Abstract
The FliG protein is essential for assembly, rotation and clockwise/counter-clockwise (CW/CCW) switching of the bacterial flagellum. About 25 copies of FliG are present in a large rotor-mounted assembly termed the ‘switch complex’, which also contains the proteins FliM and FliN. Mutational studies have identified the segments of FliG most crucial for flagellar assembly, rotation and switching. The structure of the C-terminal domain, which functions specifically in rotation, was reported previously. Here, we describe the crystal structure of a larger fragment of the FliG protein from Thermotoga maritima, which encompasses the middle and C-terminal parts of the protein (termed FliG-MC). The FliG-MC molecule consists of two compact globular domains, linked by an α-helix and an extended segment that contains a well-conserved Gly–Gly motif. Mutational studies indicate that FliM binds to both of the globular domains, and given the flexibility of the linking segment, FliM is likely to determine the relative orientation of the domains in the flagellum. We propose a model for the organization of FliG-MC molecules in the flagellum, and suggest that CW/CCW switching might occur by movement of the C-terminal domain relative to other parts of FliG, under the control of FliM.
Keywords: chemotaxis/energy transduction/flagella/molecular motors
Introduction
Bacterial flagella are turned by rotary motors that are assembled from ∼25 different kinds of proteins (Macnab, 1992, 1996; Schuster and Khan, 1994; Blair, 1995; Berry and Armitage, 1999; Berg, 2000). The motors are located in the cell membrane and obtain energy from the membrane gradient of protons (Larsen et al., 1974; Manson et al., 1977; Glagolev and Skulachev, 1978) or, in some species, sodium ions (Hirota and Imae, 1983; Kawagishi et al., 1995). Proton-fueled motors turn at speeds up to 300 revolutions per second (r.p.s.) (Lowe et al., 1987), while Na+-driven motors can rotate as fast as 1700 r.p.s. (Magariyama et al., 1994). Physiological studies have shown that the flagellar motor utilizes a mechanism with a high duty cycle (Ryu et al., 2000) to couple the flow of several hundred protons to each revolution (Meister et al., 1987; Samuel and Berg, 1995) with apparently high efficiency (Manson et al., 1980; Khan and Berg, 1983; Meister and Berg, 1987).
Mutational studies identified five flagellar proteins that are involved in rotation of the motor. MotA and MotB are membrane proteins (Dean et al., 1984; Stader et al., 1986) that function in proton conduction (Blair and Berg, 1990, 1991; Stolz and Berg, 1991) and form the stator (Chun and Parkinson, 1988; Blair et al., 1991; DeMot and Vanderleyden, 1994). The sodium-driven motors of Vibrio species contain proteins closely related to MotA and MotB, called PomA and PomB (Asai et al., 1997; Sato and Homma, 2000). Each motor contains several MotA–MotB complexes that function independently to generate torque (Block and Berg, 1984; Blair and Berg, 1988). The rotor is formed from the proteins FliG, FliM and FliN, which are essential for flagellar assembly and also function, to various extents, in rotation and clockwise/counter-clockwise (CW/CCW) switching (Yamaguchi et al., 1986a,b). FliG, particularly the C-terminal domain (FliG-C), is most important for the generation of torque (Irikura et al., 1993; Lloyd et al., 1996). This domain contains a set of conserved charged residues that are essential for motor rotation in Escherichia coli, and interact with charged residues in the stator protein MotA (Lloyd and Blair, 1997; Zhou et al., 1998a). Other parts of FliG (the N-terminal and middle domains) appear to be important for flagellar assembly and for proper switching between CW and CCW rotation (Irikura et al., 1993).
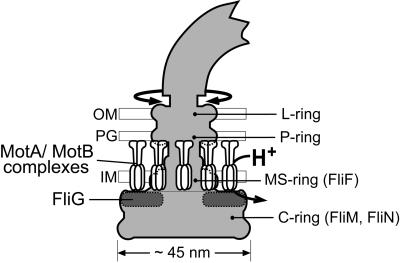
Electron microscopic images of the structure at the base of the flagellum show a set of ∼25 nm diameter, membrane-embedded rings (termed the L, P and MS-rings), attached to an ∼45 nm diameter drum-shaped structure (termed the C-ring) that extends into the cytoplasm (Aizawa et al., 1985; Khan et al., 1992; Francis et al., 1994) (Figure 1). The MS-ring is located in the cytoplasmic membrane, and is formed from ∼25 copies of the integral membrane protein FliF (Jones et al., 1990; Sosinsky et al., 1992; Ueno et al., 1992). If sufficiently gentle purification methods are used, the basal structures also contain the FliG, FliM and FliN proteins. FliN forms the C-ring, probably along with FliM (Kihara et al., 1996; Zhao et al., 1996a,b). FliG binds to FliF (Francis et al., 1992; Oosawa et al., 1994; Kubori et al., 1997; Kihara et al., 2000), and also to FliM and FliN (Marykwas and Berg, 1996; Marykwas et al., 1996; Tang et al., 1996; Toker and Macnab, 1997), thus forming a junction between the MS-ring and the C-ring. MotA and MotB are not found in basal structures as they are currently purified, but freeze–fracture electron micrographs of E.coli membranes show rings of membrane-embedded particles surrounding depressions left by the MS-rings (Khan et al., 1988). These particles are not found in cells that lack MotA or MotB, and are presumably the MotA–MotB complexes.
Fig. 1. Schematic diagram of the E.coli flagellum, showing the approximate location of FliG. The shape of the rotor (shaded) is based on EM reconstructions of Francis et al. (1994). The stator is formed from multiple MotA–MotB complexes with probable composition (MotA)4(MotB)2 (Chun and Parkinson, 1988; DeMot and Vanderleyden, 1994; Sato and Homma, 2000; Braun and Blair, 2001). The placement of MotA–MotB complexes is based on freeze–fracture micrographs of Khan et al. (1988). IM, inner membrane; PG, peptidoglycan; OM, outer membrane.
High-resolution structural data on the motor proteins is needed to provide a framework for understanding the data from physiological, mutational and biochemical studies, and to guide experiments to probe the mechanism. We recently reported the 2.3 Å resolution structure of the C-terminal domain of FliG (FliG-C) from the thermophile Thermotoga maritima (Lloyd et al., 1999). The structure of FliG-C showed that the functionally important charged residues are clustered along a ridge on the domain. We proposed that in the flagellum this ridge is pointed towards the stator, and is oriented to allow the charged groups to interact sequentially with charged groups on MotA as the rotor turns. Because it appeared that a single orientation of the domain would not allow all of the charge–charge interactions suggested by the mutational studies, we proposed that CW and CCW rotation might utilize different subsets of charged groups on FliG, brought into play by two different orientations of the domain.
