Abstract
The mitochondrial (mt) plasmid mp1 of Chenopodium album replicates by a rolling-circle (RC) mechanism initiated at two double-stranded replication origins (dso1 and dso2). Two-dimensional gel electrophoresis and electron microscopy of early mp1 replication intermediates revealed novel spots. Ribonucleotide (R)-loops were identified at dso1, which function as a precursor for the RCs in vivo and in vitro. Bacteriophage T4-like networks of highly branched mp1 concatemers with up to 20 monomer units were mapped and shown to be mainly formed by replicating, invading, recombining and resolving molecules. A new model is proposed in which concatemers were separated into single units by a ‘snap-back’ mechanism and homologous recombination. dso1 is a recombination hotspot, with sequence homology to bacterial Xer recombination cores. mp1 is a unique eukaryotic plasmid that expresses features of phages like T4 and could serve as a model system for replication and maintenance of DNA concatemers.
Keywords: 2D-loop/dso/invasion/phage T4/snap-back
Introduction
Until recently, the formation of multimeric head-to-tail DNA molecules (concatemers) was considered to be limited to double-stranded (ds) DNA phages (Mosig, 1998; Kreuzer, 2000), certain bacterial plasmids (Viret et al., 1991) and some animal viruses (deLange and McFadden, 1990). Four mechanisms were initially proposed as models for the production of concatemers: (i) sigma (σ) or rolling-circle replication (RCR) as known from phage λ (Takahashi, 1975), the replication cycle of single-stranded (ss) Escherichia coli phages (prototyped by φX174) and bacterial RC plasmids (prototyped by pT181) (Khan, 1997; del Solar et al., 1998); (ii) a replication mode involving the production of highly branched molecules such as that of phage T4 (Mosig, 1998; Kreuzer, 2000); (iii) the replication of certain linear ds genomes forming concatemers to replicate their 3′ ends, such as phage T7 (Watson, 1972); and (iv) linear dsDNA genomes with hairpin loops and terminal repeats (e.g. poxviruses) that are involved in the generation of branched concatemers (deLange and McFadden, 1990).
After replication, the concatemers need to be separated into unit-length molecules. This occurs during packaging, when viral DNA is recognized at specific sequences (cos or pac) and cut by terminases (Becker and Murialdo, 1990; Black, 1995; Catalano et al., 1995). Models have been developed to explain the processing of phage concatemers, such as the nicking model (Watson, 1972) or the double-strand breakage model (Fujisawa and Morita, 1997). The resolution of poxvirus telomeres may proceed by conservative strand exchange of the concatemer junctions (deLange and McFadden, 1990). In contrast, the concatemers that were recently detected in the mitochondria of protists, yeast and plants have a widely unknown mechanism of replication and resolution (Maleszka et al., 1991; Lockshon et al., 1995; Backert et al., 1996; Preiser et al., 1996; Oldenburg and Bendich, 2001). Consistent with a prokaryotic origin of mitochondria, mtRNA polymerases of yeast and plants are homologous to phage T7, T3 and SP6 single-subunit RNA polymerases (Hedtke et al., 1997). These phage-type RNA polymerases might be involved in mtDNA replication (Schinkel and Tabak, 1989).
The eukaryotic plasmid mp1 (1309 bp) from the mitochondria of the dicot Chenopodium album (L.) shares unique features of RCR (Backert et al., 1996). During logarithmic growth stage, σ-like RCs were identified similar to classical phage λ replication. mp1 contains two double-stranded origins (dso) with homology to those of bacterial RC plasmids and produces single-strand copies of only one plasmid strand, but a typical RC initiator protein is not encoded by mp1 (Backert et al., 1997, 1998). However, a predominant nicking site (dso1) and a less often used region 180 bp away (dso2) have been identified that share the 5′ TAAG/GG nicking motif (Backert et al., 1997). The ends of σ-structures and linear mp1 molecules constituted either dso1 or dso2 (Backert et al., 1996). Besides the identification of RCs, the presence of mp1 recombination intermediates was a common feature both in vivo (Backert et al., 1997) and in vitro (Backert et al., 1998), but their nature remained unknown.
The aim of the present study was to investigate early replicating mp1 intermediates and to elucidate the nature of the recombination structures. Novel mp1 molecules produced in vivo and in vitro, including ribonucleotide (R)-loops and highly branched structures, were analysed on two-dimensional electrophoretic (2-DE) gels and studied by electron microscopy (EM). Based on these observations, a new model for resolving DNA concatemers is predicted.
