of E2F. (A) Cells carrying inducible dominant-negative Brg-1 (dnBrg-1) (B05-1) or inducible dominant-negative Brm (dnBrm) (H-16) were transfected with E2CAT and E2F1. Expression of the dominant-negative Brm or Brg-1 was confirmed by the immunoblot using a Flag antibody (lanes 4 and 8). Expression of transfected E2F (lanes 2–4, 6, 7 and 8) and prohibitin (lanes 3, 4, 7 and 8) was confirmed by immunoblot. Immunoblot of the same cell extracts with the p38 antibody confirmed equal loading. (B) CAT assay. Co-transfection of a prohibitin expression vector repressed the E2F-induced transcription (lanes 3 and 7) in the presence of tetracycline (absence of dnBrg-1 and dnBrm). Induction of dnBrg-1 and dnBrm expression by withdrawal of tetracycline from the medium reversed the prohibitin- mediated E2F repression (lanes 4 and 8). Quantitation of CAT activity: the radioactivity recovered from the TLC plate was measured and the relative activity was calculated based on ‘E2CAT + E2F1 + tetracycline’ as ‘100’.
To test whether Brg-1 or Brm is required similarly for prohibitin-mediated growth suppression, a prohibitin expression vector was stably transfected into the H-16 (for dnBrm) and B05-1 (for dnBrg-1) cell lines. As shown in Table I, transfection of control vectors (pBabe.puro and pCDNA3) resulted in ∼200 colonies, whether or not tetracycline (and dnBrm or dnBrg-1) were present. Ectopic expression of prohibitin dramatically reduced the number of colonies to ∼40 in B05-1 cells and 50 in H-16 cells, as expected when tetracycline was present and expression of dnBrg-1 or dnBrm was suppressed. Induction of the expression of dnBrg-1 or dnBrm by withdrawal of tetracycline from the medium (confirmed by immunoblot) completely released the prohibitin-induced repression of colony formation. This result demonstrated that prohibitin requires Brm and/or Brg-1 for growth suppression, as well as for inhibition of E2F-mediated gene transcription.
Table I. Reversal of prohibitin-induced repression of colony formation by dnBrg-1 and dnBrma.
| Vector transfected | No. of colonies |
|||
|---|---|---|---|---|
| B05-1 (dnBrg-1) |
H-16 (dnBrm) |
|||
| Experiment 1 | Experiment 2 | Experiment 1 | Experiment 2 | |
| PCDNA3 + pBabe.puro + Tet | 234 | 225 | 203 | 209 |
| PCDNA3 + pBabe.puro – Tet | 238 | 232 | 219 | 217 |
| PCDNA3Prohibitin + pBabe.puro + Tet | 31 | 36 | 53 | 59 |
| pCDNA3Prohibitin + pBabe.puro – Tet | 224 | 215 | 198 | 205 |
aApproximately 10 000 B05-1 or H-16 cells were transfected with 2 µg of the indicated vectors. Colonies with ≥20 cells were counted after 14 days of selection in 1 µg of puromycin per ml.
Rb is required for prohibitin/Brg-1/Brm-mediated transcriptional repression
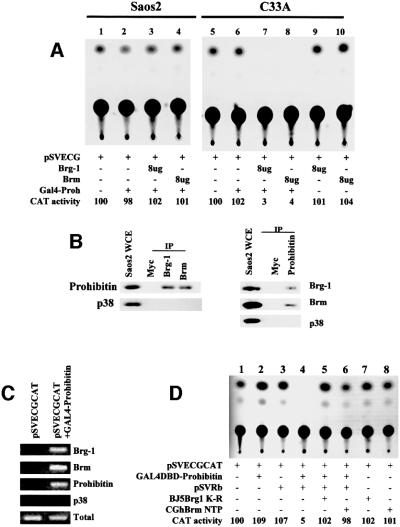
We had found previously that prohibitin did not repress E2F activity in one Rb-negative cell line, Saos2 (unpublished data), and have reported that an Rb binding-deficient prohibitin lost its ability to repress E2F (Wang et al., 1999a). We therefore tested whether Rb is necessary for prohibitin/Brg-1/Brm-mediated transcriptional repression. The pSVECGCAT reporter vector (which is driven by an SV40 promoter carrying a GAL4-binding site, and is not an E2F-responsive promoter) was transfected into Saos2 cells, which produced a basal level of CAT activity (Figure 5A, lane 1). Transfection of GAL4–prohibitin alone, or together with Brg-1 or Brm, failed to repress the pSVECGCAT (Figure 5A, lanes 2–4). To determine whether prohibitin can interact with, and recruit, Brg-1 and Brm in the Rb-null Saos2 cells, immunoprecipitation/immunoblot and CHIP assays were performed. As shown in Figure 5B, prohibitin was detected in the Brg-1 and Brm immunoprecipitates, and Brg-1 and Brm were also detected in the reciprocal prohibitin immunoprecipitates using Saos2 cell lysates, demonstrating that the association between prohibitin and Brg-1/Brm remains intact in the Rb-null Saos2 cells. In the CHIP assay, transfection of prohibitin produced a product when Brg-1 or Brm antibodies were used in the CHIP/PCR, indicating that Brg-1 and Brm were recruited to the transfected pSVECGCAT promoter in Saos2 cells (Figure 5C). We next tested whether the failure of repression of pSVECGCAT expression by prohibitin/Brg-1/Brm was due to the lack of Rb in the Saos2 cell line. As shown in Figure 5D, lanes 2 and 3, expression of Rb or GAL4–prohibitin alone in Saos2 cells did not repress the activity of pSVECGCAT, which does not contain an E2F-responsive promoter. Co-transfection of Rb and GAL4–prohibitin, however, dramatically repressed the activity of the pSVECGCAT reporter in Saos2 cells (Figure 5D, lane 4). This reconstituted, Rb-dependent repression was released by co-transfection of dominant-negative Brg-1 (BJ5Brg1 K-R) or dominant-negative Brm (CGhBrm NTP) (Figure 5D, lanes 5–8), indicating that Rb is required for prohibitin/Brg-1/Brm-mediated transcription repression in Saos2 cells.
