Abstract
The MinE protein functions as a topological specificity factor in determining the site of septal placement in Escherichia coli. MinE assembles into a membrane-associated ring structure near midcell and directs the localization of MinD and MinC into a membrane- associated polar zone that undergoes a characteristic pole-to-pole oscillation cycle. Single (green fluorescent protein) and double label (yellow fluorescent protein/cyan fluorescent protein) fluorescence labeling experiments showed that mutational alteration of a site on the α-face of MinE led to a failure to assemble the MinE ring, associated with loss of the ability to support a normal pattern of division site placement. The absence of the MinE ring did not prevent the assembly and disassembly of the MinD polar zone. Mutant cells lacking the MinE ring were characterized by the growth of MinD polar zones past their normal arrest point near midcell. The results suggested that the MinE ring acts as a stop-growth mechanism to prevent the MinCD polar zone from extending beyond the midcell division site.
Keywords: cell division/CFP/minicell/septation/YFP
Introduction
In most organisms, the site of cell division is placed at the midpoint of the cell. This is required to permit the equal distribution of cellular components into the two daughter cells. Aberrant placement of the division site can result in unequal distribution of genetic material and fragmentation of chromosomes due to a guillotine effect of the eccentrically placed division septum (Niki et al., 1991). In Escherichia coli, the placement of the division site at midcell is accomplished by the cooperative action of the MinC, MinD and MinE proteins (de Boer et al., 1989). The three proteins ensure that division is limited to midcell by preventing septation at alternative division sites that can be located elsewhere along the cell cylinder.
The Min proteins play different roles in the division site selection process [summarized in Rothfield et al. (1999)]. MinC is a non-specific division inhibitor that is capable of blocking cell division at all potential division sites when expressed alone, and is responsible for preventing septation specifically at aberrant sites when accompanied by MinD and MinE. MinD is a peripheral membrane protein that is required for the membrane association of MinC and MinE. MinE is a topological specificity factor that imparts site specificity to the membrane-associated MinC division inhibitor, preventing MinC from blocking division at midcell while permitting it to block septation at unwanted sites elsewhere along the cell. MinE appears to carry out this function by directing the membrane localization pattern and dynamic behavior of MinC and MinD.
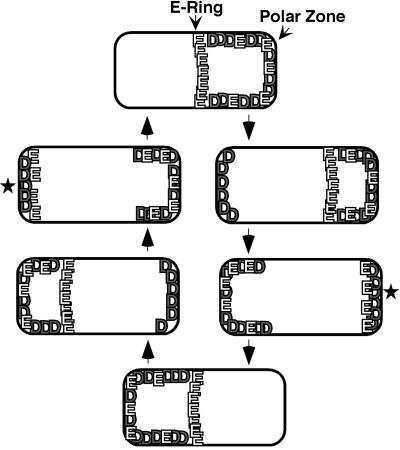
In the absence of MinE, the MinC and MinD proteins are associated with the membrane around the entire periphery of the cell. When MinE is also present, MinD and MinC are redistributed to one end of the cell where they form a membrane-associated polar zone that extends, at its fullest extent, from the cell pole to a position near midcell (Figure 1) (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a,b; Rowland et al., 2000). Coincident with formation of the MinC/MinD polar zone, MinE assembles into a ring-like structure near midcell. Most of the remaining membrane-associated MinE assembles into a polar zone at one end of the cell (Raskin and de Boer, 1997) whose appearance resembles that of the MinC/MinD polar zone. The MinE ring and the MinC/MinD/MinE polar zones undergo a repetitive cycle in which the structures oscillate rapidly from pole to pole many times within each division cycle (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a,b; Rowland et al., 2000; Fu et al., 2001; Hale et al., 2001).
Fig. 1. Dynamic behavior of MinD polar zone and MinE ring and polar zone. See text for details. The star indicates that the new MinD polar zone grows from the end of the cell toward midcell as shown previously (Fu et al., 2001; Hale et al., 2001), and that the MinE polar zone co-assembles with the MinD polar zone although this is still conjectural. MinC (not shown in the figure), undergoes pole-to-pole oscillation together with MinD (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a), although the details of assembly and disassembly of the MinC polar zones are not yet known in as much detail.
The pole-to-pole movement involves the assembly of the polar zone and E-ring at one end of the cell, followed by their disassembly and subsequent reassembly at the other end of the cell (Figure 1) (Fu et al., 2001; Hale et al., 2001). During the assembly stage, the new polar zone grows from the cell pole until it reaches a position near midcell, where zonal growth stops and a new E-ring appears at the leading edge of the polar zone. During the disassembly stage, the polar zone and E-ring retract from midcell back to the original pole. As a result of this pattern, the time-averaged concentration of the MinC division inhibitor is low near midcell and progressively higher toward the cell pole. This allows the division inhibitor to prevent aberrant septation events away from midcell while not interfering with division at the normal midcell division site. The rapid pole-to-pole oscillation permits the MinC division inhibitor to act at both poles, presumably because the total time that the ends of the cell are covered by the division inhibitor is sufficient to prevent assembly of the division machinery at these locations.
Little is known about the mechanisms responsible for formation of the MinC/MinD/MinE polar zone or the E-ring, or for the pole-to-pole oscillation of the three proteins. It has been suggested that the role of the E-ring during the oscillation cycle is to cause disassembly of the MinD polar zone, by activating the ATPase activity of MinD molecules at the medial edge of the polar zone (Hu and Lutkenhaus, 2001). The present study deals with the topological specificity function of MinE, which is responsible for formation of the E-ring near midcell and for the effects of MinE on the localization pattern of the other Min proteins.
The C-terminal region of MinE (MinE31–88; the MinE topological specificity domain, MinETSD) is required for its topological specificity function (Zhao et al., 1995; Zhang et al., 1998). Partial or complete deletion of the MinETSD leads to loss of topological specificity so that septation frequently occurs near the cell poles, giving rise to chromosomeless minicells (Pichoff et al., 1995; Zhao et al., 1995).
The MinETSD is a unique dimeric structure with an upper surface (the α-face) that is formed by a pair of antiparallel α-helices, and a lower surface (the β-face) that comprises four antiparallel β-strands (King et al., 2000). Previous studies showed that mutational alteration of either of two amino acid residues on the α-face (Asp45 and Val49) leads to loss of the ability of MinE to prevent minicelling when minE is coordinately expressed with minC and minD (King et al., 2000), thereby implicating these residues in the topological specificity function of MinE.
Here we attempt to answer two questions. What site within the three-dimensional structure of MinE is responsible for its ability to assemble into the E-ring? What role does the E-ring play in the localization and dynamic behavior of the MinD polar zone? We show that Asp45 and Val49 define a site on the α-surface of MinE that is required for formation of MinE rings but is not required for formation of MinD polar zones. Thus, these two integral parts of the topological specificity function of MinE can be dissociated. The absence of E-rings did not prevent the assembly or disassembly of the MinD polar zones but did interfere with the spatial and temporal regulation of the assembly/disassembly cycle. The results suggested a primary role for the MinE ring in the arrest of growth of the MinD polar zone that normally prevents the polar zone from extending beyond the normal midcell division site. These observations have important implications for an understanding of the interdependent roles of the Min proteins in coordinating the localization and dynamic behavior of these membrane-associated structures and in ensuring proper placement of the division site.
