Abstract
The lac repressor–operator system is a model system for understanding protein–DNA interactions and allosteric mechanisms in gene regulation. Despite the wealth of biochemical data provided by extensive mutations of both repressor and operator, the specific recognition mechanism of the natural lac operators by lac repressor has remained elusive. Here we present the first high-resolution structure of a dimer of the DNA-binding domain of lac repressor bound to its natural operator O1. The global positioning of the dimer on the operator is dramatically asymmetric, which results in a different pattern of specific contacts between the two sites. Specific recognition is accomplished by a combination of elongation and twist by 48° of the right lac subunit relative to the left one, significant rearrangement of many side chains as well as sequence-dependent deformability of the DNA. The set of recognition mechanisms involved in the lac repressor–operator system is unique among other protein–DNA complexes and presents a nice example of the adaptability that both proteins and DNA exhibit in the context of their mutual interaction.
Keywords: asymmetric DNA binding/DNA deformation/lac repressor/natural lac operator/NMR structure
Introduction
The mechanism by which genetic regulatory proteins discern specific target DNA sequences remains a major area of inquiry. The lac repressor protein, which is a prototype for transcription regulation, controls the expression of lactose metabolic genes by binding to its cognate operator sequence O1 (Figure 1). Effective down-regulation of transcription is enhanced further by lac repressor binding to two auxiliary operators O2 and O3 (Oehler et al., 1990) (Figure 1). The intact repressor can be viewed as a dimer of dimers, where each dimer can bind one lac operator sequence with its two N-terminal DNA-binding domains (DBDs) or headpieces. The DBD is connected to the core domain via a hinge region (residues 50–58), which has been shown to modulate the biological function of lac repressor (Falcon and Matthews, 2000; Kalodimos et al., 2001). Studies of isolated headpiece indicated that this region retains its ability for site-specific binding, yet it binds DNA with significantly lower affinity than the intact protein (Ogata and Gilbert, 1978).
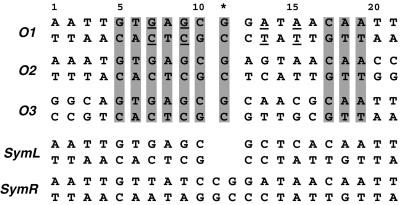
Fig. 1. Sequences of the naturally occurring and two symmetric lac operators. The three natural operators O1, O2 and O3 are aligned with the numbering referred to in the text and figures. The asterisk denotes the central base pair. The bases conserved in all natural operators are highlighted. The two binding sites within each natural operator are asymmetric and are referred to as the left (bp 1–10) and right (bp 12–21). In the O1 operator, the asymmetric regions between the two sites relative to the central base pair are underlined. The SymL operator is a palindrome of the left site of the O1 sequence with the central G:C base pair deleted, and is the sequence used in previous structural studies. The SymR operator is a palindrome of the right site of O1 including the central base pair.
lac repressor–operator interactions have been the subject of intensive study over the past decades due to their profound biochemical and biotechnological interest (for a review see Bell and Lewis, 2001a). Both the protein and DNA components have been mutated extensively in attempts to deduce the details of recognition. So far, all of the detailed structural studies (Lewis et al., 1996; Spronk et al., 1999a; Bell and Lewis, 2000) have employed a fully symmetric ‘ideal’ operator, which lacks the central base pair and is a palindrome of the left half-site of O1 (SymL operator; Figure 1). This fragment binds the repressor with the highest affinity (Sadler et al., 1983; Simons et al., 1984) and has given the best results in both crystallographic and NMR studies. The structural data have revealed the mode of lac repressor binding to DNA: the headpiece binds to the major groove of each half-site of the operator, while the hinge helices dimerize and protrude into the minor groove at the centre of the operator, thereby introducing a significant kink in the DNA. However, the absence of the central G:C base pair in the symmetric operator and the intrinsic asymmetry of the two half-sites in the natural operator suggest the possibility of significant differences in the way lac repressor binds to the natural operator sequences. The lack of structural data has prevented assessment of the importance of the naturally occurring sequence deviations from symmetry.
Within the lac operon, each of the three operator sites is pseudo-palindromic. The 2-fold symmetry is broken by variations in sequence between the two half-sites and by a central G:C base pair that separates the two half-sites (Figure 1). Relative to the complex with the SymL operator, lac repressor could accommodate binding to the natural operator in two ways: (i) as a rigid protein dimer, which implies that due to the extra central base pair a very different sequence is recognized in the left and right half-sites; or (ii) by elongation and rotation of the protein so that the consensus base pairs are recognized in both half-sites, while maintaining the hinge helix interactions. However, this latter option would entail a considerable change in conformation in one protein monomer with respect to the other. Although extensively studied, the specific mode of lac repressor binding to its natural operator has been controversial. Methylation and DNase protection studies (Ogata and Gilbert, 1979; Betz et al., 1986), operator constitutive mutations (Gilbert et al., 1975) and spectroscopic studies (Rastinejad et al., 1993; Kalodimos et al., 2001) have demonstrated symmetric as well as asymmetric contacts. Previous binding footprint studies indicated that the central base pair shifts the pattern of binding, and the right half-site shifts further away from the centre of symmetry as compared with the left site (Horton et al., 1997). In contrast, more recent biochemical studies have suggested a model in which the repressor accommodates binding to the natural operator sequence by shifting the position of the complete right headpiece by 1 bp closer to the centre (Spronk et al., 1999b). This model was supported further by a recent crystallographic analysis of a dimeric lac repressor bound to the natural operator O1, which showed that the hinge helices bind to the minor groove between the left half-site and the central G:C base pair (Bell and Lewis, 2001b). However, due to the low resolution and thermal motion of the DNA in the crystals, the interactions of lac repressor with the bases of the DNA could not be established. Previous NMR studies were also hindered by the intrinsic asymmetry of the natural operator sequence, as well as by the relatively weak affinity of the isolated DBD. Thus, a detailed view of the specific binding mode of the repressor to the natural operators is still lacking.
In the present work, we report the first high-resolution three-dimensional structure of a dimer of the DBD (residues 1–62) of lac repressor bound to its natural operator O1. Towards this end, we have taken advantage of the high affinity for DNA of a recently reported dimeric lac HP62 mutant, which was shown to bind the natural operator with an affinity comparable to that of the intact lac repressor (∼pM range) (Kalodimos et al., 2001). The V52C mutation was designed so as to link two lac headpieces by means of a disulfide bond, and biochemical experiments showed DNA-binding parameters comparable with those of the intact repressor. The close packing of the hinge helices when lac repressor binds to the natural operator (Bell and Lewis, 2001b) validates the use of this construct, which has been corroborated further by NMR analysis (Kalodimos et al., 2001). The numerous genetic and biochemical studies performed on both lac repressor and operator sequences are now complemented by the detailed structural analysis of the dimeric lac repressor DBD complexed to the natural operator O1, with the overall goal of understanding the specific recognition mode.
