Abstract
We have identified a novel 200 kDa nuclear protein that activates the proteasome. The protein, which we call PA200, has been purified to homogeneity from bovine testis and has been shown to activate proteasomal hydrolysis of peptides, but not proteins. Following γ-irradiation of HeLa cells the uniform nuclear distribution of PA200 changes to a strikingly punctate pattern, a behavior characteristic of many DNA repair proteins. Homologs of PA200 are present in worms, plants and yeast. Others have shown that mutation of yeast PA200 results in hypersensitivity to bleomycin, and exposure of yeast to DNA damaging agents induces the PA200 message. Taken together, these findings implicate PA200 in DNA repair, possibly by recruiting proteasomes to double strand breaks.
Keywords: DNA repair/double strand breaks/nucleus/PA200/proteasome activator
Introduction
The 20S proteasome is found in organisms from all three kingdoms and is the major proteolytic enzyme in the cytoplasm and nuclei of eukaryotic cells (Baumeister et al., 1998). Because the proteasome appears to be freely diffusible within these compartments (Reits et al., 1997), it presumably contacts a large number of critical proteins. Any protease with direct access to important cellular components must be tightly regulated, and the proteasome is well designed to prevent indiscriminate degradation of potential substrates. Its 14 α-subunits and 14 β-subunits are arranged in four heptameric rings stacked upon one another (αββα) to form a cylindrical particle (Grziwa et al., 1991; Lowe et al., 1995). The catalytic subunits are located in the two central β rings with their active sites facing an internal chamber separated from cellular proteins by flanking antechambers (Bochtler et al., 1999). Access to the proteasome’s active sites is restricted even further by the α rings at each end of the cylinder since N-terminal regions of the α-subunits interact extensively to seal the central proteolytic chamber from the external solvent (Groll et al., 2000).
The proteasome would be a useless enzyme were there not mechanisms for promoting the transfer of protein and peptide substrates to its sequestered active sites. Two particles have been identified that bind the ends of the proteasome and activate it. One of these is the 19S regulatory complex, a particle containing 18 different subunits that associates with the 20S proteasome to form the 26S proteasome, the ATP-dependent protease that degrades ubiquitylated proteins (Rechsteiner, 1998; Voges et al., 1999). The other proteasome activators are heptameric rings known as 11S REG or PA28 (Dubiel et al., 1992; Ma et al., 1992). These rings promote the degradation of peptides and some poorly folded small proteins, but they do not, in general, stimulate destruction of intact proteins (Rechsteiner et al., 2000).
Previously, we identified two activated forms of the 20S proteasome in rabbit reticulocyte lysate (Hoffman et al., 1992). One of these was a 20S proteasome–PA28 complex; the other consisted of a 200 kDa protein bound to the 20S proteasome. Here we present experiments demonstrating that the 200 kDa protein purified from bovine testis is a proteasome activator, hence the name PA200. Like the PA28α/β and PA28γ proteasome activators, PA200 stimulates hydrolysis of small fluorogenic peptides, but not folded proteins. Its abundance in testis, its nuclear relocalization following γ-irradiation and the fact that mutation of yeast PA200 results in bleomycin hypersensitivity indicate that PA200 plays a role in DNA repair.
Results
The sequence of human PA200 and its homologs
We sequenced three Lys-C peptides from the 200 kDa protein bound to rabbit reticulocyte 20S proteasomes; all three map onto the hypothetical translation product of a human cDNA (KIAA0077) from KG1 myeloid cells (see Figure 1). As the protein encoded by KIAA0077 appeared to be incomplete, we searched human expressed sequence tag (EST) libraries for N-terminal extensions. This analysis revealed the likely presence of an additional 45 residues at the N-terminus of human PA200. Examining mouse genomic sequences and ESTs, we arrived at a hypothetical mouse protein that is >90% identical to human PA200 over the entire 1843 amino acids. The mouse and human sequences diverge markedly just N-terminal to the postulated initiating methionine, so we consider the sequence in Figure 1 to be that of the full-length, human PA200 protein.
Fig. 1. Amino acid sequence of human PA200. The amino acid sequence shown above was deduced from a human cDNA (KIAA0077) and analysis of human and mouse ESTs or genomes. The three peptides sequenced from rabbit reticulocyte PA200 are shown in blue, the three peptides (NH, MR, CT) used to generate antibodies are shown in green, the putative nuclear targeting sequence is highlighted in red, and yellow denotes 10 potential ATM phosphorylation sites (S/T-Q).
