Abstract
ABI1, a protein phosphatase 2C, is a key component of signal transduction in Arabidopsis. It regulates diverse responses to the phytohormone abscisic acid (ABA) such as stomatal closure, seed dormancy and inhibition of vegetative growth. By analysing proteins capable of interacting with ABI1, we have identified the homeodomain protein ATHB6 as a regulator of the ABA signal pathway. Critical for interaction between ATHB6 and ABI1 is an intact protein phosphatase domain and the N-terminal domain of ATHB6 containing the DNA-binding site. ATHB6 recognizes a cis-element present in its promoter, which encompasses the core motif (CAATTATTA) that mediated ATHB6- and ABA-dependent gene expression in protoplasts. In addition, transgenic plants containing a luciferase gene controlled by the ATHB6 promoter documented a strong ABA-inducible expression of the reporter which was abrogated in the ABA-insensitive abi1 mutant. Arabidopsis plants with constitutive expression of the transcriptional regulator revealed ABA insensitivity in a subset of ABI1-dependent responses. Thus, the homeodomain protein ATHB6 seems to represent a negative regulator of the ABA signal pathway and to act downstream of ABI1.
Keywords: ABI1/abscisic acid/ATHB6/homeodomain-leucine zipper protein/PP2C
Introduction
The phytohormone abscisic acid (ABA) plays a central role in regulating plant responses to adverse environmental cues including water shortage, high osmolarity and low temperature (Leung and Giraudat, 1998). Those stress situations generally result in an elevation of the plant’s ABA levels, which leads to physiological adaptations comprising stomatal closure, growth inhibition and differential gene regulation for metabolic and developmental adjustment. Stomatal closure is induced by ABA-mediated osmoregulation of guard cells via ion fluxes through cation and anion channels localized at the plasmalemma and tonoplast (Schroeder et al., 2001). H2O2 (Pei et al., 2000), cyclic ADP-ribose (Wu et al., 1997; Leckie et al., 1998) and phospholipid-derived signals (Lemtiri-Chlieh et al., 2000; Ng et al., 2001) seem to contribute in the relay of the ABA stimulus, which generally leads to increases and oscillations of cytosolic Ca2+ levels (Staxen et al., 1999; Allen et al., 2001). In addition, protein kinases (Mori and Muto, 1997; Li et al., 2000) and protein phosphatases are involved in ABA-triggered stomatal closing (Grabov et al., 1997; Allen et al., 1999).
ABA functions as a key regulator of differential gene expression during adaptation to low water potentials and in developmental processes such as seed maturation (Busk and Pages, 1998). A plethora of transcriptional regulators emerged as targets of ABA signalling events and comprise members of the basic leucine zipper (bZIP) class (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Uno et al., 2000), AP2 (Finkelstein et al., 1998), basic helix–loop–helix (bHLH) (Abe et al., 1997), B3 (Suzuki et al., 1997) and homeodomain (HD) proteins containing a leucine zipper (HD-Zip) (Söderman et al., 1996, 1999). The G-box-like motif (C/TACGT GGC) acts as an ABA-responsive regulatory element (ABRE) with which specific bZIP transcription factors interact (Uno et al., 2000).
The ABA signal relay results in diverse physiological responses that are controlled by the central regulators ABI1 and ABI2 (Himmelbach et al., 1998; Leung and Giraudat, 1998) as well as AtPP2CA (Tähtiharju and Palva, 2001). The three proteins belong to the strictly Mg2+-dependent class 2C of serine/threonine protein phosphatases (PP2Cs). The Arabidopsis mutants abi1 and abi2 reveal a dominant ABA-insensitive phenotype due to an identical mutation of the PP2Cs within the catalytic domain that renders abi1 (ABI1Gly180Asp) and abi2 (ABI2Gly168Asp) deficient in catalytic activity (Leung et al., 1997; Leube et al., 1998; Rodriguez et al., 1998). Intragenic revertants of abi1 and abi2 with a complete loss of PP2C activity revealed an ABA-hypersensitive phenotype (Gosti et al., 1999; Merlot et al., 2001). This finding, transient expression studies (Sheen, 1998) and analysis of transgenic antisense plants (Tähtiharju and Palva, 2001) characterized PP2Cs as negative regulators of the ABA signal pathway. While AtPP2CA has been shown to interact with the K+-channel protein AKT3 (Vranová et al., 2001), no intracellular regulators or substrates of ABI1 and ABI2 have been identified.
In this study, we present the analysis of the transcriptional regulator ATHB6 as a target of ABI1 that links PP2C with gene regulation. ATHB6 belongs to the plant-specific HD-Zip class. Gene expression of ATHB6 is up-regulated by ABA and during drought stress (Söderman et al., 1999). Our analysis now reveals physical interaction of ATHB6 with ABI1. The interaction between ATHB6 and ABI1 positively correlated with the PP2C activity of the ABI1 catalytic domain and was completely abolished in a point-mutated, catalytically inactive ABI1. Stable expression analysis documented a >2000-fold ABA-mediated induction of reporter expression under the control of the ATHB6 promoter that was dependent on ABI1. The transcription factor targets the pseudopalindromic core motif (CAATTATTA) present in its own promoter that mediated ATHB6- and ABA-dependent gene expression in a transient system. Arabidopsis lines mimicking the ABA-induced state of ATHB6 expression by ectopic expression of ATHB6 displayed a reduced sensitivity towards ABA during seed germination and stomatal closure. The finding argues for a role for ATHB6 in adjusting hormonal sensitivity. In addition, the analyses reveal a transcriptional regulator as a novel interaction partner for PP2C.
Results
Identification of ATHB6 as an interaction partner of ABI1
In order to identify proteins interacting with ABI1, the yeast two-hybrid system was used (Bartel and Fields, 1995). Preliminary experiments revealed that ABI1 fused to the GAL4 DNA-binding domain (DB) resulted in activation of lacZ reporter expression (data not shown). N-terminal truncation of ABI1 (positions 1–120), however, reduced the potential to activate transcription to low background levels (Figure 1A). The modified ABI1 was used as a bait to screen an Arabidopsis expression library containing random cDNAs fused to the GAL4 activation domain (AD). From 2.5 × 106 colonies screened, two positive clones were isolated. In both cases, lacZ activation and histidine autotrophy were dependent on the expression of the cDNA fusion. DNA sequence analysis revealed that both clones were encoding N-terminally deleted versions of the HD-Zip protein ATHB6 (amino acids 44–311). In order to clarify the specificity of the interaction, we examined the full-length cDNA of ATHB6 and of two structurally related Arabidopsis HD-Zip proteins, ATHB5 and ATHB7 (Söderman et al., 1996, 1999), as GAL4 AD fusions in the yeast system (Figure 1A). The activation of the reporter gene was evident in the presence of ABI1 and ATHB6, while the other HD-Zip proteins revealed only background levels, indicative of a specific recognition of ATHB6 by ABI1.