Other domains of the FliG protein, not previously crystallized, are important for assembly of the flagellum and for controlling CW/CCW switching. Here, we report the structure of a larger fragment of FliG that includes both the middle and C-terminal domains (FliG-MC). The FliG-MC molecule consists of two compact domains joined by an α-helix and a Gly-containing segment that is presumably flexible. Mutational data indicate that FliM binds to both domains of FliG-MC, thereby suggesting that FliM might control the relative orientation of the domains in the flagellum. FliM is the rotor protein most important for CW/CCW switching (Sockett et al., 1992; Welch et al., 1993); we propose that switching occurs by FliM-regulated movement of the C-terminal domain of FliG relative to other parts of the protein.
Results and discussion
Structure determination
In a previous report we described the structure of a C-terminal fragment of FliG (FliG-C) consisting of residues 235–335, which functions specifically in rotation of the motor (Lloyd et al., 1999). For that study we used FliG-C from T.maritima, which proved more amenable to crystallization than the protein from E.coli. The full-length protein was polydisperse, prompting us to test other FliG fragments. A construct that encodes residues 104–335 of T.maritima FliG (FliG-MC) was overexpressed in E.coli, purified and formed crystals that diffracted to 2.8 Å resolution.
Data were collected at three wavelengths from a crystal of selenomethionine-substituted protein, and used to obtain experimental phases to 3.3 Å resolution (Table I). The structure was built into the solvent-flattened 3.3 Å electron density map, and refined against the native 2.8 Å data to an R-factor (Rfree) of 25.5% (28.0%), with good geometry (Table II). The density maps were clearly defined throughout the structure except for eleven residues at the N-terminus and eight residues at the C-terminus. The refined model consists of residues 115–327 of FliG, five water molecules and two calcium ions. The calcium ions are coordinated by side-chains that are not conserved in FliG proteins from other species, and so their presence probably reflects the high concentration of CaCl2 used in crystallization (200–400 mM) rather than an essential role for Ca2+ in FliG function.
Table I. Data collection statistics.
| Native | Edge | Peak | Remote | |
|---|---|---|---|---|
| Wavelength (Å) | 0.99981 | 0.97935 | 0.97906 | 0.93222 |
| Resolution (Å) | 35 to 2.8 | 30 to 3.3 | 30 to 3.3 | 30 to 3.3 |
| Highest resolution shell (Å) | 2.9–2.8 | 3.42–3.3 | 3.42–3.3 | 3.42–3.3 |
| Observed reflections | 179 161 | 186 454 | 160 380 | 159 388 |
| Unique reflections | 12 745 | 7871 | 7955 | 7847 |
| Completeness (%)a | 96.0 (90.1) | 98.0 (99.7) | 97.9 (94.2) | 98.3 (99.6) |
| Rsym (%)b | 8.4 (23.5) | 12.3 (42.0) | 11.7 (30.6) | 10.4 (32.6) |
| Average I/σ (I) | 12.3 (3.6) | 11.3 (3.6) | 10.8 (4.7) | 13.0 (4.4) |
| Mosaicity (°) | 0.808 | 0.663 | 0.725 | 0.712 |
aValues in parentheses refer to data in the highest resolution shell.
bRsym = 100 × Σ|I – <I>|/ΣI, where I is the intensity of the individual measurement and <I> is the average intensity from multiple observations.
Table II. Refinement statistics.
| Resolution range (Å)a | 35 to 2.8 (2.9–2.8) |
| No. of reflections in working set | 11 237 |
| No. of reflections in free-R set | 991 |
| No. of protein atomsb | 1702 |
| No. of solvent molecules | 5 |
| No. of metal sites (Ca2+) | 2 |
| R-factor (%)c | 25.5 (28.7) |
| Free-R factor (%)d | 28.0 (36.4) |
| R.m.s.d. (bond lengths) (Å) | 0.016 |
| R.m.s.d. (bond angles) (°) | 1.972 |
| No. of φ, ψ angles (%)e | |
| Most favored | 98 |
| Additional allowed | 2 |
aValues in parentheses refer to data in the highest resolution shell.
bNon-hydrogen atoms only.
cR-factor = 100 × Σ(||Fobs| – |Fcalc||)/Σ|Fobs|.
dRfree is the R-factor for a selected subset of reflections (8.1%) that were not included in the refinement calculations.
eStereochemistry was assessed with PROCHECK (Laskowski et al., 1993).
Structure of FliG-MC
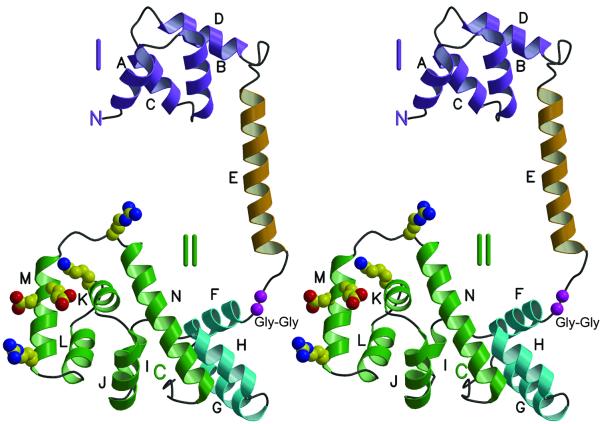
The FliG-MC molecule has overall dimensions 75 × 40 × 30 Å, and consists of two compact, mainly helical domains separated by an α-helix and a segment with extended, nonregular secondary structure (Figures 2 and 3). Domain I (residues 115–170) is formed by four helices (A–D) arranged in a right-handed super-helix. Domain II (residues 198–327) is composed of nine helices (F–N), several of which are also coiled into right-handed super-helices. Helices F–H comprise a right-handed coil of helices that form one end of domain II. Helices I–N are folded into the shape of a triangular prism, as reported previously (Lloyd et al., 1999). The structure of helices I–N overlaps well with the previous structure of FliG-C, with a root-mean-square deviation (r.m.s.d.) of 1.14 Å for main-chain atoms from residue 245 to residue 320. The largest differences occur in surface loops, probably caused by differences in crystal contacts. Major structural features of the domain, including the endpoints of the helices and the overall fold, are the same as reported earlier for the shorter construct.