Results and discussion
Structure determination of the complex
We have solved the solution structure of a 28 kDa complex between the dimeric lac HP62-V52C (residues 1–62) and the natural operator O1. The operator sequence used for the structural studies is a 23mer (5′-GAATTGTGA GCGGATAACAATTT-3′) flanking one extra base pair in each site of O1. The structure of the protein–DNA complex was solved by heteronuclear double and triple resonance NMR spectroscopy using 15N- and 13C-labelled protein and unlabelled oligonucleotide. The high stability of the complex ensured that it exists in slow exchange on the NMR chemical shift time scale. Due to the asymmetry of the complex imposed by the intrinsic asymmetry of the operator sequence, each residue of the protein should give rise to its own signal. Indeed, ∼124 signals from the protein backbone show up in the 1H–15N HSQC spectrum of the complex, indicating that the two subunits are found in different environments when bound to DNA and no symmetry relationship is present (Kalodimos et al., 2001).
The structure calculation protocol used for the dimeric lac HP62–O1 operator complex consisted first of calculation of the protein dimer alone in the complex, then its docking onto the DNA, and a final refinement step in explicit solvent (see Materials and methods). A total of 100 conformers were calculated, and the 20 of lowest energy were selected (Figure 2). The structure was determined on the basis of 2563 experimental NMR restraints (Table I). Due to the high stability of the complex, a large number of intermolecular restraints were collected (254), which permitted the fine positioning of the dimeric lac DBD onto the operator sequence. For residues 4–59, the backbone root mean square deviation (r.m.s.d.) from the mean structure is ∼0.36 Å within each protein monomer and 0.54 Å for the dimer. Elements of secondary structure were defined on the basis of nuclear Overhauser effect (NOE) connectivities, hydrogen–deuterium (H/D) exchange data and analysis using the program PROCHECK (Laskowski et al., 1996). Detailed H/D data were also used to identify intermolecular hydrogen bonds from the protein backbone to the operator. A summary of the structural and restraints statistics is given in Table I.
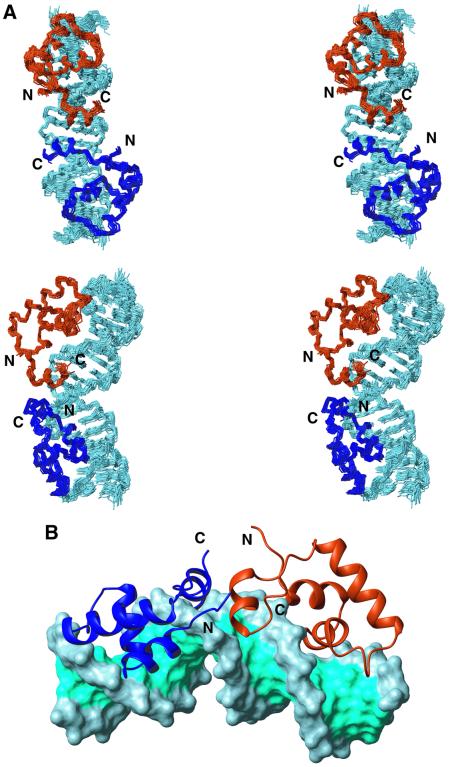
Fig. 2. Three-dimensional structure of the dimeric lac HP62–O1 operator complex. (A) Side-by-side stereo view of the ensemble of the final 20 structural conformers depicted in two different views (rotated by 90°). The left and right lac DBD subunits are coloured dark blue and dark orange, respectively, while DNA is displayed in light blue. The O1 operator is bent by ∼36°, with the kink centred between bp 10 and 11. For clarity, only the backbone atoms of residues Val4–Lys59 and DNA heavy atoms of bp 2–20 are shown. (B) Schematic diagram of the structure of the complex. A ribbon diagram of the protein is shown bound to the solvent-accessible surface of the operator. The protein is coloured as in (A), whereas the major and minor grooves of the operator are coloured cyan and the ribose phosphate backbone light blue.
Table I. Structural statistics of 20 final structures of the dimeric lac HP62–O1 operator complexa.
| R.m.s.d. (Å) with respect to meanb (backbone/heavy) | |
| lac HP62 (left subunit) | 0.34 ± 0.09/0.85 ± 0.24 |
| lac HP62 (right subunit) | 0.38 ± 0.10/0.86 ± 0.26 |
| lac HP62 (dimer) | 0.48 ± 0.13/0.93 ± 0.29 |
| O1 operator | 0.67 ± 0.37/0.62 ± 0.30 |
|
lac HP62–O1 operator |
0.62 ± 0.29/0.82 ± 0.34 |
| No. of experimental restraints | |
| Protein (dimer) | |
| Intraresidue NOEs | 514 |
| Sequential NOEs (|i – j| = 1) | 419 |
| Medium range NOEs (1<|i – j|<4) | 260 |
| Long-range NOEs (|i – j|>4) | 246 |
| Interprotein | 28 |
| Total NOEs | 1467 |
| Dihedral angle restraints | 162 |
| DNA NOEs | 680 |
| Protein–DNA NOEs | 245 |
| Protein–DNA hydrogen bonds |
9 |
| Restraint violationsc | |
| NOE distances with violations >0.3 Å | 2.9 ± 1.8 |
| Dihedrals with violations >5° |
4.2 ± 1.9 |
| R.m.s.d. for experimental restraints | |
| All distance restraints (2401) (Å) | 0.06 ± 0.01 |
| Torsion angles (162) (°) |
4 ± 2 |
| CNS energies from SAd | |
| Evdw (kcal/mol) | –1096 ± 10 |
|
Eelec (kcal/mol) |
–3077 ± 62 |
| R.m.s.d. (Å) from idealized covalent geometry | |
| Bonds (°) | 0.003 ± 0.00 |
| Angles (°) | 0.54 ± 0.01 |
| Impropers (°) |
0.51 ± 0.01 |
| Ramachandran analysis | |
| Residues in the favoured region (%) | 85.1 |
| Residues in additional allowed regions (%) | 13.8 |
| Residues in generously allowed regions (%) | 0.6 |
| Residues in disallowed regions (%) | 0.6 |
aBased on 20 structures, obtained by simulated annealing in CNS followed by refinement in explicit water using NOE distance restraints, dihedral angle restraints, bonds, angles, impropers, dihedral angle, van der Waals and electrostatic energy terms (see Materials and methods).
bResidues 3–59 for the protein and bp 2–21 for the operator are taken into account.
cNo distances were violated by >0.5 Å.
dThe non-bonded energies were calculated with the OPLS parameters using a 11.5 Å cut-off.
Structure description
The N-terminal domain of the lac repressor complexed to DNA consists of four α-helices running from residues 6 to 13, 17 to 24, 32 to 45 and 51 to 57. The α-helical content is the same in both the left and right protein subunits. The recognition helix of both headpieces makes extensive contacts with the major groove of the natural operator, while the hinge helices penetrate, as expected, into the minor groove between bp 10 and 11, thereby introducing a kink of ∼36° in the DNA. The structure of the left site of the complex is similar to the structure of the lac headpiece bound to the SymL operator, especially in the minor groove region (Spronk et al., 1999a). However, the higher affinity of the dimeric lac HP62-V52C for DNA allowed us to collect a larger number of NOEs, which, coupled with detailed H/D experiments, now give a more detailed, and slightly different, view of the protein–DNA contacts in the left site of the complex.