Results
Phenotypes of MinE α-face mutants
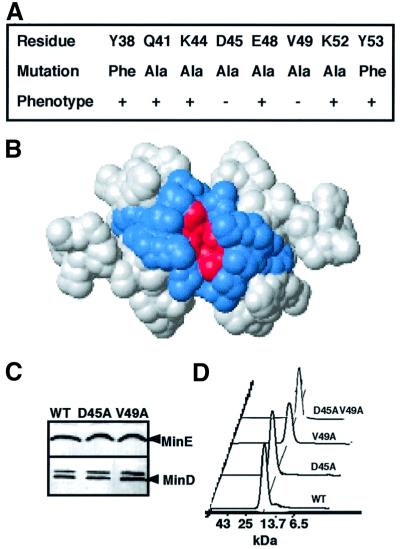
To determine whether additional residues on the α-face other than Asp45 and Val49 are implicated in the topological specificity function of MinE, we individually mutated all of the surface-exposed residues of the α-face (Figure 2A). The mutated MinETSD was substituted for the wild-type TSD in plasmid pSY1083 (Plac-minC minD minE). A classical minicelling phenotype (minicells and short filaments of 4–8 cell lengths) was observed when the strains were grown in the presence of 0.01% glucose to repress expression of minCDE. Induction of the wild-type minCDE operon by growth in 0 or 10 µM isopropyl-β-d-thiogalactopyranoside (IPTG) restored the wild-type division pattern (>98% of cells were normal cell length), with growth in 10 µM IPTG being most effective (<1% minicells). Therefore, in subsequent experiments, both with unlabeled and labeled MinD and MinE, cells were induced by growth in 10 µM IPTG. Plasmids containing minE mutations that resulted in loss of the topological specificity function were identified by loss of the ability to correct the minicelling phenotype of a ΔminCDE strain. minED45A and minEV49A were the only α-face mutations that led to loss of the ability of MinE to restore the wild-type cell division phenotype (Figure 2A). The minED45A and minEV49A mutants grown under these conditions showed a lower frequency of minicelling (13 and 6%, respectively) than cells in which Plac-minCDE was completely repressed by growth in 1% glucose (26–30% minicells).
Fig. 2. Mutations on the α-face of the MinE topological specificity domain. (A) Surface-exposed α-face residues in wild-type MinE (top row) and in mutant MinE proteins (middle row). Phenotypic effects of minE alleles were determined by their effects on cell division phenotype (see Materials and methods). +, wild-type phenotype; –, minicelling phenotype. (B) Location of mutated residues. Surface view of the MinETSD (MinE31–88), looking down at the α-helical face. Mutated residues (Asp45 and Val49 in A) associated with a minicelling phenotype are indicated in red; those with no effect on division phenotype are shown in blue. (C) Western blot analysis. Cell extracts were analyzed from strain RC1 (ΔminCDE) containing pSY1083 (WT), pYLS4 (D45A) or pYLS40 (V49A) from the cultures used to determine cell division phenotype (A). Upper panel, anti-MinE; lower panel, anti-MinD. (D) Size exclusion chromatography of MinETSD. Samples (D45AV49A, V49A, D45A and WT, top to bottom; 0.4 mM) were applied to the Superdex 75 column. Elution positions of molecular weight marker proteins are indicated on the horizontal axis, and absorbance at 220 or 276 nm on the vertical axis. The predicted molecular weights of the MinE samples are 14.09–14.24 kDa. Similar results were obtained when 2 and 10 µM samples were analyzed.
The possibility that the minicelling phenotypes of the D45A and V49A mutants were due to changes in cellular concentrations of the mutant proteins or in the ratio of MinE/MinD was excluded by quantitative western blot analysis (Figure 2C). The possibility that the phenotypes were due to a perturbation of the overall structure of the TSD was excluded by two-dimensional NMR analysis that showed no change in folding pattern (King et al., 2000). The possibility that the phenotypes were due to defects in MinE dimerization was excluded in two ways. First, multiangle laser light scattering (MALLS) analysis of the MinETSD showed that the MinEWT, MinED45A, MinEV49A and MinED45A/V49A proteins (loading concentration 400 µM) were monodisperse with weight-average molecular masses of 13.5 ± 0.2, 14.5 ± 0.4, 13.3 ± 0.2 and 13.6 ± 0.2 kDa, respectively. All of these masses are close to the predicted dimer molecular masses of 14.09–14.24 kDa. Secondly, size exclusion chromatography (Figure 2D) showed that the mutant and wild-type MinE proteins eluted with the same retention time over a wide range of concentrations (2–400 µM), indicating that the Kd for dimerization is not significantly different for any of the mutants compared with wild-type MinE.
Effect of D45A and V49A mutations on formation of MinE rings
Studies of MinE–green fluorescent protein (GFP) fusion proteins in fixed cells provided evidence that the loss of topological specificity of septal placement in the α-face mutants was accompanied by a defect in formation or stability of the MinE ring. The E-ring is defined by an annular accumulation of a high concentration of MinE molecules that can be visualized as a fluorescent ring at the medial edge of the MinE polar zone (Raskin and de Boer, 1997). The high concentration of MinE in the ring is believed to be responsible for the role of the E-ring in MinCDE function.
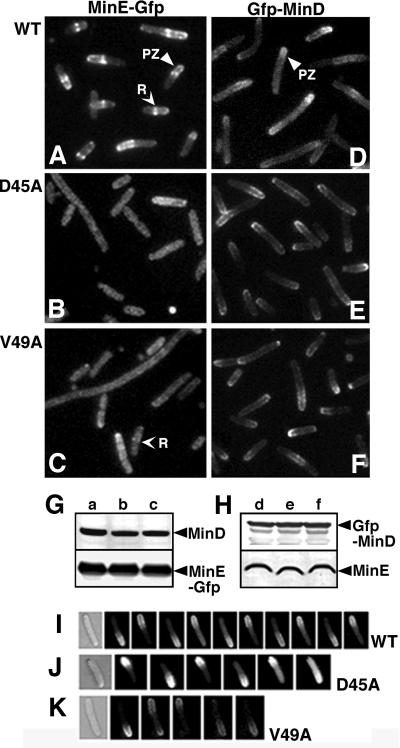
When Plac-minC minD minE::gfp containing the wild-type minE allele, in a ΔminCDE host, was induced for 2 h, MinE rings were visible as bright fluorescent bands extending across the width of the cylinder or paired fluorescent dots on either side of the cylinder (R in Figure 3A). MinE polar zones were visible in the same cells as zones of membrane-associated fluorescence that extended from the E-ring to the nearest pole (PZ in Figure 3A). The E-rings and polar zones were present in ∼80% of cells.
Fig. 3. Cellular localization of GFP–MinD and MinE–GFP. (A–F) Fluorescence micrographs of fixed cells of strain RC1 (ΔminCDE) containing: (A) pSY1083G (Plac-minC minD minE::gfp); (B) pYLS4G (Plac-minC minD minED45A::gfp); (C) pYLS40G (Plac-minC minD minEV49A::gfp); (D) pFX9 (Plac-gfp::minD minE); (E) pYLS33 (Plac-gfp::minD minED45A); and (F) pYLS46 (Plac-gfp:: minD minEV49A). PZ, polar zone; R, MinE ring. (G and H) Western blot analysis of cells from the experiments described in (A–F). Top panel, anti-MinD; bottom panel, anti-MinE. (I–K) Time-lapse images of living cells of RC1 containing: (I) pFX9 (WT); (J) pYLS33 (D45A); and (K) pYLS46 (V49A). Time intervals were 20, 30 and 20 s, respectively.
In contrast, when wild-type minE::gfp was replaced by minED45A::gfp (Figure 3B), E-rings were absent from most cells, with faint ring-like structures visible in 0.5–1% of the cells. When wild-type minE was replaced by minEV49A, 10–15% of the cells showed faint ring-like structures (R in Figure 3C), indicating that the effect of the V49A mutation on E-ring formation was less marked than that of the D45A mutation. When the minC minD minED45A::gfp cells were grown in the presence of inducer overnight and then diluted to permit resumption of exponential growth (see Materials and methods), the results were the same except that ∼2% of cells now had faint E-rings. Mutation of α-face residue K52, which did not lead to a minicelling phenotype, did not prevent formation of MinE rings and polar zones (data not shown). Western blot analysis (Figure 3G) showed that the concentrations of MinE–GFP and MinD were similar in the wild-type and mutant cells, excluding the possibility that the failure to see E-rings was due to decreased expression or more rapid degradation of the mutant MinE–GFP proteins. The minED45A::gfp and minEV49A::gfp cells showed the same minicelling phenotype as the corresponding strains without the gfp label.