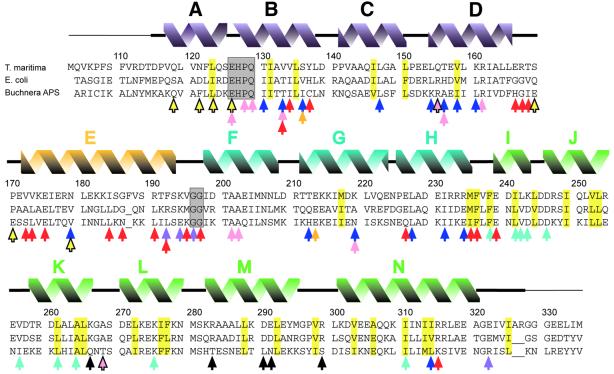
Fig. 2. Alignment of FliG-MC sequences from T.maritima, E.coli and Buchnera APS. The initiating Met in the T.maritima protein studied here replaces a Leu in the native protein. Secondary structural elements seen in T.maritima FliG-MC are shown above. A thin line denotes disordered segments not included in the refined model. Residues that form the cores of domains I and II are highlighted in yellow. Conserved EHPQ and GG motifs discussed in the text are in gray boxes. Black arrows indicate charged residues important for motor rotation in E.coli (Lloyd and Blair, 1997). Colored arrows indicate positions of mutations giving various phenotypes. Turquoise, nonmotile but flagellate (Irikura et al., 1993; Lloyd et al., 1996). Pink, nonflagellate (in E.coli) (Lloyd et al., 1996; Zhou et al., 1998b). Blue, weakened binding to FliM in a yeast two-hybrid assay (Marykwas and Berg, 1996). Red, CW motor bias (Irikura et al., 1993; Lloyd and Blair, 1997). Yellow with black outline, CCW motor bias (Irikura et al., 1993). Orange, either CW or CCW motor bias depending on the substitution (Irikura et al., 1993). Purple, suppressors of mutations in the stator protein MotB (Garza et al., 1996). Most of the point mutations that gave nonflagellate phenotype in E.coli (pink arrows) gave an immotile but flagellate phenotype in Salmonella (Irikura et al., 1993); two exceptions that gave nonflagellate phenotype in both species are indicated by pink arrows with black outline. Sequences are from DDBJ/EMBL/GenBank.
Fig. 3. Stereoview ribbon representation of the FliG-MC structure. Helices are labeled A through N, and are colored according to domain. Purple, domain I. Gold, inter-domain helix. Green, the part of domain II whose structure was reported previously (Lloyd et al., 1999). Turquoise, the other part of domain II. Charged residues that interact with the stator and are important for motor rotation in E.coli are shown in standard atom colors. A well-conserved Gly–Gly sequence that should confer flexibility between the domains is shown in magenta.
Visual inspection showed that the coil-of-helices motif recurred within the FliG-MC structure. By using the DALI server (Holm and Sander, 1997) to identify regions of internal structural similarity, helices B–D were found to align well with helices F–H (overall r.m.s.d. of 2.9 Å for the Cα backbone), and also with helices J–L (overall deviation 2.1 Å) (Figure 4A). A search of the DALI database showed that this three-helix motif resembles the armadillo (ARM) repeat found in β-catenin and in a variety of other proteins of diverse function (Huber et al., 1997) (Figure 4B). In spite of the similarity in three-dimensional structure, ARM repeats show high variability at the amino-acid level. This is also true in the case of FliG, where the repeating structural motif is not accompanied by obvious similarities in sequence. It is typical for multiple ARM motifs to pack together to form a right-handed coil of α-helices, as seen in FliG-MC (Huber et al., 1997).
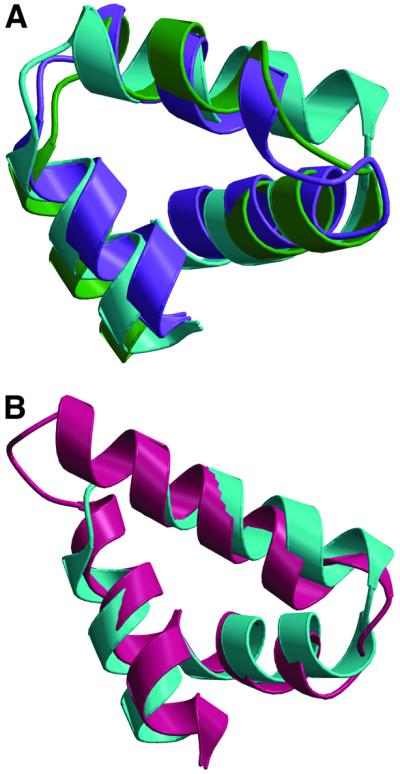
Fig. 4. Superpositions of structural elements in FliG. (A) Overlay of helices B–D of FliG-MC (purple) with helices F–H (turquoise) and helices J–L (green). (B) Overlay of helices F–H of FliG-MC (turquoise) with residues 397–444 of β-catenin (protein data bank accession code 2BCT; Huber et al., 1997).
The structure is stabilized by conserved residues
It is important to establish that the structure we have determined is a good model for the FliG-MC proteins from E.coli and Salmonella, the species for which the most data are available from mutational and physiological studies. The core of domain I is largely formed from the side-chains of six residues (Leu123, Ile131, Leu135, Ile146, Leu150 and Val158; highlighted in Figure 2), whose hydrophobic character is well conserved in the ∼40 FliG sequences in the NCBI database. (Residues with mixed polar/nonpolar character, such as Thr or Gln, are found in a few cases at positions 131, 146 and 158.) The core of domain II is formed from the side-chains of about two dozen hydrophobic residues (Met217, Met235, Phe236, Phe238, Ile241, Leu244, Ile249, Val252, Leu253, Leu261, Ala264, Leu265, Leu272, Ile276, Phe277, Leu288, Leu292, Val298, Val303, Ala306, Ile310, Ile313, Ile314 and Ile325; see Figure 2). Hydrophobic character is well conserved at these positions in the FliG sequences in the database. (Residues with mixed character sometimes occur in positions where the polar groups in the side-chain could reach the protein surface.) Patterns of sequence conservation thus indicate that the overall fold should be similar in FliG proteins from other species. This is consistent with our earlier observation that a 93-residue segment from the C-terminus of T.maritima FliG can function in place of the corresponding part of the E.coli protein (Lloyd et al., 1999).
Flexible linker between domains I and II
The two domains of FliG-MC are connected by helix E and a segment with fairly extended secondary structure (residues 193–197) that contains two consecutive Gly residues. The Gly–Gly motif is well conserved, with just a few species having a single Gly (at position 197), and only certain species of α-proteobacteria (Caulobacter crescentus, Rhodopseudomonas palustrus and Magnestospirillum magnetotacticum) having non-Gly residues in both positions. The Gly–Gly linker will allow considerable relative movement of the two globular domains in solution, and since the domains do not contact each other directly their orientations in the X-ray structure must be determined primarily by crystal contacts. Both of the globular domains, and also the linking helix, make contacts with other FliG molecules in the crystal.