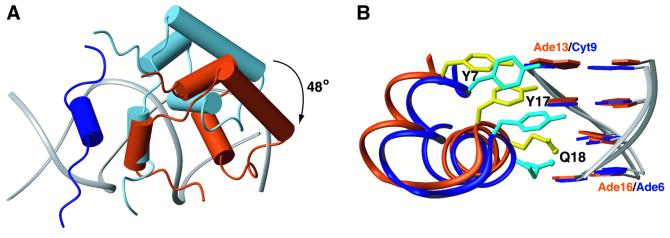
Remarkably, the right protein subunit adopts an alternative conformation in order to recognize specifically the right half-site of the natural operator (Figures 2 and 3). Although the hinge helix binds to the minor groove between bp 10 and 11, the three-helical domain shifts by 1 bp further away from the centre. In order to form an optimum interface with the right half-site of the natural operator, the right lac headpiece undergoes a 48° rotation relative to the left monomeric site. This is the result of the shift by 1 bp (the helix twist of B-DNA is 36°) and an additional 12° rotation of the recognition helix needed for maximizing the interaction with the right half-site sequence. Therefore, the two protein subunits align in a very different way with respect to the centre of the operator in order to achieve optimum juxtaposition of the protein–DNA interface in the two sites.
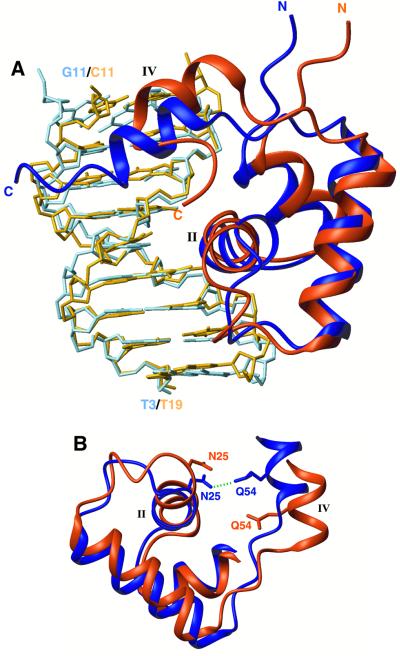
Fig. 3. Superposition of the two O1 operator-bound protein subunits. The left lac headpiece is coloured dark blue and the left half-site of the operator sequence (bp 3–11) is in light blue, whereas the right lac headpiece is coloured dark orange and the right half-site of the operator sequence (bp 11–19) is in light orange. (A) The two structures have been overlaid on their DNA backbone with respect to their approximate dyad-symmetric sequence (i.e. bp 3–11 are overlaid with bp 19–11). The hinge helix in the right site moves by ∼3.4 Å closer to the centre of the sequence and the recognition helix rotates by 12° relative to the left one. (B) In the left site, the hinge helix is stabilized through a hydrogen bond (green dashed line) between Gln54 and Asn25. The extension of the loop linking the third helix with the hinge helix in the right site results in the disruption of this critical contact, thereby diminishing the stability of the right hinge helix.
If the natural lac operator O1 is viewed as two halves, with an approximate dyad axis through the central base pair, left site to right site differences occur at bp 7 (G:C) versus 15 (A:T), and 9 (G:C) versus 13 (A:T) (Figure 1). Previous biochemical and structural studies have demonstrated that formation of the hinge helices, which is a prerequisite for the stabilization of the protein–DNA complex, requires both protein–protein and protein–DNA contacts (Spronk et al., 1996; Kalodimos et al., 2001). Therefore, the right headpiece should move closer to the centre of the natural operator, relative to the SymL operator, so that its hinge helix can interact with its left mate and strengthen the dimer interface. A recent low-resolution crystallographic analysis (Bell and Lewis, 2001b) showed that indeed the right hinge helix packs against the left one and both bind between Cyt10 and Gua11. If, however, the right headpiece moved as a rigid body to bind to a position shifted by 1 bp closer to the centre, then it would recognize a completely different sequence and not the sequence with approximate dyad symmetry to the left site. As the present results demonstrate, the lac repressor accommodates binding by a dramatic alteration of its DBD conformation, so that differences in the protein–DNA contacts between the two sites are kept to a minimum. lac repressor’s ability to accommodate conformational variations is due to the high flexibility of its DBD, as was demonstrated recently (Kalodimos et al., 2002).
The conformation adopted by the right lac headpiece results in an extension of the loop linking the third and the hinge helix in the right subunit (Figure 3B). This has some important implications regarding the stability of the complex. In the left subunit, Asn25 makes a key hydrogen bond via its side chain CO to the side chain NH2 of the hinge helix residue Gln54, thus providing a critical link between the core of the lac headpiece and the hinge helix (Figure 3B). This link is expected to contribute significantly to the stability of the hinge helix and thus of the dimer interface, and is also present in the highly homologous pur repressor (Schumacher et al., 1994). In the right lac headpiece subunit, however, these two groups are far apart, due to the structural rearrangement, rendering the formation of the corresponding hydrogen bond impossible (Figure 3B). This may explain the lower stability of the hinge helices in the complex with the natural operator as compared with that with the SymL operator. Therefore, when lac repressor binds to right symmetrized DNA fragments, the hinge helices would be expected to be even more unstable. Interestingly, in the complex of lac repressor with the SymR operator (Figure 1), the hinge helices are not well ordered and exist at equilibrium between α-helical and random coil conformations (Kalodimos et al., 2001). The alternative conformations that lac repressor adopts when bound to different sequences may also strongly affect the allosteric response to inducer molecules (Falcon and Matthews, 2001).
The total solvent-accessible surface area buried upon DNA binding is equal for the two sites (∼2220 Å2 each). Comparison of the bound monomers with the structure of free lac DBD (Slijper et al., 1996) shows that significant conformational changes occur upon binding. Apart from the ordering of the hinge region to an α-helix, the orientation of the helices within the helix–turn–helix (HTH) domain changes by ∼11°. The backbone r.m.s.d. between the free and the bound conformation for the first three helices is 1.3 Å, whereas for heavy atoms it increases to 2.4 Å due to the significant local adjustments of the configuration of the side chains (see below). Overlay of the first 45 residues of the two O1 operator-bound lac headpieces results in a 1.2 and 2.1 Å r.m.s.d. for backbone and heavy atoms, respectively. When all four helices are included, then the r.m.s.d. increases to 2.4 and 3.4 Å for backbone and heavy atoms, respectively.