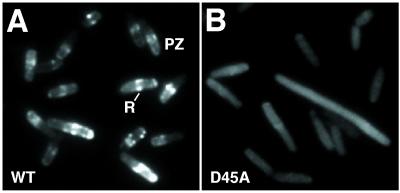
In unfixed minEWT::gfp cells, ∼90% of the cells con tained clear MinE rings (Figure 4A), whereas faint MinE rings were visible in ∼6% of minED45A::gfp cells (Figure 4B). Similar results were obtained when minEWT:: gfp and minED45A::gfp were expressed from single copy lysogens by growth of RC1(λSY1083G) and RC1 (λYLS4G) in 10 or 50 µM IPTG. Most cells contained E-rings in the minEWT::gfp culture whereas <2% of cells contained E-rings in the minED45A::gfp culture.
Fig. 4. MinE–GFP localization in living cells. Fluorescence micrographs of unfixed cells grown as described in Figure 3A and B. (A) RC1/pSY1083G (ΔminCDE/Plac-minC minD minE::gfp. (B) RC1/pYLS4G (ΔminCDE /Plac-minC minD minED45A::gfp).
These results indicated that the D45A and V49A mutations are associated with defects in formation or stability of the MinE ring and polar zone.
Effect of D45A and V49A mutations on MinD polar zones
In contrast to their effect on MinE localization, the minED45A and minEV49A mutations did not interfere with formation of MinD polar zones in cells that expressed gfp::minD and minE (Figure 3E and F). The results were similar in fixed and unfixed cells. Nearly all cells contained MinD polar zones that resembled those in cells that expressed wild-type minE (Figure 3D). However, the polar zones in the mutant cells were longer than those in cells expressing wild-type MinE (Figure 5P), extending well past midcell in most cells, as discussed further below. Western immunoblots showed that the concentrations of GFP–MinD, MinED45A and MinEV49A were similar to the concentrations of GFP–MinD and MinE in cells expressing the wild-type minE allele (Figure 3H).
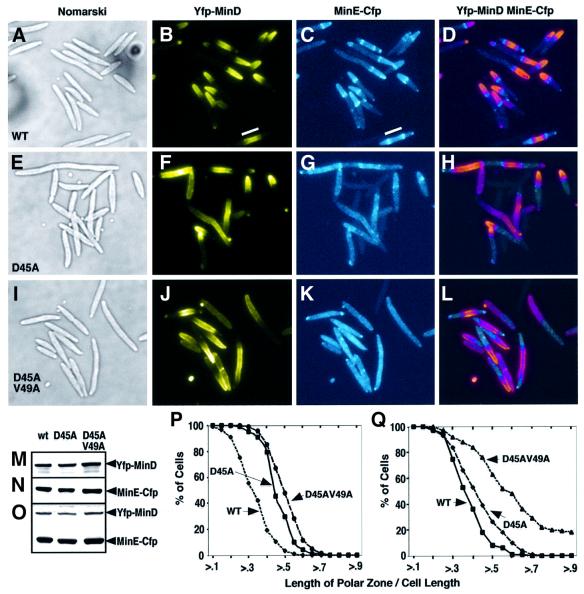
Fig. 5. Fluorescence microscopy of YFP–MinD and MinE–CFP in doubly labeled cells, induced as described in Materials and methods. (A–L) Nomarski (A, E and I) and fluorescence microscopy of fixed cells of strain RC1 containing: (A–D) pYLS68 (Plac-yfp::minD minE::cfp); (E–H) pYLS69 (Plac-yfp::minD minED45A::cfp); and (I–L) pYLS77 (Plac-yfp::minD minED45A/V49A::cfp). YFP fluorescence is shown in (B), (F) and (J); CFP fluorescence is shown in (C), (G) and (K); an overlay of YFP and CFP panels is shown in (D), (H) and (L), to show the relationship of the MinE ring to the edge of the MinD polar zone. In the overlay, MinE–CFP appears light blue, and YFP–MinD appears pink. An internal YFP–MinD/MinE–CFP zone flanked by MinE rings in a short filament is indicated by the white line (B and C). (M–O) Western blot analysis of cells from the experiments described above. Top panel, anti-MinD; middle panel, anti-MinE; bottom panel, anti-GFP. Anti-GFP antibodies also recognize the YFP and CFP moieties of the fusion proteins. (P and Q) Relationship between the length of MinD polar zones and cell length expressed as a ratio of the length of the polar zone to cell length. GFP or YFP fluorescence was monitored. (P) Cells were grown for 2 h in the presence of 10 µM IPTG as in Figure 3. WT = RC1/pFX9 (ΔminCDE /Plac-gfp::minD minE); D45A = RC1/pYLS33 (ΔminCDE /Plac-gfp::minD minED45A); D45AV49A = RC1/pYLS92 (ΔminCDE /Plac-yfp::minD minED45A/V49A). (Q) Cells were induced overnight and grown for an additional 2–3 h in the presence of IPTG as in (A–L). WT = RC1/pYLS68; D45A = RC1/pYLS69; D45AV49A = RC1/pYLS77.
MinC did not change the distribution pattern of GFP–MinD in cells where minC and gfp::minD minE were co-expressed in trans under control of Para and Plac, respectively, with wild-type, minE D45A or minE V49A (data not shown). This was consistent with previous reports that MinC is not required for formation of MinD polar zones.
The GFP–MinD polar zones in the minED45A and minEV49A mutant cells underwent cycles of pole-to-pole oscillation that resembled those in cells that expressed the wild-type minE allele (Figure 3I–K). The mean oscillation rate (Kosc, in cycles per min) was slower in the minED45A and minEV49A cells than in minEWT cells (minEWT, Kosc = 1.25 ± 0.24, n = 8; minED45A, Kosc = 0.96 ± 0.26, n = 8; minEV49A, Kosc = 0.59 ± 0.22, n = 6).
Simultaneous visualization of MinD and MinE in individual cells
The GFP labeling experiments described in the preceding section suggested that formation and oscillation of MinD polar zones might not require the presence of the MinE ring. However, because it is not possible to visualize MinE and MinD separately in the same cell when both proteins are labeled with GFP, the possibility could not be excluded that the gfp::minD minED45A cells that showed fluorescent GFP–MinD polar zones might have contained MinE rings despite the fact that MinE–GFP rings were not observed in most cells that co-expressed unlabeled minD and minED45A::gfp. To circumvent this problem, we utilized mutant GFP proteins with different absorption and emission spectra [yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP), see Materials and methods]. In these experiments, the host cell contained a deletion of the chromosomal copy of minCDE, and yfp::minD and minE::cfp were co-expressed from Plac-yfp::minD minE::cfp, making it possible to visualize both proteins within the same cell.
To observe significant fluorescence from MinE–CFP, it was necessary to grow cells for longer than the 2–4 h period that is used for visualization of GFP- and YFP-labeled proteins. This probably explains previous failures to utilize CFP successfully in double-label studies in E.coli. Cells were therefore grown overnight in the presence of inducer and then diluted to permit resumption of exponential growth before examination (see Materials and methods). Western blot analysis showed that the cellular concentrations of the labeled wild-type and mutant proteins were approximately the same under the 2 h and overnight induction protocols, indicating that a steady-state level had been achieved in most cells with the shorter growth period. The longer growth required for use of CFP might indicate a difference in folding efficiency or rate of chromophore formation between GFP variants since chromophore formation appears to be the rate-limiting step in generating fluorescent GFP proteins (Heim et al., 1994; Davis et al., 1994).
