Abstract
During the elongation cycle of protein biosynthesis, the specific amino acid coded for by the mRNA is delivered by a complex that is comprised of the cognate aminoacyl-tRNA, elongation factor Tu and GTP. As this ternary complex binds to the ribosome, the anticodon end of the tRNA reaches the decoding center in the 30S subunit. Here we present the cryo- electron microscopy (EM) study of an Escherichia coli 70S ribosome-bound ternary complex stalled with an antibiotic, kirromycin. In the cryo-EM map the anticodon arm of the tRNA presents a new conformation that appears to facilitate the initial codon–anticodon interaction. Furthermore, the elbow region of the tRNA is seen to contact the GTPase-associated center on the 50S subunit of the ribosome, suggesting an active role of the tRNA in the transmission of the signal prompting the GTP hydrolysis upon codon recognition.
Keywords: decoding/electron microscopy/elongation factor Tu/ribosome/tRNA
Introduction
In protein biosynthesis the decoding process is the fundamental step that assures the accuracy of the translation from mRNA into polypeptide. The codon in the mRNA has to be correctly recognized by the anticodon of the cognate tRNA, leading to its incorporation, while incorrect matches must lead to rejection. Elongation factor Tu (EF-Tu) plays a crucial role in this function of delivering the aminoacyl-tRNA (aa-tRNA) as part of the ternary complex (aa-tRNA·EF-Tu·GTP). When this ternary complex binds to the ribosome, the tRNA is bound in the A/T state, as its anticodon is able to interact with the mRNA in the A-site while its acceptor end is still bound to the factor (Moazed and Noller, 1989). In this way, the codon–anticodon match can be tested before the new amino acid is incorporated. When a cognate tRNA is present, EF-Tu catalyzes the hydrolysis of GTP and changes its conformation (Berchtold et al., 1993; Kjeldgaard et al., 1993; Polekhina et al., 1996), thereby reducing its affinity for the ribosome and the tRNA (Dell et al., 1990). The remaining EF-Tu·GDP binary complex leaves the ribosome, so that the tRNA is free to move its CCA end towards the peptidyltransferase center (PTC), allowing the polypeptide to be elongated by the new amino acid residue (Kaziro, 1978). The transition of tRNA from the A/T site to A-site is known as accommodation.
Several important questions have remained unanswered. First, how the anticodon of the aa-tRNA recognizes the codon in the mRNA while it is delivered by EF-Tu at a very different angle compared with the orientation assumed in the final A-site. Secondly, how this codon–anticodon recognition acts as a signal to trigger the GTP hydrolysis and the release of EF-Tu. And finally, what mechanism is employed that makes possible a tRNA rotation towards the PTC while maintaining the codon– anticodon pairing. As a signal for GTP hydrolysis, one possibility is that the cognate tRNA–mRNA interaction in the decoding center promotes conformational changes in the ribosome (Geigenmüller and Nierhaus, 1990; Powers and Noller, 1994; Lodmell and Dahlberg, 1997) that are communicated through the inter-subunit bridges to the GTPase-associated center of the 50S subunit (Rodnina and Wintermeyer, 2001). Another possibility that has been discussed is that a conformational change in the tRNA following codon–anticodon pairing (Moras et al., 1985, 1986) might act as the signal (Rodnina et al., 1994; Yarus and Smith, 1995).
The ribosome-bound EF-Tu ternary complex has previously been visualized by cryo-electron microscopy (EM) techniques in the presence of the antibiotic kirromycin (Stark et al., 1997; Agrawal et al., 2000b). Kirromycin is known to block the conformational changes in EF-Tu that follow GTP hydrolysis (Wolf et al., 1977; Parmeggiani and Stewart, 1985). As a result of this inhibition, the factor remains bound to the ribosome in a conformation that is thought to be an intermediate between the GTP and the GDP states. This previous study suggested that EF-Tu binds to the base of L7/L12 stalk of the 50S subunit, close to the universally conserved α-sarcin–ricin loop (SRL) of the 23S rRNA, and showed the anticodon loop of the tRNA interacting with the decoding region. One of the puzzles posed by the results of previous cryo-EM studies is that the orientation of the tRNA deduced does not facilitate codon–anticodon interaction.
In the present work we have employed cryo-EM to study a complex of 70S ribosomes with P-site tRNA and EF-Tu ternary complex carrying cognate aa-tRNA and GDP in the presence of kirromycin. The first three-dimensional (3D) reconstruction of the whole set of single particles (see Agrawal et al., 2000b) showed low (∼50%) density in the region of the ternary complex. Computation of a difference map revealed the overall binding position of the complex, but a detailed interpretation of binding sites was not possible. We have now applied a supervised classification strategy based on the similarity of these images with distinct 3D references. One of the resultant 3D maps shows the ternary complex with much higher definition than seen in the earlier reports (Stark et al., 1997; Agrawal et al., 2000b). Most importantly, EF-Tu and the tRNA are now distinctly recognizable, and the tRNA is seen to interact with the base of the L7/L12 stalk and with protein S12. Detailed fitting, supported by quantitative measures, reveals that the tRNA structure is different from the reported X-ray structures and from the structure deduced in the former cryo-EM work (Stark et al., 1997) due to a change of orientation in the anticodon arm, towards an orientation that facilitates codon–anticodon interaction. Furthermore, the T-loop side of the tRNA is seen to interact with the 58-nucleotide segment of the 23S rRNA that forms a complex with protein L11 (Wimberly et al., 1999), a region known as the GTPase-associated center (GAC). The results bring new insights in understanding the incorporation of tRNA into the ribosome and strongly implicate tRNA as an active player in this process.
Results
3D reconstruction and analysis of the cryo-EM maps
A set of 22 905 projections was selected by a previously described quasi-automated method (Lata et al., 1995). By image processing techniques (see Materials and methods), a first map was calculated. The map showed the presence of extra density located in the inter-subunit space, which we attributed to the ternary complex. However, the extra density was fragmented, and could only be seen by rendering the volume at lowered density threshold. This initial poor representation of the putative ternary complex can be understood either as the result of averaging a population of ribosomes with low occupancy of the factor, or as a consequence of merging particles where the complex exists in different conformations, or as a combination of both effects. Evidence that low occupancy is a major factor comes from the filter-binding assay (see Materials and methods), which estimated ∼50% occupancy for the ternary complex.
In order to separate the heterogeneous particle set into more homogeneous subsets, we have used a supervised classification method (details described in the Supplementary data, available at The EMBO Journal Online), where the images are classified according to their similarity to distinct references (see Frank, 1996). Briefly, our strategy for separation makes use of the resemblance that the ternary complex and EF-G share in their architecture and their overall binding position on the ribosome, and the fact that the projection of ribosomes carrying the ternary complex would show more similarity with an EF-G bound ribosome than with a ribosome with an empty A-site. After the selection, new 3D reconstructions were computed. Application of the same method of classification to a control set of empty ribosomes did not show a mass at the A-site in the reconstruction from the subset having highest similarity with the EF-G bound reference (see Supplementary data).