Fig. 5. Rb is required for the prohibitin/Brg-1/Brm-mediated transcriptional repression. (A) Saos2 cells (lacking Rb) and C33A cells (Brg-1 and Brm deficient, and expressing a mutant Rb that cannot bind to Brg-1) were transfected with pSVECGCAT. The basal CAT activity was not affected by the co-transfection of GAL4–prohibitin, Brg-1 and Brm in Saos2 cells (lanes 1–4). The activity of pSVECGCAT was repressed when prohibitin was co-transfected with Brg-1 or Brm in C33A cells, while prohibitin, Brg-1 or Brm alone did not affect the activity (lanes 5–10). (B) Saos2 cell lysate was immunoprecipitated using either Brg-1, Brm or prohibitin antibodies followed by immunoblotting using prohibitin or Brg-1 and Brm antibodies. A c-Myc antibody was used as a negative control for the immunoprecipitation and a p38 antibody for the immunoblot analysis. (C) CHIP assay using the same Saos2 cell extracts shown in (A), which were transfected with pSVECGCAT and GAL4–prohibitin as indicated. The DNA recovered from immunoprecipitation by Brg-1, Brm, prohibitin or p38 (as negative control) antibodies was amplified by PCR, using primers against a region of the CAT gene. Co-transfection of the GAL4–prohibitin expression vector resulted in a PCR product using DNA recovered from the Brg-1, Brm or prohibitin immunoprecipitates, indicating that GAL4–prohibitin, Brg-1 and Brm are recruited to this promoter, which carries a GAL4-binding site. Direct PCR using the cell extracts without immunoprecipitation detected comparable amounts of the CAT gene from both extracts tested, demonstrating the equal transfection efficiency. PCR failed to detect the CAT gene in the p38 immunoprecipitates, indicating the specificity of the CHIP assay. (D) Saos2 cells were transfected with pSVECGCAT. Co-transfection of GAL4–prohibitin or Rb alone failed to repress the transcriptional activity of the reporter (lanes 1, 2 and 3). CAT activity was repressed by co-transfection of GAL4–prohibitin plus Rb expression vectors (lane 4), and this repression was reversed by co-transfection of dominant-negative Brg-1 (BJ5Brg1 K-R) or dominant-negative Brm (CGBrm NTP) (lanes 5–8).
Interestingly, co-transfection of Brg-1 or Brm with GAL4–prohibitin did repress pSVECGCAT activity in the Brg-1/Brm-deficient C33A cells (Figure 5A, lanes 5–10). C33A cells express a mutant Rb protein, carrying a four amino acid deletion in the B pocket domain, impairing its ability to bind to E1A and Brg-1, and rendering it non-functional in repressing E2F-mediated transcription (Scheffner et al., 1991; Dunaief et al., 1994). In a CHIP assay using C33A cells, transfection of prohibitin produced a product when Brg-1 or Brm antibodies were used in the CHIP/PCR, indicating that Brg-1 and Brm were recruited to the transfected pSVECGCAT promoter in cells containing a mutant Rb incapable of binding cells to Brg-1 (Dunaief et al., 1994), further suggesting that prohibitin interacts with Brg-1/Brm independently of Rb (data not shown).
SV40T reverses prohibitin-mediated E2F transcriptional repression through interruption of the prohibitin-mediated recruitment of Brg-1 and Brm
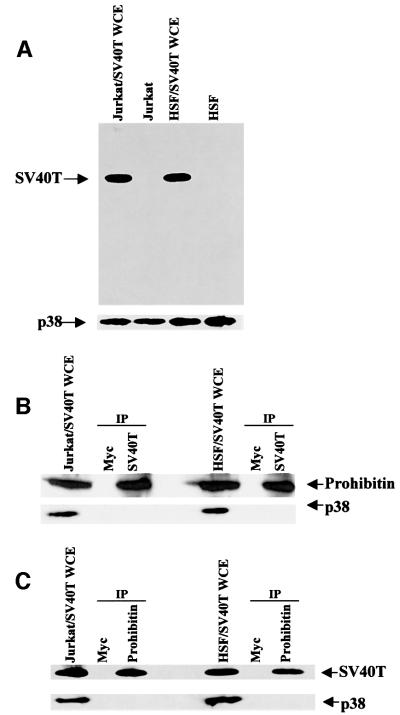
We speculated that prohibitin, as a modulator of E2F, may be targeted by viral oncoproteins, such as SV40T, which are known to inactivate Rb (Brehm et al., 1999). This possibility was tested first by E2F-dependent transcriptional assays. Jurkat and T47D cells were transfected with E2CAT and E2F1. Co-expression of prohibitin or Rb resulted in profound suppression of E2F-induced transcription (Figure 6A, lanes 3 and 6). The prohibitin- or Rb-mediated repression of E2F-dependent transcription was fully released, however, by the co-expression of SV40T (Figure 6A, lanes 4 and 7). In contrast, co-expression of E1A released Rb-mediated repression of E2F (lane 5), but had no effect on prohibitin-mediated repression (lane 8), as we have reported previously (Wang et al., 1999b). We demonstrated above that prohibitin recruits Brg-1 or Brm to E2F-responsive promoters for transcriptional repression. We therefore tested whether SV40T affects this recruitment by performing CHIP assays (Luo et al., 1998; Luo and Dean, 1999). We first performed the CHIP assay in the transient transfection system described above. The DNA recovered from immunoprecipitations using the Brg-1 or Brm antibodies was amplified by PCR, using primers specific for the CAT gene. Transfection of a prohibitin expression vector resulted in the amplification of a band in the PCRs using DNA recovered from the Brg-1 and Brm immunoprecipitates, indicating that Brg-1 and Brm are recruited to this E2F-responsive promoter (Figure 6B). Co-expression of SV40T, however, resulted in no such PCR product, demonstrating that the recruitment of Brg-1 or Brm to the E2F-responsive promoter is blocked by SV40T (Figure 6B). Interestingly, SV40T expression failed to affect the generation of PCR products from DNA recovered by prohibitin immunoprecipitations (Figure 6B), suggesting that the association between E2F1 or prohibitin with the transfected promoter was intact even though SV40T was present. To test whether SV40T interacts with prohibitin in vivo, immunoprecipitation/immunoblot analysis was performed using Jurkat or HSF (a primary human fibroblast cell line; Wang et al., 1998) cells transfected with SV40T. As shown in Figure 7A, an SV40T antibody, Ab-2, detected one band specific to SV40T-transfected cells, demonstrating the expression of SV40T in the transfected Jurkat and HSF cells, but not in the parental cells. Immunoprecipitations were performed using the SV40T or prohibitin antibody, followed by immunoblot analysis using either the prohibitin or SV40T antibody. These immunoprecipitation/immunoblot experiments indicated an association between prohibitin and ectopically expressed SV40T in vivo (Figure 7B and C).