Results
2-DE reveals novel plasmid mp1 intermediates
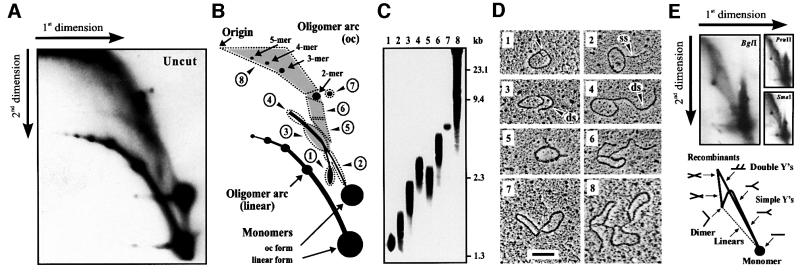
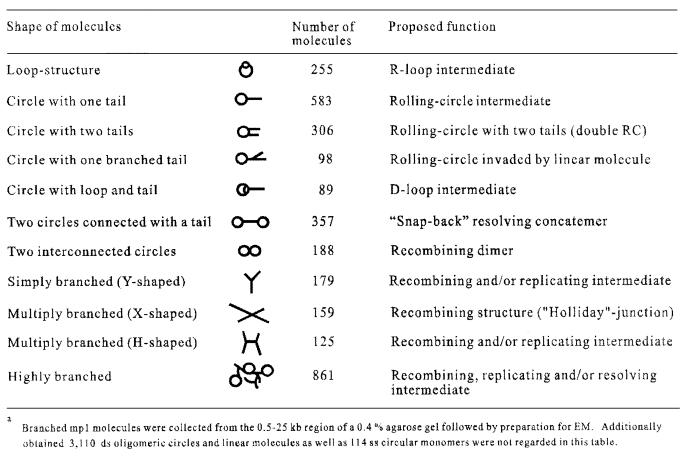
Two powerful techniques, 2-DE and EM, were combined to map a complete set of intermediates produced during replication and recombination of mp1 in the early stages of cell growth. For this purpose, mitochondria were prepared from C.album 12 h after transfer of cells into new medium, when most of the mtDNA undergoes replication (Oldenburg and Bendich, 2001). Uncut mtDNA was separated on 2-DE gels, blotted by denaturing transfer, and then hybridized with mp1 DNA (Figure 1A). To visualize better the zone containing branched forms, the supercoiled and ssDNA monomers were allowed to run from the gel. The strongest signals were at the positions of the open circular (oc), linear and supercoiled forms of the monomer, as well as at two curves representing linear and oc oligomers up to a 5mer (Figure 1B). A series of novel spots have been detected that were gently recovered from a gel (Figure 1C) and subjected to EM. From each fraction, 100 molecules were randomly photographed and classified (Figure 1D). Surprisingly, spot 1 contained theta (θ)-like mp1 monomers with a small loop (100–150 bases). Above this spot, a curve between the linear and circular molecules was detected, which extended past the linear dimer. These signals mainly originated from spot 1 and in part from the circular monomer. EM analysis of both arcs (fraction 2) revealed RC monomers, with one growing single-stranded or double-stranded tail from ∼0.1 up to 0.5 kb. Circles with one growing double-stranded tail (0.5–1 kb) were observed in fraction 3, while RCs with longer tails (1–4 kb) originated from the extending arc (fraction 4). Careful examination revealed variable single-stranded regions (50–200 bases) at the circle–tail junction and the tip of the tail in 82% of the RCs. Circles with two short (0.1–0.3 kb) double-stranded tails have been observed in fraction 5. Fraction 6 revealed the presence of circles with two extending double-stranded tails (0.4–1.0 kb). Spot 7 contained recombining mp1 molecules, and highly branched concatemers were found in a large zone of the gel shaded in grey (fraction 8). Quantitative analysis of the mp1 hybridization signals obtained in Figure 1A revealed the following distribution: 37.5% linear molecules (mono- and oligomers), 34% circles (mono- and oligomers), 21% branched concatemers, 7% σ-like molecules and 1.5% θ-like monomers. These results are in good agreement with a quantitative EM analysis of 3200 total mp1 molecules that were electrophoretically separated from chromosomal mtDNA and gently recovered from the 1–25 kb zone of an agarose gel (Table I).
Fig. 1. 2-DE and EM analysis of intact plasmid mp1 in vivo replication and recombination intermediates. (A) Autoradiography of a 2-DE gel probed with mp1 DNA. Hybridization signals are explained in (B). Signals of unknown nature are marked with dashed lines and arrowheads. (C) Molecules from regions 1–8 were gently recovered from a gel. Enrichment of accumulated replication and recombination intermediates is shown. (D) One hundred molecules from each fraction were analysed by EM (for details see text). Bar = 0.5 kb. (E) Autoradiography of 2-DE gels of plasmid mp1 digested with the indicated restriction endonucleases. Hybridization patterns are explained schematically.
Table I. EM categorisation of 3200 early in vivo plasmid mp1 replication and recombination intermediatesa.
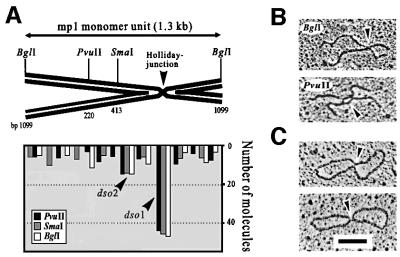
To identify mp1 replication and recombination intermediates, plasmid DNA from the 1–25 kb region was digested with restriction endonucleases with a single cutting site in mp1, and separated by 2-DE. mp1 hybridization patterns revealed a continuous arc of growing Y-shaped replication intermediates expanding from the linear monomer to the dimer, plus a spike of double Ys and recombining X-molecules (Figure 1E). These pictures were compatible with σ-like but not with θ-like replication intermediates according to the model of Brewer and Fangman (1987) (Figure 1E, bottom). Almost identical 2-DE patterns were observed for BglI, PvuII and SmaI digests, respectively, thus not allowing 2-DE mapping of branched mp1 forms. To map mp1 replication and recombination intermediates in the following experiments, individual fractions 1–8 were cut with the same set of enzymes and prepared for EM.
EM mapping of mp1 rolling circles
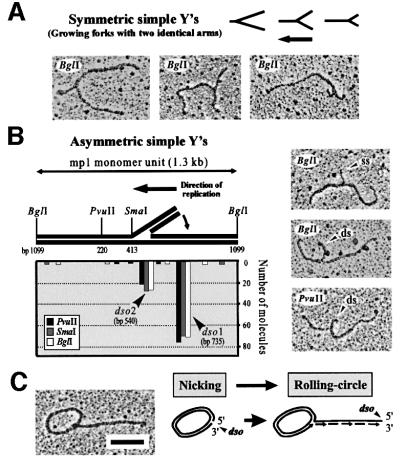
In a first experiment, σ-molecules with one growing tail in fractions 2, 3 and 4 were combined and analysed in three parallel samples digested with BglI, PvuII and SmaI, respectively. EM of these samples revealed symmetric and asymmetric simple Ys. The lengths of the arms and the branch points were carefully measured as 100 molecules from each digest. The growing symmetric simple Ys originated from the large σ-structures because the position of branch points was highly variable, as expected from moving replication forks (Figure 2A). The asymmetric simple Ys contained short single-stranded or double-stranded tails and, therefore, may originate from strand displacement in early RCs. To reveal whether these tails have specific ends, the tails were projected onto the linearized monomer and the origin was deduced from two possible orientations (Figure 2B). In agreement with a previous study (Backert et al., 1996), the origin determined for the majority (74%) of these Y-shaped molecules was at position 735 of the mp1 map (dso1). The origin of 22% of the Y-shaped molecules was determined to be 200 bp downstream from dso1, correlating with the location of dso2. The direction of replication initiated at dso1 or dso2 was identical (Figure 2B, arrow). This supports a RC model of mp1 replication initiated at two alternating origins (Figure 2C).