The final 3D cryo-maps are depicted in Figure 1A (map 1) and B (map 2). Final map 1 (Figure 1A) was reconstructed from particles more similar to an empty ribosome. It is clear that the ternary complex occupancy is poor. In contrast, the occupancy is large in final map 2 (Figure 1B), a reconstruction obtained from those particles most similar to an EF-G-bound ribosome. In this latter map (Figure 1B) the extra density (in red) is a continuous mass with a volume large enough to accommodate the whole ternary complex.
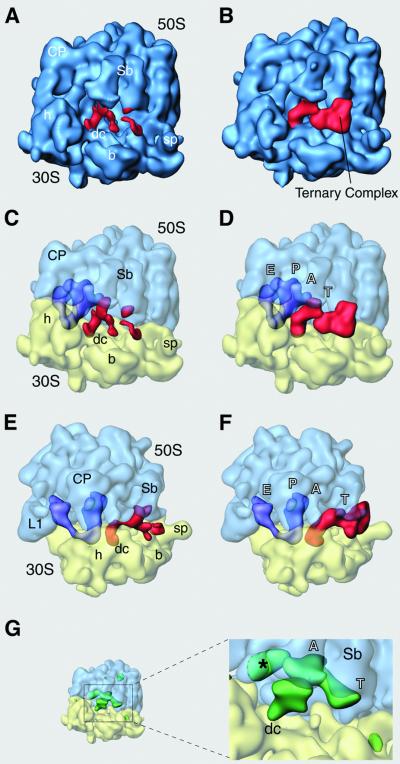
Fig. 1. Cryo-EM maps resulting from the classification, and their analysis. Map 1 (A, C and E) and map 2 (B, D and F) are depicted in two orientations related by a 90° rotation around a vertical axis in the plane [(A–D) show the side views, (E) and (F) the top views of the 70S ribosome). Side views are rotated in the plane by 90° from their conventional presentation (see Supplementary figure S1) to maintain continuity in the tRNA positions with (E) and (F)]. In (C)–(F) the ribosomal subunits are represented as semi-transparent surface (blue for the 50S and yellow for the 30S subunit), so that the positions of P- and E-site tRNAs (purple) and the mass attributable to the ternary complex (red) can be seen more clearly. (G) Representation of the difference map (see Materials and methods) calculated for the cryo-EM map 1, shown in green. Lobe of mass labeled with an asterisk is attributed to a conformational change in the L1 region. The ribosome is shown in same orientation as in (C). Landmarks are as follows: CP, central protuberance; Sb, L7/L12 stalk base; sp, spur; b, body; h, head; dc, decoding center; L1, stalk of L1 protein. Labels in tRNA positions: T, T-site within the ternary complex; A, aminoacyl site; P, peptidyl site; E, exit site.
We isolated the densities corresponding to tRNAs and tRNA–EF-Tu complex bound to the ribosome (see Materials and methods). This separation shows that both the maps carry P- and E-site tRNAs (purple in Figure 1), and the main difference relates to the presence of the ternary complex (red). [It should be noted that in all tRNA binding experiments (e.g. Cate et al., 1999; Agrawal et al., 2000a), a mass of density corresponding to the E-site tRNA is always observed.] In map 1 (Figure 1A, C and E) the putative ternary complex is under-represented and only few parts of the tRNA and the EF-Tu can be seen. Some density clearly appears at the expected binding site of the anticodon loop of the A-site tRNA on the 30S subunit, but the rest of the tRNA is not visible directly in the volume. In a calculated difference map (see Materials and methods) for this reconstruction (in green in Figure 1G) no major difference in the ribosome conformation is detected, but there is a large mass that covers the position of an A-site tRNA reaching from the decoding site in the 30S subunit to the PTC in the 50S subunit. The region that should correspond to the CCA end of the tRNA extends below the base of L7/L12 stalk, toward the region that EF-Tu binds to.
In map 2 (Figure 1B, D and F) the picture of the ternary complex (in red) changes drastically, and its full mass is now represented, together with the masses of P- and E-site tRNAs (purple). The anticodon arm of the tRNA is situated in the decoding region and the acceptor arm is still bound to the factor. No density attributable to an acceptor arm connected to the A-site in the 50S subunit is visible. In this reconstruction, both tRNA and protein interact with the base of the L7/L12 stalk, where contact points important for the activity of the elongation factors are located. The interactions between the ternary complex and the ribosome have been studied using this map.
Views of the mass attributable to the ternary complex from the solvent (Figure 2A) and from side of the inter-subunit space (Figure 2B) reveal a structure distinct enough to permit visual identification of tRNA and the domains of the protein factor. To understand the nature of the elements within the complex, the X-ray structure of the Phe-tRNAPhe·EF-Tu·GTP analog complex from Thermus aquaticus (Nissen et al., 1995) was filtered to the same resolution and displayed in similar orientation (Figure 2C and D). The tRNA and EF-Tu were colored separately for the purpose of clarity. We note that the structures come from different organisms (EF-Tu from T.aquaticus and from Escherichia coli share 68% sequence identity), the complexes have different guanine nucleotides bound, and while our cryo-EM map shows the ternary complex bound to the ribosome, the X-ray structure represents the complex in the close packing of a 3D crystal. Nevertheless, the overall mass distribution and the shape of the ribosome-bound ternary complex in our cryo-EM map and in the ternary complex crystal structure are strikingly similar.
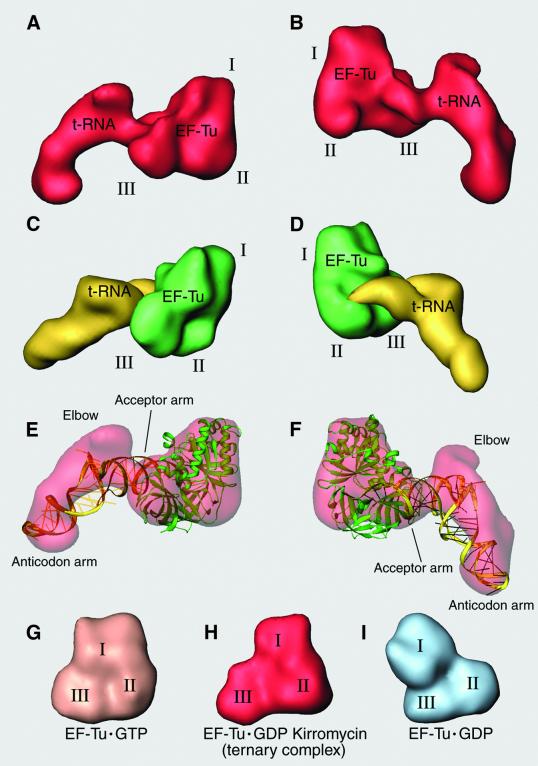
Fig. 2. Comparison of cryo-EM density and X-ray structure of the ternary complex. (A and B) Ternary complex density, isolated from the cryo-EM map: (A) as seen from the solvent side; (B) as seen from the intersubunit space side. (C and D) Equivalent orientations of the X-ray crystal structure from T.aquaticus ternary complex (Nissen et al., 1995), filtered to the resolution of the cryo-EM map. EF-Tu is shown in green, aa-tRNA in yellow. (E and F) Ribbons representation of the fitting of the ternary complex crystal structure into the cryo-EM density. (G–I) EF-Tu from T.aquaticus in the GTP (Kjeldgaard et al., 1993) and GDP (Polekhina et al., 1996) states are shown next to the EF-Tu·GDP·kirromycin inferred from the cryo-EM map at the same resolution.