Fig. 6. SV40T releases prohibitin-mediated E2F repression. (A) CAT assay. Jurkat and T47D cells were transfected with E2CAT and E2F1. The prohibitin- and Rb-mediated repression of E2F-dependent transcription (lanes 3 and 6) was released by co-transfection of an SV40T expression vector (lanes 4 and 7). Co-transfection of an E1A expression vector released Rb-mediated repression of E2F (lane 8), but had no effect on prohibitin-mediated repression (lane 5). Quantitation of CAT activity: the radioactivity of the TLC plate was measured and the relative activity was calculated based on ‘E2CAT + E2F1’ as ‘100’. (B) CHIP assay using extracts from Jurkat and T47D cells, which were transfected with the E2CAT, prohibitin and/or SV40T expression vectors as indicated. The DNA recovered from immunoprecipitation by Brg-1 or Brm antibodies was amplified by PCR, using primers against a region of the CAT gene. Transfection of a prohibitin expression vector resulted in a product in the PCR using DNA recovered from the Brg-1 and Brm immunoprecipitates, indicating that Brg-1 or Brm are recruited to this E2F-responsive promoter. Co-transfection of SV40T, however, generated no such PCR product, demonstrating that the recruitment of Brg-1 or Brm by prohibitin is blocked by SV40T. PCR using DNA recovered from the prohibitin immunoprecipitate detected the CAT gene in both prohibitin-transfected extracts, including extracts from cells that were co-transfected with SV40T, in which the prohibitin-mediated transcriptional repression was released. PCR using the DNA from the E2F immunoprecipitate showed equal association of E2F on the promoter of the transfected E2CAT. PCR failed to detect the CAT gene in the p38 immunoprecipitates, indicating the specificity of the CHIP assay.
Fig. 7. SV40T interacts with prohibitin in vivo. (A) Jurkat and HSF cells were transfected with an SV40T expression vector, and SV40T expression were confirmed by immunoblotting. Immunoblotting with a p38 antibody served as a loading control. (B and C) Immuno precipitation was performed using either an SV40T antibody or a prohibitin antibody, followed by immunoblot analysis using a prohibitin or SV40T antibody. A Myc antibody served as a control for the immunoprecipitations, and a p38 antibody as a control in the immunoblot experiments.
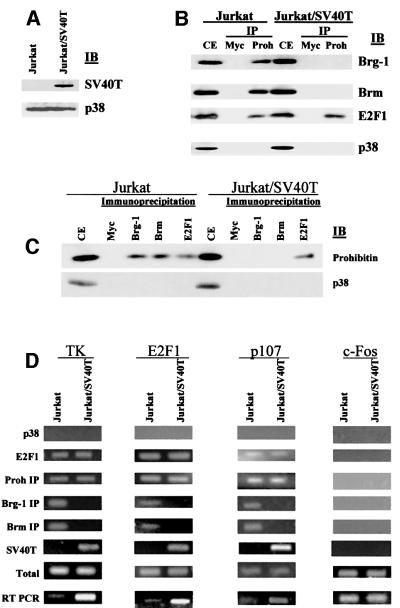
We extended our study to test whether SV40T affected the recruitment of Brg-1 and Brm to native E2F-responsive promoters by using Jurkat cells and Jurkat cells stably transfected with SV40T (designated Jurkat/SV40T). The presence of SV40T in the Jurkat/SV40T cells was again confirmed by immunoblot analysis (Figure 8A). We first tested the effect of SV40T expression on the association between prohibitin and Brg-1 or Brm by the immunoprecipitation/immunoblot analyses. As shown in Figure 8B, Brg-1 and Brm were found in the prohibitin immunoprecipitates but not in the control Myc precipitates, confirming the association of prohibitin with Brg-1 and Brm. However, Brg-1 and Brm were not detected in the prohibitin immunoprecipitates from the Jurkat/SV40T cell extract, although the total amounts of Brg-1 and Brm proteins in the extracts remained the same as in the Jurkat cells (Figure 8B, Jurkat/SV40T, prohibitin, Brg-1, Brm), indicating that SV40T blocked the interaction of prohibitin with its co-repressors, Brg-1 and Brm, in vivo. Interestingly, the association between prohibitin and E2F1 was not affected by SV40T (Figure 8B). These immunoprecipitation/immunoblot assays were repeated in a reciprocal fashion, with Brg-1- and Brm-associated complexes being immunoprecipitated first. Brg-1- and Brm-associated prohibitin was again detected by immunoblotting of these complexes in the control Jurkat cell extract but was not detected in the Jurkat/SV40T cells. The prohibitin and E2F1 association was detected in both Jurkat and Jurkat/SV40T cells. The p38 antibody (as a control) failed to detect a band in any immunoprecipitates, demonstrating specificity. We next assayed for the recruitment of Brg-1 and Brm to native E2F-responsive promoters in Jurkat and Jurkat/SV40T. As shown in Figure 8D, a CHIP assay using Jurkat or Jurkat/SV40T (stably transfected with SV40T) cell extracts was performed to test three E2F-responsive promoters [thymidine kinase (TK), E2F1 and p107] and a non-E2F-responsive promoter, c-Fos (as a control). The DNA recovered from immunoprecipitates by the indicated antibodies was amplified by PCR using primers against a region