Study of RC1/pYLS68 (ΔminCDE/Plac-yfp::minD minEWT::cfp) confirmed that MinD polar zones and MinE rings and polar zones were present in nearly all cells that expressed wild-type minE (Figure 5B and C). The MinD polar zones and MinE rings and polar zones (Figure 6A and B) underwent the characteristic pole-to-pole movement that has been described previously in single-labeled GFP–MinD and MinE–GFP cells (Raskin and de Boer, 1999b; Fu et al., 2001; Hale et al., 2001). The MinD oscillation rate (in cycles per min) in the yfp::minD minEWT::cfp double-labeled cells (Kosc = 0.27 ± 0.18, n = 25) was slower than in singly labeled gfp::minD minEWT cells (Kosc = 1.25 ± 0.24, n = 8). The reason for the slower oscillation rate in the doubly labeled cells is not known. Similar results were obtained by Hale et al. (2001) in comparing MinD oscillation rates in gfp::minD minEWT::bfp double-labeled cells and gfp::minD minEWT single-labeled cells. In the GFP/BFP double-label experiments, the weakness of the blue fluorescent protein (BFP) fluorescence signal precluded the observation of both labeled proteins in the same cell.
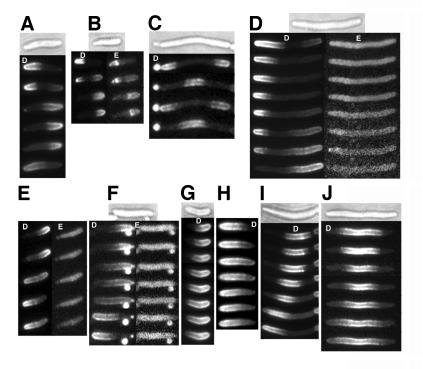
Fig. 6. Localization of YFP–MinD and MinE–CFP in living cells. Time-lapse fluorescence micrographs of strain RC1 containing: (A–C) pYLS68 (Plac-yfp::minD minE::cfp) or (D–J) pYLS77 (Plac-yfp::minD minED45A/V49A::cfp). White letters: D, YFP–MinD; E, MinE–CFP. Nomarski images are also shown. Time intervals between images were: (A–F) 1 min; (G) 2 min; (H) 1.5 min; (I) 1.75 min; (J) 1 min. In (B) and (D–F), MinE–CFP images (on the right) were acquired 10 s after the corresponding YFP–MinD images.
MinD and MinE localization in minED45A cells
When wild-type minE was replaced by minED45A [in pYLS69 (Plac-yfp::minD minED45A::cfp)], MinE rings were visible in ∼40% of fixed cells (Figure 5G), although the rings were less distinct and more difficult to visualize than in the case of wild-type E-ring. In the same cells, MinD polar zones were present in almost all cells (Figure 5F). The concentrations of MinD and MinE were similar in the wild-type and minED45A cells (Figure 5M–O). These observations indicated that the MinED45A protein is less efficient than wild-type MinE in forming E-rings, but is still capable of fully supporting the formation of MinD polar zones.
It was interesting that MinE–CFP rings were visualized in the doubly labeled minED45A::cfp cells in larger numbers than in the singly labeled minED45A::gfp cells when both were grown under the same conditions. This does not reflect differences in fluorescence emission of GFP and CFP (Clontech Living Colors User Manual). It also did not result from differences in MinE concentration as determined by immunoblot analysis. The increased number of E-rings in the yfp::minD minED45A::cfp cells might reflect interaction between the YFP and CFP moieties since GFP is capable of self-interaction (Clontech Living Colors User Manual) and YFP and CFP are derived from GFP (Cormack et al., 1996; Tsien, 1998). These secondary MinD–MinED45A interactions could have stabilized nascent or unstable MinED45A rings in the mutant cells if ring assembly depends on MinE–MinD interactions.
MinD and MinE localization in minED45A/V49A cells
The Asp45 and Val49 residues form a four amino acid cluster on the α-face of the MinE dimer (Figure 2B). The studies of the minED45A and minEV49A single mutants suggested that the Asp45Val49 cluster might identify a localized site that plays a role in formation of the MinE ring. To determine whether complete loss of the site might give a more pronounced phenotype than seen in the minED45A and minEV49A single mutants, we constructed a minE double mutant in which MinE residues Asp45 and Val49 were both replaced by alanine. When minED45A/V49A::gfp was expressed in the presence of minC and minD, the cells showed a minicelling pattern that was similar to that of cells expressing minED45A:: gfp or minED45A (Figure 3e in King et al., 2000). As discussed above, MALLS and gel filtration analysis showed that the D45A/V49A mutation did not alter the oligomerization state of MinE. The localization patterns of MinED45A/V49A–CFP and YFP–MinD from the doubly labeled plasmid pYLS77 (Plac-yfp::minD minED45A/V49A:: cfp) were then determined.
As shown in Figure 5K, the minED45A/V49A cells contained no detectable E-rings or MinE polar zones in fixed cells under conditions where E-rings and MinE polar zones were present in nearly all cells that expressed wild-type MinE (Figure 5C). The MinED45A/V49A–CFP fluorescence was distributed diffusely throughout the cell, with no evidence of a peripheral pattern that would suggest membrane localization of the MinED45A/V49A protein. This suggests that all or most of the MinED45A/V49A was located in the cytoplasm although some MinE may have been present in the membrane at low levels that could not be discriminated by fluorescence microscopy. Strikingly, despite the absence of E-rings and MinE polar zones, MinD polar zones were present in most of the minED45A/V49A cells (Figure 5J). Similar results were obtained in living cells (Figure 6D–F); MinE–CFP rings and polar zones were not detectable whereas YFP–MinD polar zones were present in nearly all cells. The absence of E-rings and presence of MinD polar zones in the minED45A/V49A mutant cells in the double-label experiments was confirmed in single label experiments with pYLS92 (Plac-yfp::minD minED45A/V49A) and pYLS76G (Plac-minC minD minED45A/V49A::gfp).
Immunoblot analysis using anti-MinD, anti-MinE and anti-GFP antibody (Figure 5M–O) showed that the cellular concentrations of YFP–MinD and MinED45A/V49A–CFP were similar to those of YFP–MinD and MinE–CFP in the parallel experiments with wild-type MinE. This excluded the possibility that the absence of visible MinED45A/V49A rings and polar zones reflected differences in expression or turnover of the wild-type and mutant proteins. We conclude that MinE rings and polar zones are not required for formation of MinD polar zones.
The GFP and CFP moieties in minE::gfp and minE::cfp both affected the cellular concentration of MinE. This led to an increase in MinE/MinD ratios as compared with cells that expressed unlabeled minE (Figures 2C, 3G and H, and 5O). The reason is unknown, although it could reflect an effect of the GFP/CFP moieties on the turnover rate of MinE. The effect on MinE concentration was the same for the wild-type and mutant proteins (Figures 3 and 5), indicating that it was not responsible for the altered localization and changes in dynamic behavior of MinE and MinD in the mutant strains.
The MinD polar zones were significantly longer in the doubly labeled minED45A and minED45A/V49A cells than in minEWT cells (Figure 5Q). In the presence of wild-type MinE, 82% of the polar zones were shorter than half the cell length, whereas in the presence of MinED45A and MinED45A/V49A, respectively, 40 and 76% of the polar zones in YFP/CFP-labeled cells extended beyond the midpoint of the cell. The MinD polar zones were also significantly longer in minED45A and minED45A/V49A singly labeled cells, where only MinD was labeled with the fluorescent probe (Figure 5P); 45 and 68% of polar zones extended beyond midcell in minED45A and minED45A/V49A cells, respectively, whereas only 10% of polar zones extended beyond midcell in the presence of wild-type MinE. The distribution should represent cells at all stages of the cycle of polar zone assembly and disassembly.