The result of the rigid-body fitting of the X-ray structure form of the ternary complex into the cryo-EM map is presented in Figure 2E and F. Clearly, the domains of the elongation factor have not undergone a large rearrangement in the interaction with the ribosome, but the tRNA has. In the cryo-EM map, the acceptor arm of the tRNA that is held by EF-Tu apparently maintains its binding with the protein as in the crystal structure, but the elbow of the tRNA has moved significantly from its position in the crystal structure. However, in both versions of the tRNA, the anticodon loops are in the same position due to an apparent kink in the anticodon arm. In this way, the T and D loops forming the elbow of the tRNA in our cryo-EM map move toward the base of the L7/L12 stalk of the ribosome, while the two ends of the L-shaped structure remain in the same place, changes that imply a large internal reorganization of the tRNA structure.
Docking of tRNA and EF-Tu X-ray structures
The E.coli EF-Tu has been divided into three domains: domain I, the guanine nucleotide binding domain that accounts for the first 204 amino acids of the N-terminus; domain II, residues 205–298; and domain III, the C-terminal domain that covers positions 299–393. In the X-ray structure of T.aquaticus EF-Tu, the constellation of these domains changes significantly from the GTP (Figure 2G; from Kjeldgaard et al., 1993) to the GDP (Figure 2I; from Polekhina et al., 1996) states. It is clear that the architecture of EF-Tu within the kirromycin-stalled ternary complex (Figure 2H) resembles more closely the EF-Tu·GTP state in both, binary (Figure 2G) and ternary complexes (Figure 2C and D) than the GDP state (Figure 2I). This similarity with the GTP state is revealed by computation of cross-correlation coefficients between the crystal structures and the cryo-EM density (Table I). For E.coli, the X-ray structure was calculated in its GDP state (Song et al., 1999), and in order to obtain an acceptable fit, domain I needed to be moved significantly from its original position, close to domain III, to the position where the contacts between these two domains of the protein are minimized and enough room is created to accommodate the tRNA acceptor arm in the space available between them (Figure 3). Those changes produce a much better fit (Table I) and establish a GTP-state architecture.
Table I. Correlation between the fitted X-ray structures and the cryo-EM density.
| X-ray structure | Rigid body fitting | Flexible fitting |
|---|---|---|
| T.aquaticus EF-Tu·GDP | 0.51 | – |
| T.aquaticus EF-Tu·GTP | 0.78 | – |
| E.coli EF-Tu·GDP | 0.52 | 0.81 |
| Phe-tRNAPhe·EF-Tu·GTP | 0.69 | 0.83 |
| Phe-tRNAPhe | 0.77 | 0.81a |
aValue when the anticodon tip of the A-site tRNA was used for the fitting.
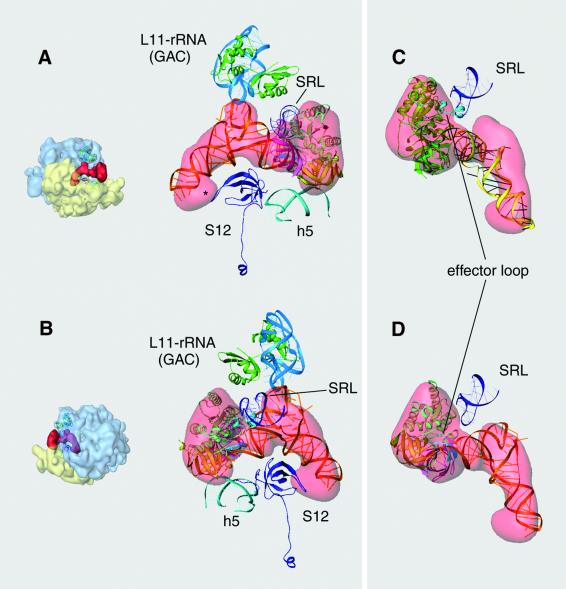
Fig. 3. Docking of EF-Tu and aa-tRNA into the cryo-EM density of the ternary complex: (A) as seen from the solvent side; (B) as seen from the intersubunit side. (Views are similar, but not identical to those in Figure 2A and B.) Fitted atomic coordinates of E.coli EF-Tu (Song et al., 1999) and Phe-tRNAPhe [(from the ternary complex of T.aquaticus (Nissen et al., 1995)] are shown inside the semi-transparent cryo-EM density of the ternary complex. The domains of EF-Tu were fitted independently (domain I shown in green, domain II in yellow and domain III pink). The switch-I region within domain I is highlighted in cyan. The dotted line in (A) indicates the place of the kink in the anticodon arm of the tRNA. Orientations of the ribosome are shown as thumbnails on the left. SRL, α-sarcin–ricin loop; L11–rRNA (GAC), protein L11 and the segment of 58 nucleotides of the 23S rRNA, also known as the GTPase-associated center; S12, protein S12 from the small subunit; h5, helix 5 from the 16S rRNA; dc, decoding center in the A-site of the 30S subunit.
In the case of the tRNA, the Phe-tRNAPhe within the ternary complex from T.aquaticus in the presence of a non-hydrolyzable GTP analog (Nissen et al., 1995) was used. It was not possible to get a full adjustment. The best docking was achieved by using the putative T and D loops in the elbow region of the L-shaped molecule as the main guide. The cryo-EM mass describes the above-mentioned change in the shape of the anticodon arm (‘kinked’ appearance; the position of the kink is indicated in Figure 3A by a dotted line), and this distortion causes the anticodon loop of the atomic coordinates of the Phe-tRNAPhe (Nissen et al., 1995) to lie partially outside the density mass (Figure 3A).
Inside domain I of EF-Tu there is a segment of the sequence that overlaps with the fitted tRNA structure in the CCA end (Figure 3B). This part of the protein corresponds to a single turn of an α-helix known as the switch-I region (depicted in cyan in the figures), a putative ribosomal interaction site that undergoes a dramatic conversion in conformation to a β structure in the transition from the GTP to the GDP state (Abel et al., 1996). Obviously, either the position of this part of the protein or the position of the tRNA must be changed in order to avoid the overlap.
Interaction of the ternary complex with the ribosome
Fitting of X-ray structures into the cryo-EM map reveals three of the most conserved regions in the ribosome that are present in the immediate vicinity of the ternary complex: the decoding site on the 30S subunit (see Ogle et al., 2001); the L11–rRNA complex (Wimberly et al., 1999); and the SRL (Wool et al., 1992) at the base of the stalk of the large subunit. These ribosomal components were fitted in both subunits to study their interactions with Phe-tRNAPhe and with EF-Tu. The results of this analysis are shown in Figures 4 and 5. According to the positions found, some of the interactions can now be described.
Fig. 4. Interaction of EF-Tu and aa-tRNA with the ribosome. (A and B) Ribbons representation of the docked EF-Tu and aa-tRNA within the ternary complex. (C and D) Focus on the interaction between the α-sarcin–ricin loop (SRL) and the effector loop within domain I of EF-Tu (cyan). In (C) the coordinates of the whole ternary complex from T.aquaticus with a GTP analog (Nissen et al., 1995) were used for the fitting, while in (D) the crystal structure of EF-Tu from E.coli bound to GDP (Song et al., 1999) was used. Orientation of the ribosomes for (A) and (B) are shown as thumbnails on the left. Labeling is the same as in Figure 5.