Homologs of mammalian PA200 are present in the worm Caenorhabditis elegans, the plant Arabidopsis thaliana and the yeast Saccharomyces cerevisiae, but they are much less similar to human PA200 with identities ranging from 29% for worm PA200 to 17% for the yeast protein (see Supplementary figure S1 available at The EMBO Journal Online). We have not found a PA200 homolog in the published Drosophila melanogaster or Schizosaccharomyces pombe genomes. The sequence of human PA200 contains a bipartite nuclear targeting sequence (shown in red in Figure 1), which is consistent with its nuclear localization (see below). However, an equivalent nuclear targeting sequence is not present in worm, plant or yeast PA200. Larger structural motifs in the PROSITE database, such as ring fingers, WD repeats, etc., are not evident in any of the homologs, although each contains a number of potential protein kinase C and casein kinase II phosphorylation sites as well as potential sites, shown yellow in Figure 1, for the important ‘DNA repair kinases’ ATM (ataxia telangiectasia mutated) and DNA-PK (Kastan and Lim, 2000).
Antibodies to PA200
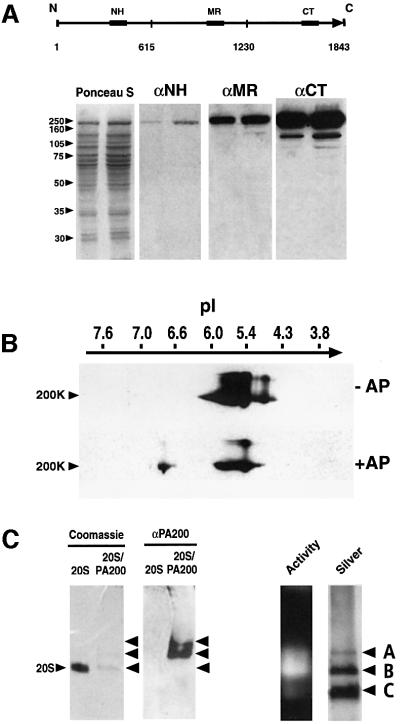
Antibodies to PA200 were generated by immunizing rabbits with synthetic peptides from three regions of the protein, and a 200 kDa protein was detected by western blot analysis of a fraction from bovine testis using each antiserum (Figure 2A). The single 200 kDa protein resolved into a number of isoelectric variants following 2D PAGE (Figure 2B). All of the variants were more acidic than the pI of 6.8 expected for mammalian PA200 based on the human or mouse sequences. To determine whether the acidic forms of PA200 result from phosphorylation, we treated a fraction enriched in bovine testicular PA200 with calf intestinal alkaline phosphatase and found that the most acidic form of PA200 was converted to a species with the expected pI of 6.8. Surprisingly, several intermediate isoelectric variants were unaffected by phosphatase treatment (see Figure 2B). Antibodies to PA200 also recognize PA200–proteasome complexes separated on native gels (Figure 2C). Anti-PA200 did not react with the free 20S proteasome (denoted ‘C’ in Figure 2C). However, the antisera did identify two slower migrating forms of the enzyme (denoted ‘A’ and ‘B’ in Figure 2C). Fluorogenic substrate overlays demonstrated that the slower migrating proteasome–PA200 complexes are more active than the free 20S proteasome (see right panel of Figure 2C).
Fig. 2. Anti-peptide antibodies to PA200. (A) Western blot analysis of PA200 separated by SDS–PAGE. Samples (7 and 14 µg) of partially purified bovine testis PA200 were separated on 10% polyacrylamide gels and transferred to nitrocellulose. Pairs of lanes were then probed with a 1:1000 dilution of antiserum to each of the immunizing peptides, the location of which is shown schematically above. (B) Identification of PA200 isoforms by 2D PAGE. PA200 resolves into a series of isoelectric variants, at least one of which is phosphorylated based on alkaline phosphatase (AP) treatment. (C) Recognition by anti-PA200 of PA200–proteasome complexes separated on native gels. Proteasome– PA200 complexes and purified 20S proteasomes were separated in adjacent lanes of a 4.5% native acrylamide gel. A portion of the gel was stained directly with Coomassie Brilliant Blue (left) and an equivalent pair of lanes were transferred to nitrocellulose for western blotting with anti-PA200 (αPA200). Similar native gel analyses were performed in which proteasome activity was monitored by sLLVY-MCA over lay (activity) prior to fixation and silver staining of the same lane (far right). The species denoted A, B and C are thought to be PA200– proteasome–PA200 complexes, PA200–proteasome complexes and free 20S proteasomes, respectively.