Fig. 2. Mapping of σ-like mp1 replication intermediates with one tail. Plasmid molecules from fractions 2, 3 and 4 were combined and linearized with BglI, PvuI and SmaI, respectively. EM investigation of 100 molecules each revealed symmetric simple Ys (A) and asymmetric simple Ys (B). The location of the origins dso1 and dso2 was deduced as described in the text. The x-axis corresponds to positions on the plasmid map as indicated in the model above. Examples are shown in the right panel. (C) Example and model of an uncut mp1 RC. Bar = 0.5 kb.
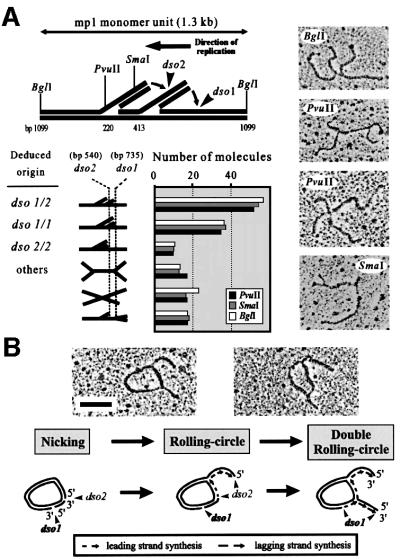
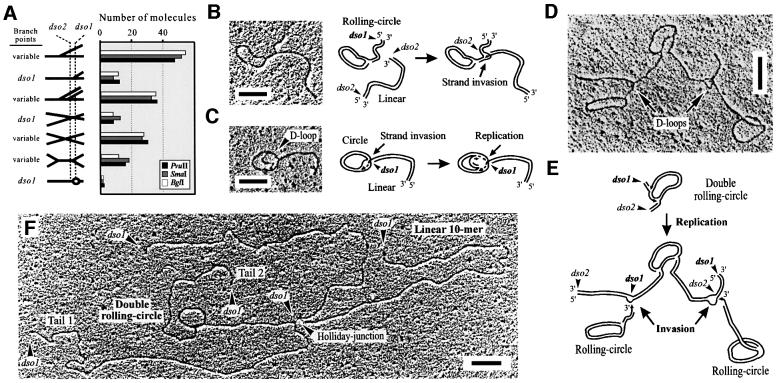
Next, fractions 5 and 6 containing σ-molecules with two growing tails (double RCs) were combined and digested. EM revealed symmetric and asymmetric double Ys (Figure 3A). The branch points in the symmetric double Ys were variable, indicating ongoing replication in two moving replication forks per molecule. The detection of a few X-shaped molecules (12%) also suggested the presence of recombining intermediates. However, the majority of asymmetric double Ys with two short tails were mapped to either dso1/dso1, dso2/dso2 or dso1/dso2, indicating that restarts can occur at the dsos and that both origins can be used simultaneously for the initiation of RCR. In agreement with these data, the ends of the tails in uncut double RCs fall together in either one origin, dso1 or dso2 (142 molecules), or are separated by 0.2 kb (164 molecules), which corresponds to the distance between the dsos (Table I). Two representative double RCs are shown and explained schematically (Figure 3B).
Fig. 3. Mapping of σ-like mp1 intermediates with two tails (double RCs). (A) Plasmid DNA from fractions 5 and 6 was combined and linearized with BglI, PvuI and SmaI, respectively. EM of 150 molecules in each digest revealed various double Ys. The location of the origins dso1 and dso2 was mapped. Examples are shown in the right panel. (B) Uncut double RCs initiated at dso1 and dso2 are explained in the model. Bar = 0.5 kb.
Identification of in vivo R-loops
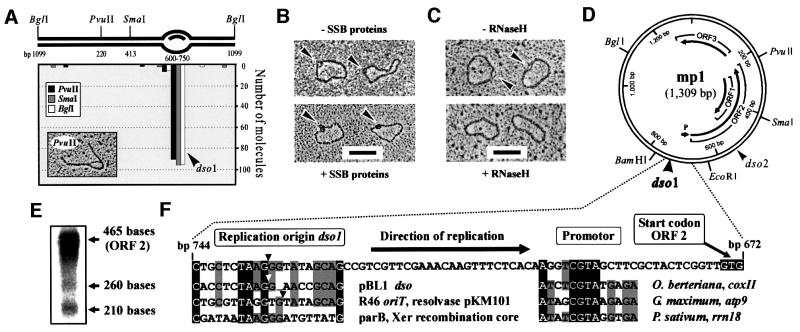
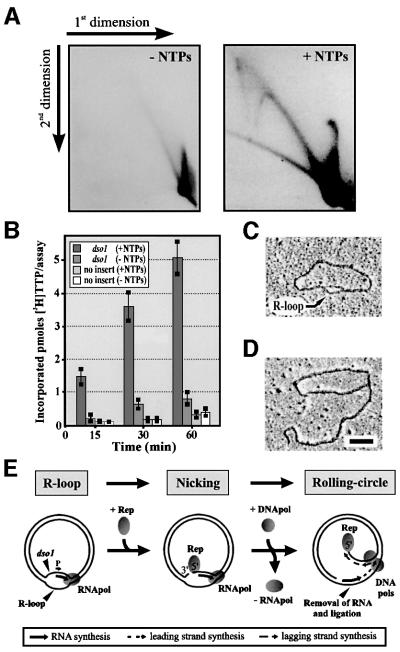
To map the position of the loop in θ-like mp1 molecules, fraction 1 was prepared for EM after digestion with BglI, PvuII and SmaI. Surprisingly, this loop was located at position 600–750 of the plasmid map, correlating to dso1 (Figure 4A). To characterize this loop, θ molecules were incubated with SSB proteins from E.coli and prepared for EM. Binding of two to three SSB protein particles at this loop demonstrated that it contains at least one single strand (Figure 4B). This observation suggested two possibilities: (i) a displacement (D)-loop; or (ii) an R-loop. Incubation with RNaseH, an RNase that specifically hydrolyses RNA molecules in RNA/DNA hybrids, led to the disappearance of the loops in 92% of the molecules (Figure 4C). These loops also disappeared by treatment with a RNace-it ribonuclease cocktail (RNaseA and non-specific RNase from Aspergillus), but not with λ-exonuclease (data not shown). This indicated that the loop represents an R-loop. R-loops were also observed in linearized mp1 and did not exceed 150 bases (Figure 4A). Thus, artificial R-loops with sizes of up to 600 bases that were observed in supercoiled but not in linear plasmids can be excluded (Richardson, 1975). The only possible open reading frame (ORF) in this region that could be involved in the initiation of replication is ORF2 (Figure 4D). Northern blot analysis of in vivo-transcribed mt mRNA indicated abundant transcripts of ORF2 (Figure 4E). The genetic organization of dso1 and ORF2 with a promotor consensus motif is shown in Figure 4F.