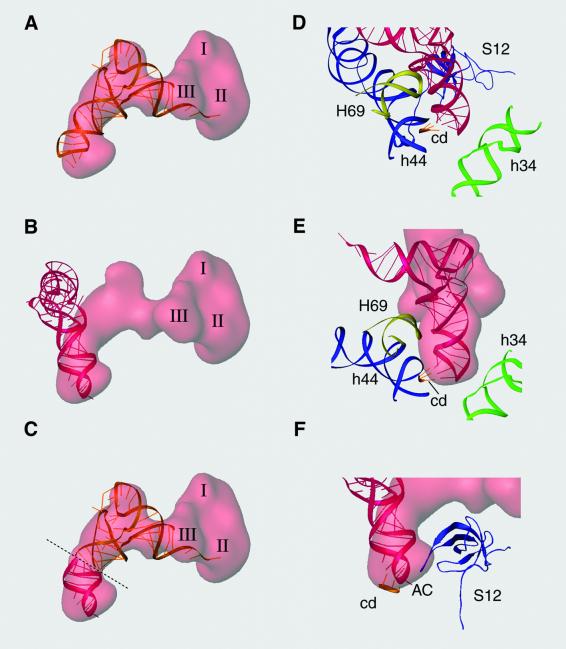
Fig. 5. Interaction in the decoding center and accommodation of the aa-tRNA. (A–C) Semi-transparent representation of the ternary complex density from the cryo-EM map showing the fitted tRNA (gold) and the A-site tRNA (red) with corresponding mRNA codon (Yusupova et al., 2001). (C) A tRNA construct in which the anticodon position, up to the kink, is adopted from (B) and the rest of the tRNA from (A). (D–F) Interaction in the anticodon loop of the tRNA in the decoding site. H69, helix 69 from 23S rRNA; h44, helix 44 from 16S rRNA; h34, helix 34 from 16S rRNA; cd, A-site codon in the mRNA; AC, anticodon loop; S12, protein S12 in the 30S subunit.
Turning first to the 50S subunit, the T loop of the aa-tRNA closely interacts with the region of the 58-nucleotide rRNA segment that binds to the L11 protein (Figure 4A and B). This portion of the ribosome is known as the GTPase-associated center (GAC) due to the fact that antibiotics that bind to it strongly affect the activity of EF-G and EF-Tu (Vazquez et al., 1979). The GAC is seen to interact with the T arm of the tRNA through a loop, around positions 1060–1075 in the 23S rRNA, which protrudes between the C- and the N-terminal domains of L11. In this loop, a single transversion in the residue 1067 impairs the function of the elongation factors (Saarma et al., 1997). Residues around position 2660 of the rRNA (within the SRL) were defined as binding site for EF-Tu by footprinting experiments (Moazed et al., 1988). The fitting leaves a distance between the SRL and EF-Tu large enough (10–15 Å) to be uncertain about the existence of a real binding contact. However, the effector loop region inside domain I of EF-Tu switches from an α-helical to a β-sheet structure and moves within the protein in the transition between GTP and GDP states (Abel et al., 1996). In Figure 4C and D, those two different positions for the effector loop are represented. When the coordinates of the EF-Tu·GTP form are used (Figure 4C), this switch-I region interacts with the SRL and avoids the earlier mentioned steric conflict with the CCA tip of the tRNA that the GDP state shows (Figure 4D).
In the 30S subunit the ternary complex forms three contacts, one with domain II of EF-Tu and two with the aa-tRNA. Domain II contacts helix 5 of 16S rRNA; the acceptor arm of the tRNA interacts with the protein S12; and the anticodon loop reaches the decoding center. In Figure 4A and B it can be seen that protein S12 extends into a loop that goes close to the decoding center. This loop includes the conserved PNSA (Pro-Asn-Ser-Ala) amino acid sequence at positions 48–51. The mass in the region of the anticodon tip of the tRNA is unexpectedly wide, but the additional mass could be explained as the result of a conformational change of this loop of S12 such that it reaches the density in immediate vicinity to the tRNA (labeled with an asterisk in Figure 4A).
Particular interest is focused on the codon–anticodon interaction and in the dynamic transition of the tRNA from the T- to the A-site. The rigid-body fitting of the tRNA structure inside the cryo-EM map leaves the anticodon loop partially outside the density (Figure 5A). Evidently, the tRNA crystal structure is different from the structure of the tRNA in the transition state observed here, and hence cannot be used to study the interactions in the region. However, it is important for understanding the process of tRNA accommodation that the anticodon portion of the tRNA interacting with the mRNA in the A-site (Agrawal et al., 2000a; Yusupova et al., 2001) partially overlaps the cryo-EM density of the tRNA up to the observed kink, and that the correlation coefficient is improved in the docking (Table I). The position of this A-site tRNA was deduced by the alignment of the P- and E-site tRNAs from the X-ray structure (Yusupova et al., 2001) with those from our density map. Figure 5C shows a merged representation of both tRNAs where the kink in the anticodon arm separates an A-site anticodon loop (red) from the rest of the tRNA still residing in the T-site (gold).
We have used the accommodated position of the tRNA to represent the interactions in the decoding center. In Figure 5B the density identified with the anticodon is broadened on both sides. The extra mass not filled with the tRNA structure might be understood to be a result of contributions from the tight interactions of the anticodon end with the surrounding ribosomal elements (Figure 5D). This way, the parts of helix 69 from 23S rRNA and helix 44 from 16S rRNA, which interact tightly with the codon of the mRNA (Figure 5E), and as well as a putative contribution due to a conformational change of S12 (Figure 5F), would be displayed as a compact mass at the resolution of our map.
Discussion
Ternary complex in the presence of kirromycin
The biggest problem that arose in our sample was its heterogeneity. Taking into account the occupancy of the ternary complex that the filter binding assays determined, it was reasonable to expect a significant fraction of the population of ribosomes to be without the factor. The classification produced two 3D reconstructions with different features. One of the maps, which showed very low occupancy, has to be interpreted with caution because the set of individual particles that it comes from represents, in all likelihood, a mixture of factor-free and factor-bound ribosomes. However, it shows an interesting presence of some densities in the calculated difference map that we attribute to the mass of tRNAs in the process of moving to the A-site from the initial position where they were bound to EF-Tu. This interpretation requires the assumption that kirromycin fails to block the conformational change of EF-Tu at least in a fraction of the ribosomes.