Distribution of PA200 in mouse tissues
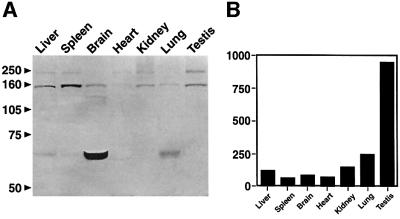
Because PA200 is not abundant in rabbit reticulocyte lysate, we surveyed several mouse organs for the protein by western blotting. Each of the anti-peptide antibodies identified two immunoreactive bands with apparent molecular weights of 200 and 160 kDa in SDS–PAGE samples from most organs. The 200 kDa species was most abundant in testis, whereas in the other organs the 160 kDa protein was generally more reactive (Figure 3A). A third immunoreactive species with a molecular weight of 60 kDa, present in liver and lung, was especially prominent in brain. The 60 kDa species only reacted with one of the three anti-peptide antibodies (anti-MR), but it was not detected by pre-immune serum, suggesting that it may be a shortened version of PA200, perhaps a proteolytically processed species or a splicing variant.
Fig. 3. (A) Tissue distribution of PA200. Mouse organs were homogenized in 0.25%Triton X-100, 10 mM Tris pH 7.5, 1 mM DTT. Samples (20 µg) were resolved on 10% SDS–PAGE gels, transferred to nitrocellulose, and PA200 was detected by western blotting. Note that full-length PA200 is most abundant in testis, whereas the 160 kDa species is enriched in spleen and the 60 kDa immunoreactive protein is greatly enriched in brain. (B) PA200 mRNA levels in various human tissues and cell lines.The sequence of human PA200 was evaluated by a high-throughput gene expression profiling resource (Su et al., 2002; publicly available at http://expression.gnf.org).
Proteasome activation by purified bovine testis PA200
In view of the relative abundance of PA200 in mouse testis and message profiles showing PA200 mRNA to be enriched in human testis (Figure 3B), we selected bovine testis for the purification of the molecule. DEAE, size exclusion and Mono Q chromatography followed by glycerol gradient centrifugation (see Supplementary figures S2–S5) resulted in full purification of PA200 (Figure 4). Proteasome activation was then examined using the purified protein. Eukaryotic proteasomes contain three catalytic subunits known as the chymotrypsin-like (CT), the trypsin-like (T) and the peptidylglutamyl peptide hydrolyzing (PGPH) activities. To determine whether PA200 activates all three catalytic subunits, we measured proteasomal hydrolysis of six fluorogenic peptides in the presence or absence of PA200. As can be seen in the upper portion of Table I, PA200 stimulated cleavage of all six peptides with greater activation for the PGPH substrates LLE-βNA and IETD-MCA. This pattern of activation differs from that seen by the 11S REG (PA28α/β) or the 19S regulatory complex (RC) (see Discussion). We also asked whether PA200 stimulated the degradation of [125I]casein or ubiquitin (Ub)–[125I]lysozyme conjugates. The lower portion of Table I shows that PA200 inhibited degradation of [125I]casein slightly and had no effect on the degradation of Ub–lysozyme conjugates. Proteasome activation by PA200 or the lack thereof was unchanged in the presence of 1 mM ATP (data not shown). Thus, PA200 is a protein that activates proteasomal cleavage of peptides in an energy-independent manner.
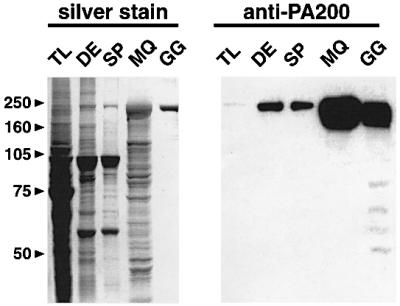
Fig. 4. Purification of bovine testicular PA200. Fractions obtained during the purification of PA200 (see Supplementary data) were analyzed by SDS–PAGE and western blotting. Samples from testis lysate (TL), DEAE-pool (DE), a sizing pool (SP), a Mono Q pool (MQ) and the final glycerol gradient (GG) were separated on 10% SDS–PAGE gels and either silver stained (left) or transferred to nitrocellulose and probed with anti-PA200 (right).