Fig. 1. Interaction of ATHB6 with ABI1 in the yeast two-hybrid system and in vitro. (A) ABI1 fused to the GAL4 DNA-binding domain (DB fusion) was analysed for interaction with fusions of ATHB6, ATHB5 and ATHB7 to the GAL4 activation domain (AD fusion), respectively. Cells transformed with empty vectors for AD and DB fusion (C) were used as control. The β-galactosidase activity and the standard deviation are presented as the relative values of three independent experiments. (B) The in vitro binding of ATHB6 and ABI1 protein was performed by affinity interaction. ATHB6 fused to MBP (MBP–ATHB6) or MBP as a control were immobilized on amylose resins. Binding of ABI1 was analysed by chromatography of radiolabelled ABI1 on a column containing the charged resins. Fractions were collected and quantified for radioactivity. Elution of the ATHB6–ABI1 complex was initiated by administration of maltose-containing solution indicated by an arrow, and 32% of applied ABI1 was recovered subsequently (continuous line). ABI1 chromatography on MBP-charged resin yielded a recovery of 3.8% of applied ABI1 in the elution fractions (dashed line). (C) Purified proteins used in the in vitro binding assay were separated by SDS–PAGE and visualized by silver staining. In addition, MBP and MBP–ATHB6 were identified by immunodetection using antibodies directed against MBP, and the radiolabelled ABI1 protein fraction was analysed by autoradiography. The positions and molecular weight of protein size markers are indicated on the left.
Subsequently, the physical interaction of both proteins was corroborated by in vitro binding experiments. In this analysis, radiolabelled ABI1 was tested for interaction with ATHB6 during affinity chromatography (Figure 1B and C). ATHB6 was tethered via a maltose-binding protein fusion (MBP–ATHB6) to amylose beads, and binding of radiolabelled ABI1 was determined by recovery of radiolabel after several washing steps and elution of bound protein complexes. A total of 32% of applied ABI1 was recovered in elution fractions containing ATHB6. In control experiments providing only MBP as interaction partner for ABI1, recovery of ABI1 in the elution fraction was <4%. Similar results were obtained with pull-down assays of ABI1 and MBP–ATHB6. Recovery of radiolabelled ABI1 from amylose beads yielded 4.8% after extensive washing steps, while only background levels of ABI1 (0.3%) were detected in a control experiment by replacing MBP–ATHB6 with MBP. These in vitro data validate the in vivo analysis and support a bona fide interaction.
Protein domains critical for ABI1 and ATHB6 interaction
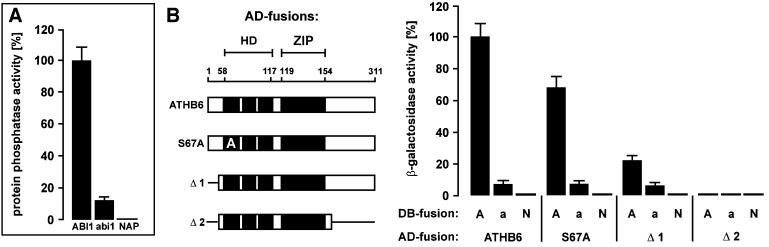
Recently, the interaction of a PP2A with an HD protein has been characterized to reflect a substrate–enzyme interference (Berry and Gehring, 2000). In order to examine the role of the catalytic PP2C domain in the interaction with ATHB6, two point-mutated forms of ABI1 were tested in the yeast system: abi1 and a non-active protein phosphatase (NAP). The abi1 (ABI1Gly180Asp) contains a glycine residue within the catalytic domain exchanged for aspartic acid that interferes with Mg2+ binding and renders the PP2C strongly diminished in enzymatic activity (Leube et al., 1998). NAP (ABI1Asp177Ala) lacks an aspartic acid residue within the catalytic cleft of PP2Cs essential for Mg2+ co-ordination (Das et al., 1996). The enzymatic activity of abi1 and NAP were 13 and 0% of that of wild-type protein, respectively (Figure 2A). The yeast analysis yielded a strongly reduced level of ATHB6-dependent lacZ activation for abi1 compared with ABI1, with a residual level of 7% (Figure 2B). In agreement with this observation, elimination of the PP2C activity in NAP resulted in a complete failure to transactivate the reporter in the interaction analysis (Figure 2B). The experiments document the requirement for either a functional catalytic domain of ABI1 or its intact topology for binding of ATHB6.
Fig. 2. Dependence of ABI1 and ATHB6 interaction on the functional catalytic domain of PP2C. (A) Specific protein phosphatase activity of purified ABI1, and both point-mutated forms abi1 (ABI1Gly180Asp) and non-active ABI1Asp177Ala (NAP) were measured and expressed as relative activities (n = 3, ± SD). (B) The homeodomain (HD) and leucine zipper (ZIP) regions of the AD fusion for ATHB6 (amino acids 1–311) and mutant versions thereof [Δ1 (amino acids 44–311), Δ2 (amino acids 44–217) and S67A (ATHB6Ser67Ala)] are presented schematically (left panel). They were analysed for binding to DB fusions of ABI1 (A), abi1 (a) and NAP (N) in the yeast two-hybrid system. The β-galactosidase activities for combination with the empty AD vector were subtracted as background from the values presented.
In order to identify contact sites of ATHB6 with ABI1, several modified versions of ATHB6 were examined for interaction (Figure 2B). Deletion of the N-terminal part (positions 1–43 in Δ1) reduced the lacZ reporter activity to a residual level of 25%. Additional deletion of the DNA-contacting α-helix 3 region of the HD (Gehring et al., 1994) had no further effect (data not shown), while additional truncation of the C-terminus (Δ2, Figure 2B) completely abolished the reporter gene activation to background levels. Throughout the analysis, interaction levels with abi1 and NAP fusions were reduced in comparison with ABI1, corresponding to the diminished enzymatic activity of the mutated PP2Cs. In conclusion, the N- and C-terminal parts of ATHB6 seem to constitute major determinants for ABI1 interaction.
In view of the above results, it is conceivable that ATHB6 serves as a substrate of ABI1. N-terminal phosphorylation in the proximity of the DNA-binding sites frequently fulfils a regulatory function of transcription factors (Hunter and Karin, 1992) and seems also to regulate transcriptional ABA response factors (Uno et al., 2000). In silico analysis of ATHB6 for putative serine/threonine phosphorylation sites revealed a prominent target serine residue (S67) in the consensus motif KRRLSINQV (Blom et al., 1999) immediately adjacent to HD α-helix 1. Mutation of the predicted phosphorylation site to an alanine residue (S67A) and subsequent analysis of the modified ATHB6 for protein interaction with ABI1 revealed a moderate reduction of the interaction by ∼30% (Figure 2B).