The good representation of the ternary complex in the high-occupancy map suggests the existence of a uniform subset of particles, and the inferred structure for the ternary complex has to be understood as the most abundant conformation of the complex in the sample, i.e. the conformation corresponding to the bottleneck in the inhibition of the decoding by the antibiotic. The kirromycin binding site in EF-Tu is formed by residues of domains I and III (Abdulkarim et al., 1994; Mesters et al., 1994), situated at a crucial place to stall the rearrangements of the interdomain positions that the change from GTP to GDP forms requires (Berchtold et al., 1993; Kjeldaarg et al., 1993; Polekhina et al., 1996; Song et al., 1999). Evidently, as our fitting shows, the structure that EF-Tu·GDP·kirromycin displays in the cryo-EM map is closer to a GTP constellation of domains. Furthermore, we have seen that the switch-I region (termed effector loop) inside the domain G of EF-Tu cannot be fitted properly into the isolated density when the coordinates from the E.coli factor in its GDP form are used. The effector loop is known to move and modify the secondary structure upon the change of the guanine nucleotide, and it is considered a putative interaction region with the ribosome (Song et al., 1999). By using the X-ray structure from the whole ternary complex with a GTP analog (Nissen et al., 1995) it is seen that the effector loop contacts the SRL from the ribosome as in the case of EF-G (Wriggers et al., 2000), and avoids a clash with the docked tRNA. Altogether, the data indicate that the ternary complex density in this work represents a stalled structure where the factor has been caught in an intermediate conformation, which is similar to the GTP form of the X-ray structure.
Interaction of the ternary complex with proteins and rRNA in the entrance channel of the ribosome
Previously, the only ribosome-binding region defined for EF-Tu was the above-mentioned SRL (Hausner et al., 1987; Moazed et al., 1988) interaction that in our map, as noted above, appears to include the switch-I region of domain I of EF-Tu. In the same work, chemical probes revealed that EF-G binds to the GAC in domain II of 23S rRNA, but no interaction from EF-Tu was detected in this region. On the other hand, the rRNA segment that folds with the L11 protein is known to have a direct influence on the GTPase activity of both factors when they are bound to the ribosome: the GAC is the binding site for antibiotics such as thiostrepton and micrococcin that affect the GTPase activity not only of EF-G, but also of EF-Tu (Vazquez, 1979). Even a single change of the nucleotide in position 1067 of the 23S rRNA (located within the GAC) was found to impair the function of both elongation factors (Saarma et al., 1997). In our cryo-EM map there is a clear interaction between the ternary complex and the GAC, but the interaction involves the tRNA instead of EF-Tu. More precisely, it is the T-loop region of the tRNA, the part of the nucleic acid that contacts this region. In the experiments probing 23S rRNA protection by the ternary complex against chemical modification (Moazed et al., 1988), a tRNA–GAC interaction was not observed, but neither did P- or A-site tRNAs give any effect in the 23S rRNA labeling pattern. The discrepancy of the results seems to be related to the reduced sensitivity of the technique for tRNA–rRNA interactions rather than to a real difference in the sample.
When comparing the overall structure of the ternary complex with that of EF-G, one is struck by their similarity (Nissen et al., 2000) and finds that individual domains of EF-G are mimicking parts of the ternary complex. In the case of the tRNA within the ternary complex, the domains in EF-G that build up a more or less compact structure are domains III, IV and V. Domain IV goes into the decoding site in the 30S subunit, the same place the anticodon of the tRNA binds to. Domain V of EF-G is the substitute for the T region of the tRNA. It is this domain that binds to the GAC in the case of EF-G (Agrawal et al., 1998, 1999, 2001). According to our present findings, the molecular mimicry goes beyond this, and now includes the fact that the interaction of protein S12 with the acceptor arm of the tRNA inside the ternary complex closely parallels its interaction with domain III of EF-G (Agrawal et al., 1998). In our fitting the protein is also close to the anticodon loop of the tRNA in the decoding site.
In the previous cryo-EM reconstruction of a ribosome– ternary complex (Stark et al., 1997), where the ternary complex density was seen without requiring any classification, a connection between EF-Tu and the base of the L7/L12 stalk close to the L11–rRNA complex was shown at the same place where the arc-like connection was observed in the EF-G·GDP·ribosome complex (Agrawal et al., 1998, 1999, 2000b). In our new map this connection cannot be seen. An interpretation of the binding in this region was made that implicated L7/L12 in the interaction (Stark et al., 1997). However, this and other differences in fine features can be caused by the difference in resolution of the recon structions [using the same criteria as defined in Orlova et al. (1997), the resolution of our map is 11 Å, as compared with the 18 Å resolution reported by those authors].
An active role for the tRNA
The incorporation of tRNAs into the A-site in the ribosome by EF-Tu follows a sequence of closely related steps: initial binding of the ternary complex to the ribosome; codon recognition; GTP hydrolysis; release of the EF-Tu·GDP accompanied by the accommodation of the tRNA in the A-site; and formation of a new peptide (Rodnina et al., 1994). Two main questions that remained unanswered have been how the codon–anticodon interaction triggers the GTP hydrolysis by EF-Tu and how the conformational changes after this event facilitate both the exit of the factor and the accommodation of the tRNA. In the present work, the ribosome-bound ternary complex was blocked with kirromycin, so that the structure visualized in our cryo-EM map represents an intermediate state after the GTP hydrolysis and before the completion of the accommodation process. The antibiotic kirromycin is known to stimulate the GTP hydrolysis by EF-Tu (Wolf et al., 1977) and we cannot distinguish between a GTPase stimulation by the antibiotic from an activation due the codon–anticodon recognition. Nevertheless, the cognate aa-tRNA shows a clear interaction with the decoding center, and we assume that the anticodon-pairing signal has been triggered.
The elbow in the L shape of the tRNA that comprises the D- and T-loop regions moves significantly toward the ribosome, and the T loop contacts the GAC of the 23S rRNA. Evidence of this movement, together with the evidence that cognate codon recognition is not sufficient for the GTPase activation and that an intact tRNA is required for the transmission of the signal (Piepenburg et al., 2000), points directly to the role of tRNA as the communicator that translates a cognate interaction in the decoding center into a signal for GTPase activation in the EF-Tu interaction with the ribosome. It is necessary to point out that in the previous cryo-EM work (Stark et al., 1997) a change in the relative position between the tRNA and EF-Tu was described, but no changes were detected in the structure of the aa-tRNA, nor did the study detect any interaction between the tRNA and the GAC.
In dynamic studies of the interaction of the ternary complex with the ribosome, Rodnina et al. (1994) found that the D loop of the tRNA, labeled with proflavin, changed its structure after codon–anticodon recognition and before GTP hydrolysis. These changes were deduced from a rapid increase of fluorescence followed by a slow decrease, reflecting two different levels of exposure of the label. In that work, the same experiment in the presence of kirromycin revealed an initial rapid increase of the signal, while the slow decrease was completely suppressed. Thus, the change that the tRNA structure undergoes upon codon recognition is not reversed when the complex is bound to the antibiotic. We conjecture that the structural change in the tRNA deduced in the dynamic experiments (Rodnina et al., 1994) is captured by the cryo-EM map we have obtained, where the tRNA shows a distorted structure and reaches the GAC in the 50S subunit, apparently sending the signal for the GTP hydrolysis.
The idea of the tRNA having an active role in translation is not new and has been postulated in several works by Woese (1970) (and more recently Woese, 2001), a concept that is at odds with the currently prevailing view of tRNAs as mere adaptors during the process.