specific for each of the three E2F-responsive promoters, TK, E2F1 and p107, and the non-E2F-responsive promoter, c-Fos. An amplified product was detected in the CHIP assay of the E2F-responsive promoters using the DNA recovered from Brg-1 or Brm immunoprecipitates from the Jurkat cell extract, indicating an association of Brg-1 or Brm with the E2F-responsive promoters. No such products were detected using the DNA recovered from the Brg-1 or Brm immunoprecipitates from the Jurkat/SV40T cell extract for the same E2F-responsive promoters, demonstrating that the association of Brg-1 or Brm with these E2F-responsive promoters was blocked by the presence of SV40T (Figure 8D). CHIP assays of the E2F promoters using prohibitin or E2F-1 antibodies produced equal amounts of PCR products in both Jurkat and Jurkat/SV40T cells, indicating that the association between prohibitin or E2F-1 proteins with these promoters was not affected by the presence of SV40T (Figure 8D, E2F1 IP, prohibitin IP). CHIP assays using an SV40T antibody (Ab-2) produced a PCR product in all the SV40T-transfected cells, demonstrating the association of SV40T with the tested E2F-responsive promoters (Figure 8D, SV40T). CHIP assays using a non-related antibody, p38, produced no PCR products in any of the assays, demonstrating the specificity of the CHIP assay. PCR using DNA directly isolated from the cell extracts generated products in all the samples tested, which serves as a positive control of the PCR (Figure 8D, Total). RT–PCR demonstrated an increase in transcript levels of these E2F-responsive promoters in the Jurkat/SV40T cells relative to the Jurkat cells lacking SV40T (Figure 8D, RT PCR). CHIP assay of the non-E2F-responsive gene c-Fos demonstrated no association with E2F1, prohibitin, Brg-1 or Brm, and RT–PCR analyses showed no differences in the levels of c-fos transcripts between the cell lines, demonstrating the specificity of the assays (Figure 8D, c-Fos).
Fig. 8. SV40T blocks prohibitin-mediated recruitment of Brg-1 and Brm to native E2F-responsive promoters. (A) Immunoblot (IB) analysis showing the presence of the SV40T proteins in the Jurkat/SV40T cell line. As a loading control, equal levels of p38 protein were detected in both Jurkat and Jurkat/SV40T cells. (B) Jurkat or Jurkat/SV40T cell extracts were immunoprecipitated (IP) using Myc (as a negative control) or prohibitin (Proh) antibodies, followed by IB analysis using Brg-1, Brm or E2F1 antibodies. (C) Jurkat and Jurkat/SV40T cell extracts were immunoprecipitated using Myc (negative control), Brg-1, Brm or E2F1 antibodies, followed by IB analysis using the prohibitin antibody or a control antibody (p38). (D) CHIP assay using Jurkat or Jurkat/SV40T cell extracts as indicated. The DNA recovered from the immunoprecipitate by the indicated antibodies was amplified by PCR using primers against a region on one of three E2F-responsive promoters (TK, E2F1 or p107) and one non-E2F-responsive promoter (c-Fos) as a control. An amplified band was detected in the CHIP assay of the E2F-responsive promoters, but not of the c-Fos promoter, using the DNA recovered from Brg-1 or Brm immunoprecipitates from the Jurkat cell extract, indicating the specific association of Brg-1 or Brm with the E2F-responsive promoters. There was no PCR band detected using the DNA recovered from the Brg-1 or Brm immunoprecipitates from the Jurkat/SV40T cell extract for the same E2F-responsive promoters, demonstrating that the association of Brg-1 or Brm with the E2F-responsive promoters is blocked by the presence of SV40T (demonstrated in the CHIP assay using the SV40T antibody, Ab-2, as indicated). CHIP assay of the E2F promoters using prohibitin or E2F 1 antibodies resulted in an equal amount of PCR products using extracts from both Jurkat and Jurkat/SV40T cells, indicating that the associations between prohibitin or E2F1 proteins and the promoters were not affected by the presence of SV40T. CHIP assay using a non-related antibody against p38 generated no PCR band in any other reactions, demonstrating the specificity of the CHIP. PCR using DNA directly isolated from the cell extracts resulted in bands in all the lanes tested, serving as a positive control of the PCR (Total). RT–PCR assays demonstrated a relative increase in transcription of the E2F-responsive genes in the Jurkat/SV40T cells, compared with the parental Jurkat cells. Results from the control CHIP assay and RT–PCR of the non-E2F-responsive gene, c-Fos, confirmed the specificity of the assays.