Fig. 4. Identification of in vivo R-loops in plasmid mp1. (A) EM mapping of 100 θ-like mp1 molecules. The x-axis corresponds to the plasmid map as indicated. (B) EM of uncut mp1 molecules with loops (arrowheads) that were incubated with SSB proteins. (C) Treatment of these molecules (arrowheads) with RNaseH resulted in the removal of the loops. Bar = 0.5 kb. (D) Physical and genetic map of mp1 depicting three putative ORFs. Arrowheads label the origins dso1 and dso2. (E) Northern blotting revealed three major transcripts with the sizes of ORF1–3 using mp1 DNA as probe. (F) Genetic organization of dso1 and ORF2 with a plant mt promotor consensus motif 5′ CGTA (Tracy and Stern, 1995; Binder et al., 1996). An alignment of dso1 to dsos of bacterial RC plasmids (Backert et al., 1998) and a Xer recombination core (Hodgman et al., 1998) is presented. Identical nucleotides are shaded in black and at least three identical residues are shaded in grey. Arrowheads indicate the RC nicking site.
R-loops are precursors of rolling circles
To identify the R-loop as the initial structure of mp1 RCR, an in vitro replication system was utilized. A cloned 148 bp fragment containing dso1, the ORF2 promotor and the first 44 nucleotides of ORF2 was incubated with a mitochondrial replication complex in the presence or absence of ribonucleoside triphosphates (NTPs). Efficient replication was observed only in the presence of NTPs with typical 2-DE gel patterns of RC concatemers (Figure 5A). A time course showed that incorporation of methyl-[3H]-TTP in the dso-containing construct was linear and time dependent, whereas in vitro replication in the absence of NTPs was low and comparable to the empty vector control (Figure 5B). This suggested that R-loops at dso1 may also be formed in vitro. To test this, in vitro replication intermediates of the origin-containing construct were prepared for EM, and revealed the presence of R-loops at dso1 after 10 min (Figure 5C) and subsequently RCs after 30 min (Figure 5D). EM of in vitro replication products without NTPs revealed no R-loops, with only very little random displacement synthesis, similar to the vector control (data not shown). This clearly excluded the possibility that lagging strand synthesis (via ribonucleotide primers) at the displaced strand was blocked. Instead, these observations collectively suggest that R-loops of ORF2 were essential in the initiation of RCR (Figure 5E).
Fig. 5. Analysis of early mp1 replication by an in vitro replication assay. (A) 2-DE of a construct harbouring the dso1 origin sequence, which was incubated in vitro for 60 min with a mitochondrial enzyme complex in the presence or absence of 10 µM NTPs. (B) Time course of mp1 in vitro replication using the dso1 construct and vector as a control. The results are the means of three experiments. (C) EM analysis of in vitro replication intermediates revealed R-loops (arrow) after 10 min. (D) Typical RCs were observed after 30 min. Bar = 0.5 kb. (E) Hypothetical model for the initiation of mp1 RCR involving an R-loop transcribed from ORF2 (see text).
dso1 is a recombination hotspot
In another experiment, the recombining mp1 molecules observed in fraction 7 were digested and prepared for EM. This fraction exclusively revealed symmetric X-shaped molecules, indicating the presence of stable ‘Holliday’ junctions in recombining dimers or two monomers (Figure 6, arrowheads). Measuring revealed that the junction in almost half of these molecules also mapped to dso1. This demonstrated that dso1 could be used in both the initiation of mp1 RCR and recombination. Interestingly, the dso1 nicking sequence 5′ TAAG/GG is present in bacterial Xer recombination cores (Figure 4F). Recombining mp1 intermediates were compatible with the strand invasion mechanism (Smith, 1988; Mosig, 1998). No intermediates consistent with the resection-annealing model have been found and no molecules typical of artificial branch migration were observed.
Fig. 6. EM mapping of in vivo mp1 recombination intermediates. (A) DNA from fraction 8 was cut with BglI, PvuI and SmaI. EM of 100 molecules in each digest revealed X-shaped mp1 recombinants that mapped to dso1. Examples of cut (B) and uncut mp1 dimers (C) containing recombination junctions (arrowheads). Bar = 0.5 kb.
Categorization and mapping of complex mp1 molecules
The highly complex mp1 molecules extracted from fraction 8 of a 2-DE gel were cut and prepared for EM as described above. This study predominantly revealed symmetric simple Ys, double Ys and X-shaped molecules (Figure 7A). Measurement of the X-structures showed a similar distribution as shown in Figure 6, with 32% of molecules mapping to dso1. The fork position in ∼22% of the simple Ys also mapped to dso1, but the majority of simple Ys, double Ys and X-forms contained variable fork positions and, therefore, originated from the branched concatemers that can be explained as replication/recombination intermediates (Figure 7A). In particular, various events such as multiple replication forks and/or recombination junctions, and DNA end invasions must be considered for the generation of these highly complex structures. According to their shape, a high number (2362) of uncut mp1 molecules were categorized into specific groups: (i) double RCs; (ii) RCs with a branched tail; (iii) two circles connected by a tail; (iv) recombining oligomers; (v) Y-structures; (vi) X-structures; (vii) H-structures; and (viii) highly branched molecules (Table I).