Protein–DNA contacts
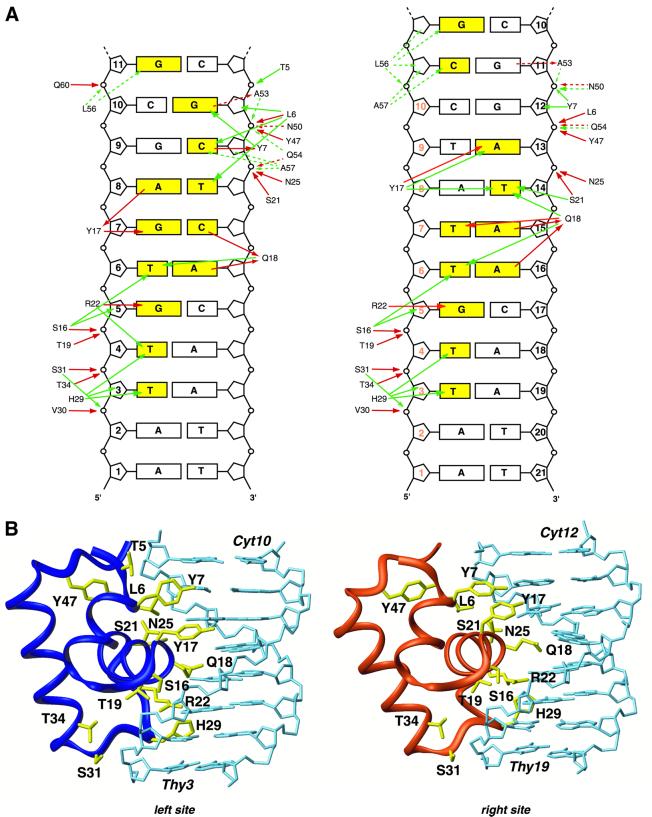
The intermolecular contacts observed in both sites of the complex are summarized in Figure 4. As can be seen, most of the base pairs are contacted in a specific manner through the side chains of residues of both the HTH domain and the hinge helices. With respect to the left site complex, many contacts have been addressed in previous studies (Chuprina et al., 1993; Spronk et al., 1999a). However, the higher number of NOEs collected in the present work, in conjunction with H/D experiments, reveals additional and important DNA interactions. First of all, lac repressor makes extensive hydrogen bonding contacts to the sugar phosphate through many residues located in the HTH domain. More specifically, Leu6, Ser16, Thr19, Ser21, Asn25, Val30, Ser31 and Thr34 hydrogen-bond to the DNA backbone either with their side chains or with their backbone, or with both. Remarkably, the right headpiece subunit retains all these non-specific interactions with the sugar phosphate backbone of the DNA despite the alternative conformation it assumes (Figure 4). Thus, the flexibility in the DBD allows lac repressor to accommodate conformational variations while retaining high affinity by maintaining sugar phosphate backbone interactions.
Fig. 4. Comparison of the protein–DNA contacts in the two half-sites of the dimeric lac HP62–O1 operator complex. (A) Schematic summary of the DNA contacts in the left and right half-site. The bases that are recognized specifically by the lac repressor are coloured yellow. The solid and dashed lines indicate interactions in the major and minor grooves, respectively. Red and green lines indicate hydrogen bonding and hydrophobic contacts, respectively. In the right site, the numbering displayed in orange denotes the symmetry-related base pairs in the left half-site. (B) View of the interactions of the left and right lac globular subdomains (residues 1–49) with their corresponding half-sites of the O1 operator. Residues involved in protein–DNA interactions are shown in yellow.
Sequence-specific protein–DNA contacts are provided mostly by residues of the HTH domain. Leu6, the first residue of the first helix, makes extensive apolar contacts with the bases of Thy8 and Cyt9, whereas Tyr7 contributes to specificity by a direct hydrogen bond from its side chain oxygen atom to the H4 proton of Cyt9 and strong apolar contacts to the base of Gua10. Interestingly, these intermolecular contacts are not conserved in the right half-site of the complex. Therefore, the first helix residues do not confer any specificity to the recognition of the right site sequence, since they are involved only in non-specific contacts (Figure 4B).
The second helix residues are responsible for extensive recognition of the major groove of the operator. In contrast to previous structural studies, a large number of intermolecular NOEs have now been collected for this region. Ser16, Tyr17, Gln18, Thr19, Ser21 and Arg22 give in total 60 protein–DNA NOE restraints, resulting in their position being very well defined. In both sites, the recognition helix is anchored to DNA through hydrogen bonds to sugar phosphates formed by Ser16, Thr19, Ser21 and Asn25, which align the helix in a proper way across the major groove. In the left site, the hydroxyl group of Tyr17 hydrogen-bonds to both O6 and N7 atoms of Gua7, while it accepts a hydrogen bond from the H6 proton of Ade8. Gln18, a key residue in the recognition process, confers specificity by accepting hydrogen bonds from both Ade6 and Cyt7. Previous genetic data have indicated the important role of Arg22 for interacting with DNA. The side chain of Arg22 is poised to interact favourably with the base of Gua5, which is in agreement with mutational data (Sartorius et al., 1989). Additionally, Thy4 and Thy6 are also recognized specifically by strong apolar contacts through the side chains of Ser16, Gln18 and Arg22.
As pointed out above, the right lac headpiece undergoes a 48° rotation, relative to the left one, so as to contact the DNA sequence with approximate dyad symmetry to the left site. However, two base pairs are still different; G:C base pairs at positions 7 and 9 now become A:T (at positions 13 and 15). The G:C base pair at position 9 is recognized through specific contacts by Leu6 and Tyr7; these contacts are missing in the right site. Interestingly, the A:T base pair at position 13 is recognized by Tyr17, the side chain of which hydrogen-bonds to the N7 atom of Ade13. This contact is achieved by an alteration of the side chain conformation of Tyr7 and Tyr17 in the right site, in which they form an aromatic ring-stacking interaction that reorients the two tyrosines relative to the left site (Figure 4B). In the left half-site, Thy8 is contacted specifically by the methyl groups of Leu6. In the right site, however, there is a complete reorganization: the symmetry-related Thy14 is contacted at its methyl group by Tyr17, Gln18 and Ser21 in a specific manner. This side chain rearrangement, in conjunction with an additional rotation of 12° of the recognition helix relative to the left one, allows Gln18 to recognize three different bases, instead of two in the left site (Figure 4). The amino group of its side chain hydrogen-bonds to the O4 atom of Thy15, whereas the carbonyl group of its side chain accepts a hydrogen bond from the N6 atom of both Ade15 and Ade16. Arg22 is seen to participate in four possible hydrogen bonds in the structure ensemble, all of them solely with Gua17. Overall, there are surprisingly extensive structural differences between the left and right half-sites with respect to protein–DNA interactions, including a shift of the right lac headpiece by 1 bp and a rotation of 48° relative to the left one.
Tyr17 and Gln18 are the most important residues for specific recognition of the lac operator and can be changed to obtain lac repressors that recognize different operator sequences (Sartorius et al., 1989). These two residues recognize three base pairs in the left half-site (the T:G:A triplet at positions 6, 7 and 8), whereas they contact four base pairs in the right site (the quadruplet A:T:A:A at positions 13, 14, 15 and 16). It is interesting that Tyr17 contacts the A:T pair at position 13, while its left site symmetry-related G:C pair is recognized by Tyr7. Therefore, despite the global shift of the right HTH domain by 1 bp further away from the centre, the side chain of Tyr17 adjusts locally and moves towards the centre of the operator, thereby forming entirely different contacts to the operator compared with the left site. A similar rearrangement and shift is also seen for the side chain of Gln18 (Figure 4B). These asymmetric binding modes are consistent with the large chemical shift differences of these residues in the two sites. Tyr7 makes specific contacts only to the left half-site, yet it participates indirectly in the recognition of the right site by forming an aromatic cluster with Tyr17, which orients the side chain of the latter to interact favourably with the bases. The mechanisms of recognition of the natural operator O1 by the lac repressor present a good example of the adaptability that proteins exhibit towards discrimination of different operator sequences.