Discussion
Alteration or remodeling of nucleosome structure by ATP-dependent remodeling complexes is a critical step in the regulation of transcription. The human SWI/SNF (hSWI/SNF) family is composed of complexes that contain either Brg-1 or Brm as the central ATPase (Peterson and Tamkun, 1995). Recruitment of SWI/SNF complexes previously had been associated with transcriptional activation. The more recent finding of an association between Rb and Brg-1/Brm indicates that the SWI/SNF complex is also involved in transcriptional repression (Dunaief et al., 1994; Strobeck et al., 2000). The results in this report, demonstrating that prohibitin-mediated repression of E2F transcriptional activity requires Brg-1/Brm, further support the concept that SWI/SNF activity is indeed involved in some mechanisms of repression of transcriptional activity. The complete reversal of the prohibitin-induced repression of E2F activity by the expression of dominant-negative Brg-1/Brm demonstrated that Brg-1 and Brm are required for the prohibitin/E2F pathway. Our initial CHIP assays demonstrated an association between Brg-1/Brm and prohibitin on exogenously introduced E2F-responsive promoters. Although these findings were consistent with the results of the co-immunoprecipitation analyses, it was possible that these data using exogenously introduced templates, in which any chromatin structure may not be similar to native chromatin, might not reflect the interactions of ATP-dependent chromatin remodeling complexes on their physiological substrates. We therefore carried out in vivo CHIP assays on endogenous E2F-responsive promoters residing in native chromatin, and confirmed the prohibitin-dependent recruitment of Brg-1/Brm complexes to prohibitin-inhibited E2F-responsive promoters. Furthermore, the dissociation of Brg-1/Brm from prohibitin/E2F complexes by SV40T was also observed on these native E2F-responsive promoters in the in vivo CHIP assays. It is therefore plausible to hypothesize that signals which regulate the function of prohibitin on E2F-driven promoters may be able to do so through modulation of the interactions of prohibitin with Brg-1/Brm. The results of our studies with SV40T provide strong support for such a regulatory mechanism.
It was established that the Rb/E2F pathway is targeted by viral oncoproteins, such as SV40T (Harbour and Dean, 2000). As our previous findings have demonstrated that prohibitin is an important regulator of E2F, and distinct in its actions from the Rb family (Wang et al., 1999a,b), determining whether prohibitin-mediated E2F repression could also be targeted by viral oncoproteins was of considerable interest. The results presented herein demonstrate that SV40T can indeed interact with prohibitin and relieve the transcriptional repression imposed by prohibitin. Our demonstration that SV40T expression coincidentally interrupts the association between Brg-1/Brm and an E2F-responsive promoter, without affecting the association of prohibitin with the E2F-responsive promoter, is striking and suggests this dissociation as the mechanism of the reversal of E2F-dependent transcriptional inhibition. Moreover, this result is in distinct contrast to the action of SV40T on Rb-mediated gene suppression, wherein Rb itself (and presumably the associated co-repressors Brg-1 and Brm) are dissociated from the promoter by the viral oncoprotein. Our results thus define a novel type of functional interaction for SV40T with transcriptional regulators, as well as demonstrating a new level of regulation in Brg-1/Brm co-repressor–transcription complex interactions. In addition, these results further support a causal linkage between the recruitment of Brg-1 and Brm and transcriptional repression by prohibitin, and suggest that the interaction of prohibitin and Brg-1/Brm is targeted by SV40T. The in vivo CHIP assay results from the endogenous cellular E2F-responsive promoters not only confirm that prohibitin is a target of SV40T, as indicated in the transient expression experiments, but also demonstrate that this regulation occurs on native chromatin. It is possible that the abnormal proliferation of normal cells induced by SV40T is mediated in part through its action on prohibitin-mediated cell cycle control.
We have found previously that prohibitin can bind to, and inhibit all the members of the E2F family (Wang et al., 1999a,b). It is possible that prohibitin tethers to the E2F-responsive promoter through the associated E2Fs without itself affecting the promoter activity, and that the repressive effects of prohibitin require the recruitment of co-repressors, such as Brg-1/Brm. In response to certain anti-mitotic signals, prohibitin molecules already associated with E2F-responsive promoters might recruit Brg-1/Brm, resulting in repression of transcription. Conversely, mitotic or oncogenic signals might dissociate Brg-1/Brm from prohibitin, while maintaining a co-repressor-free prohibitin/E2F complex, which would enable transcription. One potential advantage of such a model is that prohibitin would already be in place at E2F promoters, allowing a more rapid response to appropriate signals than if recruitment were required.
As we have reported previously, prohibitin interacts with all members of the Rb family (Wang et al., 1999a). One major difference in the mechanism of E2F repression by prohibitin compared with the repression by Rb family members is that prohibitin-mediated repression cannot be reversed by adenovirus E1A protein and/or the stress-signaling kinase p38, while E1A and p38 can release Rb-mediated E2F repression, as has been established by ourselves and others (Adams and Kaelin, 1995; Wang et al., 1999b). The involvement of the SWI/SNF family in Rb-mediated growth regulation was demonstrated recently (Strobeck et al., 2000). Our discovery of the requirement for Brg-1/Brm in prohibitin-mediated E2F repression and growth suppression may provide a basis for the differential regulation of Rb and prohibitin by various signals, including viral oncoproteins. It is also possible that in the normal situation, Rb independently recruits co-repressors for repression of E2F-responsive promoters. Signals through E1A or p38 release this Rb-mediated repression by dissociating Rb from E2F. When prohibitin is present in the Rb complex, however, reagents such as E1A may still affect Rb, but ‘activated’ prohibitin/Brg-1/Brm remains able to inhibit E2F. SV40T, however, is able to affect both Rb- and prohibitin-mediated repression of E2F by dissociating Rb from E2F and Brg-1/Brm from prohibitin, allowing E2F activation.
GAL4–prohibitin failed to repress transcription of a transfected GAL4-binding site/SV40 promoter-driven reporter gene in Saos2 cells, which lack the Rb protein, although both Brg-1 and Brm were still recruited to the promoter. Ectopic expression of wild-type Rb reconstituted the transcriptional repression. We hypothesize that the recruitment of a wild-type Rb protein by prohibitin to a promoter (in this case, a promoter lacking an E2F-binding site) may ‘activate’ the prohibitin/Brg-1/Brm repressor complex. Alternatively, wild-type Rb may carry out the functional repression itself, once recruited by prohibitin to the promoter of the transfected reporter. The presence of prohibitin may also facilitate the Rb-mediated transcriptional repression, perhaps by recruiting more Brg-1/Brm (or other co-repressors) and/or stabilizing the Rb– repressor complex.