Characterization of ATHB6 as a transcriptional activator
The major DNA interaction site of HD proteins is formed by α-helix 3 of the homeobox, which frequently establishes contacts to AT-rich cis-elements (Gehring et al., 1994). Analysis of the plant ATHB1 protein characterized the 9 bp pseudopalindromic core sequence (CAATTA TTG) as a recognition element (Sessa et al., 1993). The primary structure of ATHB1 and ATHB6 within the α-helix 3 (residues 42–58 of HD) is identical and points to a similar contact site. Analysis of the promoter region of ATHB6 revealed a sequence (CAATTATTA) almost identical to the ATHB1-binding motif, which is located at position –620 upstream of the predicted transcriptional start site. A 30mer oligonucleotide (oligo α) of the promoter region encompassing the AT-rich sequence was analysed for ATHB6 binding by gel retardation assays. Purified MBP–ATHB6 fusion protein specifically formed a complex with oligo α in the analysis (Figure 3). The complex between MBP–ATHB6 and oligo α was destabilized by increasing the salt concentration of the sample solution indicative of a non-covalent interaction. No binding was observed in controls using either oligo β, which was identical in sequence to oligo α except for a single base substitution at the invariable core motif, or exchanging MBP–ATHB6 for MBP. In addition, ATHB6 phosphorylation by protein kinase A (PKA) reduced binding to the cis-element >10-fold in vitro, as revealed by gel retardation assay (Figure 3). Thus, binding of ATHB6 to the promoter fragment oligo α in vitro reflects specific interaction via the CAATTATTA sequence. The finding suggests phosphorylation-dependent interference of ATHB6 with its own promoter containing the binding site.

Fig. 3. Specific binding of ATHB6 to a cis-element present in the ATHB6 promoter. The 30mer oligomeric DNA fragment (oligo α) contains a putative cis-element located between position –638 and –609 of the ATHB6 promoter. The core interaction motif (bold) is mutated in oligo β by the transition of A to G (underlined). ATHB6 fused to MBP and MBP as control were analysed for specific DNA binding to oligo α versus oligo β. Complexes of ATHB6 and DNA (arrowhead) were tested in the presence of increasing ionic strength (0, 50, 100 and 500 mM KCl). PKA-phosphorylated MBP–ATHB6 was tested for binding to oligo α in the absence of KCl. Complexes were separated from unbound radiolabelled DNA by EMSA and visualized by autoradiography.
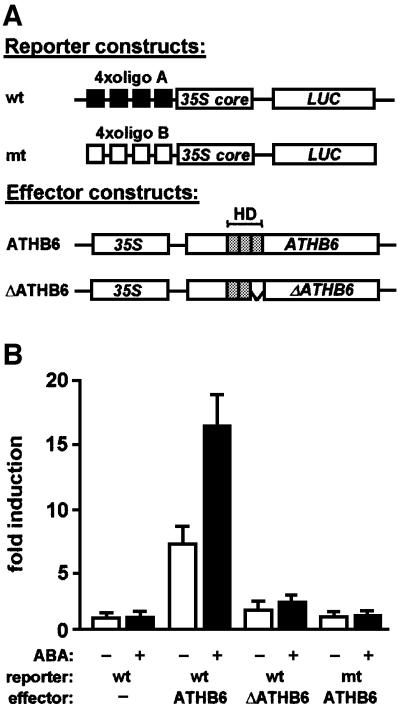
In order to elucidate the role of ATHB6 in targeting the cis-element, four binding sites were fused in tandem orientation to a minimal –46 cauliflower mosaic virus (CaMV) 35S promoter that controls expression of firefly luciferase (LUC). Regulation of the reporter gene by ATHB6 in a transient expression system was expected to define the role of the HD protein as a transcriptional activator or repressor. Arabidopsis protoplasts were transfected with both the promoter–LUC reporter and effector DNA, providing constitutive expression of ATHB6 (Figure 4A). In addition, the cells were co-transfected with an aequorin reporter gene for standardization of LUC expression. Subsequently, protoplasts were incubated in the absence or presence of ABA (30 µM). Analysis of LUC activity revealed a 7-fold induction of reporter expression by co-expression of ATHB6 (Figure 4B). In the presence of ABA and ectopically expressed ATHB6, normalized LUC activity increased 17-fold compared with the control. The corresponding reporter construct with the point-mutated binding sites (CAATTGTTA) yielded only background levels irrespective of ABA and ectopically expressed ATHB6. Likewise, deletion of the HD α-helix 3 of ATHB6 (ΔATHB6) resulted in background levels of reporter expression. Thus, ATHB6 represents an activator of transcription in these analyses that specifically recognizes a binding site present in its own promoter region.
Fig. 4. Requirement for a functional cis-element and ATHB6 for ABA-mediated promoter activation in Arabidopsis protoplasts. (A) The reporter constructs (wt and mutant) contain four tandemly orientated copies of the ATHB6-binding sequence (CAATTATTA, oligo A) or the point-mutated sequence (CAATTGTTA, oligo B), respectively, fused upstream to the CaMV 35S core promoter (35S core) that controls expression of firefly luciferase (LUC). They are represented schematically together with the effector constructs that allow constitutive expression of ATHB6 and ΔATHB6 under control of the 35S promoter (35S). In ΔATHB6, the α-helix 3 (amino acids 96–117) of the homeodomain (HD) essential for DNA binding was deleted. The positions of HD helices are indicated by shaded boxes. (B) The ATHB6-dependent activation of reporter constructs was tested in a transient gene expression system. Transfected Arabidopsis protoplasts were incubated for 24 h in the presence (+) or absence (–) of 30 µM ABA and analysed for expression of the LUC reporter. Co-transfection of a constitutively expressed aequorin gene allowed normalization of expression in independent experiments. The induction of LUC activity from wild-type and mutant reporter constructs by different effectors is shown relative to the expression from wild-type reporter in the absence of ectopically expressed effector. The data reflect the results of three independent transfections ± SD.
Functional analysis of ATHB6 in transgenic Arabidopsis
The previous results imply a role for ATHB6 as a transcriptional regulator. Due to its interaction with ABI1, a key regulator of ABA responses, ATHB6 possibly represents a master switch to ABA-specific developmental adaptations. The reported ABI1-dependent up-regulation of ATHB6 mRNA abundance (Söderman et al., 1999) may result from ATHB6 interaction with its own promoter. In consequence, ATHB6 accumulation could mediate or modify ABA responses.