Flexibility of tRNA: codon recognition and accommodation
By proposing that the relative movement of parts of the tRNA acts as signal after the codon–anticodon recognition we assume that the tRNA possesses flexibility. Indeed, in our data, the tRNA appears distorted and we have been unable to fit any atomic structure of tRNA from the CCA end all the way up to the anticodon loop without leaving significant parts of the nucleic acid outside the map. The change in the structure of the tRNA occurs during the interaction with the ribosome following cognate tRNA–mRNA recognition (Rodnina et al., 1995). The question is, can the codon–anticodon interaction promote structural changes in the aa-tRNA? Interesting results were introduced with the crystal structure of a yeast tRNAAsp (Dumas et al., 1985). In the 3D crystals the tRNA molecules were interacting via their GUC anticodon sequence with a slight mismatch at the uridine position, and the structure was considered a model for the effects of a codon–anticodon binding on the tRNA (Moras et al., 1986). The comparison with the yeast tRNAPhe structure revealed a more rigid anticodon loop and a gain in flexibility of the T and D loops in tRNAAsp versus tRNAPhe. Studies with tRNA mutants have shown that tRNAs with altered D and T arm interactions have different degrees of stability on the ribosome and favor near-cognate recognition (reviewed in Yarus and Smith, 1995). These authors proposed a general ‘waggle’ theory that attributes some role to the flexibility and structural reorganization of the elbow region in the tRNA during the decoding process. Thus, there are data that suggest conformational changes in the D and T arms of the tRNA after the pairing of the anticodon loop with the com plementary RNA sequence, and indicate that the disruption of this region has direct effects on the accuracy of the decoding process. The present work brings direct evidence of a structural modification in the tRNA within the ternary complex after the codon–anticodon pairing.
In the delivery of the aa-tRNA to the ribosome it moves from the A/T position to the position in the A-site. The tRNA structure inferred from the density map shows a better fit in the anticodon loop if we use the position of a full A-site tRNA interacting with the mRNA (Yusupova et al., 2001) than the tRNA structure within the ternary complex crystal structure (Nissen et al., 1995). The kink that the anticodon arm shows represents a modification from the known structures of tRNAs that could explain the phase mismatch between an anticodon loop already accommodated and an acceptor arm still residing in the T site. The kirromycin has stalled the complex at the stage where the tRNA has moved toward the ribosome to contact the GAC. This interaction could trigger the GTPase activity of EF-Tu, and the subsequent liberation of the acceptor arm of the tRNA from the binding with the factor could allow the acceptor arm to swing and reach the PTC. Furthermore, the contacts of the tRNA with two different parts of protein S12 would connect the movement of the acceptor arm with the previously described reorganization of the decoding site, particularly the helix 44 of 16S rRNA (VanLoock et al., 2000; see also Carter et al., 2001), in a way that both changes could be synchronized. In any event, the flexibility of the anticodon arm of tRNA would have an interesting role as a mechanism to absorb the temporary discrepancy between the positions of acceptor and anticodon arms of the tRNA in the transition toward the A-site position.
A 5′-stacked anticodon loop?
After the submission of the present work, Simonson and Lake (2002) presented the ‘transorientation hypothesis’, where they revive the idea that the tRNA might flip between a 5′-stacked and a 3′-stacked anticodon loop structure (Woese, 1970), and that these two conformations might facilitate both the codon recognition and the accommodation process. All the X-ray-determined structures for tRNAs are in the 3′-stacked form, and no direct structural evidence for the existence of the 5′-stacked form has been reported. Nevertheless, the authors of this hypothesis constructed an energy-minimized model for a tRNA with a 5′-stacked anticodon loop. We have used this model to see if its conformation is compatible with our cryo-EM density (details shown in Supplementary data). By using the anticodon of this modeled tRNA we can fill the whole density of our cryo-EM map satisfactorily. However, in the best fit, the codon and the anticodon run in the same 3′→5′direction, showing a parallel orientation rather than the antiparallel one required for base pairing (S.M.Stagg and S.C.Harvey, personal communication). In order to accomplish the codon recognition, the whole tRNA would have to be rotated by about 180° around the anticodon arm. Thus, given the antiparallel base pairing requirements, and the known orientation of the mRNA, our map is not compatible with such a tRNA structure.
Materials and methods
Preparation of 70S·aa-tRNA·EF-Tu·GDP·kirromycin complex
Escherichia coli ribosomes (10 pmol) were incubated with 15 pmol of MF-mRNA (Triana-Alonso et al., 1995) and 14 pmol of [35S]fMet-tRNAfMet (575 d.p.m./pmol) at 37°C for 10 min in 30 µl of a buffer containing 20 mM HEPES–KOH pH 7.5, 6 mM MgCl2, 150 mM NH4Cl, 4 mM 2-mercaptoethanol, 0.05 mM spermine and 2 mM spermidine according to Blaha et al. (2000). Three microliters of a 100 mM kirromycin solution were added to the reaction mixture and the incubation was continued for another 5 min. Separately, another incubation with 14 pmol of [14C]Phe-tRNAPhe (996 d.p.m./pmol), 20 pmol of EF-Tu, and 8 µl of a 2.5 mM solution of [α-32P]GTP (150 d.p.m./pmol) was carried out for 10 min at the same temperature. The samples with the 70S ribosomes and with the ternary complex were mixed and another 10 min incubation at the same temperature was performed before the cryo-grids were prepared. The occupancy of individual components on the ribosome was measured by filter-binding assays in parallel incubation mixtures, and was found to be ∼60% for both, the P-site fMet-tRNAfMet and the A-site Phe-tRNAPhe. The radioactivity contributed by the α-32P moiety of the GDP served to calculate a ∼50% occupancy for the elongation factor.
Electron microscopy and image processing
The grids for cryo-EM were prepared following standard procedures (Wagenknecht et al., 1988). Micrographs were taken following low-dose procedures on a Philips EM420 at 120 kV and a magnification of ×52 000. The micrographs were digitized with a step size of 14 µm on a Zeiss/Imaging scanner (Z/I Imaging Corporation, Huntsville, AL), corresponding to 2.69 Å on the object scale. The 3D reconstructions were calculated using 3D projection alignment procedures (Penczek et al., 1994; Frank et al., 2000). The volumes were CTF-corrected in defocus groups, with an average of ∼1000 individual images per group (Frank and Penczek, 1995) and the resolution was estimated by the Fourier shell correlation with a 0.5 threshold (Malhotra et al., 1998). X-ray solution scattering data for the E.coli ribosome were used to correct the Fourier amplitudes of the maps (Gabashvili et al., 2000). Difference maps were calculated by subtracting an E.coli 70S ribosome reference volume that had both P- and E-sites occupied with tRNAs (M.Valle, unpublished). Before the subtraction, the maps were normalized taking into account their average density and their variability (σ) and in the rendering only significant values (>2σ) were displayed. The isolation of tRNAs and ternary complex within the volumes was performed by masking out the ribosomal subunits and selecting the remaining masses by clustering procedures.