The MinD polar zones in living minED45A/V49A cells were capable of moving from pole to pole in the absence of a visible E-ring (Figure 6D–F), but the pattern of polar zone assembly and disassembly was significantly perturbed. In the presence of wild-type MinE, the growth of the MinD polar zone almost always stops when the edge of the zone approaches midcell (Fu et al., 2001; Hale et al., 2001) (Figure 1). The polar zone then retracts back toward the original pole during the polar zone disassembly phase of the cycle. In contrast, in the presence of MinED45A/V49A, the MinD polar zone often continued to grow for a considerable distance beyond midcell (Figure 6F–H) before beginning to retract. In some cases, the switch from polar zone assembly to disassembly was characterized by stuttering, in which the medial edge underwent several short periods of retraction and regrowth before continuing to disassemble toward the cell pole. In many cells, the MinD polar zone grew until it reached the opposite pole (Figure 6F–H) so that the usual pole-to-pole oscillation did not occur. In these cases, the YFP–MinD was distributed peripherally around the entire surface of the cell, indicating that it remained membrane associated. After a highly variable time, the continuous zone of membrane-associated MinD began to disassemble, sometimes from the original pole (Figure 6F) and sometimes from the distal pole.
We conclude from these results that loss of the E-ring does not prevent the growth of the MinD polar zone nor its later unidirectional disassembly. However, the results indicate that loss of the E-ring does interfere with the arrest of polar zone growth that normally occurs when the leading edge of the polar zone reaches midcell, leading to delays in initiation of the disassembly stage and in interference with the coordination of the assembly and disassembly processes.
Internal MinD zones
Additional evidence that the absence of E-rings in minED45A/V49A cells is associated with loss of the normal regulation of growth of membrane-associated MinD zones came from study of internal MinD zones. In cells that co-express minD and wild-type minE, membrane-associated MinD zones are present along the body of the short filaments that accumulate in minicelling cultures (Figures 5B and 6C) and in cells that are blocked in cell division (Raskin and de Boer, 1999b). Formation of the internal MinD zones requires MinE. In the presence of wild-type MinE, each internal MinD zone is flanked by two MinE rings (Figure 5C and D). The internal zones undergo repetitive cycles of assembly, disassembly and reassembly that lead to their oscillating presence at adjacent intervals along the cell (Figure 6C).
This pattern was altered significantly when wild-type MinE was replaced by the mutant MinED45A/V49A protein. The flanking E-rings were absent (Figure 5K and L) and the internal MinD zones in the minED45A/V49A cells usually failed to stop growing when the zones achieved their normal size. As a result, the zones were often much longer than in wild-type cells. A variety of assembly/disassembly patterns were observed. In some cases, the internal zone appeared to drift unidirectionally within the membrane (Figure 6I). This pattern suggested that assembly took place at one end of a zone while disassembly occurred at the other end. This differed from the normal situation, where assembly and disassembly of internal MinD zones are symmetrical processes in which the two ends of the zone are growing or shrinking coordinately at the same time. Sometimes, the MinD zone underwent a period of symmetric retraction, where the zone shrank from both ends, followed by a period of growth from both ends (Figure 6J). This differed from the usual situation in which the zone continues to shrink until it disappears as part of the oscillation sequence (Raskin and de Boer, 1999b). We interpret all of these patterns as manifestations of a failure of the MinED45A/V49A protein to regulate zonal growth of the internal MinD zones properly and thereby coordinate the assembly and disassembly stages of the MinD cycle.
Rate of polar zone disassembly
To determine whether the absence of E-rings affected the rate of zonal disassembly, we measured the rate of shrinkage of the MinD polar and internal zones during their disassembly stages in minEWT and minED45A/V49A cells. This showed that the rate of zonal disassembly was slower in minED45A/V49A cells (16.2 ± 8.0 nm/s, n = 65) than in minEWT cells (21.7 ± 9.9 nm/s, n = 69; P = 0.0003).
Quantitation of MinD and MinE
It has been reported previously in separate studies that the cellular concentrations of MinE (Zhao et al., 1995) and MinD (de Boer et al., 1991) in E.coli UT481 were ∼200 and 3000 molecules per cell, respectively. We have re-examined the concentrations of the two proteins within the same cell extracts, using a different anti-MinE antibody (anti-MinD2–19) and more rigorously excluding possible protein degradation during sample preparation, in cells growing exponentially at 30 and 37°C. In strain MC1000, the min+ parent of the Δmin host strain used in the present studies, the concentrations of MinE and MinD were ∼1400 and 2000 molecules per cell, respectively, under all conditions. The results were similar in strain UT481. Thus, MinE and MinD were present at approximately equivalent concentrations, and the previous suggestion that MinD is normally present in large excess over MinE was apparently wrong, at least for cells grown and analyzed under these conditions (see Materials and methods).
Discussion
The specificity of division site placement is mediated by the pole-to-pole oscillation of the MinD/MinC polar zone and the MinE ring. In the present work, we began to dissect the details of this complex system by the study of mutations that uncoupled MinD polar zone formation from E-ring assembly. For this purpose, we used a double-label technique that made it possible, for the first time, to visualize MinD and MinE independently within the same cell.
The Asp45Val49 site
The mutagenesis studies identified a site on the surface of the TSD of MinE (the Asp45Val49 site) that is required for the normal process of formation of the MinE ring and polar zone. Because of the dimeric structure of MinE, Asp45 and Val49 define a contiguous four amino acid cluster that lies within a small cleft near the center of the α-helical face of the TSD (Figure 2B). Within the Asp45Val49 site, the entire Asp45 side chain is exposed on the surface, whereas only one of the terminal γ-methyl groups of Val49 is fully surface exposed. This probably explains the greater effect of the minED45A mutation than the minEV49A mutation on E-ring formation. Complete loss of visible E-rings and MinE polar zones required that both residues be mutated, supporting the idea that Asp45 and Val49 form part of a single functional locus.
These observations suggest that the Asp45Val49 site normally binds to a complementary site on another protein, possibly MinD or MinE itself, in an interaction that is required for assembly or stability of the MinE ring and polar zone. The interaction could be required directly for E-ring assembly if interactions between MinE molecules within the E-ring or between MinE and the edge of the MinD/MinE polar zone are involved in E-ring structure. This would explain the absence of E-rings in minED45A/V49A cells although it would not explain directly the absence of MinE polar zones. Alternatively, the Asp45Val49 site might only be involved in assembly of the MinE polar zone. If the E-ring is assembled normally on the free edge of a MinE or MinE/MinD polar zone, then an inability of minED45/V49A cells to form MinE polar zones would be sufficient to explain the failure to assemble the MinE ring.
Mechanism of polar zone assembly and disassembly
Strikingly, as discussed below, the absence of visible E-rings and MinE polar zones in the minED45A/V49A cells did not prevent the assembly and disassembly of MinD polar zones, although it significantly perturbed the temporal and spatial regulation of these processes.
In the present discussion, we define three stages in the dynamic behavior of the MinD polar zone: (i) assembly of the MinD polar zone; (ii) arrest of polar zone assembly near midcell, coupled to formation of the E-ring; and (iii) disassembly of the polar zone, associated with movement of the E-ring from midcell toward the pole. Normally, this sequence of events occurs repetitively, first at one pole and then at the opposite pole, in an oscillatory fashion. In this sequence of events, MinE is required for at least two events, assembly of the MinD polar zone and formation of the E-ring.
(i) Assembly of the MinD polar zone. It is not known why MinE is required for formation of the MinD polar zone. This function remained intact in the MinED45/V49A protein. The present study provides direct confirmation of previous suggestions (Raskin and de Boer, 1999b; Fu et al., 2001) that the MinD and MinE polar zones are located at the same end of the cell and oscillate synchronously. The similar appearance and location of the MinD and MinE polar zones and the fact that formation of the MinD and MinE zones each requires the co-expression of minE and minD (Raskin and de Boer, 1999a,b; Rowland et al., 2000) are compatible with a simple model in which the polar zone is formed by co-assembly of the two proteins into a common MinD/MinE polar zone. The observation in minED45A/V49A cells that the MinD polar zone can be assembled in the absence of visible MinE polar zones argues against the obligatory co-assembly idea, although it does not exclude the possibility that MinE and MinD do co-assemble and the Asp45Val49 site is required for the stability of MinE within the zone after the co-assembly event. Alternatively, MinE (and the MinED45A/V49A protein) may act on the assembly process only at the initial step, where MinD initiates the process of zonal assembly at the cell pole. In the latter scenario, there would be no need for MinE to participate in the subsequent entry of MinD molecules into the polar zone, thereby explaining the presence of MinD polar zones in the absence of MinE zones in the minED45A/V49A cells.