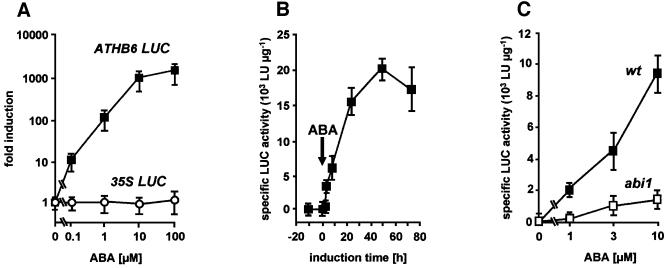
A first step to test the idea was the generation of suitable transgenic plants to define more clearly the effect of ABA and ABI1 on the promoter activity of ATHB6. An Arabidopsis reporter line was established that expressed the LUC gene under the control of the ATHB6 promoter. The reporter analysis documented a remarkable dependence of LUC activity present in the transgenic line on the presence of exogenous ABA (Figure 5A). Reporter expression increased with increasing ABA concentrations, and induction levels beyond a factor of 2000 were recorded. The maximal expression observed (1.7 × 104 LU/µg) even surpassed the expression level of the reporter under the control of the strong 35S promoter (1.3 × 104 LU/µg). Reporter activation was detectable within 4 h of ABA addition (10 µM), reaching half-maximal level after ∼12 h (Figure 5B). For analysis of ABI1-dependent reporter activation, the reporter line was crossed with the dominant abi1 mutant and wild-type Arabidopsis. To avoid segregational variation of ABA sensitivities of the different parental lines, the analysis was performed with bulked F2 seedlings. The expression of LUC in seedlings of the abi1 cross was reduced at all ABA concentrations examined by a factor of at least 4 compared with those of the wild-type cross (Figure 5C). The Mendelian segregation of abi1 results in a 25% fraction of seedlings lacking the dominant mutant trait. Thus, the reporter activity in the abi1 cross observed probably stems from non-mutant seedlings and, therefore, the mutant abi1 protein appears to block activation of the ATHB6 promoter efficiently.
Fig. 5. ABA- and ABI1-dependent activation of the ATHB6 promoter. The regulation of the ATHB6 promoter was analysed by stable integration of an ATHB6 promoter driving LUC expression in Arabidopsis. (A) The specific LUC activity of seedlings homozygous for the ATHB6 reporter construct and the 35S LUC control was determined from extracts 24 h after ABA administration, respectively. Values are expressed as fold induction of specific LUC activity relative to the untreated control. (B) The time-dependent increase of specific LUC activity in the transgenic reporter line by 10 µM ABA. The arrow signifies the time point of challenge to exogenous ABA. (C) The ABI1 dependence of the ABA-mediated reporter activation was studied 24 h after ABA addition in bulked F2 seedlings from crosses of the reporter line to the ABA-insensitive abi1 mutant (abi1) and to the wild-type (wt) of the same ecotype to avoid ecotype-specific differences in ABA sensitivity. The Mendelian segregation of the dominant mutant trait results in a quarter of the seedlings of the F2 population expressing LUC in a wt ABI1 background. Specific activity is expressed as light units (LU) per µg of protein. The standard deviation among four independent experiments is indicated.
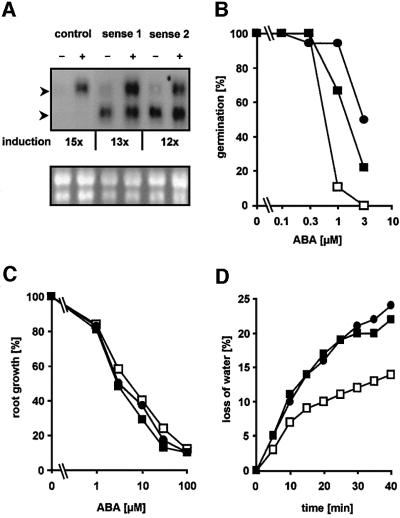
The strong ABA-dependent induction of the ATHB6 promoter with a strength comparable to that of the 35S promoter prompted us to generate plants with a pre-induced state of ATHB6. Transgenic plants were generated that constitutively express the ATHB6 gene under the control of the 35S promoter in order to subsequently analyse ABA-dependent gene regulation and ABA responses. Two independent lines homozygous for the transgene were randomly selected. Northern blot analysis confirmed the presence of the ectopic transcript discernible by its shorter length of the 5′- and 3′-untranslated mRNA sequences compared with the endogenous ATHB6 message (Figure 6A).
Fig. 6. Analysis of ABA responses in Arabidopsis plants with ectopic expression of ATHB6. (A) Northern blot analysis for ATHB6-specific RNA abundance in two transgenic Arabidopsis lines (sense 1 and sense 2) ectopically expressing ATHB6, and in control plants in which the ATHB6 structural gene had been replaced by the GUS coding sequence (control). Total RNA (20 µg) of seedlings treated with (+) or without (–) 10 µM ABA for 24 h was separated by electrophoresis and probed for the presence of ATHB6 transcripts (upper panel). The position of endogenous ATHB6 and ectopically expressed mRNA is indicated by the upper and lower arrowhead, respectively. The transcripts differ in size due to deletions in the untranslated leader sequences of the ATHB6 RNA constitutively expressed from the 35S promoter. The level of endogenous transcript induction by ABA is stated below and is corrected for equal RNA loading, which was determined by imaging and quantifying rRNA bands of the RNA samples stained with ethidium bromide (lower panel). (B) Fraction of seeds germinating in the presence of ABA after 5 days determined from a total of 80 seeds. (C) Five-day-old seedlings grown under sterile conditions were transferred on solid medium with various ABA concentrations. Root growth within 4 days of transfer was determined (n = 30, SD ± 14%). In the absence of ABA, root growth equalled 15 ± 1, 14 ± 2 and 14 ± 1 mm for the control, sense 1 and sense 2 lines, respectively. (D) Stomatal response measured by water loss of excised leaves. Leaves of a comparable developing stage (n = 5) from 4-week-old plants were excised and the loss of the fresh weight was measured at ambient conditions. The values are indicated for the control (open squares), as well as for the transgenic sense 1 (closed squares) and sense 2 (closed circles) lines.
Interestingly, overexpression of the ATHB6 transgene did not interfere with regulation of the endogenous transcript. The expression level of the endogenous ATHB6 gene in the absence of exogenous ABA was low but appeared not to be altered in the overexpressing lines. ABA treatment (10 µM) resulted in comparable induction levels of the endogenous ATHB6 transcript in constitutively expressing lines and in control plants by a factor of ∼12 and 15, respectively. Additional analysis of mRNA abundance for the ABA-regulated genes rd29b and rab18 (Busk and Pages, 1998) failed to reveal deregulated expression in the ATHB6 lines irrespective of the ABA concentration (data not shown). The level of the ectopically expressed transcript remained unchanged. Therefore, constitutive expression of ATHB6 per se did not affect endogenous ATHB6 regulation.
The ATHB6 transgenic lines were analysed for the physiological responses of ABA-mediated inhibition of both germination and vegetative growth as well as for control of stomatal aperture (Figure 6B–D). ABA action on seed germination and vegetative growth involves different signalling components (Himmelbach et al., 1998). All three physiological responses are easy to score and provide a means to characterize deregulated signalling components by interference with the ABA response spectrum.
Both ATHB6 transgenic seed batches and control seed material were examined for inhibition of germination by ABA. Clearly, seeds of the overexpression lines were more insensitive to ABA than the controls (Figure 6B). Half-maximal inhibition by exogenous ABA was observed at 0.6 µM ABA in seeds of the control, while the IC50 value for seed batches of both ATHB6 lines was shifted by a factor of 3 and 5 to higher ABA concentrations. Interestingly, inhibition of vegetative growth by ABA as analysed by root expansion was identical in all lines tested (Figure 6C).