Fig. 7. EM of highly complex in vivo mp1 replication and recombination intermediates. (A) Molecules from fraction 8 were linearized with BglI, PvuI and SmaI, respectively. EM of 150 molecules mainly revealed symmetric simple Ys and double Ys. Examples are shown in the right panel. (B–F) Examples and explanation of uncut complex mp1 concatemers. (B) Open circle with a branched double-stranded tail of 1.3 kb. (C) Open circle with a 1.1 kb tail and a D-loop. (D) Highly branched mp1 molecule (total size 8.9 kb). (E) Model for the origin of this structure. The upper molecule may represent a double RC initiated at dso1 and dso2. Both RC tails are invaded by two other RCs (bottom). (F) Highly branched mp1 molecule (total size is 26.3 kb) that represents a double RC (reinitiated at dso1), recombining with a linear 10mer at dso1. Bars = 0.5 kb.
The presence of two mp1 origins (dso1 and dso2) that replicated unidirectionally in the same direction enabled measurement of circles with several branchings. This study generated interesting insights into the nature of these complex structures. First, the observation of RCs with a branched tail (Figure 7B, 98 molecules) or circles containing a D-loop connected to a tail (Figure 7C, 89 molecules) supported the idea of strand invasion events in mp1. Secondly, recombinative invasion could trigger replication because the D-loops contained growing dsDNA stretches (Figure 7C–E, arrows; 289 molecules). Thirdly, measurement of the highly complex concatemers suggested that in 72% of these molecules the origin structure was a double RC that initiated either at one or both dsos. The tails of double RCs were subject to intermolecular recombination and/or invasion at dso1 or dso2 (Figure 7D–F). Similar observations were made for 860 other mp1 molecules (Table I), suggesting that these structures are not artefacts but represent recombining, invading and replicating plasmid concatemers similar to phage T4 (Mosig, 1998; Kreuzer, 2000).
Processing of mp1 concatemers
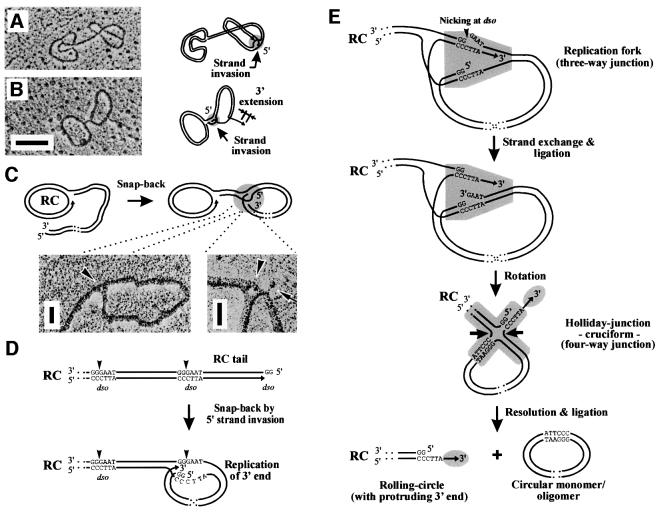
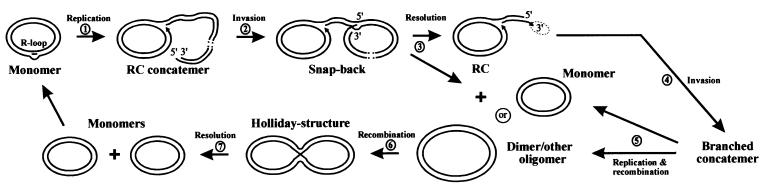
The formation of DNA concatemers requires an enzymatic machinery (e.g. a terminase and a debranching enzyme) that has not yet been reported for any mitochondrial genome. In this study, 357 uncut molecules were measured in which two or more mp1 monomeric circles appeared to be interconnected by a double-stranded tail (Figure 8A and B; Table I). These molecules suggest a mechanism for the resolution of head-to-tail concatemers. A new model proposed here invokes that the 5′ end of the tail invaded the next dso sequence of the same tail (1.3 kb away) to form a snap-back structure, a term describing the annealing of a palindromic ssDNA into a double-stranded form. This model is supported by the following observations. First, the fork position of 22% of all simple Ys in fraction 8 matched to dso1 (Figure 7A). Secondly, EM investigation of these snap-back junctions at high magnification revealed interconnected DNA strands in triangle structures (Figure 8C; arrowheads). After invasion, a replication fork may form and extension of the 3′ end begins for full replication of the dso (Figure 8D). The replicated dso sequence is nicked again, followed by strand exchange (Figure 8E, top). This leads to conversion of the replication fork into a Holliday junction, thereby forming protruding 3′ ends (Figure 8E, grey circles). Thirdly, in about half of the snap-back junctions, variable ssDNA ‘whiskers’ have indeed been detected that exceeded mp1 monomer size by ∼50–200 bases (Figure 8B and C, arrows). Finally, this junction can easily be resolved by the recombination machinery to release replicated circular monomer/oligomer units (Figure 8E, bottom). The remaining RC structure contains a free 3′ end at the tip of the tail, which may allow invasion of other molecules to trigger replication (see Figures 7E and 9).