(ii) Arrest of MinD polar zone assembly and formation of the MinE ring. In cells that express wild-type minE, the growth of the MinD zone is arrested near midcell. This occurs at approximately the same time that the E-ring becomes visible at the leading edge of the polar zone (Figure 1), although the exact timing of E-ring appearance has not been established rigorously. As discussed further below, the present results suggest that the arrest of MinD polar zone growth may be a direct result of E-ring formation. The mechanism that temporally couples E-ring formation to the arrival of the MinD polar zone at midcell is not known.
(iii) Disassembly of the MinD polar zone. The stage of polar zone disassembly is defined by the progressive poleward shrinkage of the MinD polar zone. Disassembly of the polar zone begins after the MinE ring is formed, and the E-ring normally remains close to the medial edge of the zone during the disassembly process. The close association of the MinE ring with the edge of the MinD polar zone during the disassembly phase has suggested that the high concentration of MinE in the E-ring might play a direct role in the disassembly process. Based on the demonstration that MinE can activate the ATPase activity of MinD in vitro, it was proposed that the E-ring is responsible for the disassembly of the polar zone and that this is accomplished by the MinE-mediated activation of the ATPase activity of MinD molecules at the edge of the polar zone (Hu and Lutkenhaus, 2001). However, the present study shows that the progressive disassembly of MinD polar zones and internal zones can take place in the absence of a visible E-ring.
Role of MinE and the E-ring
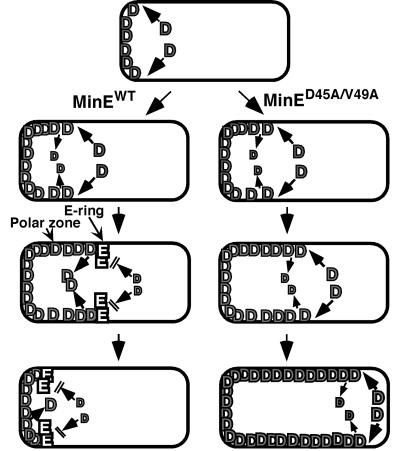
Instead of a primary role for the E-ring in disassembly of the MinD polar zone, we propose that the major role of the E-ring is to prevent addition of new MinD molecules at the leading edge of the polar zone, thereby blocking further growth of the zone (Figure 7). In this view, the high concentration of MinE in the E-ring provides a stop signal that terminates the polar zone assembly phase when the polar zone approaches midcell, thereby triggering the transition from polar zone assembly to polar zone disassembly.
Fig. 7. Suggested role of the E-ring in regulating growth of the MinD polar zone. The MinD polar zone grows by addition of new MinD molecules at the medial edge of the zone at a rate that exceeds the rate of spontaneous release of MinD molecules from the polar zone. When the polar zone approaches midcell, the E-ring assembles at its leading edge, and the E-ring then blocks subsequent entry of MinD molecules. In the absence of addition of MinD molecules, the polar zone shrinks, due to spontaneous desorption of MinD. The rate of MinD desorption may also be increased by the presence of the E-ring, as suggested by the decreased rate of shrinkage of the MinD zones in minED45A/V49A cells that lacked MinE rings (see Results). In the absence of the MinE ring, MinD molecules continue to be added to the growing edge of the polar zone, permitting the zone to grow past the midcell division site. MinE is not shown within the polar zone but, in wild-type cells, MinE within the polar zone presumably behaves like MinD. See text for further discussion.
This would explain the temporal coupling of the arrest of growth of the MinD polar zone to the appearance of the E-ring in cells that contain wild-type MinE. It would also explain the observation that the absence of visible E-rings in minED45A and minED45A/V49A cells is associated with loss of regulation of growth of the MinD polar zone, leading to the continued growth of the zone beyond the normal stop point near midcell (Figure 5P and Q). The idea that the role of the E-ring is to terminate the zonal growth stage is also consistent with the observed failure of the internal MinD zones that are present in long minED45A/V49A cells to stop growing when the zones reach their normal size and with the loss of the normally ordered cycles of assembly and disassembly of the internal MinD zones in filamentous cells.
All of the observations on the patterns of assembly of polar and internal MinD zones in cells that lack E-rings can be explained by a failure to terminate zonal assembly at the proper time, leading to an apparently random choice of timing of the switch from assembly mode to disassembly mode. The possibility that other defects may also be present cannot be excluded.
Mutational alteration of the Asp45Val49 site leads to a minicelling phenotype. This presumably results from the defect in arrest of polar zone growth that frequently permits the zone to extend beyond the normal midcell stop point. This was reflected by the increased length of the polar zones in minED45A and minED45A/V49A cells. In both wild-type and mutant cells, new polar zones usually fail to initiate at the free cell pole until the polar zone at the other end of the cell stops growing and begins to retract, perhaps because polar zone disassembly is required to provide sufficient free MinD molecules to initiate assembly of the new zone. In many of the mutant cells, the opposite pole is free of MinD for relatively long periods of time because of the prolonged period of polar zone growth. Because the distribution of MinC normally depends on the distribution of MinD (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a), we speculate that the opposite pole also remains free of the MinC division inhibitor for a long enough time to initiate the polar division process that gives rise to minicells. Establishment of the basis of minicelling in the minE mutants must await studies of the simultaneous localization of MinC and MinD in the mutant cells.
Despite the absence of E-rings, minED45A/V49A cells were still able to disassemble the polar and internal MinD zones, although the onset of the disassembly stage was delayed and irregular. The disassembly process was polarized in both wild-type and mutant cells, with fluorescence disappearing progressively from the end of the zone rather than decaying randomly along the length of the zone. This suggests that the basic mechanism of disassembly is likely to be similar in the two cases.
How might disassembly of the membrane-associated MinD zones take place in the absence of the E-ring? It is possible that the assembly/disassembly transition can occur spontaneously in the absence of the E-ring, although inefficiently, due to random fluctuation of the state of the leading edge of the polar zone that prevents further addition of MinD molecules. Disassembly might then occur due to the spontaneous desorption of MinD molecules that normally is counterbalanced by the higher rate of addition at the growing edge of the zone (Figure 7). Desorption might, for example, be catalyzed by the endogenous ATPase activity of MinD (de Boer et al., 1991). Since a later fluctuation could restore the state of the leading edge, this could explain why shrinking MinD zones in minED45A/V49A cells sometimes prematurely resume their growth. If this were correct, the only role of the E-ring would be to give temporal and spatial specificity to the triggering of the assembly/disassembly transition and to ensure completion of the disassembly phase. There presently is no suggested mechanism for the predicated spontaneous fluctuation of the state of the leading edge of the polar zone.
On the other hand, MinE might still play a role in the disassembly process even in the absence of a visible E-ring. It has been suggested by Hu and Lutkenhaus (2001) that the MinE ring is responsible for driving polar zone disassembly, acting by activating the ATPase activity of MinD molecules at the edge of the polar zone. While the present work shows that the high concentration of MinE in the E-ring is not needed for the disassembly process (Figure 6D), disassembly might still be driven by MinE-modulated activation of MinD ATPase. In wild-type cells, the high density of MinE within the E-ring provides a high local MinE concentration at the leading edge of the polar zone. In contrast, in minED45A/V49A cells, the MinE molecules are distributed diffusely within the cell so that the local concentration of MinE in the vicinity of the leading edge of the polar zone is substantially lower than in wild-type cells. Because of this, the probability of productive interaction of MinE molecules with the edge of the MinD polar zone will be much lower in the mutant cells. If arrest of polar zone assembly requires the binding of a critical number of MinE molecules at the edge of the zone, the lower probability of interaction would lead to the delayed and irregular arrest of MinD polar zone assembly that was observed in the minED45A/V49A cells. The MinE molecules associated with the leading edge that caused arrest of polar zone assembly might also be sufficient to drive the disassembly process, as suggested for the MinE ring by Hu and Lutkenhaus (2001). The slower rate of polar zone disassembly in mutant cells that lack visible E-rings would be consistent with this. The disassembly process could be catalyzed by a MinE function that resides in another region of the protein, perhaps the N-terminal region, or could reflect a low level of residual activity of the mutated C-terminal TSD. In view of the suggestion that activation of the MinD ATPase activity by MinE drives the polar zone disassembly process, it will be of interest to see whether the MinED45A/V49A protein is still capable of activating the ATPase activity of MinD. Further work will be needed to distinguish between these and other possibilities.