The analysis of stomatal regulation, however, supports a role for ATHB6 in vegetative responses. Transpiration is controlled by stomatal aperture, which is reduced by ABA, and altered stomatal responses of ABA-insensitive or -hypersensitve mutants to ABA are mirrored in enhanced or reduced water loss of detached leaves, respectively (Meyer et al., 1994; Pei et al., 1998). Leaves of the same age and size either from ATHB6-overexpressing plants or from control transformed lines were tested (Figure 6D). Surprisingly, the rate of water loss in leaves from both ATHB6-overexpressing lines was increased ∼2-fold compared with the control and reached a level comparable to the severe wilty mutant abi1 (Meyer et al., 1994). Microscopic examination of stomatal pores of detached leaves corroborated the observation. Immediately after leaf excision, stomatal aperture of the ATHB6 lines averaged 2.8 ± 0.7 and 3.0 ± 0.7 µm (n = 30), which changed to 2.5 ± 0.5 and 2.5 ± 0.3 µm, respectively, within 30 min. In the wild-type control and in the abi1 mutant, stomatal pore widths were initially 3.0 ± 0.6 and 4.1 ± 1.1 µm (n = 30), respectively, which closed to 0.5 ± 0.2 and 2.5 ± 0.5 µm after 30 min of leaf detachment. In conclusion, the deregulated expression of ATHB6 decreased the sensitivity towards ABA in two out of three responses that are controlled by ABI1.
Discussion
Homeobox proteins represent master control switches involved in developmental processes and are ubiquitous in higher organisms. They function as transcriptional regulators that are characterized by an evolutionarily conserved HD responsible for specific DNA binding (Gehring et al., 1994). In plants, two major classes have been identified: the HD class represented by KNOTTED1 (Vollbrecht et al., 1991) and the family of HD-Zip proteins (Schena and Davis, 1992). The HD-Zip proteins are characterized by an additional leucine zipper motif adjacent to the HD which facilitates homo- and heterodimerization of the transcriptional regulators (Johannesson et al., 2001). Functional characterization of some members supports a role for HD-Zip proteins as key regulators of adaptational responses including developmental adjustment to environmental cues such as light, pathogens and water stress (Mayda et al., 1999; Steindler et al., 1999). Drought, high osmolarity and ABA induce gene expression of HD-Zip ATHB6 (Söderman et al., 1999), ATHB7 (Söderman et al., 1996) and ATHB12 (Lee et al., 2001) from Arabidopsis.
Physical interaction of ABI1 and ATHB6
PP2As have been recognized to target HD transcription factors such as the human HOX11 (Kawabe et al., 1997) and SCR from Drosophila (Berry and Gehring, 2000). PP2C targeting of a transcription factor represents a novel paradigm. The specificity of the interaction between ABI1 and ATHB6 is reflected by in vivo analysis in yeast and transgenic plants as well as by in vitro binding assays. It is noteworthy that protein–protein interaction analysis with ABI1 and transcriptional regulators involved in ABA responses, such as ABI3, ABI4 and ABI5, failed to reveal binding to the PP2C (Nakamura et al., 2001). The analysis of ABI1 and ATHB6 in the yeast two-hybrid system disclosed the necessity of a functional protein phosphatase domain for interaction. In addition, a critical serine residue within the homeobox of ATHB6 and additional structural features located outside the HD-Zip motif contribute to the interaction. Truncation of the N-terminal region up to the HD and deletion of the C-terminal part negatively affected binding to ABI1. It is possible that these regions are essential for structural integrity. Deletion of the HD α-helix 3 required for DNA binding, however, did not affect protein–protein interaction, and indicates that ABI1 does not recognize the topology of the DNA-binding domain of ATHB6.
The ability of ABI1 to interact with ATHB6 in yeast was strictly correlated with the PP2C activity of ABI1. Reduction of enzymatic activity by a single point mutation reduced the interaction of ABI1 with the transcription factor to a similar extent. Hence, complete loss of PP2C activity completely abolished the interaction. These analyses point to the function of ATHB6 as a substrate of ABI1 or, at least, to a specific recognition of the transcription factor via the catalytic centre of ABI1. Provided the HD transcriptional regulator is phosphorylated in yeast adjacent to or within the HD, as observed for PHO2 (Liu et al., 2000), the differences in PP2C and ATHB6 interaction could reflect phosphorylation-dependent interactions. Indeed, modifying a predicted phosphorylation site within the HD of ATHB6 (ATHB6Ser67Ala) negatively affected the interaction with ABI1; however, it did not completely abolish binding. The serine residue could form a topological determinant and/or a phosphorylation site important for interaction. Phosphorylation of transcription factors has been shown to regulate DNA binding (Hunter and Karin, 1992). Interestingly, ATHB6 phosphorylation by PKA in vitro abolished specific binding to the cis-element. At this stage, any information on phosphorylation of HD proteins in higher plants is lacking. A paradigm for such a regulational level in plants is the phosphorylation of an HD protein during fertilization of the unicellular alga Chlamydomonas (O’Connell et al., 1999). In animals, the phosphorylation status regulates protein association, DNA-binding ability or subcellular compartmentation (Whitmarsh and Davies, 2000). Such cellular consequences of phosphorylation/dephosphorylation of plant HD-Zip proteins in planta have to be expected. The analysis of the intracellular localization of ATHB6 fused to GUS revealed a clear nuclear compartmentation of the HD protein irrespective of ABA levels (data not shown, see Supplementary data available at The EMBO Journal Online). This finding suggests that ATHB6 is not regulated by different compartmentation, and the interaction with ABI1 necessitates the transport of the cytosolic PP2C into the nucleus, reminiscent of human PP2Cγ (Murray et al., 1999).
ATHB6 as a transcriptional regulator
It is an attractive hypothesis that ATHB6 functions as a specific transcription factor of the ABA response. The predicted serine phosphorylation site within the DNA-binding domain of ATHB6 represents a determinant for protein–protein interaction and could also form a regulation site for protein–DNA interaction, as exemplified by the soybean transcription factor g/HBF-1 (Dröge-Laser et al., 1997). Contact between HD proteins and DNA is established primarily through the α-helix 3 of the homeobox domain that targets the major groove of the DNA helix (Gehring et al., 1994). The homology of the DNA binding α-helix 3 of ATHB6 to the corresponding region of ATHB1 (Sessa et al., 1993) led to the identification of an ATHB1-similar binding site within the ATHB6 promoter. Binding of ATHB6 to a 30mer oligonucleotide promoter element containing the sequence CAATTATTA was demonstrated by electrophoretic mobility shift assays (EMSAs) and supported by transient gene expression studies. The sequence is similar to the binding site CAATTATTG of ATHB1 and ATHB2 (Sessa et al., 1993; Aoyama et al., 1995; Steindler et al., 1999). Specific targeting of the binding site by ATHB6 is revealed by the complete prevention of interaction in the presence of a single point mutation within the recognition sequence both in vitro and in vivo. The binding sites fused to a minimal promoter were sufficient to mediate ATHB6- and ABA-dependent activation of gene expression. Reporter expression was induced ∼7-fold by ectopic expression of ATHB6, and the additional presence of ABA further stimulated the expression to a 17-fold induction level while in the absence of ectopic ATHB6 little or no induction was observed. The mode of ABA action is not clear. It could either enhance ATHB6 activity, e.g. via ABI1, and/or activate additional transcriptional regulator(s) that depend on ATHB6 for stimulation of promoter activity. Clearly, the ABA-mediated activation was fully dependent on a functional DNA-binding domain of ATHB6 as well as on the correct ATHB6-binding sequence. The analyses define a novel cis-element regulated by the ABA signal. The results support a transcriptionally activating role of the HD-Zip protein for the chimeric promoter and suggest a feedback regulation of the ATHB6 promoter.