The loop following the recognition helix is also involved in interactions with the operator, in agreement with previous studies (Spronk et al., 1999a). The side chain of His29 makes extensive hydrophobic contacts with Thy3 and Thy4. The backbone of Val30 and Ser31 and the side chain of Thr34 hydrogen-bond to the DNA phosphates. Exactly the same set of interactions is also observed in the right half-site involving the A:T base pairs at positions 18 and 19. Just beyond this point, only Tyr47 contacts DNA, with a hydrogen bond to the DNA backbone. This interaction is also present in the right site and its importance is reflected in its intolerance to mutations and its highly conserved nature in the LacI family of repressors (Weickert and Adhya, 1992; Markiewicz et al., 1994).
The left hinge helix makes extensive contacts to the minor groove that are very similar to those seen in the complex with the SymL operator. Asn50 and Gln54 hydrogen-bond to the DNA backbone, whereas Ala53, Leu56 and Ala57 are involved in extensive hydrophobic interactions with the bases at the centre of the operator. A similar set of interactions is also observed for the right hinge helix, with the exception of the backbone of Asn50 that does not hydrogen-bond to DNA. The bending mechanism is the same as in the SymL operator; the side chain of Leu56 of both subunits protrudes into the minor groove and pries open the DNA.
DNA conformation and recognition through sequence-dependent conformation
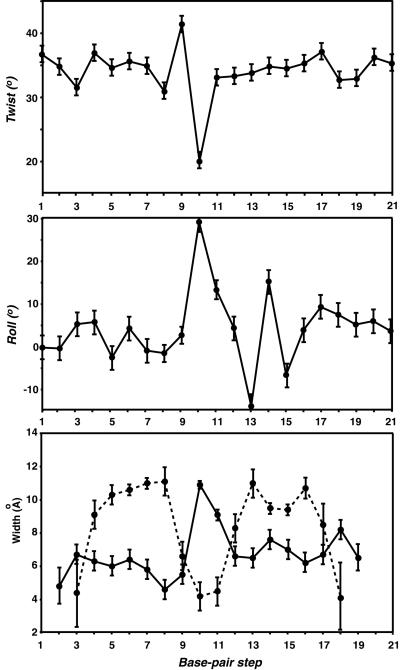
Understanding the role DNA plays in facilitating the association of DNA-binding proteins is necessary for understanding how sequence specificity is accomplished. The observed parameters of the O1 operator bound to the lac repressor DBD (base pair roll, twist, and major and minor groove width) are summarized in Figure 5. The natural lac operator O1 is bent globally by ∼36° (the kink is located between positions 10 and 11), some 9° less than the SymL operator. This is in agreement with gel shift mobility experiments which showed a lower degree of bending for the natural operator (Spronk et al., 1999b; Kalodimos et al., 2001). The kink is reflected in increases in the roll and twist angles of the central base pairs and in deviations of the major and minor groove widths and depths, compared with standard B-DNA values (Figure 5). All parameters in the left half-site of the operator sequence are normal for undistorted B-DNA. However, the base pairs of the triplet A:T:A in the right half-site show significant local deformations, with base pair rolls that deviate significantly from the averaged values of B-DNA. In addition, the major groove in this region is narrower by ∼2 Å compared with its symmetry-related region in the left half-site. The conformation of the A:T:A triplet differs significantly from that of its symmetry-related region in the left site, where a G:A:G triplet exists, and imparts asymmetry by providing unique contacts to the HTH domain of the lac headpiece. The specific contacts to base pairs provided by Tyr7, Tyr17 and Gln18 are very different between the two half-sites. Apparently, the base pairs at positions 13, 14 and 15 should deform significantly for optimum interaction with the side chains of the residues located in the recognition helix. Therefore, appropriate recognition of the right half-site requires that the lac repressor assumes an alternative conformation in conjunction with the sequence-dependent ability of the half-site to adopt the required conformation upon repressor binding.
Fig. 5. Averaged helical parameters of the lac operator O1 in complex with the dimeric lac HP62. Twist, roll and width of the major (dashed line) and minor (solid line) grooves are shown.
Analysis of the mutational data provides interesting insights into the role played by DNA. None of the naturally occurring lac operators adhere to the idealized palindromic sequence. Alignment of the three operators O1, O2 and O3 reveals that bp 5–9 are absolutely conserved (Figure 1). The importance of this region is also consistent with mutational data, which showed that any base pair substitution results in very low affinity for the lac repressor (Lehming et al., 1987). The central CpG step, where the hinge helices bind, is also very sensitive to mutations. The central sequences in the natural operators may play a key role in bending and structural flexibility to generate the DNA-binding conformation necessary for high-affinity protein binding. Even a purine to purine change in this central sequence abolishes high-affinity complex formation. This may be due to the fact that CpG steps have higher than average roll angles, resulting in enhanced flexibility and the ability to adopt greater positive rolls (Dickerson, 1998).
In contrast, the right half-site sequence at positions 12–16, where the recognition helix binds, has no consensus. Mutational analysis has shown that alterations in the right site sequence content are far less deleterious compared with the left site (Sadler et al., 1983; Betz et al., 1986). Thymine at positions 18 and 19 is conserved in the right half-site within the natural operators, the methyl groups of which make favourable van der Waals contacts with the side chain of His29. In contrast, in the left half-site, this region does not show strong consensus; in fact, substitutions at positions 3 and 4 are much less detrimental to protein binding compared with the base pairs at positions 5–10 (Lehming et al., 1987). The only unique and consistent sequence similarity between the two half-sites of the natural operators is the G:C pair at position 5 (position 17 in the right site). Any mutation at this position results in a complex being >100-fold less stable (Lehming et al., 1987). This base pair is contacted specifically, in both sites, by Arg22, which in general appears to be a very favourable contact (Pabo and Nekludova, 2000).
How can the diminished stability of the complex with the O1 operator compared with that with the SymL operator be explained? Operators with single base pair mutations that make the right site of the natural operator more symmetric than the left bind this operator with lower affinity than the natural operator. The doubly substituted operator, with two identical left halves reflected about the central base pair, is an even weaker binding site (Betz et al., 1986). In each case, the left mutation is more deleterious to repressor binding than the right mutation (Sadler et al., 1983), suggesting a greater overall contribution to binding by the left operator half. However, the base pairs that provide maximal repressor affinity at given positions in the left site of the operator do not improve binding of repressor when symmetrically introduced into the right side; in fact they appear to be detrimental to binding. Therefore, reduction in the affinity of the right half-site cannot be ascribed to ‘incorrect’ base pairs in this sequence, particularly those of A:T pairs at positions 13 and 15, since symmetrization gives rise to significant loss of affinity. This result might appear intriguing since, as the present data demonstrate, the lac repressor is capable of reaching out 1 bp further away from the centre so that it could optimize its binding to the right half-site using the same mechanism for recognizing the left half-site. A plausible explanation based on the present results could be that distortions in the backbone and base stacking propagated from the central G:C generate a conformation that is less favourable for lac repressor recognition and binding. Thus, the naturally occurring right-site lac operator sequences must compensate for binding loss generated by the presence of the central G:C base pair. As our results demonstrate, in the O1 operator this is accomplished by significant distortions of the base pairs at positions 13, 14 and 15. Our suggestions are in close agreement with recent biochemical data demonstrating that subtle variations in DNA sequence can lead to structural changes that are detected by the lac repressor and are reflected in large alterations in affinity and/or allostery (Falcon and Matthews, 2000, 2001).