Flexibility between the two domains may be amplified by unravelling of helix E, which is stabilized by contacts with other FliG molecules in the crystal but could be flexible in solution. This ∼20-residue helix is highly amphipathic, displaying the hydrophobic side-chains of Val172, Val173, Ile176, Leu180, Ile184, Phe187, Val188 and Phe192 on one face, and polar residues on the opposite face. The residues listed above are conserved as hydrophobic, except that residues 187, 188 and 192 are polar in the FliG proteins from some species. Residues 173 and 176 are close to Val133 and Tyr137 on domain I, and may interact with these residues to stabilize the position of helix E. Resides 172, 180 and 184 do not form obvious contacts with other parts of the FliG-MC molecule. They might interact with the N-terminal part of FliG (the part missing from FliG-MC), or with another protein of the flagellum.
Interactions with the stator
Three charged residues of FliG (corresponding to Arg283, Asp290 and Glu291 in T.maritima) were shown to be very important for motor rotation in E.coli, and two others (Lys266 and Arg299) were found to be moderately important (Lloyd and Blair, 1997). These residues function redundantly, and charge is their most critical feature. These charged residues are located along a prominent ridge on the C-terminal domain, with the three more important residues all on helix M (Figure 3). As discussed previously (Lloyd et al., 1999), in the flagellum this charge-bearing ridge on FliG is presumably positioned to allow interactions with charged groups in the stator protein MotA (Zhou and Blair, 1997; Zhou et al., 1998a). The precise orientation of the domain vis-à-vis the stator, and the precise role of these rotor–stator interactions in rotation, are not known.
In the previously determined structure of FliG-C (Lloyd et al., 1999), all of the functionally important charged groups were exposed to solvent. The location of the Lys266 side-chain was somewhat uncertain, however, because its conformation differed in the two FliG-C molecules of the asymmetric unit, with one more exposed to solvent than the other. In the FliG-MC structure, the side-chain of Lys266 adopts the less-exposed conformation. In this conformation, Lys266 is hydrogen bonded to backbone carbonyl groups of residues 292 and 296 near the end of helix M, and is not involved in lattice contacts. In the more exposed conformation seen previously, Lys266 forms a hydrogen bond with the terminal carboxyl of a symmetry related molecule, which might stabilize it in the more outward position. The less-exposed conformation of Lys266 thus appears more probable. The hydrogen bonding to the backbone raises the possibility that mutations in Lys266 might impair function by altering or destabilizing a region of local structure, rather than by directly affecting electrostatic interactions with the stator.
FliG sequences are presently known from ∼40 species. Examination of these sequences shows that in cases where a species is motile and contains only a single FliG protein, the charged residues discussed above are well conserved. Some species have two genes that resemble fliG, and in those cases the charged residues are better conserved in one of the FliG proteins than in the other. The FliG protein from Buchnera is exceptional in showing no conservation of the charged residues implicated in rotor–stator interactions (Figure 2). Buchnera is an obligate intracellular symbiont of aphids that provides its host with certain essential amino acids (Shigenobu et al., 2000), and is presumably immotile. Its chromosome has genes for all flagellar proteins except the filament protein flagellin and the stator proteins MotA and MotB. The flagellar apparatus of Buchnera thus lacks specifically the parts that function in propulsion, while retaining the parts for other functions such as flagellum-specific export. Overall, the pattern of sequence conservation in FliG proteins in the database reinforces the view that the charged residues on the ridge of domain II function specifically in rotation.
Mutations that affect flagellar assembly
In Salmonella, most FliG mutations that disrupt flagellar assembly involve frameshifts or other changes that eliminate most of the protein (Irikura et al., 1993). Exceptions were the two point mutations Arg154→Pro and Ala266→Asp, at positions corresponding to Gln155 and Ala268 in the T.maritima protein (Figure 5B). Gln155 is located in an α-helix where the Pro replacement would disrupt local secondary structure. The side-chain of Ala268 is directed towards the core of the protein where the Asp substitution could alter the structure; this residue is conserved as small (Ala, Ser, Cys) in most FliG proteins.
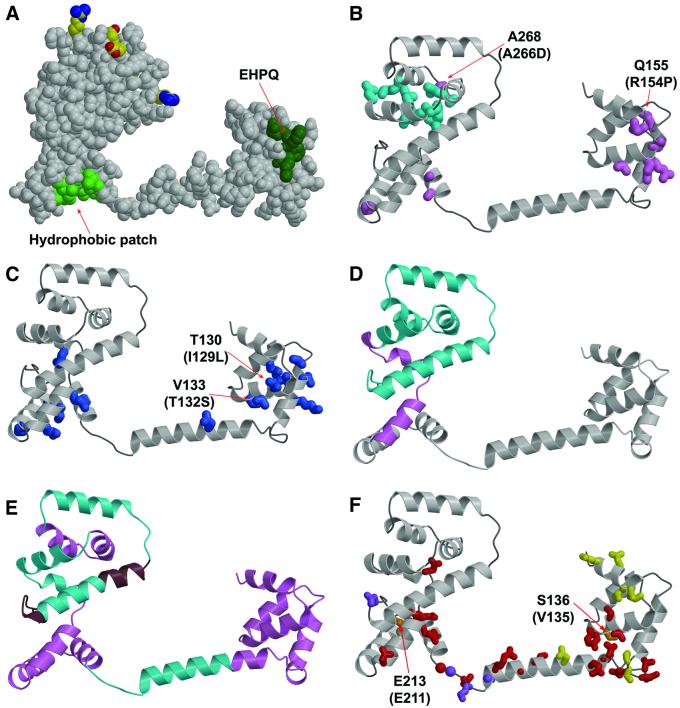
Fig. 5. Mapping of mutations and deletions to the structure. (A) Conserved surface features. Dark green, EHPQ motif in domain I. Light green, surface hydrophobic patch on domain II. Charged residues that interact with the stator are shown in standard atom colors. (B) Turquoise, mutations that eliminate motor rotation but allow flagellar assembly. Pink, mutations that disrupt flagellar assembly in E.coli. Mutations indicated by the labels (Q155 and A268) also disrupt flagellar assembly in Salmonella (compare with the legend to Figure 2; the mutational changes in Salmonella are given in parentheses). (C) Blue, mutations that weaken binding to FliM in the yeast two-hybrid assay (Marykwas and Berg, 1996). Two mutations that involved relatively minor changes in surface-exposed side-chains are indicated, with the mutational changes in the E.coli protein given in parentheses. These two mutations are adjacent to the EHPQ motif. (D) Turquoise, an 86-residue C-terminal segment that can be deleted while still allowing flagellar assembly and binding of FliG to FliM and FliN. Pink, a 19-residue segment whose further deletion prevents flagellar assembly and weakens binding to FliM and FliN (Tang et al., 1996). (E) Turquoise, segments of the protein in which 10-residue deletions prevent motor rotation but allow flagellar assembly. Pink, segments of the protein in which 10-residue deletions prevent flagellar assembly. Brown, segments of the protein in which 10-residue deletions allow both assembly and rotation. (F) Red, mutations giving CW motor bias (Irikura et al., 1993; Lloyd and Blair, 1997). Yellow, mutations giving CCW motor bias (Irikura et al., 1993). Orange, two positions that, depending on the substitution, give either CW or CCW motor bias; the residue numbers for T.maritima and Salmonella are shown (Salmonella in parentheses) (Irikura et al., 1993). Purple, mutations that suppress motility defects caused by mutations in the stator protein MotB (Garza et al., 1996).