Materials and methods
Strains, growth conditions and nomenclature
The E.coli strains used in this study were RC1 (ΔminCDE; Rowland et al., 2000) and DH5α (Bethesda Research Laboratories). Strains were grown in LB medium and the required antibiotics were present in all cultures. The nomenclature of minE alleles and gene products indicates the change in the protein product (e.g. minED45A indicates a mutant allele in which amino acid 45 has been changed from aspartate to alanine; MinE31–88 indicates a MinE fragment containing amino acids 31–88).
Site-directed mutagenesis
To introduce changes in the target amino acids of MinE, complementary 25–35 bp primer pairs were designed to introduce point mutations into minE in a PCR using plasmid pSY1083 (Plac-minC minD minE) as template, in a reaction catalyzed by Pfuturbo DNA polymerase as described by Stratagene. This yielded the complete plasmid containing the desired minE mutation. The PCR products were treated with DpnI to remove parental plasmids (Nelson and McClelland, 1992) and then transformed into DH5α. The plasmids were sequenced to confirm the presence of the desired base substitutions. In the case of mutations that were associated with phenotypic changes, including pYLS4 (Plac-minC minD minED45A) and pYLS40 (Plac-minC minD minEV49A), the presence of the desired mutations in minE and the absence of mutations in minC and minD in the same plasmid were confirmed by DNA sequencing.
Plasmids and bacteriophages
For a list of the plasmids used in this study, see Table I. Plasmids pSY1083G (Plac-minC minD minE::gfp), pYLS4G (Plac-minC minD minED45A::gfp) and pYLS40G (Plac-minC minD minEV49A::gfp) were obtained by treating pSY1083 (Fu et al., 2001), pYLS4 and pYLS40, respectively, with BamHI followed by religation to generate in-frame minE::gfp fusions. pYLS33 (Plac-gfp::minD minED45A) was constructed by a three-fragment ligation between the EcoRI–XmnI fragment from pFX9 (Rowland et al., 2000) that contains gfp::minD, an XmnI–XbaI fragment from pYLS4C, containing minED45A, that was blunt-ended at the XbaI end by treatment with Klenow fragment and deoxynucleoside triphosphates, and the EcoRI–HindIII fragment from the pMLB1113 vector (de Boer et al., 1989) in which the HindIII end of the fragment was blunt-ended. pYLS46 (Plac-gfp::minD minEV49A) was constructed in a three-fragment ligation between the EcoRI–XmnI fragment from pFX9, the EcoRI–HindIII fragment from the pMLB1113 vector and the XmnI–HindIII fragment containing minEV49A that was obtained by PCR from pYLS40. pFX40 (Plac-yfp::minD minE) was obtained by replacing the gfp cassette between the EcoRI and XbaI sites of pFX9 with yfpmut2, obtained by PCR from pKL184 (kindly provided by Cormack et al., 1996). pFX56 (Plac-minC minD minE::cfp) was obtained by replacing the gfp cassette between the BamHI and HindIII sites of pSY1083G with cfp, obtained by PCR from pJCL61 [kindly provided by R.Tsien (Tsien, 1998)]. For the construction of pYLS68 (Plac-yfp::minD minE::cfp), a PCR was performed using pFX40 as template to obtain the yfp::minD minE fragment; the PCR also introduced a BamHI site to replace the stop codon of minE. The PCR product was digested with EcoRI and BamHI and used to replace the EcoRI–BamHI fragment in pFX56; this generated an in-frame minE::cfp fusion. Plasmid pYLS69 (Plac-yfp::minD minED45A::cfp) was made by replacing the XbaI–BamHI fragment in pYLS68 with the XbaI–BamHI fragment containing minD minED45A that was obtained in a PCR using pYLS33 as template. pYLS76G (Plac-minC minD minED45A/V49A::gfp) was constructed in a three-fragment ligation between the large EcoRI–BamHI fragment from pSY1083, the 1.5 kb EcoRI–XmnI fragment from pSY1083 and the XmnI–BamHI fragment containing minED45A/V49A obtained by PCR from pYLS77. pYLS77 (Plac-yfp::minD minED45A/V49A::cfp) was made by introducing the V49A mutation into pYLS69 using mutagenic PCR as described above. pYLS92 (Plac-yfp:: minD minED45A/V49A) was made by ligating the large XmnI–HindIII fragment from pFX40 to the XmnI–HindIII fragment containing minED45A/V49A that was obtained by PCR from pYLS77.
Table I. List of plasmids.
| Genotypea |
minE alleles |
|||
|---|---|---|---|---|
| minEWT | minED45A | minEV49A | minED45A/V49A | |
| Plac-minC minD minE | pSY1083b | pYLS4 | pYLS40 | |
| Plac-minC minD minE::gfp | pSY1083Gb | pYLS4G | pYLS40G | pYLS76G |
| Plac-gfp::minD minE | pFX9c | pYLS33 | pYLS46 | pYLS92d |
| Plac-yfp::minD minE::cfp | pYLS68 | pYLS69 | pYLS77 | |
aThe presence of the desired mutations and the absence of min mutations elsewhere were confirmed by sequencing the entire min locus of the plasmids.
dContains yfp in place of gfp.
Bacteriophages λSY1083G (Plac-minC minD minE::gfp) and λYLS4G (Plac-minC minD minED45A::gfp) were constructed by crossing the minC minD minE::gfp segments from pSY1083G and pYLS4G into λNT5 (de Boer et al., 1989).
Determination of cell division phenotype
The effects of the mutant and wild-type minE alleles in plasmids pSY1083, pYLS4 and pYLS40 were studied in a ΔminCDE strain (RC1). The plasmid-containing strains were grown in the presence of 0.25% glucose for 3–4 h to an OD600 nm of ∼0.4. Cells were then washed and resuspended in fresh medium that contained 0.01% glucose or 10 µM IPTG. The cultures were grown at 37°C to OD600 nm 0.4–0.5. Unfixed cells or cells fixed in 0.5% glutaraldehyde for 30 min at room temperature were examined by phase contrast microscopy to determine the cell division phenotype as described previously (de Boer et al., 1989).
Fluorescence microscopy
For study of GFP fusion proteins, cultures were grown overnight at 37°C in the presence of 0.25% glucose and then diluted 100-fold into the same medium and grown for 2 h at 30°C unless otherwise indicated. Cells were washed twice, resuspended in the presence of 10 µM IPTG and grown at 30°C for an additional 2 h. Cells were examined on glass slides without fixation or were fixed in 0.2% glutaraldehyde/2% formaldehyde for 20 min at room temperature. For study of YFP- and CFP-labeled proteins, or for study of GFP-labeled proteins after long-term induction, cultures were grown in 0.25% glucose at 30°C until exponential phase was reached, and then diluted 5000-fold into medium containing 10 µM IPTG. After overnight growth at 30°C, the cultures were diluted 50 times in the same medium and were grown for 2–3 h at 30°C. Cells were then prepared and examined as described above.