Furthermore, mutational studies have shown that changes at position 12 did little to reduce or enhance repressor affinity, the protein recognizing the sequence quite well regardless of the base pair in this position (Betz et al., 1986). This is in agreement with the present structural data, which show that there is no specific contact to this base pair. Actually, one could argue, based on the shift of the right headpiece further away from the centre, that position 12 in the operator represents an extra base pair. However, an operator with position 12 merely deleted presents a very weak repressor target, >150 times weaker than the wild-type sequence (Betz et al., 1986). Overall, the present data indicate that optimizing protein binding is not simply a matter of providing two of the sequence preferential half operator sites, but that other factors, such as the relative rotational orientation of half operator sites within the context of the DNA helix, play a significant role. For example, the SymL operator binds to the repressor ∼8 times more tightly than wild-type in vitro, but only ∼2 times more tightly in vivo, where presumably the operator plasmids are under tension and the operator segments are underwound (Sadler et al., 1983). The present results contribute to an evolving view of the importance of sequence-dependent conformational flexibility of the DNA for protein recognition and affinity (Koudelka, 1998; Lefstin and Yamamoto, 1998).
Comparison with other asymmetric protein–DNA complexes
Pseudo-dyad-related sequences are found commonly as the DNA target sites of both prokaryotic and eukaryotic multimeric gene regulatory proteins. Structural data on these complexes would be very interesting, since they may reveal how proteins are able to recognize different DNA sequences simultaneously. However, in NMR studies, it has been a common strategy to use symmetric sequences to force identical and symmetric protein–DNA interactions in order to reduce the assignment task and the number of NOESY cross-peaks by 2-fold. The present structure is the first of an asymmetric protein–DNA complex solved by NMR spectroscopy. Similarly, only a small set of crystal structures of dimeric proteins bound to asymmetric DNA target sites has been obtained, as in most cases altered sequences with idealized symmetry for improved crystallization and diffraction have been used.
Crystallographic analysis of the glucocorticoid receptor DBD bound to target DNA with improper spacing between the half-sites demonstrated that the DNA-induced dimer fixes the separation of the subunits’ recognition surfaces so that one subunit interacts specifically with the consensus target half-site and the other contacts what is effectively a non-specific site, making many fewer contacts to the DNA bases (Luisi et al., 1991; Gewirth and Sigler, 1995). This is due to more favourable protein–protein than protein–half-site interactions, which force the right subunit to shift as a rigid body closer to the centre of the operator. Comparison of the structure of the human oestrogen receptor DBD complexed to a consensus binding site with that containing a non-consensus DNA target reveals that recognition of the non-consensus sequence is achieved by the rearrangement of a lysine side chain (Schwabe et al., 1995). Rearrangement of amino acid side chains between the protein–DNA interface at the consensus and non-consensus sequences, accompanied by displacement of the phosphate backbone of DNA, have also been seen in the structures of 434 repressor complexed with different DNA targets (Rodgers and Harrison, 1993), as well as in the case of the NF-κB p65 transcription factor (Chen et al., 2000). In all these cases, however, the protein subunits align roughly symmetrically on the DNA half-sites. lac repressor adopts a novel mode for recognizing its pseudo-symmetric natural operator sequence, which is in fact a combination of the strategies employed by the above-mentioned proteins. The dimer interface, which is essential for formation of a high stability complex, is always formed by packing of the two hinge helices. If the spacing of the two headpieces is too large, then complex formation is abolished (Spronk et al., 1999b). In the O1 operator, both hinge helices bind to the minor groove between bp 10 and 11, thereby ensuring that the dimer interface remains tight. However, the right three-helical domain rotates by 48° relative to the left one and shifts further away from the centre by ∼3.4 Å. Additionally, extensive differences are observed in the protein–DNA interface as well as significant rearrangement of the side chain packing of residues located in the HTH domain (Figure 4B). This dramatic structural rearrangement of the protein, accompanied by significant local deformation of the DNA (Figure 5), suggests that proteins can adapt to recognize different DNA sequences by small- to large-scale rearrangement of side chains and/or entire domains.
Conclusions
The present report describes in detail the specific recognition mechanism of the natural lac operator O1 by lac repressor, a system with a profound biochemical and biotechnological interest. Recognition of the operator involves four distinct mechanisms, which are summarized schematically in Figure 6: (i) a hinge helix–minor groove interaction dictated by protein–protein interactions and the stronger binding of the left headpiece; (ii) elongation of the right headpiece so that the three α-helical domain shifts by 1 bp further away from the centre and rotates by 48° relative to the left one; (iii) significant rearrangement of the side chains of the residues located in the HTH domain; and (iv) a sequence-dependent deformation of the operator. This mode of recognition is unique among other protein–DNA systems. Our results underscore how proteins can take advantage of the intrinsic flexibility of its DBD to recognize different DNA targets, as well as the essential role that DNA sequence plays in the association of the repressor with the operator through sequence-dependent deformability that contributes significantly to binding and sequence discrimination. The numerous genetic and biochemical studies performed in both lac repressor and its natural operator sequences are now complemented by the detailed structural analysis. These combined efforts have led to a better understanding of the mechanisms a protein can use in order to recognize different DNA targets.
Fig. 6. Distinct recognition mechanisms involved in lac repressor interaction with the left and right operator sites. (A) The left hinge helix is displayed in dark blue. If the lac repressor recognized the right half-site in a similar way to the left site, then it would adopt the conformation displayed in light blue. However, recognition of the right half-site is accomplished by a 48° rotation and a shift of ∼3.4 Å further away from the centre of the operator of the three α-helical domain, whereas the hinge helices still pack together. The calculated conformation of the right subunit of lac repressor’s DBD is displayed in dark orange. (B) Specific recognition of the two sites also requires significant rearrangement of the side chain conformations (the three most important residues, Y7, Y17 and Q18, are depicted) as well as a sequence-dependent deformation of the operator. Dark blue and dark orange denote the left and right site, respectively.
Materials and methods
Expression and preparation of the dimeric lac HP62-V52C protein and its complex with the natural lac operator O1
The HP62-V52C mutant was amplified from the corresponding Lac I genes by PCR and expressed in Escherichia coli using a T7 polymerase-based system. The protein was purified as described earlier (Kalodimos et al., 2001).
Uniformly 13C/15N- and 15N-labelled proteins were grown in BIOEXPRESS-CN 1000 (CIL) medium. The natural lac operator O1 fragment was purchased from Carl Roth GmbH (Germany) and purified further on a Q-Sepharose (Pharmacia) column. For complex formation, the protein was mixed with an equimolar amount of the operator and dissolved in 0.01 M KPi buffer pH 6.0 containing 0.02 M KCl. All samples were concentrated using Cenricon concentrators (Amicon) and dissolved in 95% H2O/5% D2O. Trace amounts of NaN3 were added as a preservative.