Flagellar assembly is more easily disrupted in E.coli, as many point mutations give a nonflagellate phenotype (Lloyd et al., 1996) (Figure 5B). Five such mutations (in residues 126, 128, 129, 133 and 161) are clustered together on the surface of domain I, overlapping a patch of four well conserved residues (EHPQ126–129; colored dark green in Figure 5A) that are exposed to solvent and do not have obvious roles in stabilizing the structure. Most FliG proteins conform to this EHPQ sequence, with only a few differing at one, or at most two, positions. Three other point mutations giving nonflagellate phenotype in E.coli (in residues 201, 202 and 219) are on the bottom of domain II, overlapping a sizable patch of surface hydrophobic residues. This hydrophobic patch is formed from the side-chains of ∼9 residues (Ile198, Ala201, Ile204, Met205, Leu208, Ile216, Leu220, Leu227 and Ile231, colored light green in Figure 5A), whose hydrophobic character is well conserved. These clusters of assembly-disrupting mutations identify regions of FliG likely to interact with other flagellar proteins.
Data from a yeast two-hybrid assay (Marykwas and Berg, 1996) indicate that the EHPQ motif in domain I and hydrophobic patch in domain II are important for binding to FliM (Figure 5C). Several mutations that weaken binding to FliM occur in and around the hydrophobic patch on domain II. They also occur on the surface of domain I, including two that are adjacent to the EHPQ motif and involve only mild changes to the side-chain (in E.coli the mutations were Ile129→Leu and Thr132→Ser, corresponding to positions 130 and 133 in T.maritima). Just one mutation affecting binding to FliM was found on the inter-domain helix.
Apart from the region of the hydrophobic patch, domain II does not appear to make contacts essential for flagellar assembly. Several mutations in the upper part of the domain prevent rotation, but permit normal flagellar assembly, in both E.coli and Salmonella (Figure 5B). A large deletion that removes the upper part of domain II (residues 246–331 in the E.coli protein) gave a FliG fragment that was stable, sufficient for flagellar assembly, and able to bind FliM and FliN in co-precipitation experiments. Deletion of 19 additional residues gave a stable FliG fragment that did not support flagellar assembly, and did not bind to FliM or FliN (Tang et al., 1996). This critical 19-residue segment is near the bottom of domain II, where it contains one of the residues of the surface hydrophobic patch (Ile231) and should be essential for organizing most of the other residues of the patch (Figure 5D).
Smaller deletions give further evidence that domain I and the hydrophobic patch of domain II are important for flagellar assembly (Kihara et al., 2000). Ten-residue deletions in domain I or the hydrophobic patch of domain II prevented flagellar assembly, whereas deletions in most other parts of the protein did not (Figure 5E). Two 10-residue deletions in the middle of domain II did disrupt flagellar assembly, even though a much larger deletion encompassing those segments did not (as described above). These deletions might alter the domain so that it cannot be properly incorporated into the flagellum. Most of the 10-residue deletions that allowed assembly did prevent rotation, consistent with the importance of a properly positioned C-terminal domain for rotation. Rotation persisted in only two 10-residue deletions, in segments that are C-terminal to the charge-bearing ridge (Figure 5E) and evidently not essential for the proper positioning of the ridge.
Mutations that affect CW/CCW motor bias
Several FliG mutations are known that affect the CW/CCW bias of the motors in Salmonella (Irikura et al., 1993) or E.coli (Lloyd and Blair, 1997). Most such mutations are in the part of the protein crystallized here, with just a few CW-bias mutations occurring immediately N-terminal to FliG-MC, in positions between residues 95 and 110. The bias-altering mutations do not disrupt flagellar assembly, and most do not fall directly in regions implicated in binding to FliM, one exception being a mild (Glu→Asp) mutation in residue 126 (Figure 5F). The mutations that give CW motor bias (red) fall on the lower parts of the molecule, in both of the globular domains and also the inter-domain helix. The mutations giving CCW bias (yellow) are distributed differently, occurring on the top as well as the bottom of domain I, in only one position in the inter-domain helix, and not in domain II.
Several CW and CCW mutations are found near the junction of domain I and helix E, where they might alter the relationship between the domain and the helix (CW mutations at residues 133, 134, 137, 166, 167, 168, 172, 173; CCW mutations at 169, 170). Three of the CW-bias mutations in domain II are in places (residues 226, 235 and 315) where they might alter the relationship between the upper and lower parts of that domain. Mutations in two positions (136 and 213; orange) gave either CW- or CCW-bias, depending upon the substitution. One of these is near the junction between domain I and helix E, and the other is at the interface between the upper and lower parts of domain II. Overall, the mutational data are consistent with the idea that switching involves relative movement of FliG domains.
FliG mutations that alter the relationship of rotor and stator
The proper relationship of the rotor and stator is critical for rotation of the flagellar motor, and certain mutations in the stator protein MotB disrupt motility by altering the rotor–stator relationship (Blair et al., 1991; Garza et al., 1996). These MotB mutations can be suppressed by compensating mutations in FliG that evidently restore a working alignment (Garza et al., 1996). These suppressor mutations in FliG occur in four positions, one centered on the surface of domain II, in residue 322, and three in the linking segment between helix E and domain II (Phe192, Lys194 and Gly196 in T.maritima; Figure 5F). All four suppressing mutations give an aberrantly CW motor bias, in both mutant and wild-type backgrounds. The struc tural basis of suppression by the residue-322 mutation (Glu320→Lys in E.coli) is not obvious. The occurrence of three suppressor mutations in the linker is intriguing given the possibility that this segment functions to allow relative movement of domains I and II, and indeed prompted Garza et al. (1996) to propose the existence of a hinge in this part of the molecule. One mutation in the linker (Gly194→Ser in E.coli; corresponding to position 196 in T.maritima) made CW/CCW switching slower than normal, a strong indication that the linker has a role in facilitating the transition between CW and CCW states.