Microscopy was performed with an Olympus BX60 microscope using Chroma filter sets 41071 for visualizing GFP, 41029 for visualizing YFP and 31044v2 for visualizing CFP. Images were collected using a Hamamatsu Orca digital camera (model C4742-95) and Openlab software (Improvision). Images were processed using Adobe PhotoShop. The same exposure times and other acquisition and processing parameters were used on all samples from each set of experiments. It was observed that the CFP fluorescence in E.coli was photobleached much more rapidly than YFP when repeated observations were made (see Figure 6B, D and F). In measurement of GFP–MinD oscillation cycles, one cycle is defined as the time from appearance of a polar zone at one end of the cell to the reappearance of a new polar zone at the same end, and the oscillation rate (Kosc) is expressed in cycles per min. In time-lapse experiments, images were captured every 20–30 s when one label (GFP or YFP) was monitored, and at 1 min intervals when CFP and YFP were both monitored in the same experiment. Cells were observed at 20–30 s intervals even when images were not captured, to verify that significant changes did not occur during the intervals. For each series, every second or third image is shown in the figures. Rates of disassembly of MinD zones were determined by measuring the change in length of the MinD zone during its shrinkage stage, as a function of time. P-values were derived using the Student’s t-test. The length of polar zones was measured from the tip of the cell to the edge of the polar zone. In the figures showing the ratio of polar zone length to cell length, 100–150 cells were measured for each curve.
Western blot analysis
Cell extracts were prepared by suspending cells in SDS–PAGE solubilization buffer and heating in a boiling water bath for 5 min. Western blot analysis was performed on 10 µg of cell extract as described previously (Zhang et al., 1998), using polyclonal antibodies directed against MinE2–19 (Zhang et al., 1998), MinD (Rowland et al., 2000) or GFP (Clontech). The blots shown within each figure were performed at the same time with the same batch of diluted antibody so that band intensities can be compared directly. Blots that are shown in different figures were done at different times, so that band intensities from figure to figure cannot be compared quantitatively. Quantitation was performed by comparing the intensities of the MinD or MinE band from western blots on several aliquots of the SDS extract with standard curves prepared from purified MinD and MinE on the same gel. Bovine serum albumin (BSA) was used as a standard to establish protein concentrations of purified MinE and MinD using the Bio-Rad (Bradford) assay reagent; when MinE and MinD concentrations were based on the Pierce BCA protein assay, the calculated MinE/MinD ratios in the extracts were similar, although the calculated cellular concentrations of MinE and MinD were ∼30% lower.
Gel filtration and multiangle laser light scattering analysis
For gel filtration analysis, MinE31–88 fragments, prepared as described previously (King et al., 1999), were concentrated and exchanged into 20 mM sodium phosphate pH 7.0, 0.1 M NaCl, 1 mM EDTA, 1 mM tris(2-carboxyethyl)phosphine (TCEP) using a YM-3 Centriprep device (Millipore). The samples were then applied to a Superdex 75 column (HR10/30, Pharmacia) and eluted in the same buffer (flow rate 0.5 ml/min). Eluted proteins were detected by absorbance at 220 or 276 nm. Molecular weight standard proteins were run separately to calibrate the column. MALLS was performed using a miniDAWN Tristar light scattering instrument and Optilab DSP interferometric refractometer (Wyatt Technology Corporation). Analyses were performed as described previously (Folta-Stogniew and Williams, 1999) except that size exclusion chromatography was performed on a Superdex 75 HR 10/30 column using 20 mM HEPES, 150 mM NaCl, 1 mM EDTA pH 8.0 as the elution buffer. MinE samples were prepared in elution buffer containing TCEP as reducing agent. Weight-average molecular weights were determined using the Debye fitting method (ASTRA software; Wyatt Technology Corporation) as described previously (Folta-Stogniew and Williams, 1999).
Acknowledgments
Acknowledgements
We acknowledge the help and advice of M.J.Osborn throughout this work and the assistance of J.Clive with statistical methods. We thank Scott Robson for assistance with the MALLS measurements. This work was supported by US National Institutes of Health grants GM-60632 to L.R. and GM-48583 to G.F.K.
References
- Cormack B.P., Valdivia,R.H. and Falkow,S. (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene, 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Davis D.F., Ward,W.W. and Cutler,M.W. (1994) Posttranslational chromophore formation in recombinant GFP from E.coli requires oxygen. In Campbell,A.K., Kricka,L.J. and Stanley,P.E. (eds), Bioluminescence and Chemiluminescence: Fundamentals and Applied Aspects. Proceedings of the 8th International Symposium on Bioluminescence and Chemiluminescence, Cambridge. Wiley, New York, NY, pp. 569–599. [Google Scholar]
- de Boer P.A.J., Crossley,R.E. and Rothfield,L.I. (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E.coli. Cell, 56, 641–649. [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Crossley,R.E., Hand,A.R. and Rothfield,L.I. (1991) The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J., 10, 4371–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta-Stogniew E. and Williams,K.R. (1999) Determination of molecular masses of proteins in solution: implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Tech., 10, 51–63. [PMC free article] [PubMed] [Google Scholar]
- Fu X., Shih,Y.-L., Zhang,Y. and Rothfield,L.I. (2001) The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the E.coli division cycle. Proc. Natl Acad. Sci. USA, 98, 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C.A., Meinhardt,H. and de Boer,P.A. (2001) Dynamic localization cycle of the cell division regulator minE in Escherichia coli. EMBO J., 20, 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R., Prasher,D.C. and Tsien,R.Y. (1994) Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl Acad. Sci. USA, 91, 12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. and Lutkenhaus,J. (1999) Topological regulation in Escherichia coli involves rapid pole-to-pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol., 34, 82–90. [DOI] [PubMed] [Google Scholar]
- Hu Z. and Lutkenhaus,J. (2001) Topological regulation of cell division in E.coli: spatiotemporal oscillation of MinD occurs through stimulation of its ATPase activity by MinE and phospholipid. Mol. Cell, 7, 1337–1343. [DOI] [PubMed] [Google Scholar]
- King G.F., Rowland,S.L., Pan,B., Mackay,J., Mullen,G. and Rothfield,L.I. (1999) The dimerization and topological specificity functions of MinE reside in a structurally autonomous C-terminal domain. Mol. Microbiol., 31, 1161–1169. [DOI] [PubMed] [Google Scholar]
- King G.F., Shih,Y.-L., Maciejewski,M.W., Bains,N.P.S., Pan,B., Rowland,S., Mullen,G.P. and Rothfield,L.I. (2000) Structural basis for the topological specificity function of MinE. Nat. Struct. Biol., 7, 1013–1017. [DOI] [PubMed] [Google Scholar]
- Nelson M. and McClelland,M. (1992) Use of DNA methyltransferase/endonuclease enzyme combinations for megabase mapping of chromosomes. Methods Enzymol., 216, 279–303. [DOI] [PubMed] [Google Scholar]
- Niki H., Jaffé,A., Imamura,R., Ogura,T. and Hiraga,S. (1991) The new gene mukB codes for a 177 kD protein with coiled-coil domains involved in chromosome partition in E.coli. EMBO J., 10, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S., Vollrath,B., Touriol,C. and Bouché,J.-P. (1995) Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol. Microbiol., 18, 321–329. [DOI] [PubMed] [Google Scholar]
- Raskin D. and de Boer,P. (1997) The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E.coli. Cell, 91, 685–694. [DOI] [PubMed] [Google Scholar]
- Raskin D. and de Boer,P. (1999a) MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol., 181, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A. (1999b) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of E.coli. Proc. Natl Acad. Sci. USA, 96, 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L., Justice,S. and Garcia-Lara,J. (1999) Bacterial cell division. Annu. Rev. Genet., 33, 423–448. [DOI] [PubMed] [Google Scholar]
- Rowland S.L., Fu,X., Sayed,M.A., Zhang,Y., Cook,W.R. and Rothfield,L.I. (2000) Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J. Bacteriol., 182, 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R.Y. (1998) The green fluorescent protein. Annu. Rev. Biochem., 67, 509–544. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Rowland,S., King,G. and Rothfield,L. (1998) Relation of the oligomeric structure of MinE to its topological specificity function. Mol. Microbiol., 30, 265–273. [DOI] [PubMed] [Google Scholar]
- Zhao C.-R., de Boer,P. and Rothfield,L. (1995) Proper placement of the E.coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl Acad. Sci. USA, 92, 4313–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]