NMR spectroscopy
All NMR spectra were recorded on Bruker DRX750 and DRX600 spectrometers equipped with triple resonance gradient probes at 315 K. Sequential assignement of the 1H, 13C and 15N protein chemical shifts was achieved by means of through-bond heteronuclear scalar correlations along the backbone and the side chains using conventional three-dimensional pulse sequences (Cavanagh et al., 1996). DNA assignments were obtained from 2D-NOE and simultaneous 13C–15N double-half filter NOE experiments recorded on a sample containing a 1:1 complex of 13C–15N-dimeric HP62-V52C protein and unlabelled operator, using conventional sequential assignment methodology for nucleic acids (Wijmenga and van Buuren, 1998). Interproton distance restraints within the protein were derived from three-dimensional 13C- and 15N-separated NOE experiments. Protein–DNA interactions were assigned from two-dimensional time-shared 13C–15N double-half filter, 2D-NOE and 3D-NOESY-HSQC experiments. Amide proton exchange rates were determined from the time course of the peak intensities in a series of 1H–15N HSQC spectra after dissolving lyophilized samples in D2O (Kalodimos et al., 2002). All spectra were processed using the NMRPipe software package (Delaglio et al., 1995) and analysed with NMRView (Johnson and Blevins, 1994).
Structure calculations
All calculations were performed with CNS (Brünger et al., 1998) using the ARIA set-up and protocols (Linge and Nilges, 1999). Approximate interproton distance restraints were grouped into four distance ranges, 1.8–2.8, 1.8–3.4, 1.8–5.0 and 1.8–6.0 Å, corresponding to strong, medium, weak and very weak NOEs, respectively. Watson–Crick base pairing was maintained in the DNA by the following hydrogen bond restraints (r): for G:C pairs rG(N1)–C(N3) = 2.95 ± 0.2 Å, rG(N2)–C(O2) = 2.86 ± 0.2 Å and rG(O6)–C(N4) = 2.91 ± 0.2 Å; for A:T base pairs rA(N1)–T(N3) = 2.82 ± 0.2 Å and rA(N6)–T(O4) = 2.95 ± 0.2 Å. Loose torsion angle restraints were used to alleviate problems associated with mirror images: α = –70 ± 50, β = 180 ± 50, γ = 60 ± 35, ε = 180 ± 50 and ζ = –85 ± 50 (Huang et al., 2000). Weak planarity restraints were also included during the simulated annealing protocol. The 13Cα, 13Cβ, 13C′, Hα, 15N and NH chemical shifts of 116 residues served as input for the TALOS program (Cornilescu et al., 1999) to extract Φ and Ψ angles. The structure of the complex was calculated in two phases. First the structure of the dimeric lac HP62-V52C alone was calculated following the standard Cartesian space simulated annealing set-up and protocols in ARIA. The best 100 structures in terms of restraint energies subsequently were refined in explicit water using the OPLS parameters (Jorgensen and Tirado-Rives, 1988) and the TIP3P water model (Jorgensen et al., 1992). Those 100 structures were then docked onto a B-DNA following a torsion angle dynamics (TAD) simulated annealing protocol. The protein was positioned randomly within a 10 Å cube at a 50 Å distance from the DNA. For the high temperature (5000 steps at 5000 K) and the first cooling stage (from 5000 to 500 K in 10 000 steps), both protein and DNA were treated as semi-rigid bodies: the backbone of the core (residues 6–45) of the two lac headpieces and of the two hinge helices (residues 51–57) and the entire DNA except the central three base pairs were defined as rigid bodies in TAD. This was followed by Cartesian space refinement: in the first cooling stage (from 1000 to 500 K in 10 000 steps), position restraints were applied on the DNA atoms with a force constant of 20 kcal/Å2/mol, then, in the final refinement (from 500 to 50 K in 10 000 steps), both protein and DNA were completely free to move. The NOE restraint force constants were set initially to 50 and 5 kcal/Å2/mol intra- and intermolecular NOEs, respectively. The intermolecular force constant was scaled up to 10 kcal/Å2/mol during the first TAD cooling stage. The resulting complex structures were refined in explicit water with all NOE force constants set to 50 kcal/Å2/mol using electrostatic and Lennard–Jones non-bonded energy terms. A modified version of the PARALLHDG5.2 parameter set was used for the protein (M.Williams and A.Bonvin, personal communication), and the DNA parameters were taken from the dna-rna-allatom parameter set from the CNS distribution. The best 20 structures in terms of restraint energies were selected for analysis. Structural DNA parameters were analysed using the program CURVES (Lavery and Sklenar, 1988). Structure figures were generated using the program MOLMOL (Koradi et al., 1996). The chemical shifts of the protein–DNA complex have been deposited in the BMRB (accession number 5345). The final structures have been deposited in the PDB (accession code 1L1M).
Acknowledgments
Acknowledgements
We thank Dr G.Folkers for his assistance in the early phase of this project. This work was supported financially by NOW-CW. R.K.S. is a recipient of a PhD fellowship from CNPq (Brazil).
References
- Bell C.E. and Lewis,M. (2000) A closer view of the conformation of the Lac repressor bound to operator. Nat. Struct. Biol., 7, 209–214. [DOI] [PubMed] [Google Scholar]
- Bell C.E. and Lewis,M. (2001a) The Lac repressor: a second generation of structural and functional studies. Curr. Opin. Struct. Biol., 11, 19–25. [DOI] [PubMed] [Google Scholar]
- Bell C.E. and Lewis,M. (2001b) Crystallographic analysis of Lac repressor bound to natural operator O1. J. Mol. Biol., 312, 921–926. [DOI] [PubMed] [Google Scholar]
- Betz J.L., Sasmor,H.M., Buck,F., Insley,M.Y. and Caruthers,M.H. (1986) Base substitution mutants of the lac operator: in vivo and in vitro affinities for lac repressor. Gene, 50, 123–132. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cavanagh J., Fairbrother, WJ., Palmer,A.G.,III and Skelton,N.J. (1996) Protein NMR Spectroscopy. Academic Press, San Diego, CA.
- Chen Y.Q., Sengchanthalangsy,L.L., Hackett,A. and Ghosh,G. (2000) NF-κB p65 (RelA) homodimer uses distinct mechanisms to recognize DNA targets. Structure Fold. Des., 8, 419–428. [DOI] [PubMed] [Google Scholar]
- Chuprina V.P., Rullmann,J.A., Lamerichs,R.M., van Boom,J.H., Boelens,R. and Kaptein,R. (1993) Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics. J. Mol. Biol., 234, 446–462. [DOI] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio,F. and Bax,A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek,S., Vuister,G., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX PIPES. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Dickerson R.E. (1998) DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res., 26, 1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon C.M. and Matthews,K.S. (2000) Operator DNA sequence variation enhances high affinity binding by hinge helix mutants of lactose repressor protein. Biochemistry, 39, 11074–11083. [DOI] [PubMed] [Google Scholar]
- Falcon C.M. and Matthews,K.S. (2001) Engineered disulfide linking the hinge regions within lactose repressor dimer increases operator affinity, decreases sequence selectivity and alters allostery. Biochemistry, 40, 15650–15659. [DOI] [PubMed] [Google Scholar]
- Gewirth D.T. and Sigler,P.B. (1995) The basis for half-site specificity explored through a non-cognate steroid receptor–DNA complex. Nat. Struct. Biol., 2, 386–394. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Gralla,J., Majors,J. and Maxam,A. (1975) Lactose operator sequences and the action of lac repressor. In Sund,H. and Blauer,G. (eds), Protein–Ligand Interactions. De Gruyter, Berlin, Germany, pp. 93–210.