The analysis of transgenic Arabidopsis plants revealed a more complex regulation of ATHB6 expression. The ectopic expression of ATHB6 in these lines did not interfere with the accumulation of endogenous ATHB6 transcript irrespective of the ABA signal. The surprising observation indicates an additional negative regulation of the ATHB6 promoter which is not abolished by ectopic ATHB6 expression. It underlines the importance of further components required for regulation of the ATHB6 promoter. In silico analysis revealed additional promoter elements including a MYB recognition sequence (Abe et al., 1997) and ABRE located downstream of the ATHB6-specific motif (Busk and Pages, 1998; Uno et al., 2000). A concerted action of transcriptional regulators is conceivable via interference with the different regulatory elements. The possibility of heterodimerization of ATHB6 with other HD-Zip proteins in planta is supported by in vitro analysis of ATHB6 complex formation with ATHB5, which have a similar expression pattern in Arabidopsis (Johannesson et al., 2001). The ABA-dependent activation of the ATHB6 promoter in our analysis approximately equalled expression from the strong 35S promoter. Such regulatory dynamics are unique, and the physiological role of such high expression of a transcriptional regulator remains mysterious. So far, it is not known whether the observed induction also leads to a corresponding accumulation of ATHB6 protein. Overexpression of ATHB6 fused to GUS, however, resulted in the expected hyperaccumulation of the fusion protein. In view of the capacity of ATHB6 to heterodimerize, the massive accumulation of the specific transcription factor could compete out other complexes of transcriptional regulators and thereby mediate adaptional responses to ABA. The induced state of ATHB6 expression was mimicked by constitutive expression of the ATHB6 transcript with the idea to look for physiological consequences of ATHB6 up-regulation in transgenic plants.
Functional role in ABA responses
Physiological analysis clearly demonstrated ATHB6-mediated alterations of ABA responses in transgenic Arabidopsis lines. Ectopic expression of ATHB6 reduced the sensitivity of the plants towards ABA 3- to 5-fold during germination, reminiscent of ABA-insensitive mutants (Finkelstein et al., 1998; Himmelbach et al., 1998). In addition, the ATHB6-expressing lines were affected in the regulation of water status similarly to the wilty mutants abi1 and abi2. Again, the observed phenotypic alteration is compatible with reduced ABA sensitivity and defines ATHB6 as a negative regulator of the hormone response. The plants, however, responded indistinguishably from control lines to ABA in the regulation of vegetative growth as deduced from comparable inhibitions of root extension. The impact of constitutive ATHB6 expression on ABA action differs from that of known ABA-insensitive mutants, which are either affected in all the above-mentioned ABA responses such as abi1 and abi2 or have only an altered germination response like abi3–abi5 (Himmelbach et al., 1998). The ATHB6 transgenic lines resemble in the alteration of the response spectrum the ABA-hypersensitive era1 (Pei et al., 1998). The ‘loss-of-function’ mutant defined the farnesyltransferase ERA1 as a negative regulator of the ABA-mediated stomatal response and germination inhibition. Hence, both ABA response pathways could share more signalling components or regulators such as ERA1 and ATHB6 than the signalling chain leading to regulation of vegetative growth. Surprising in this context is the regulation of the stomatal response by the transcription factor ATHB6 (Figure 6D), which is strongly expressed in stomata (Söderman et al., 1999). A link between gene expression and regulation of ABA-triggered ion fluxes has been established recently by the identification of the mRNA cap-binding protein ABH1 as a negative regulator of ABA responses including stomatal closure (Hugouvieux et al., 2001).
The stomata of plants can respond to nanomolar concentrations of ABA with closure. During ongoing water stress, however, ABA can accumulate to millimolar concentrations (Harris et al., 1988) and, even under such situations of severe water shortage, the plant has to regulate gas exchange (Grill and Ziegler, 1998). The dynamics and action of ATHB6 induction could form a basis for incremental desensitization of the stomata to the accumulating hormonal signal by accumulating the HD-Zip protein as a negative regulator of the ABA response. The mechanism of ATHB6-mediated desensitization is not clear; however, ATHB6 induction requires the action of ABI1, a key regulator of ABA responses which targets the transcriptional regulator.
Materials and methods
Plasmid constructs
The cDNAs of ATHB5 and ATHB7 were provided by Dr Engström (University of Uppsala, Uppsala, Sweden). ATHB6 full-length cDNA was obtained by RT–PCR. Two copies of the oligonucleotide (CTAGCTAGCAATT[A/G]TTACAATT[A/G]TTAGATA) containing two wild-type (CAATTATTA, oligo A) or mutant (CAATTGTTA, oligo B) binding motifs were cloned in tandem orientation upstream of the –46 CaMV 35S core promoter (Odell et al., 1985). Details of plasmid constructs are available in the Supplementary data.
Yeast two-hybrid assays
The Arabidopsis cDNA library from 3-week-old leaves of ecotype Columbia was generated by directional insertion of cDNAs into pGAD424 (Bartel and Fields, 1995). Screening for interacting proteins and subsequent analysis were performed as described in Clontech protocols (manual PT3024-1). The library was transformed into yeast strain HF7c harbouring the GAL4-ABI1 (amino acids 121–424) bait.
Recombinant proteins and interaction assays
ABI1 and its derivatives were expressed in Escherichia coli with a C-terminal PKA recognition sequence (RRASV) (Blanar and Rutter, 1992) in front of a His6 tag (pQE60) and purified under native conditions as described by the manufacturer (Qiagen). Labelling of ABI1 and phosphorylation of MBP–ATHB6 were performed as described (Blanar and Rutter, 1992). MBP and MBP–ATHB6 fusion protein were expressed in E.coli (pMAL) and affinity purified according to the manufacturer’s instructions (New England Biolabs). The same protocol for affinity chromatography was used to tether the MBP–ATHB6 to amylose resin for analysis of interaction with PP2C (see Supplementary data).