Organization of FliG-MC in the flagellum
The information from mutational studies allows us to propose a model for the arrangement of FliG-MC molecules in the flagellum, pictured in Figure 6. The active-site ridge on domain II must be positioned to allow interactions with charged residues in the cytoplasmic domain of MotA (Zhou et al., 1998a; Lloyd et al., 1999). The yeast two-hybrid data (Marykwas and Berg, 1996) and the location of assembly-disrupting mutations in E.coli suggest that FliM binds to the hydrophobic patch on domain II and to the conserved EHPQ motif on domain I (see Figure 5). By binding to both domains of FliG, FliM could dictate their relative orientation. Switching from CCW to CW rotation occurs when the chemotactic signaling molecule phospho-CheY binds to FliM (Welch et al., 1993), presumably triggering a conformational change in FliM that is transmitted to FliG. Sequences near the N-terminus of FliG (not present in the FliG-MC molecule crystallized here) form the site of attachment to the MS-ring (the FliF protein) (Kihara et al., 2000). Given this N-terminal attachment to the MS-ring, and the non-involvement of most of domain II in flagellar assembly, it seems likely that domain I remains essentially fixed relative to the MS-ring, while domain II undergoes movements under the control of FliM. These movements could alter the position or orientation of the charged-residue ridge to reverse the direction of forces produced at the rotor–stator interface, and thus reverse the direction of rotation.
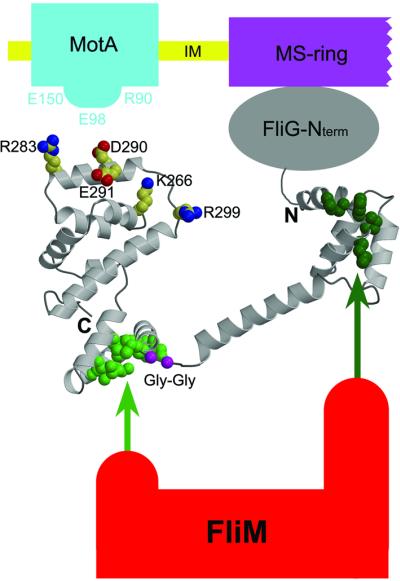
Fig. 6. Proposed arrangement of FliG-MC relative to other proteins of the flagellum. The charge-bearing ridge on the C-terminal domain is oriented to allow interactions with conserved charged residues of MotA. FliM binds to both domains of FliG-MC, most likely through the EHPQ motif in domain I (dark green) and the surface hydrophobic patch on domain II (light green). (FliM–FliG binding interactions are indicated by the arrows.) The flexible Gly–Gly linker (magenta) allows relative movement of the domains. We propose that CW/CCW switching occurs by movement of the C-terminal domain of FliG relative to the rest of the protein, under the control of FliM (see the text).
Materials and methods
Cloning and protein expression
A fragment of the T.maritima fliG gene encoding residues 104–334 of the protein was obtained by PCR amplification from a plasmid (pSL49) carrying the full-length gene, and cloned into the NdeI and BamHI sites of the vector pAED4 (Doering, 1992). Primer sequences were based on the T.maritima genome sequence (Nelson et al., 1999). The residue 104–334 protein fragment (FliG-MC) was expressed in codon-plus (RIL) cells (Stratagene), a derivative of strain BL21(DE3) (Studier and Moffatt, 1986), cultured in Luria–Bertani medium. Selenomethionine substituted protein (Se FliG-MC) was expressed in strain B834(DE3) cultured in 2× M9 medium supplemented with 40 µg/ml seleno-l-methionine (Ramakrishnan et al., 1993). The cells were grown overnight at 37°C to an OD600 of ∼1.5, then induced with 600 µM isopropyl-β-d-thiogalactopyranoside for 3 h. The cells were collected by centrifugation, frozen in liquid nitrogen, then stored at –70°C.
Protein purification
Frozen cells were thawed, resuspended in lysis buffer (50 mM Tris–HCl pH 8.0, 5 mM EDTA), and protease inhibitors were added to the following concentrations: 170 µg/ml phenylmethylsulfonyl fluoride, 0.7 µg/ml pepstatin A, 0.5 µg/ml leupeptin, 2 µg/ml aprotinin. The cells were sonicated, passed through a French pressure cell, then sonicated again. Samples were centrifuged at 20 000 g for 30 min at 4°C. The cold supernatant was mixed with three volumes of boiling lysis buffer and held at 85°C for 15 min. The lysate was cooled in an ice-water bath and the denatured proteins were pelleted by centrifugation at 100 000 g for 1 h at 4°C. The clarified lysate (typically ∼200 ml) was loaded directly onto a Q–Sepharose (Pharmacia) column, which was washed with ∼500 ml of 50 mM Tris–HCl pH 8.0, then developed with a 0→1 M NaCl gradient in the same buffer. Fractions containing FliG were pooled, and saturated (NH4)2SO4 was added to give a final concentration of 0.5 M (10% saturation). The solution was loaded onto a phenyl– Sepharose column (Pharmacia) equilibrated in 50 mM Tris–HCl pH 8.0, 0.5 M (NH4)2SO4, and proteins were eluted with a 0.5→0 M (NH4)2SO4 gradient. Fractions containing FliG were pooled, concentrated by ultrafiltration, and loaded onto a Superdex-200 size-exclusion column (Pharmacia). The column was run in 50 mM Tris–HCl pH 8.0, 200 mM NaCl. The peak fractions were pooled, dialyzed into a buffer containing 10 mM Tris–HCl pH 8.0, 10 mM NaCl, then concentrated by ultrafiltration to 10 mg/ml.
Crystallization and data collection
FliG crystallized at room temperature in sitting drop trays with a well solution containing 8–10% (v/v) isopropanol, 200–400 mM CaCl2, 100 mM sodium acetate pH 4.4–5.0 and 1% pluronic F-68. The drops were set up with a 5:2 ratio of well solution to protein. Native crystals grew to full size in ∼12 h; Se FliG-MC crystals grew in about 1 week. Crystals used for data collection typically had dimensions 200 × 20 × 20 µm.
Se FliG-MC crystals were cooled by plunging into liquid nitrogen after a brief (∼10 s) soak in 10% isopropanol, 100 mM sodium acetate pH 5.0, 200 mM CaCl2, 1% pluronic F-68, and 15% glycerol. Native crystals were treated likewise, except that the cryoprotectant consisted of 8% isopropanol, 100 mM sodium acetate pH 4.6, 400 mM CaCl2, 1% pluronic F-68 and 15% 1,4-butanediol. Native FliG-MC and Se FliG-MC data were collected at 100 K on a charged-coupled device Quantum 4 detector (ADSC) at SSRL beamline 9–2. Data were processed (Table I) using DENZO and SCALEPACK (Otwinowski, 1993). The crystals belong to space group P6422 with unit cell dimensions a = b = 113.94 Å, c = 128.83 Å, α = β = 90° and γ = 120°. There is one molecule per asymmetric unit, and the solvent content is ∼65%.