To distinguish among these possible mechanisms, we utilized a cell line (C33A) containing a mutant Rb which cannot mediate transcriptional repression by itself. The same GAL4–prohibitin vector that failed to repress the pSVECG reporter in Saos2 cells did repress transcription of the same exogenous reporter gene construct when Brg-1 and Brm were co-transfected into C33A cells (which are also Brg-1/Brm deficient). The ‘non-functional’ Rb expressed in C33A is lacking four amino acids from the B pocket domain, resulting in loss of ability to complex with adenovirus E1A in vitro (Scheffner et al., 1991) and inability to bind Brg-1 (Dunaief et al., 1994). These data further support the hypothesis that prohibitin recruits Brg-1/Brm independently of Rb, and that Rb recruited by prohibitin to the Gal4–SV40 promoter–reporter construct is not the direct effector of transcriptional repression. The ‘non-functional’ Rb protein in C33A cells may still be able to ‘activate’ the repressor activity of the prohibitin/Brg-1/Brm complex, despite having no intrinsic ability to repress E2F-mediated transcription independently of prohibitin/Brg-1/Brm due to the loss of Brg-1 binding ability. The molecular mechanism whereby Rb activates transcriptional repression by the prohibitin/Brg-1/Brm complex is under investigation. Comparison of the relative roles of Brg-1 and Brm in prohibitin- versus Rb-mediated E2F repression and growth suppression will facilitate our understanding of the differential regulation of cell proliferation by viral oncoproteins and other signals.
Future studies will also assess Brg-1/Brm activity in prohibitin-mediated E2F regulation of transcription during cell cycle progression, apoptosis and differentiation, as well as the prohibitin/Brg-1/Brm association in response to the other signaling events that are known to require prohibitin, such as IgM- and Raf-1-initiated signals (Wang et al., 1999a,b). Finally, a mechanistic understanding of the functional association of Brg-1/Brm with prohibitin and different members of the E2F family on different native promoters will elucidate more precisely the mechanisms of prohibitin-mediated repression of E2F and growth suppression.
Materials and methods
Cell lines, vectors and transfections
SW13, HSF and C33A cells were maintained in Dulbecco’s modified Eagle’s medium, and Jurkat and T47D cells were grown in RPMI medium, both containing 10% fetal bovine serum (FBS). The B05-1 and H-16 cell lines (the generous gifts of Dr Anthony N.Imbalzano) are 3T3 cells stably transfected with dominant-negative mutants of Brg-1 or Brm (deficient in the ATP-binding site), controlled by a tetracycline-responsive promoter. Upon removal of tetracycline from the culture media, the expression of dnBrg-1 or dnBrm protein is induced, as confirmed by immunoblot analysis (de La Serna et al., 2000). Transient transfections of H-16, B05-1, SW13 and T47D cells were carried out using the calcium phosphate DNA precipitation method, according to standard protocols. Both Jurkat and T47D cells are Brg-1/Brm positive, while SW13 cells are Brg-1/Brm negative.
A total of 2 µg of plasmid vectors was used in all transfections for reporter analyses, unless noted otherwise; a total of 8 µg of the vectors was used when extracts had to be prepared from the transfected cells for biochemical analysis. A 1 µg aliquot of a pSV-βGal vector was included as internal control in all transfections, and the β-galactosidase activity varied only slightly (<5%) within each experiment. In all cases, representative CAT assay results from multiple experiments are shown. The total amount of DNA used in each transfection was normalized with salmon sperm DNA.
The pSVECG CAT reporter was a kind gift from Dr Joseph Nevins. pCGhBrm was kindly provided by Dr Stephen P.Goff (Columbia University). pBJ5Brg-1, pBJ5Brg1 K-R (dnBrg-1) (Khavari et al., 1993) and pCGhBrm NTP (dnBrm) (Muchardt and Yaniv, 1993) were the generous gift of Dr Anthony N.Imbalzano (University of Massachusetts, Worcester, MA). The E2CAT vector contains the CAT gene driven by the adenovirus E2 promoter, which contains two E2F-binding sites. pCR3.1GAL4DBD-prohibitin was generated by fusing a full-length mouse prohibitin cDNA to the C-terminus of the GAL4 DNA-binding domain, using SOE-PCR and inserting the product into the pCR3.1 vector (Invitrogen Corporation). A full-length E2F1 expression vector (pDCE2F1) was used to induce E2CAT in transfections. PCDNA3prohibitin has been described previously (Wang et al., 1999b). PW2ori-TDL, an expression vector for SV40T, was the gift of Dr Kathy Rundell (Northwestern University). Stable transfections were performed on 35 mm diameter dishes, using ∼10 000 cells per dish, and subjected to selection in the appropriate antibiotic for 14 days. The total amount of DNA transfected in each sample was normalized with salmon sperm DNA. Cells were fixed and stained with crystal violet, and colonies consisting of >20 cells were scored.
CHIP assays
Cells were treated with formaldehyde at a final concentration of 1% in phosphate-buffered saline (PBS) for 10 min at room temperature. Cells were then washed with PBS and re-suspended in lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.1, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM pepstatin A, 1 mM aprotinin] and sonicated. The samples were centrifuged at 14 000 r.p.m. at 4°C for 5 min. One-third of the supernatant was used to perform a control PCR to verify equivalence of the total amount of plasmid transfected (or the total amount of genomic DNA) among the samples. One-third of the supernatant was immunoprecipitated with a control antibody, or antibodies to Brg-1 or Brm, in a buffer containing 0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.1 and 150 mM NaCl. The antibody-bound complexes were recovered on protein A or protein G beads and the DNA was released by heating to 65°C for 4 h. The DNA was re-suspended in 200 µl of water and treated with 40 µg of proteinase K at 37°C for 30 min, follow by phenol/chloroform extraction and ethanol precipitation. PCR was carried out using 20–100 ng (500–1000 ng for genomic DNA) of DNA as template.