Gel retardation assay
The analysis of DNA-binding properties of MBP–ATHB6 and MBP was performed with double-stranded oligo α (GCATATAATCCAATTATTACAACTTTAACA) identical to the ATHB6 promoter sequence –620 to –628 relative to the transcription start site. Oligo β (GCATATAATCCAATTGTTACAACTTTAACA) was used as a control. The oligos were labelled by filling in 7 bp overhangs with Klenow polymerase using [α-32P]dATP (3000 Ci/mmol). Binding assays were performed with 100 ng of MBP or MBP–ATHB6, oligo α or β (5000 c.p.m.), 750 ng of poly(dI–dC) and 1 µg of bovine serum albumin in a total volume of 15 µl of buffer consisting of 10 mM HEPES pH 7.5, 8 mM MgCl2, 1 mM dithiothreitol, 4 mM spermidine and various concentrations of KCl for adjustment of ionic strength. The components were incubated for 15 min at 4°C. Subsequently, protein–DNA complexes were separated from free oligos by electrophoresis in a 5% polyacrylamide gel and analysed in a phosphorimager (Fuji BAS-1800).
Transient expression and LUC measurements
Isolation of Arabidopsis mesophyll protoplasts from 4-week-old Arabidopsis RLD plants and polyethyleneglycol (PEG)-mediated DNA transfection were performed as described (Abel and Theologis, 1994). For transfections of 5 × 105 protoplasts, plasmid DNAs of aequorin standard (75 µg), reporter (25 µg) and effector (50 µg) were used. LUC and aequorin activity were assayed in a luminometer (Berthold) with a 90 s integration period of light emission.
Transgenic plants and analysis
The transformation of Agrobacterium tumefaciens strain MP90 and Arabidopsis plants ecotype RLD was carried out as described (Meyer et al., 1994). The conditions for the ABA response assays were as mentioned previously (Rodriguez et al., 1998). Stomatal aperture was determined in epidermal peels of Arabidopsis generated immediately prior to microscopic examination (Zeiss Axioskop). The epidermis was immobilized on translucent adhesive tape and mounted on a microscopic slide for digital photography (Nikon Coolpix 990). The stomatal opening was measured by using imaging software (Corel Draw8).
The ABA regulation of the ATHB6 promoter was analysed in a homozygous RLD::ATHB6LUC reporter line. For analysis of the ATHB6 promoter in the abi1 background, F2 seedlings of a cross between the reporter line and ecotype Landsberg-erecta and abi1 (Landsberg-erecta) were used. Fourteen-day-old plantlets (∼30 per data point) were cultivated in nutrient solution as described (Söderman et al., 1999) and challenged with various ABA concentrations and for different time periods. Extracts were analysed for specific LUC activity. In wild-type plants, no LUC activity was found.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr P.Engström (University of Uppsala, Sweden) for providing cDNA for ATHB-5 and -7. The financial support of the DFG and Fonds der Chemischen Industrie is gratefully acknowledged.
References
- Abe H., Yamaguchi-Shinozaki,K., Urao,T., Iwasaki,T., Hosokawa,D. and Shinozaki,K. (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell, 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S. and Theologis,A. (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J., 5, 421–427. [DOI] [PubMed] [Google Scholar]
- Allen G.J., Kuchitsu,K., Chu,S., Murata,Y. and Schroeder,J. (1999) Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell, 11, 1785–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G.J., Chu,S., Harrington,C.L., Schumacher,K., Hoffmann,T., Tang,Y.Y., Grill,E. and Schroeder,J.I. (2001) A defined range of guard cell calcium oscillation parameters encodes stomatal movements. Nature, 411, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Aoyama T., Dong,C., Wu,Y., Carabelli,M., Sessa,G., Ruberti,I., Morelli,G. and Chua,N.H. (1995) Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell, 7, 1773–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P.L. and Fields,S. (1995) Analyzing protein–protein interactions using two-hybrid system. Methods Enzymol., 254, 241–263. [DOI] [PubMed] [Google Scholar]
- Berry M. and Gehring,W. (2000) Phosphorylation status of the SCR homeodomain determines its functional activity: essential role for protein phosphatase 2A,B′. EMBO J., 19, 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanar M.A. and Rutter,W.J. (1992) Interaction cloning: identification of a helix–loop–helix zipper protein that interacts with c-Fos. Science, 256, 1014–1018. [DOI] [PubMed] [Google Scholar]
- Blom N., Gammeltoft,S. and Brunak,S. (1999) Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol., 294, 1351–1362. [DOI] [PubMed] [Google Scholar]
- Busk P.K. and Pages,M. (1998) Regulation of abscisic acid-induced transcription. Plant Mol. Biol., 37, 425–435. [DOI] [PubMed] [Google Scholar]
- Das A., Helps,N., Cohen,P. and Barford,D. (1996) Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 Å resolution. EMBO J., 15, 6798–6809. [PMC free article] [PubMed] [Google Scholar]
- Dröge-Laser W., Kaiser,A., Lindsay,W., Halkier,B., Loake,G., Doerner,P., Dixon,R. and Lamb,C. (1997) Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses. EMBO J., 16, 726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R. and Lynch,T.J. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell, 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Wang,M., Lynch,T.J., Rao,S. and Goodman,H.M. (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell, 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W.J., Affolter,M. and Bürglin,T. (1994) Homeodomain proteins. Annu. Rev. Biochem., 63, 487–526. [DOI] [PubMed] [Google Scholar]
- Grill E. and Ziegler,H. (1998) A plant’s dilemma. Science, 282, 252–253. [DOI] [PubMed] [Google Scholar]
- Gosti F., Beaudoin,N., Serizet,C., Webb,A.R., Vartanian,N. and Giraudat,J. (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell, 11, 1897–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A., Leung,J., Giraudat,J. and Blatt,M.R. (1997) Alteration of anion channel kinetics in wild-type and abi1-1 transgenic Nicotiana benthamiana guard cells by abscisic acid. Plant J., 12, 203–213. [DOI] [PubMed] [Google Scholar]
- Harris M.J., Outlaw,W.H., Mertens,R. and Weiler,E.W. (1988) Water-stress-induced changes in the abscisic acid content of guard cells and other cells from Vicia faba L. leaves as determined by enzyme-amplified immunoassay. Proc. Natl Acad. Sci. USA, 85, 2584–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A., Iten,M. and Grill,E. (1998) Signalling of abscisic acid to regulate plant growth. Philos. Trans. R. Soc. Lond. B Biol. Sci., 353, 1439–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V., Kwak,J.M. and Schroeder,J. (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell, 106, 477–487. [DOI] [PubMed] [Google Scholar]
- Hunter T. and Karin,M. (1992) The regulation of transcription by phosphorylation. Cell, 70, 375–387. [DOI] [PubMed] [Google Scholar]
- Johannesson H., Wang,Y. and Engström,P. (2001) DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol. Biol., 45, 63–73. [DOI] [PubMed] [Google Scholar]