Fitting of atomic structures
After docking of X-ray structures into the cryo-EM volume using program O (Jones et al., 1991); the visualization of the density maps was performed in IRIS Explorer (Numerical Algorithms Groups, Inc., Downers Grove, IL). EF-Tu domains were taken from the E.coli crystal structure (PDB code 1EFC, from Song et al., 1999) and aa-tRNA or the whole ternary complex (PDB code 1TTT, from Nissen et al., 1995) as the initial coordinates for fitting. The EF-TU·GTP (PDB code 1EFT, from Kjeldgaard et al., 1993) and EF-TU·GDP (PDB code 1TUI, from Polekhina et al., 1996) were also used. The coordinates of the ribosomal proteins and rRNAs were taken from published atomic structures (PDB code 1FFK for the 50S subunit, from Ban et al., 2000; PDB code 1FJF for the 30S, from Wimberly et al., 2000). For the final rendering of fitted structures, the Ribbons package (Carson, 1997) was used. For computation of the cross-correlation coefficient, the fitted PDB coordinates were converted into electron densities, which were low-pass filtered to the resolution of the cryo-EM map.
Coordinates
The atomic coordinates of E.coli EF-Tu (GDP form; Song et al., 1999) and yeast Phe-tRNAPhe (Nissen et al., 1995), independently fitted into the cryo-EM map, have been deposited in the Protein Data Bank (http://www.rcsb.org) with the accession code 1LS2. In addition, the anticodon loop region of the tRNA, fitted separately into the cryo-EM map, has been deposited with the accession code 1LU3.
Supplementary data
A description of the supervised classification method is provided as Supplementary data, available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Christian Spahn for helpful suggestions. We are grateful to Michael Watters and Yu Chen for the preparation of the illustrations. The work was supported by National Institutes of Health Grants R37 GM29169, R01 GM55440, P41 RR01219 and NSF BIR 9219043 (to J.F.), and R01 GM61576 (to R.K.A.).
References
- Abdulkarim F., Liljas,L. and Hughes,D. (1994) Mutations to kirromycin resistance occur in the interface of domains I and II of EF-Tu–GTP. FEBS Lett., 352, 118–122. [DOI] [PubMed] [Google Scholar]
- Abel K.M., Yoder,M.D., Hilgenfeld,R. and Jurnak,F. (1996) An α to β conformational switch in EF-Tu. Structure, 4, 1153–1159. [DOI] [PubMed] [Google Scholar]
- Agrawal R.K., Penzeck,P., Grassucci,R.A. and Frank,J. (1998) Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl Acad. Sci. USA, 95, 6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Heagle,A.B., Penzeck,P., Grassucci,R.A. and Frank,J. (1999) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol., 6, 643–647. [DOI] [PubMed] [Google Scholar]
- Agrawal R.K., Spahn,C.M.T., Penzeck,P., Grassucci,R.A., Nierhaus,K.H. and Frank,J. (2000a) Visualization of tRNA movements on the Escherichia coli ribosome during the elongation cycle. J. Cell Biol., 150, 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Heagle,A.B. and Frank,J. (2000b) Studies of elongation factor G-dependent tRNA translocation by three-dimensional cryo-electron microscopy. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 53–62.
- Agrawal R.K., Linde,J., Sengupta,J., Nierhaus,K.H. and Frank,J. (2001) Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation. J. Mol. Biol., 311, 777–787. [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- Berchtold H., Reshetnikova,L., Reiser,C.O., Schirmer,N.K., Sprinzl,M. and Hilgenfeld,R. (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature, 365, 126–132. [DOI] [PubMed] [Google Scholar]
- Blaha G., Stelzl,U., Spahn,C.M.T., Agrawal,R.K., Frank,J. and Nierhaus,K.H. (2000) Preparation of functional ribosomal complexes and the effect of buffer conditions on tRNA positions observed by cryoelectron microscopy. Methods Enzymol., 317, 292–309. [DOI] [PubMed] [Google Scholar]
- Carson M. (1997) Ribbons. Methods Enzymol., 277, 493–505. [PubMed] [Google Scholar]
- Carter A.P., Clemons,W.M., Brodersen,D.E., Morgan-Warren,R.J., Hartsch,T., Wimberly,B.T. and Ramakrishnan,V. (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science, 291, 498–501. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Yusupov,M.M., Yusupova,G.Z., Earnest,T.N. and Noller,H.F. (1999) X-ray structures of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- Dell V.A., Miller,D.L. and Johnson,A.E. (1990) Effects of nucleotide- and aurodox-induced changes in elongation factor Tu conformation upon its interaction with aminoacyl-tRNA. A fluorescence study. Biochemistry, 29, 1757–1763. [DOI] [PubMed] [Google Scholar]
- Dumas P., Ebel,J.P., Giegé,R., Moras,D., Thierry,J.C. and Westhof,E. (1985) Crystal structure of yeast tRNAAsp: atomic coordinates. Biochimie, 67, 597–606. [DOI] [PubMed] [Google Scholar]
- Dvorak D., Kidson,C. and Winzor,D.J. (1978) Conformational changes in tRNA: consequences of aminoacylation and codon–anticodon recognition. FEBS Lett., 90, 187–188. [DOI] [PubMed] [Google Scholar]
- Frank J. (1990) Classification of macromolecular assemblies studied as ‘single particles’. Rev. Biophys., 23, 281–329. [DOI] [PubMed] [Google Scholar]
- Frank J. (1996) Three-dimensional Electron Microscopy of Macro molecular Assemblies. Academic Press, San Diego, CA.
- Frank J. and Penczek,P. (1995) On the correction of the contrast transfer function in biological electron microscopy. Optik, 98, 125–129. [Google Scholar]
- Frank J., Penczek,P., Agrawal,R.K., Grassucci,R.A. and Heagle,A.B. (2000) Three-dimensional cryo-electron microscopy of ribosomes. Methods Enzymol., 317, 276–291. [DOI] [PubMed] [Google Scholar]
- Gabashvili I.S., Agrawal,R.K., Spahn,C.M.T., Grassucci,R.A., Svergun,D.I., Frank,J. and Penczek,P. (2000) Solution structure of E. coli 70S ribosome at 11.5 Å resolution. Cell, 100, 537–549. [DOI] [PubMed] [Google Scholar]
- Geigenmüller U. and Nierhaus,K.H. (1990) Significance of the third tRNA binding site, the E site, on E.coli ribosomes for the accuracy of translation: an occupied E site prevents the binding of non-cognate aminoacyl-tranfer RNA to the A site. EMBO J., 9, 4527–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner T.P., Atmadja,J. and Nierhaus,K.H. (1987) Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie, 69, 911–923. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zhou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kaziro Y. (1978) The role of guanosine-5′-triphosphate in polypeptide chain elongation. Biochim. Biophys. Acta, 505, 95–127. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nissen,P., Thirup,S. and Nyborg,J. (1993) The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure, 1, 35–50. [DOI] [PubMed] [Google Scholar]
- Lata K.R., Penczek,P. and Frank,J. (1995) Automatic particle picking from electron micrographs. Ultramicroscopy, 58, 381–391. [DOI] [PubMed] [Google Scholar]
- Lodmell J.S. and Dahlberg,A.E. (1997) A conformational switch in Escherichia coli 16S ribosomal RNA during the decoding of messenger RNA. Science, 277, 1262–1267. [DOI] [PubMed] [Google Scholar]