Structure determination and refinement
The structure was determined to 3.3 Å resolution by the multi-wavelength anomalous diffraction (MAD) method. The FliG-MC fragment (residues 104–334) contains seven methionines, five at internal positions in the sequence and one at each terminus. The selenium sub-structure for four of the seven methionines was determined with the program SHAKE’N’BAKE (Weeks and Miller, 1999). The positions of the sites were refined and initial phases were calculated using the program SHARP (De la Fortelle and Bricogne, 1997). Density modification was performed with the program DM (Cowtan and Zhang, 1999) as provided in the CCP4 suite (Collaborative Computational Project, Number 4, 1994). The resulting 3.3 Å maps were clear and showed continuous density characteristic of α-helices through much of the model region. The program O (Jones et al., 1991) was used for model building, and refinement calculations were performed with REFMAC5 (Murshudov et al., 1999). Refinement and model building were continued against the 2.8 Å data with REFMAC5 and O, respectively. Refinement statistics are in Table II. Figures were prepared using BobScript (Esnouf, 1999), MolScript (Kraulis, 1991) and Raster3D (Merritt and Bacon, 1997). Coordinates of FliG-MC have been deposited with the RCSB Protein Data Bank, under accession No. 1LKV.
Acknowledgments
Acknowledgements
We thank F.Whitby, H.Schubert, N.-L.Chan and B.Howard for assistance and advice at many stages of this project, D.Mackay and G.Cortez for assistance with cloning and protein purification and J.S.Parkinson for helpful comments. Supported by grant R01-GM61145 and training grant 5T32-GM08537 from the US National Institutes of Health. The protein–DNA core facility at the University of Utah receives support from the National Cancer Institute (5P30 CA42014). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
References
- Aizawa S.-I., Dean,G.E., Jones,C.J., Macnab,R.M. and Yamaguchi,S. (1985) Purification and characterization of flagellar hook–basal body complex of Salmonella typhimurium. J. Bacteriol., 161, 836–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai J., Kojima,S., Kato,H., Nishioka,N., Kawagishi,I. and Homma,M. (1997) Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J. Bacteriol., 179, 5104–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H.C. (2000) Constraints on models for the flagellar rotary motor. Phil. Trans. R. Soc. Lond. B Biol. Sci., 355, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R.M. and Armitage,J.P. (1999) The bacterial flagella motor. Adv. Microb. Physiol., 41, 291–337. [DOI] [PubMed] [Google Scholar]
- Blair D.F. (1995) How bacteria sense and swim. Annu. Rev. Microbiol., 49, 489–522. [DOI] [PubMed] [Google Scholar]
- Blair D.F. and Berg,H.C. (1988) Restoration of torque in defective flagellar motors. Science, 242, 1678–1681. [DOI] [PubMed] [Google Scholar]
- Blair D.F. and Berg,H.C. (1990) The MotA protein of E.coli is a proton-conducting component of the flagellar motor. Cell, 60, 439–449. [DOI] [PubMed] [Google Scholar]
- Blair D.F. and Berg,H.C. (1991) Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J. Mol. Biol., 221, 1433–1442. [DOI] [PubMed] [Google Scholar]
- Blair D.F., Kim,D.Y. and Berg,H.C. (1991) Mutant MotB proteins in Escherichia coli.J. Bacteriol., 173, 4049–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block S.M. and Berg,H.C. (1984) Successive incorporation of force-generating units in the bacterial rotary motor. Nature, 309, 470–472. [DOI] [PubMed] [Google Scholar]
- Braun T.F. and Blair,D.F. (2001) Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H+ channels in the stator complex. Biochemistry, 40, 13051–13059. [DOI] [PubMed] [Google Scholar]
- Chun S.Y. and Parkinson,J.S. (1988) Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science, 239, 276–278. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cowtan K.D. and Zhang,K.Y. (1999) Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol., 72, 245–270. [DOI] [PubMed] [Google Scholar]
- Dean G.E., Macnab,R.M., Stader,J., Matsumura,P. and Burke,C. (1984) Gene sequence and predicted amino acid sequence of the MotA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol., 159, 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy atom parameter refinement in the MIR and MAD methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- DeMot R. and Vanderleyden,J. (1994) The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol., 12, 333–334. [DOI] [PubMed] [Google Scholar]
- Doering D.S. (1992) Functional and structural studies of a small F-actin binding domain. PhD thesis, Massachusetts Institute of Technology, Cambridge, MA.
- Esnouf R.M. (1999) Further additions to MolScript ver 1.4, including reading and contouring electron density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Francis N.R., Irikura,V.M., Yamaguchi,S., DeRosier,D.J. and Macnab,R.M. (1992) Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl Acad. Sci. USA, 89, 6304–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N.R., Sosinsky,G.E., Thomas,D. and DeRosier,D.J. (1994) Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol., 235, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Garza A.G., Biran,R., Wohlschlegel,J. and Manson,M.D. (1996) Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J. Mol. Biol., 25, 270–285. [DOI] [PubMed] [Google Scholar]
- Glagolev A.N. and Skulachev,V.P. (1978) The proton pump is a molecular engine of motile bacteria. Nature, 272, 280–282. [DOI] [PubMed] [Google Scholar]
- Hirota N. and Imae,Y. (1983) Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J. Biol. Chem., 258, 10577–10581. [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1997) Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res., 25, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A.H., Nelson,J.W. and Weis,W.I. (1997) Three-dimensional structure of the armadillo repeat region of β-catenin. Cell, 90, 871–882. [DOI] [PubMed] [Google Scholar]
- Irikura V.M., Kihara,M., Yamaguchi,S., Sockett,H. and Macnab,R.M. (1993) Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation and switching of the flagellar motor. J. Bacteriol., 17, 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.J., Macnab,R.M., Okino,H. and Aizawa,S.-I. (1990) Stoichiometric analysis of the flagellar hook–(basal body) complex of Salmonella typhimurium. J. Mol. Biol., 212, 377–387. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kawagishi I., Maekawa,Y., Atsumi,T., Homma,M. and Imae,Y. (1995) Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J. Bacteriol., 177, 5158–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. and Berg,H.C. (1983) Isotope and thermal effects in chemiosmotic coupling to the flagellar motor of Streptococcus. Cell, 32, 913–919. [DOI] [PubMed] [Google Scholar]
- Khan S., Dapice,M. and Reese,T.S. (1988) Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol., 202, 575–584. [DOI] [PubMed] [Google Scholar]
- Khan I.H., Reese,T.S. and Khan,S. (1992) The cytoplasmic component of the bacterial flagellar motor. Proc. Natl Acad. Sci. USA, 89, 5956–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M., Francis,N.R., DeRosier,D.J. and Macnab,R.M. (1996) Analysis of a FliM–FliN flagellar switch fusion mutant of Salmonella typhimurium. J. Bacteriol., 178, 4582–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M., Miller,G.U. and Macnab,R.M. (2000) Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol., 182, 3022–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MolScript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Kubori T., Yamaguchi,S. and Aizawa,S.-I. (1997) Assembly of the switch complex onto the MS-ring complex of Salmonella typhimurium does not require any other flagellar proteins. J. Bacteriol., 179, 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen S.H., Adler,J., Gargus,J.J. and Hogg,R.W. (1974) Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc. Natl Acad. Sci. USA, 71, 1239–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Moss,D.S. and Thornton,J.M. (1993) Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol., 231, 1049–1067. [DOI] [PubMed] [Google Scholar]
- Lloyd S.A. and Blair,D.F. (1997) Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol., 266, 733–744. [DOI] [PubMed] [Google Scholar]
- Lloyd S.A., Tang,H., Wang,X., Billings,S. and Blair,D.F. (1996) Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol., 178, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd S.A., Whitby,F.G., Blair,D.F. and Hill,C.P. (1999) Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature, 400, 472–475. [DOI] [PubMed] [Google Scholar]
- Lowe G., Meister,M. and Berg,H.C. (1987) Rapid rotation of flagellar bundles in swimming bacteria. Nature, 325, 637–640. [Google Scholar]
- Macnab R. (1992) Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet., 26, 129–156. [DOI] [PubMed] [Google Scholar]
- Macnab R.M. (1996) Flagella and motility. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, 2nd edn. ASM Press, Washington, DC, pp. 123–145.