Fig. 8. Model for the resolution of mp1 concatemers. (A and B) Two representative examples of mp1 in vivo circles interconnected by a double-stranded tail. Hypothetical schemes depict a snap-back mechanism using invasion of the ends of the RC tails (grey circles) to release mp1 monomer units (1.3 kb). Bar = 0.5 kb. (C) Hypothetical model and enlarged sections of snap-back junctions (arrowheads) that were recently regarded as strand-switching intermediates (Backert, 2000). The arrow shows a free ssDNA end originating from the junction. Bar = 100 bases. (D) Mp1 concatemers produced by RCR are head-to-tail molecules with specific ends (dso1 or dso2). The 5′ overhang of the RC tail may invade the next dso to form a snap-back molecule. (E) After invasion, a replication fork (shaded grey) is formed in which the 3′ end extends. Resolution of this intermediate proceeds by nicking of the dso motif and strand exchange. This results in a Holliday structure that can easily be resolved by the recombination machinery (arrows). Finally, circular mp1 mono- or oligomers are released from the replicating RC that retains a protruding 3′ end (grey circle).
Fig. 9. Hypothetical model for the replication cycle of the mt plasmid mp1. Mp1 produces long dsDNA concatemers by a unique RC mechanism that is mainly initiated at origin dso1. During processing of the concatemers into single plasmid units, RCs or linear molecules with invasive 3′ ends are produced that allow invasion of other mp1 structures similar to phage T4. This model is compatible with structures mapped in this study.
Discussion
Homologous recombination and replication are two of the most fundamental processes in living cells and are tightly interconnected (for reviews see Smith, 1988; Viret et al., 1991; Kowalczykowski et al., 1994; Kogoma, 1997; Cox, 1998). In this report, 2-DE and EM were combined to study replication and recombination intermediates of the mitochondrial plasmid mp1 from C.album. R-loops were identified as precursors of the RC structures. Structural evaluation of thousands of complex mp1 molecules resulted in a new model for the production and resolution of highly branched DNA concatemers in eukaryotes that showed similarities to the replication cycle of certain phages, bacterial RC plasmids and poxviruses.
Several lines of evidence for replication of mp1 according to a RC mechanism during the logarithmic growth stage have been found, such as the identification of σ-shaped intermediates, single-strand plasmid copies and two typical RC origins, dso1 and dso2 (Backert et al., 1996, 1998). Very early intermediates detected in this study revealed R-loops of 100–150 bases that mapped specifically to dso1, but sequence homology to known R-loops was not observed. In vitro studies demonstrated further that initiation of RCR was strongly dependent on the presence of NTPs. Consequently, RNA synthesis from ORF2 might be essential for initiation of RCR. As the ORF2 promotor is located 16 bp downstream of dso1, respective transcripts do not cross dso1. Thus, RNA synthesis could structurally support an initial intermediate by opening the dso sequence for nicking and initiation of unidirectional RCR (Figure 5E). Leading-strand replication of mp1 then initiates unidirectionally using the generated 3′ end in the DNA as primer and/or bound RNA as primer (either at the native 3′ end, an internal RNase cleavage site, or a 3′ end generated by a 3′→5′ exoribonuclease). Alternatively, RNA can be extruded from the template circle, for example when topoisomerase activity is limiting. Subsequent nicking of the template strand may convert this form into a RC.
The use of an RNA transcript to prime replication has been described in several other systems, including ColE1, phages T4 and T7, and mitochondria (Itoh and Tomizawa, 1980; Fuller and Richardson, 1985; Schinkel and Tabak, 1989; Kreuzer, 2000). Interestingly, phage-type RNA polymerases have been detected in the mitochondria that might be involved in mtDNA replication (Schinkel and Tabak, 1989; Hedtke et al., 1997). Similar to dso1 in mp1, RNA synthesis is required for initiation of phage λ replication at ori λ, but does not supply primers because it initiates as far as 95 bp downstream (Furth et al., 1982). Although the mechanism implied is still unclear, many of the late RCs also initiated at ori λ. The model proposed here can explain how RCR might be initiated in mp1 and phage λ. In contrast, R-loops have not been detected among bacterial RC plasmids (Khan, 1997; del Solar et al., 1998).
Many of the mp1 σ-structures had long tails, as known from classical phage λ replication (Takahashi, 1975) or RCR of chloroplast DNA (Williamson et al., 2001). However, linear concatemers are also produced during bacterial plasmid RCR in the absence of exonuclease V activity (Viret et al., 1991). In contrast to the situation with mp1 where replication is initiated at one or two distinct dsos, recombination-dependent replication in bacteria is not, in the main, initiated at specific origins. Investigation of highly branched mp1 concatemers showed that invasion of various plasmid molecules in other types of molecules (predominantly double RCs) strictly occurred in a dso-dependent manner. In agreement with this model, ss mtDNA stretches were detected at the tips of linear and RC molecules (Backert et al., 1996). Invasion resulted in D-loops from which replication was initiated, as initially described for phage T4. Here, free 3′ ends are produced by unreplicated ends of the linear T4 genome that invade the terminal redundancy of the same molecule, or homologous regions of others, to trigger replication (Mosig, 1998; Kreuzer, 2000). The generation of branched concatemers has also been described for vaccinia and other poxviruses (deLange and McFadden, 1990).
EM observations made with mp1 concatemers are unique so far and compatible with two hypothetical resolution mechanisms that may occur in parallel: a novel snap-back model and homologous recombination (Figure 9). The snap-back mechanism requires tandemly arrayed head-to-tail concatemer tails with specific ends (dso1 or dso2) in a RC or a linear molecule produced by RCR. The 5′ ends of the tails may invade the next dso of the tail to form a snap-back molecule. The large number of these molecules and their consistency at high pH (9–10.5) reflected their great stability during preparation. This widely excluded several other possibilities for the artificial generation of snap-back molecules (e.g. annealing and recombination of two RCs or two tails). Resolution of the snap-back structure may proceed by nicking at the dso, followed by interconversion of the replication fork into a Holliday junction. The model proposed in Figure 8 exactly reflected the structures observed by EM, and circular mp1 monomer/oligomer units can be released. The remaining RCs contain a new protruding 3′ end which allows invasion-triggered replication, similar to phage T4. In general, processing (and subsequent packaging) of ds phage concatemers occurs either precisely at a single unique sequence (e.g. cos in λ), at variable positions around a specific site (e.g. pac in P1 and P22) or at non-specific sequences such as phage T4 (Becker and Murialdo, 1990; Black, 1995; Catalano et al., 1995). However, the model of mp1 concatemer processing is novel and the dso shares no homology with cos/pac sequences.