- Horton N., Lewis,M. and Lu,P. (1997) Escherichia coli lac repressor–lac operator interaction and the influence of allosteric effectors. J. Mol. Biol., 265, 1–7. [DOI] [PubMed] [Google Scholar]
- Huang K., Luis,J.M., Donaldson,L., Lim,F.-L., Sharrocks,A.D. and Clore,M.G. (2000) Solution structure of the MEF2A–DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J., 19, 2615–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A. and Blevins,R.A. (1994) NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR, 4, 603–614. [DOI] [PubMed] [Google Scholar]
- Jorgensen W. and Tirado-Rives,J. (1988) The OPLS force field for proteins. Energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc., 110, 1657–1666. [DOI] [PubMed] [Google Scholar]
- Jorgensen W., Chandrasekhar,J. Madura,J.D., Impey,R.W. and Klein,M.L. (1992) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys., 79, 926–935. [Google Scholar]
- Kalodimos C.G., Folkers,G.E., Boelens,R. and Kaptein,R. (2001) Strong DNA binding by covalently linked dimeric Lac headpiece: evidence for the crucial role of the hinge helices. Proc. Natl Acad. Sci. USA, 98, 6039–6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalodimos C.G., Boelens,R. and Kaptein,R. (2002) A residue-specific view of the association and dissociation pathway in protein–DNA recognition. Nat. Struct. Biol., 9, 193–197. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter,M. and Wuthrich,K., (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Koudelka G.B. (1998) Recognition of DNA structure by 434 repressor. Nucleic Acids Res., 26, 669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Rullmannn,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 1, 63–91. [DOI] [PubMed] [Google Scholar]
- Lefstin J.A. and Yamamoto,K.R. (1998) Allosteric effects of DNA on transcriptional regulators. Nature, 392, 885–888. [DOI] [PubMed] [Google Scholar]
- Lehming N., Sartorius,J., Niemoller,M., Genenger,G., von Wilcken-Bergmann,B. and Muller-Hill,B. (1987) The interaction of the recognition helix of lac repressor with lac operator. EMBO J., 6, 3145–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M., Chang,G., Horton,N.C., Kercher,M.A., Pace,H.C., Schumacher,M.A., Brennan,R.G. and Lu,P. (1996) Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science, 271, 1247–1254. [DOI] [PubMed] [Google Scholar]
- Linge J.P. and Nilges,M. (1999) Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR, 13, 51–59. [DOI] [PubMed] [Google Scholar]
- Luisi B.F., Xu,W.X., Otwinowski,Z., Freedman,L.P., Yamamoto,K.R. and Sigler,P.B. (1991) Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature, 352, 497–505. [DOI] [PubMed] [Google Scholar]
- Markiewicz P., Kleina,L.G., Cruz,C., Ehret,S. and Miller,J.H. (1994) Genetic studies of the lac repressor. XIV. Analysis of 4000 altered Escherichia coli lac repressors reveals essential and non-essential residues, as well as ‘spacers’ which do not require a specific sequence. J. Mol. Biol., 240, 421–433. [DOI] [PubMed] [Google Scholar]
- Oehler S., Eismann,E.R., Kramer,H. and Muller-Hill,B. (1990) The three operators of the lac operon cooperate in repression. EMBO J., 9, 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R.T. and Gilbert,W. (1978) An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc. Natl Acad. Sci. USA, 75, 5851–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R.T. and Gilbert,W. (1979) DNA-binding site of lac repressor probed by dimethylsulfate methylation of lac operator. J. Mol. Biol., 132, 709–728. [DOI] [PubMed] [Google Scholar]
- Pabo C.O. and Nekludova,L. (2000) Geometric analysis and comparison of protein–DNA interfaces: why is there no simple code for recognition? J. Mol. Biol., 301, 597–624. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Artz,P. and Lu,P. (1993) Origin of the asymmetrical contact between lac repressor and lac operator DNA. J. Mol. Biol., 233, 389–399. [DOI] [PubMed] [Google Scholar]
- Rodgers D.W. and Harrison,S.C. (1993) The complex between phage 434 repressor DNA-binding domain and operator site OR3: structural differences between consensus and non-consensus half-sites. Structure, 1, 227–240. [DOI] [PubMed] [Google Scholar]
- Sadler J.R., Sasmor,H. and Betz,J.L. (1983) A perfectly symmetric lac operator binds the lac repressor very tightly. Proc. Natl Acad. Sci. USA, 80, 6785–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorius J., Lehming,N., Kisters,B., von Wilcken-Bergmann,B. and Muller-Hill,B. (1989) lac repressor mutants with double or triple exchanges in the recognition helix bind specifically to lac operator variants with multiple exchanges. EMBO J., 8, 1265–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M.A., Choi,K.Y., Zalkin,H. and Brennan,R.G. (1994) Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by α helices. Science, 266, 763–770. [DOI] [PubMed] [Google Scholar]
- Schwabe J.W., Chapman,L. and Rhodes,D. (1995) The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation. Structure, 3, 201–213. [DOI] [PubMed] [Google Scholar]
- Simons A., Tils,D., von Wilcken-Bergmann,B. and Muller-Hill,B. (1984) Possible ideal lac operator: Escherichia coli lac operator-like sequences from eukaryotic genomes lack the central G X C pair. Proc. Natl Acad. Sci. USA, 81, 1624–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slijper M., Bonvin,A.M., Boelens,R. and Kaptein,R. (1996) Refined structure of lac repressor headpiece (1–56) determined by relaxation matrix calculations from 2D and 3D NOE data: change of tertiary structure upon binding to the lac operator. J. Mol. Biol., 259, 761–773. [DOI] [PubMed] [Google Scholar]
- Spronk C.A., Slijper,M., van Boom,J.H., Kaptein,R. and Boelens,R. (1996) Formation of the hinge helix in the lac repressor is induced upon binding to the lac operator. Nat. Struct. Biol., 3, 916–919. [DOI] [PubMed] [Google Scholar]
- Spronk C.A., Bonvin,A.M., Radha,P.K., Melacini,G., Boelens,R. and Kaptein,R. (1999a) The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator. Structure Fold. Des., 7, 1483–1492. [DOI] [PubMed] [Google Scholar]
- Spronk C.A., Folkers,G.E., Noordman,A.M., Wechselberger,R., van den Brink,N., Boelens,R. and Kaptein,R. (1999b) Hinge-helix formation and DNA bending in various lac repressor–operator complexes. EMBO J., 18, 6472–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert M.J. and Adhya,S. (1992) A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem., 267, 15869–15874. [PubMed] [Google Scholar]
- Wijmenga S.S. and van Buuren,B.N.M. (1998) The use of NMR methods for conformational studies of nucleic acids. Prog. NMR Spectrosc., 32, 287–387. [Google Scholar]