- Malhotra A., Penczek,P., Agrawal,R.K., Gabashvili,I.S., Grassucci,R.A., Burhardt,N., Jünemann,R., Nierhaus,K.H. and Frank,J. (1998) E. coli 70S ribosome at 15Å resolution by cryo-electron microscopy: localization of fMet-tRNAfMet and fitting of L1 protein. J. Mol. Biol., 280, 103–116. [DOI] [PubMed] [Google Scholar]
- Mesters J., Zeef,L., Hilgenfeld,R., Degraaf,J., Kraal,B. and Bosch,L. (1994) The structural and functional basis for the kirromycin resistance of mutant EF-Tu species in E.coli. EMBO J., 13, 4877–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. and Noller,H.F. (1989) Intermediate states in the movement of transfer RNA in the ribosome. Nature, 342, 142–148. [DOI] [PubMed] [Google Scholar]
- Moazed D., Robertson,J.M. and Noller,H.F. (1988) Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S rRNA. Nature, 334, 362–364. [DOI] [PubMed] [Google Scholar]
- Moras D., Dock,A.C., Dumas,P., Westhof,E., Romby,P., Ebel,J.P. and Giegé,R. (1985) The structure of yeast tRNA (Asp). A model for tRNA interacting with messenger RNA. J. Biomol. Struct. Dyn., 3, 479–493. [DOI] [PubMed] [Google Scholar]
- Moras D., Dock,A.C., Dumas,P., Westhof,E., Romby,P., Ebel,J.P. and Giegé,R. (1986) Anticodon–anticodon interaction induces con formational changes in tRNA: yeast tRNAAsp, a model for tRNA-mRNA recognition. Proc. Natl Acad. Sci. USA, 83, 932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F.C. and Nyborg,J. (1995) Crystal structure of the ternary complex of the Phe-tRNAPhe, EF-TU and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldaarg,M. and Nyborg,J. (2000) Molecular mimicry. EMBO J., 19, 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen,D.E., Clemons,W.M., Tarry,M.J., Carter,A.P. and Ramakrishnan,V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science, 292, 868–869. [DOI] [PubMed] [Google Scholar]
- Orlova E.V., Dube,P., Harris,J.R., Beckman,E., Zemlin,F., Marki,J. and van Heel,M. (1997) Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 Å resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol., 271, 417–437. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A. and Stewart,G.W. (1985) Mechanism of action of kirromycin-like antibiotics. Annu. Rev. Microbiol., 39, 557–577. [DOI] [PubMed] [Google Scholar]
- Penczek P., Grassucci,R.A. and Frank,J. (1994) The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy, 53, 251–270. [DOI] [PubMed] [Google Scholar]
- Piepenburg O., Pape,T., Pleiss,J.A., Wintermeyer,W., Uhlenbeck,O.C. and Rodnina,M.V. (2000) Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry, 39, 1734–1738. [DOI] [PubMed] [Google Scholar]
- Polekhina G., Thirup,S., Kjeldgaard,M., Nissen,P., Lippman,C. and Nyborg,J. (1996) Helix unwinding in the effector region of elongation factor EF-Tu–GDP. Structure, 4, 1141–1151. [DOI] [PubMed] [Google Scholar]
- Powers T. and Noller,H.F. (1994) The 530 loop of 16S rRNA: a signal to EF-Tu? Trends Genet., 10, 27–31. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V. and Wintermeyer,W. (2001) Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanism. Annu. Rev. Biochem., 70, 415–435. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Fricke,R. and Wintermeyer,W. (1994) Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry, 33, 12267–12275. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Fricke,R., Kohn,L. and Wintermeyer,W. (1995) Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J., 14, 2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarma U., Remme,J., Ehrenberg,M. and Bilgin,N. (1997) An A to U transversion at position 1067 of 23S rRNA from Escherichia coli impairs EF-Tu and EF-G function. J. Mol. Biol., 272, 327–335. [DOI] [PubMed]
- Schwarz U., Möller,A. and Gassen,H.G. (1978) Induced structural transitions in tRNA. FEBS Lett., 90, 189. [DOI] [PubMed] [Google Scholar]
- Simonson A.B. and Lake,J.A. (2002) The transorientation hypothesis for codon recognition during protein synthesis. Nature, 416, 281–285. [DOI] [PubMed] [Google Scholar]
- Song H., Parsons,M.R., Rowsel,S., Leonard,G. and Phillips,S.E.V. (1999) Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05Å resolution. J. Mol. Biol., 285, 1245–1256. [DOI] [PubMed] [Google Scholar]
- Stark H., Rodnina,M.V., Rinke-Appel,J., Brimacombe,R., Wintermeyer,W. and van Heel,M. (1997) Visualization of elongation factor Tu on Escherichia coli ribosome. Nature, 389, 403–406. [DOI] [PubMed] [Google Scholar]
- Stark H., Rodnina,M.V., Wieden,H., van Heel,M. and Wintermeyer,W. (2000) Large scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell, 100, 301–309. [DOI] [PubMed] [Google Scholar]
- Triana-Alonso F.J., Dabrowski,M., Wadzack,J. and Nierhaus,K.H. (1995) Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J. Biol. Chem., 270, 6298–6307. [DOI] [PubMed] [Google Scholar]
- VanLoock M.S., Agrawal,R.K., Gabashvili,I.S., Qi,L., Frank,J. and Harvey,S.C. (2000) Movement of the decoding region of the 16S ribosomal RNA accompanies tRNA translocation. J. Mol. Biol., 304, 507–515. [DOI] [PubMed] [Google Scholar]
- Vazquez D. (1979) Inhibitors of Protein Synthesis. Springer Verlag, New York, NY.
- Wagenknecht T., Grassucci,R.A. and Frank,J. (1988) Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. J. Mol. Biol., 199, 137–145. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Guymon,R., McCutcheon,J.P., White,S.W. and Ramakrishnan,V. (1999) A detailed view of a ribosomal active site: the structure of the L11–RNA complex. Cell, 97, 491–502. [DOI] [PubMed] [Google Scholar]
- Wimberly B., Brodersen,D.E., Clemons,W.M., Morgan-Warren,R.J., Carter,A.P., von Rhein,C., Hartsch,T. and Ramakrishnam,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- Woese C.R. (1970) Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature, 226, 817–820. [DOI] [PubMed] [Google Scholar]
- Woese C.R. (2001) Translation: in retrospect and prospect. RNA, 7, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H., Chinali,G. and Parmeggiani,A. (1977) Mechanism of the inhibition of protein synthesis by kirromycin. Eur. J. Biochem., 75, 67–75. [DOI] [PubMed] [Google Scholar]
- Wool I.G., Gluck,A. and Endo,Y. (1992) Ribotoxin recognition of ribosomal-RNA and a proposal for the mechanism of translocation. Trends Biochem. Sci., 17, 266–269. [DOI] [PubMed] [Google Scholar]
- Wriggers W., Agrawal,R.K., Drew,D.L., McCammon,A. and Frank,J. (2000) Domains motions of EF-G bound to the ribosome: insights from a hand-shaking between multi-resolution structures. Biophys. J., 79, 1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M. and Smith,D. (1995) tRNA on the ribosome: a waggle theory. In Söll,D. and RajBahandary,U. (eds), tRNA: Structure, Biosynthesis and Function. American Society for Microbiology, Washington, DC, pp. 443–468.
- Yusupova G.Z., Yusupov,M.M., Cate,J.H.D. and Noller,H.F. (2001) The path of the messenger RNA through the ribosome. Cell, 106, 233–241. [DOI] [PubMed] [Google Scholar]