Another important finding of this work are the recombining mp1 molecules. Holliday junctions were mainly mapped to the replication origin, indicating that dso1 is a recombination hotspot. Recombining intermediates were compatible with strand invasion but not with the resection-annealing mechanism. The invasion mechanism was proposed for conservative processes in meiotic recombination, yeast transformation, or bacterial or phage T4 recombination (e.g. Smith, 1988; Cox, 1998; Mosig, 1998). Surprisingly, the dso1 nicking sequence of mp1 shows homology to core sequences of the site-specific bacterial recombination system Xer, which converts plasmid and chromosomal dimers into monomers (Sherratt et al., 1995; Hodgman et al., 1998). Thus, it is tempting to speculate that mp1 may have adapted special replication/recombination functions from ancestor bacteria or bacteriophages to resolve concatemers into monomer units after replication.
In conclusion, data presented here have demonstrated unique mechanisms of replication and recombination of plasmid mp1 isolated from eukaryotic mitochondria. The results showed that mp1 shares the features of bacteria and bacteriophages. Several types of replication could coexist, including R-loop, D-loop and RCR combined with invasion and recombination events. The replication parallel to phage T4 is especially curious because T4 displays striking affinities with eukaryotes, such as type II 3′ ends topoisomerases and DNA polymerases as well as the mobile self-splicing introns that are remarkably similar to those found in the mtDNA of microbial eukaryotes. The mechanisms proposed here could serve as a model system to study replication and maintenance of concatemeric DNA in the mitochondria.
Materials and methods
Preparation of mitochondria
Mitochondria were isolated from a suspension culture of C.album (Backert et al., 1996). In order to minimize breakage of mtDNA molecules, purified mitochondria were prepared in low melting point (LMP)–agarose (Oldenburg and Bendich, 2001). mtRNA was isolated using the RNAzol™ kit (Biotecx Laboratories, Houston, TX).
Cloning of plasmid mp1
Plasmid mp1 (DDBJ/EMBL/GenBank accession No. X58911) was linearized with BamHI and ligated into vector pGEM3zf(+) (Promega, Madison, WI). A 148-bp BamHI–EcoRI fragment containing the replication origin dso1 was subcloned, and supercoiled DNA was purified using CsCl gradients.
In vitro mp1 replication assay
mtDNA polymerase was partially purified via successive chromatography steps on DEAE–cellulose, phosphocellulose and heparin–sepharose columns, and optimized for maximal incorporation of methyl-[3H]-TTP (Backert et al., 1998). The final mtDNA polymerase activity was step eluted for retaining the activities from DNA primase, RNA polymerase and topoisomerase I in the same peak. The standard replication assay (100 µl) contained: 50 mM Tris–HCl pH 7.5, 170 mM KCl, 12 mM MgCl2, 1 µg of plasmid template, 10 µl of the mitochondrial enzyme fraction, 10 µM each of CTP, GTP, ATP and TTP, 100 µM each of dGTP, dATP and dTTP, and 1.85 MBq [α-32P]dCTP. The reactions were incubated over a time course at 37°C and the amount of incorporated radioactivity was measured (Backert et al., 1998). For analysis by 2-DE, replication products were linearized with XmnI (Promega).
Two-dimensional (neutral/neutral) gel electrophoresis
2-DE was performed according to Brewer and Fangman (1987). The DNAs were separated for 25 h at 1 V/cm in the first dimension on 0.4% agarose gels in 1× TBE. Separation in the second dimension was performed on 1.0–1.5% agarose gels in 1× TBE with 0.3 µg/ml ethidium bromide at 5 V/cm for 4 h. In vivo-replicated plasmid DNA was visualized by hybridization with mp1. Gels with [32P]dCTP-labelled in vitro replication products were dried and exposed to X-ray film.
Electron microscopy
mp1 was separated from chromosomal mtDNA using a 0.4% LMP–agarose gel without ethidium bromide. Agarose blocks containing (i) total mp1 molecules (1–25 kb) and (ii) mp1 molecules from fractions 1–8 (Figure 1) were excised from gels, digested with gelase (Biozym, Hameln, Germany) and prepared for transmission EM (E400T, Philips, Eindhoven, The Netherlands). Plasmid DNA (100 ng) was resuspended in carbonate buffer in the presence of formamide (Backert et al., 1996). For mapping experiments, mp1 was digested with PvuII, SmaI or BglI (NEB, Beverly, CA). For visualization of ssDNA, mp1 was coated with SSB protein from E.coli (Backert et al., 1996). For analysis of the mp1 loop, 50 ng of mp1 eluted from spot 1 (Figure 1) were treated with RNace-it ribonucleases (Stratagene, La Jolla, CA), RNaseH or λ-exonuclease (NEB). Internal standard was phage ΦX174 DNA (5386 bases).
Acknowledgments
Acknowledgements
I wish to thank Drs Thomas Börner, Rudi Lurz and Brent L.Nielsen for support over many years, and Drs Juan Alonso and Gisela Mosig for helpful discussions. This work was funded by a post-doctoral fellowship of the German Science Foundation to S.B.