- Manson M.D., Tedesco,P.M., Berg,H.C., Harold,F.M. and van der Drift,C. (1977) A protonmotive force drives bacterial flagella. Proc. Natl Acad. Sci. USA, 74, 3060–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson M.D., Tedesco,P.M. and Berg,H.C. (1980) Energetics of flagellar rotation in bacteria. J. Mol. Biol., 138, 541–561. [DOI] [PubMed] [Google Scholar]
- Magariyama Y., Sugiyama,S., Muramoto,K., Maekawa,Y., Kawagishi,I., Imae,Y. and Kudo,S. (1994) Very fast flagellar rotation. Nature, 371, 752. [DOI] [PubMed] [Google Scholar]
- Marykwas D.L. and Berg,H.C. (1996) A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J. Bacteriol., 178, 1289–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marykwas D.L., Schmidt,S.A. and Berg,H.C. (1996) Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J. Mol. Biol., 256, 564–576. [DOI] [PubMed] [Google Scholar]
- Meister M. and Berg,H.C. (1987) The stall torque of the bacterial flagellar motor. Biophys. J., 52, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister M., Lowe,G. and Berg,H.C. (1987) The proton flux through the bacterial flagellar motor. Cell, 49, 643–650. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A., Lebedev,A., Wilson,K.S. and Dodson,E.J. (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D, 55, 247–255. [DOI] [PubMed] [Google Scholar]
- Nelson K.E. et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]
- Oosawa K., Ueno,T. and Aizawa,S.-I. (1994) Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol., 176, 3683–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. (1993) Oscillation data reduction program. In Sawyer,L., Isaacs,N. and Bailey,S. (eds), Data Collection and Processing. SERC Daresbury Laboratory, Warrington, UK, pp. 56–62.
- Ramakrishnan V., Finch,J.T., Graziano,V., Lee,P.L. and Sweet,R.L. (1993) Crystal structure of the globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219–223. [DOI] [PubMed] [Google Scholar]
- Ryu W.S., Berry,R.M. and Berg,H.C. (2000) Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature, 403, 444–447. [DOI] [PubMed] [Google Scholar]
- Samuel A.D. and Berg,H.C. (1995) Fluctuation analysis of rotational speeds of the bacterial flagellar motor. Proc. Natl Acad. Sci. USA, 92, 3502–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K. and Homma,M. (2000) Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus.J. Biol. Chem., 275, 5718–5722. [DOI] [PubMed] [Google Scholar]
- Schuster S.C. and Khan,S. (1994) The bacterial flagellar motor. Annu. Rev. Biophys. Biomol. Struct., 23, 509–539. [DOI] [PubMed] [Google Scholar]
- Shigenobu S., Watanabe,H., Hattori,M., Sakaki,Y. and Ishikawa,H. (2000) Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature, 407, 81–86. [DOI] [PubMed] [Google Scholar]
- Sockett H., Yamaguchi,S., Kihara,M., Irikura,V.M. and Macnab,R.M. (1992) Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol., 174, 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky G.E., Francis,N.R., Derosier,D.J., Wall,J.S., Simon,M.N. and Hainfeld,J. (1992) Mass determination and estimation of subunit stoichiometry of the bacterial hook–basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proc. Natl Acad. Sci. USA, 89, 4801–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stader J., Matsumura,P., Vacante,D., Dean,G.E. and Macnab,R.M. (1986) Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J. Bacteriol., 166, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz B. and Berg,H.C. (1991) Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol., 173, 7033–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. and Moffatt,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- Tang H., Braun,T.F. and Blair,D.F. (1996) Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol., 261, 209–221. [DOI] [PubMed] [Google Scholar]
- Toker A.S. and Macnab,R.M. (1997) Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol., 273, 623–634. [DOI] [PubMed] [Google Scholar]
- Ueno T., Oosawa,K. and Aizawa,S.-I. (1992) M-ring, S-ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol., 227, 672–677. [DOI] [PubMed] [Google Scholar]
- Weeks C.M. and Miller,R. (1999) The design and implementation of SnB v 2.0. J. Appl. Cryst., 32, 120–124. [Google Scholar]
- Welch M., Oosawa,K., Aizawa,S.-I. and Eisenbach,M. (1993) Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl Acad. Sci. USA, 90, 8787–8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita,H., Ishihara,A., Aizawa,S.-I. and Macnab,R.M. (1986a) Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation and switching. J. Bacteriol., 166, 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Aizawa,S.-I., Kihara,M., Isomura,M., Jones,C.J. and Macnab,R.M. (1986b) Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol., 168, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Amsler,C.D., Matsumura,P. and Khan,S. (1996a) FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J. Bacteriol., 17, 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Pathak,N., Jaffe,H., Reese,T.S. and Khan,S. (1996b) FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol., 261, 195–208. [DOI] [PubMed] [Google Scholar]
- Zhou J. and Blair,D.F. (1997) Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol., 273, 428–439. [DOI] [PubMed] [Google Scholar]
- Zhou J., Lloyd,S.A. and Blair,D.F. (1998a) Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl Acad. Sci. USA, 95, 6436–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Sharp,L.L., Tang,H.L., Lloyd,S.A., Billings,S., Braun,T.F. and Blair,D.F. (1998b) Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol., 180, 2729–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]