The following PCR primers were used in the CHIP assay for these specific genes. (i) CAT: size of the PCR product 205 bp; 5′ primer, 5′-ACCACCGTTGATATATCC-3′; 3′ primer, 5′-TTGCCATACGGAGTTCCG-3′. (ii) Human E2F1 gene: size of the PCR product 195 bp; 5′ primer, 5′-GCAGCCAATTGTGGCGGC-3′; 3′ primer, 5′-GACGCTCACGGCCCG-3′. (iii) Human p107 gene: size of the PCR product 198 bp; 5′ primer, 5′-TCTTTCAGAATCTGAGGTAC-3′; 3′ primer, 5′-CCGACTTCTTTCTCCCTCC-3′. (iv) Human TK gene: size of the PCR product 200 bp; 5′ primer, 5′-TCCCGGATTCCTCCCACGAG-3′; 3′ primer, 5′-TGCGCCTCCGGGAAGTTCAC-3′. (v) Human cellular oncogene c-fos: size of the PCR product 209 bp; 5′ primer, 5′-TGT TGGCTGCAGCCCGCGAGCAGTTC-3′; 3′ primer, 5′-GGCGCGTGTCCTAATCTCGTGAGCAT-3′.
The following PCR primers were used in RT–PCR for these specific genes. (i) Human E2F1 gene: size of the RT–PCR product 225 bp; 5′ primer, 5′-CTTGGCCGGGGCCCCTGCGG-3′; 3′ primer, 5′-TGTGGGCCGGGGCGCCTGCG-3′. (ii) Human p107 gene: size of the RT–PCR product 363 bp; 5′ primer, 5′-TGGTGTCGCAAATGATGCCTG-3′; 3′ primer, 5′-AGGAGCTGATCCAAATGCCTG-3′. (iii) Human TK gene: size of the RT–PCR product 204 bp; 5′ primer, 5′-ATGAGCTGCATTAACCTGCCCACT-3′; 3′ primer, 5′-ATGTGTGCAGAAGCTGCTGC-3′. (iv) Human cellular oncogene c-fos: size of the PCR product 212 bp; 5′ primer, 5′-ATGATGTTCTCGGGCTTCAACGCA-3′; 3′ primer, 5′-CGTCCCGCCTGTCGGCTCCT-3′.
Immunoprecipitation and immunoblots
Monoclonal antibodies to prohibitin were purchased from NeoMarkers Inc., and anti-Rb, anti-SV40T (Ab-2) and anti-c-Myc antibodies were obtained from Oncogene Science-Calbiochem. Antibodies to Brm, Brg-1 and JNK 1 were obtained from Santa Cruz Biotechnologies. Whole-cell extracts were prepared using the protocol supplied by Santa Cruz. Portions of whole-cell extracts (50–200 µg) were treated with 5 µl of the appropriate primary antibody in a volume of 100 µl at 4°C for 1 h. A 3 mg aliquot of protein A–Sepharose or protein G–Sepharose in a 100 µl volume was added to each sample and incubated for an additional hour. The binding was performed in a buffer containing 20 mM HEPES pH 7.9, 40 mM KCl, 1 mM MgCl2, 0.1 mM EGTA, 0.1 mM EDTA, 0.1 mM dithiothreitol (DTT), 0.1 mM NaF, 0.1 mM Na3VO4, 0.5% NP-40 and 3 mg of bovine serum albumin per ml. The beads were washed six times with 600 µl of the same buffer, boiled in 20 µl of SDS sample buffer and separated on 8 or 10% polyacrylamide gels. After semi-dry transfer to supported nitrocellulose membranes, the blots were probed with the appropriate antibody. The proteins were detected by using an enhanced chemiluminescence assay system from Amersham.
Acknowledgments
Acknowledgements
We thank Dr Srikumar Chellappan for his continuous support, Dr Anthony N.Imbalzano for the kind gifts of the dnBrg-1/Brm cell lines and plasmids, and Ms Samantha Minc for her assistance. This work was partially supported by grants from the American Cancer Society (IRG-72-001-27-IRG) (S.W.) and the National Cancer Institute (CA 84193) (D.V.F.). S.W. is the recipient of the BUSM Department of Medicine Pilot Project Grant Award and an Aid for Cancer Research grant award.