References
- Backert S. (2000) Strand switching during rolling circle replication of plasmid-like DNA circles in the mitochondria of the higher plant Chenopodium album (L.). Plasmid, 43, 166–170. [DOI] [PubMed] [Google Scholar]
- Backert S., Dörfel,P., Lurz,R. and Börner,T. (1996) Rolling-circle replication in mitochondria of the higher plant Chenopodium album (L.). Mol. Cell. Biol., 16, 6285–6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Meissner,K. and Börner,T. (1997) Unique features of the mitochondrial rolling-circle plasmid mp1 from the higher plant Chenopodium album (L.). Nucleic Acids Res., 25, 582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Kunnimalaiyaan,M., Börner,T. and Nielsen,B.L. (1998) In vitro replication of mitochondrial plasmid mp1 from the higher plant Chenopodium album (L.): a remnant of bacterial rolling circle and conjugative plasmids? J. Mol. Biol., 284, 1005–1015. [DOI] [PubMed] [Google Scholar]
- Becker A. and Murialdo,H. (1990) Bacteriophage λ DNA: the beginning of the end. J. Bacteriol., 172, 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S., Marchfelder,A. and Brennicke,A (1996) Regulation of gene expression in plant mitochondria. Plant Mol. Biol., 32, 303–314. [DOI] [PubMed] [Google Scholar]
- Black L.W. (1995) DNA packaging and cutting by phage terminases: control in phage T4 heads. BioEssays, 17, 1025–1030. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman,W.L. (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- Catalano C.E., Cue,D. and Feiss,M. (1995) Virus DNA packaging: the strategy used by phage λ. Mol. Microbiol., 16, 1075–1086. [DOI] [PubMed] [Google Scholar]
- Cox M.M. (1998) A broadening view of recombinational DNA repair in bacteria. Genes Cells, 3, 65–78. [DOI] [PubMed] [Google Scholar]
- deLange A.M. and McFadden,G. (1990) The role of telomers in poxvirus DNA replication. Curr. Top. Microbiol. Immunol., 163, 71–93. [DOI] [PubMed] [Google Scholar]
- del Solar G., Giraldo,R., Ruiz-Echeverria,M.J., Espinosa,M. and Diaz-Orejas,R. (1998) Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev., 62, 434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H. and Morita,M. (1997) Phage DNA packaging. Genes Cells, 2, 537–545. [DOI] [PubMed] [Google Scholar]
- Fuller C.W. and Richardson,C.C. (1985) Initiation of DNA replication at the primary origin of bacteriophage T7 by purified proteins: initiation of bidirectional synthesis. J. Biol. Chem., 260, 3197–3206. [PubMed] [Google Scholar]
- Furth M.E., Dove,W.F. and Meyer,B.J. (1982) Specificity determinants for bacteriophage λ DNA replication. J. Mol. Biol., 154, 65–83. [DOI] [PubMed] [Google Scholar]
- Hedtke B., Börner,T. and Weihe,A. (1997) Mitochondrial and chloroplast phage-like RNA polymerases in Arabidopsis. Science, 277, 809–811. [DOI] [PubMed] [Google Scholar]
- Hodgman T.C., Griffiths,H. and Summers,D.K. (1998) Nucleoprotein architecture and ColE1 dimer resolution: a hypothesis. Mol. Microbiol., 29, 545–558. [DOI] [PubMed] [Google Scholar]
- Itoh T., and Tomizawa,J.-I. (1980) Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl Acad. Sci. USA, 77, 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.A. (1997) Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev., 61, 442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination and transcription. Microbiol. Mol. Biol. Rev., 61, 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S.C., Dixon,D.A., Eggleston,A.K., Lauder,S.D. and Rehraurer,W.M. (1994) Biochemistry of homologous recombination in E. coli. Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer K.N. (2000) Recombination-dependent DNA replication in phage T4. Trends Biochem. Sci., 25, 165–173. [DOI] [PubMed] [Google Scholar]
- Lockshon D., Zweifel,S.G., Freeman-Cook,L.L., Lorimer,H.E., Brewer,B. and Fangman,W. (1995) A role of recombination junctions in the segregation of mitochondrial DNA in yeast. Cell, 81, 947–955. [DOI] [PubMed] [Google Scholar]
- Maleszka R., Skelly,P.J. and Clark-Walker,G.D. (1991) Rolling circle replication in yeast mitochondria. EMBO J., 10, 3923–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig G. (1998) Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev. Genet., 32, 379–413. [DOI] [PubMed] [Google Scholar]
- Oldenburg D.J. and Bendich,A.J. (2001) Mitochondrial DNA from the liverwort Marchantia polymorpha: circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J. Mol. Biol., 310, 549–562. [DOI] [PubMed] [Google Scholar]
- Preiser P.R., Wilson,R.J.M., Moore,P.W., McCready,S., Hajibagheri,M.A.N., Blight,K.J., Strath,M. and Williamson,D.H. (1996) Recombination associated with replication of malarial mitochondrial DNA. EMBO J., 15, 684–693. [PMC free article] [PubMed] [Google Scholar]
- Richardson J.P. (1975) Attachment of nascent RNA molecules to superhelical DNA. J. Mol. Biol., 98, 565–579. [DOI] [PubMed] [Google Scholar]
- Schinkel A.H. and Tabak,H.F. (1989) Mitochondrial RNA polymerase: dual role in transcription and replication. Trends Genet., 5, 149–154. [DOI] [PubMed] [Google Scholar]
- Sherratt D.J., Arciszewska,L.K., Blakely,G., Colloms,S., Grant,K., Leslie,N. and McCulloch,R. (1995) Site-specific recombination and circular chromosome segregation. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 347, 37–42. [DOI] [PubMed] [Google Scholar]
- Smith G.R. (1988) Homologous recombination in procaryotes. Microbiol. Rev., 52, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S. (1975) The starting point and direction of rolling-circle replication intermediates of coliphage λ DNA. Mol. Gen. Genet., 142, 137–153. [DOI] [PubMed] [Google Scholar]
- Tracy R.L. and Stern,D.B. (1995) Mitochondrial transcription initiation: promotor structures and RNA polymerases. Curr. Genet., 28, 205–216. [DOI] [PubMed] [Google Scholar]
- Viret J.F., Bravo,A. and Alonso,J.C. (1991) Recombination-dependent concatemeric plasmid replication. Microbiol. Rev., 55, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.D. (1972) Origin of concatemeric T7 DNA. Nat. New Biol., 239, 197–201. [DOI] [PubMed] [Google Scholar]
- Williamson D.H., Denny,P.W., Moore,P.W., Sato,S., McCready,S. and Wilson,R.J. (2001) The in vivo conformation of the plastid DNA of Toxoplasma gondii: implications for replication. J. Mol. Biol., 306, 159–168. [DOI] [PubMed] [Google Scholar]