- Kawabe T., Muslin,A. and Korsmeyer,S.J. (1997) HOX11 interacts with protein phosphatases PP2A and PP1 and disrupts a G2/M cell-cycle checkpoint. Nature, 385, 454–458. [DOI] [PubMed] [Google Scholar]
- Leckie C.P., McAinsh,M.R., Allen,G.J., Sanders,D. and Hetherington,A.M. (1998) Abscisic acid-induced stomatal closure mediated by cyclic ADP-ribose. Proc. Natl Acad. Sci. USA, 95, 15837–15842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Oh,H., Cheon,C., Hwang ,I., Kim,Y. and Chun,J. (2001) Structure and expression of the Arabidopsis thaliana homeobox gene Athb-12. Biochem. Biophys. Res. Commun., 284, 133–141. [DOI] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F., MacRobbie,E.A. and Brearley,C.A. (2000) Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl Acad. Sci. USA., 97, 8687–8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leube M.P., Grill,E. and Amrhein,N. (1998) ABI1 of Arabidopsis is a protein serine/threonine phosphatase highly regulated by the proton and magnesium ion concentration. FEBS Lett., 424, 100–104. [DOI] [PubMed] [Google Scholar]
- Leung J. and Giraudat,J. (1998) Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol., 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Leung J., Merlot,S. and Giraudat,J. (1997) The Arabidopsis ABSCISIC ACID INSENSITIVE 2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell, 9, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang,X., Watson,M. and Assmann,S.M. (2000) Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science, 287, 300–303. [DOI] [PubMed] [Google Scholar]
- Liu C., Yang,Z., Yang,J., Xia,Z. and Ao,S. (2000) Regulation of the yeast transcriptional factor PHO2 activity by phosphorylation. J. Biol. Chem., 275, 31972–31978. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L. and Chua,N.H. (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol., 41, 541–547. [DOI] [PubMed] [Google Scholar]
- Mayda E., Tornero,P., Conejero,V. and Vera,P. (1999) A tomato homeobox gene (HD-zip) is involved in limiting the spread of programmed cell death. Plant J., 20, 591–600. [DOI] [PubMed] [Google Scholar]
- Merlot S., Gosti,F., Guerrier,D., Vavasseur,A. and Giraudat,J. (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J., 25, 295–303. [DOI] [PubMed] [Google Scholar]
- Meyer K., Leube,M.P. and Grill,E. (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science, 264, 1452–1455. [DOI] [PubMed] [Google Scholar]
- Mori I.C. and Muto,S. (1997) Abscisic acid activates a 48-kilodalton protein kinase in guard cell protoplasts. Plant Physiol., 113, 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.V., Kobayashi,R. and Krainer,A.R. (1999) The type 2C Ser/Thr phosphatase PP2Cγ is a pre-mRNA splicing factor. Genes Dev., 13, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Lynch,T.J. and Finkelstein,R.R. (2001) Physical inter actions between ABA response loci of Arabidopsis. Plant J., 26, 627–635. [DOI] [PubMed] [Google Scholar]
- Ng C.K., Carr,K., McAinsh,M., Powell,B. and Hetherington,A.M. (2001) Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature, 410, 596–599. [DOI] [PubMed] [Google Scholar]
- O’Connell J., Lu,M. and Snell,W. (1999) Flagellar adhesion between mt(+) and mt(–) Chlamydomonas gametes regulates phosphorylation of the mt(+)-specific homeodomain protein GSP1. J. Biol. Chem., 274, 34383–34388. [DOI] [PubMed] [Google Scholar]
- Odell J.T., Nagy,F. and Chua,N.H. (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature, 313, 810–812. [DOI] [PubMed] [Google Scholar]
- Pei Z.M., Ghassemian,M., Kwak,C., McCourt,P. and Schroeder,J.I. (1998) Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science, 282, 287–290. [DOI] [PubMed] [Google Scholar]
- Pei Z.M., Murata,Y., Benning,G., Thomine,S., Klusener,B., Allen,G.J., Grill,E. and Schroeder,J.L. (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature, 406, 731–734. [DOI] [PubMed] [Google Scholar]
- Rodriguez P.L., Benning,G. and Grill,E. (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett., 421, 185–190. [DOI] [PubMed] [Google Scholar]
- Schena M. and Davis,R.W. (1992) HD-Zip proteins: members of an Arabidopsis homeodomain superfamily. Proc. Natl Acad. Sci. USA, 89, 3894–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J.I., Kwak,J.M. and Allen,G.J. (2001) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature, 410, 327–330. [DOI] [PubMed] [Google Scholar]
- Sessa G., Morelli,G. and Ruberti,I. (1993) The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J., 12, 3507–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc. Natl Acad. Sci. USA, 95, 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderman E., Mattsson,J. and Engström,P. (1996) The Arabidopsis homeobox gene ATHB7 is induced by water deficit and abscisic acid. Plant J., 10, 375–381. [DOI] [PubMed] [Google Scholar]
- Söderman E., Hjellström,M., Fahleson,J. and Engström,P. (1999) The HD-Zip gene ATHB6 in Arabidopsis is expressed in developing leaves, roots and carpels and up-regulated by water deficit conditions. Plant Mol. Biol., 40, 1073–1083. [DOI] [PubMed] [Google Scholar]
- Staxen I., Pical,C., Montgomery,L., Gray,J., Hetherington,A.M. and McAinsh,M.R. (1999) Abscisic acid induces oscillations in guard-cell cytosolic free calcium that involve phosphoinositide-specific phospholipase C. Proc. Natl Acad. Sci. USA, 96, 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C., Matteucci,A., Sessa,G., Weimar,T., Ohgishi,M., Aoyama,T., Morelli,G. and Ruberti,I. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-Zip protein, a negative regulator of gene expression. Development, 126, 4235–4245. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Kao,C.Y. and McCarty,D.R. (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell, 9, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tähtiharju S. and Palva,T. (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J., 26, 461–470. [DOI] [PubMed] [Google Scholar]
- Uno Y., Furihata,T., Abe,H., Yoshida,R., Shinozaki,K. and Yamaguchi-Shinozaki,K. (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl Acad. Sci. USA, 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Veit,B., Sinha,N. and Hake,S. (1991) The developmental gene KNOTTED-1 is a member of a maize homeobox gene family. Nature, 350, 241–243. [DOI] [PubMed] [Google Scholar]
- Vranová E., Tähtiharju,S., Sriprang,R., Willekens,H., Heino,P., Palva,E.T., Inzé,D. and Van Camp,W. (2001) The AKT3 potassium channel protein interacts with the AtPP2CA protein phosphatase 2C. J. Exp. Bot., 52, 181–182. [PubMed] [Google Scholar]
- Whitmarsh A.J. and Davies,R.J. (2000) Regulation of transcription factor function by phosphorylation. Cell. Mol. Life Sci., 57, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Kuzma,J., Marechal,E., Graeff,R., Lee,H.C., Foster,R. and Chua,N.H. (1997) Abscisic acid signalling through cyclic ADP-ribose in plants. Science, 278, 2126–2130. [DOI] [PubMed] [Google Scholar]