References
- Adams P.D. and Kaelin,W.G.,Jr (1995) Transcriptional control by E2F. Semin. Cancer Biol., 6, 99–108. [DOI] [PubMed] [Google Scholar]
- Adams P.D. and Kaelin,W.G.,Jr (1996) The cellular effects of E2F overexpression. Curr. Top. Microbiol. Immunol., 208, 79–93. [DOI] [PubMed] [Google Scholar]
- Asamoto M. and Cohen,S.M. (1994) Prohibitin gene is overexpressed but not mutated in rat bladder carcinomas and cell lines. Cancer Lett., 83, 201–207. [DOI] [PubMed] [Google Scholar]
- Brehm A. and Kouzarides,T. (1999) Retinoblastoma protein meets chromatin. Trends Biochem. Sci., 24, 142–145. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska,E., Reid,J., Bannister,A. and Kouzarides,T. (1999) The cell cycle-regulating transcription factors E2F-RB. Br. J. Cancer, 80, Suppl. 1, 38–41. [PubMed] [Google Scholar]
- Chellappan S.P., Hiebert,S., Mudryj,M., Horowitz,J.M. and Nevins,J.R. (1991) The E2F transcription factor is a cellular target for the RB protein. Cell, 65, 1053–1061. [DOI] [PubMed] [Google Scholar]
- Coates P.J., Jamieson,D.J., Smart,K., Prescott,A.R. and Hall,P.A. (1997) The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr. Biol., 7, 607–610. [DOI] [PubMed] [Google Scholar]
- de La Serna I.L., Carlson,K.A., Hill,D.A., Guidi,C.J., Stephenson,R.O., Sif,S., Kingston,R.E. and Imbalzano,A.N. (2000) Mammalian SWI–SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol., 20, 2839–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Orco R.T., McClung,J.K., Jupe,E.R. and Liu,X.T. (1996) Prohibitin and the senescent phenotype. Exp. Gerontol., 31, 245–252. [DOI] [PubMed] [Google Scholar]
- Dunaief J.L., Strober,B.E., Guha,S., Khavari,P.A., Alin,K., Luban,J., Begemann,M., Crabtree,G.R. and Goff,S.P. (1994) The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell, 79, 119–130. [DOI] [PubMed] [Google Scholar]
- Harbour J.W. and Dean,D.C. (2000) The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev., 14, 2393–2409. [DOI] [PubMed] [Google Scholar]
- Ikonen E., Fiedler,K., Parton,R.G. and Simons,K. (1995) Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Lett., 358, 273–277. [DOI] [PubMed] [Google Scholar]
- Jaskelioff M., Gavin,I.M., Peterson,C.L. and Logie,C. (2000) SWI–SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol., 20, 3058–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari P.A., Peterson,C.L., Tamkun,J.W., Mendel,D.B. and Crabtree,G.R. (1993) BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature, 366, 170–174. [DOI] [PubMed] [Google Scholar]
- Luo R.X. and Dean,D.C. (1999) Chromatin remodeling and transcriptional regulation. J. Natl Cancer Inst., 91, 1288–1294. [DOI] [PubMed] [Google Scholar]
- Luo R.X., Postigo,A.A. and Dean,D.C. (1998) Rb interacts with histone deacetylase to repress transcription. Cell, 92, 463–473. [DOI] [PubMed] [Google Scholar]
- McClung J.K., King,R.L., Walker,L.S., Danner,D.B., Nuell,M.J., Stewart,C.A. and Dell’Orco,R.T. (1992) Expression of prohibitin, an antiproliferative protein. Exp. Gerontol., 27, 413–417. [DOI] [PubMed] [Google Scholar]
- McClung J.K., Jupe,E.R., Liu,X.T. and Dell’Orco,R.T. (1995) Prohibitin: potential role in senescence, development and tumor suppression. Exp. Gerontol., 30, 99–124. [DOI] [PubMed] [Google Scholar]
- Meloni A.R., Smith,E.J. and Nevins,J.R. (1999) A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl Acad. Sci. USA, 96, 9574–9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C. and Yaniv,M. (1993) A human homologue of Saccharo myces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J., 12, 4279–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. and Helin,K. (2000) The E2F transcription factors: key regulators of cell proliferation. Biochim. Biophys. Acta, 1470, M1–M12. [DOI] [PubMed] [Google Scholar]
- Nevins J.R. (1992) E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science, 258, 424–429. [DOI] [PubMed] [Google Scholar]
- Nevins J.R. (1998) Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ., 9, 585–593. [PubMed] [Google Scholar]
- Peterson C.L. and Tamkun,J.W. (1995) The SWI–SNF complex: a chromatin remodeling machine? Trends Biochem. Sci., 20, 143–146. [DOI] [PubMed] [Google Scholar]
- Sato T. et al. (1992) The human prohibitin gene located on chromosome 17q21 is mutated in sporadic breast cancer. Cancer Res., 52, 1643–1646. [PubMed] [Google Scholar]
- Sato T., Sakamoto,T., Takita,K., Saito,H., Okui,K. and Nakamura,Y. (1993) The human prohibitin (PHB) gene family and its somatic mutations in human tumors. Genomics, 17, 762–764. [DOI] [PubMed] [Google Scholar]
- Scheffner M., Munger,K., Byrne,J.C. and Howley,P.M. (1991) The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc. Natl Acad. Sci. USA, 88, 5523–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck M.W., Knudsen,K.E., Fribourg,A.F., DeCristofaro,M.F., Weissman,B.E., Imbalzano,A.N. and Knudsen,E.S. (2000) BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl Acad. Sci. USA, 97, 7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober B.E., Dunaief,J.L., Guha and Goff,S.P. (1996) Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol., 16, 1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D., Le Chalony,C., Muchardt,C., Yaniv,M. and Kouzarides,T. (1997) RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl Acad. Sci. USA, 94, 11268–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. and Chellappan,S. (2000) Prohibitin, a potential tumor suppressor, represses the transcriptional activity of E2F by recruiting histone deacetylase. Cold Spring Harbor Laboratory, 2000 Meeting on The Cell Cycle. May 17–21, 2000. Cold Spring Harbor Laboratory, New York, NY, p. 194.
- Wang S., Ghosh,R.N. and Chellappan,S.P. (1998) Raf-1 physically interacts with Rb and regulates its function: a link between mitogenic signaling and cell cycle regulation. Mol. Cell. Biol., 18, 7487–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Nath,N., Adlam,M. and Chellappan,S. (1999a) Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene, 18, 3501–3510. [DOI] [PubMed] [Google Scholar]
- Wang S., Nath,N., Fusaro,G. and Chellappan,S. (1999b) Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol. Cell. Biol., 19, 7447–7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Nath,N., Minden,A. and Chellappan,S. (1999c) Regulation of Rb and E2F by signal transduction cascades: divergent effects of JNK1 and p38 kinases. EMBO J., 18, 1559–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]