Table I. Proteasome activation by PA200.
| Substrate | Proteasome alone | Proteasome + PA200 | Stimulation (x-fold) |
|---|---|---|---|
| Peptide hydrolysis (F per min) | |||
| LLVY-MCA | 2.2 | 3.6 | 2 |
| LY-MCA | 0.7 | 2.8 | 4 |
| LRR-MCA | 0.9 | 2.9 | 3 |
| LSTR-MCA | 0.2 | 0.9 | 5 |
| LLE-βNA | 0.14 | 1.1 | 8 |
| IETD-MCA | 0.02 | 0.2 | 10 |
| Degradation (% per h) | |||
| [125I]casein | 22.4 | 18.0 | 0.8 |
| Ub–[125I]lysozyme | 1.2 | 1.5 | 1.2 |
Fluorogenic peptide assays were performed by pre-incubating 200 ng of purified bovine red cell proteasomes in the presence or absence of 340 ng of purified PA200 for 30 min at 37°C. Reactions were initiated by adding substrate (100 µM final) and incubating further at 37°C. Fluorescence (F) was measured at 380 ex/440 em or 335 ex/410 em in a Perkin Elmer LS-5 Spectrofluorimeter. Each entry is the average from four or more independent assays using several preparations of purified PA200. Ub–[125I]lysozyme conjugates were prepared as described (Hough et al., 1986); α-casein was radioiodinated by the chloramine-T procedure. Purified bovine proteasomes (1 µg) were pre-incubated in the presence or absence of 2.3 µg of purified PA200 for 30 min at room temperature. Duplicate reactions were initiated by the addition of radiolabeled substrate and incubated at 37°C. Samples were removed at 0, 30 and 60 min, and subjected to TCA precipitation. The distribution of 125I between supernate and pellet was determined in a Beckman Gamma 4000 counter and degradation rates were calculated from the increase in TCA soluble 125I. Ub–[125I]lysozyme conjugate reactions were carried out in the presence of 1 mM ATP. Stimulation of α-casein degradation remained unchanged in the presence or absence of ATP.
Proteasome activation is accompanied by formation of a PA200 proteasome complex
To explore the possibility that the proteasome activation presented in Table I was caused by an undetected contaminant in the final PA200 preparation, we used a native gel assay. A mixture of PA200 and 20S proteasomes or the separate components were analyzed by native gel electrophoresis. The resulting gel was overlaid with LLVY-MCA, to estimate proteasome activity before transferring the proteins to nitrocellulose for localization of PA200 by western blotting. The peptide overlay presented in Figure 5 shows proteasome activation upon addition of PA200, and it is evident from the adjacent western blot that some of the faster-migrating free PA200 molecules, which migrate as two bands (see Discussion for further explanation), became bound to the activated proteasome. We conclude that PA200 is responsible for the enhanced peptidase activity exhibited by PA200– proteasome complexes.
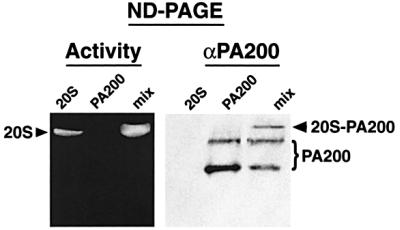
Fig. 5. Proteasome activation by purified bovine PA200. Purified PA200 or 20S proteasomes were analyzed separately or mixed prior to electrophoresis on a native gel. Peptide overlay (left) demonstrated that PA200 activated the proteasome and western blotting (right) confirmed that some of the added PA200 bound the 20S proteasome (denoted 20S–PA200) in the right panel.
Evidence that PA200 is not a proteasome-activated peptidase
Throughout the preceding sections, we have called the 200 kDa protein a proteasome activator based on the fact that purified PA200 does not hydrolyze fluorogenic peptides. However, increased cleavage of fluorogenic peptides by PA200–proteasome complexes would also result if PA200 were a proteasome-activated peptidase. Protease inhibitors were used to address this possibility. Unlike virtually all other proteases, the proteasome is sensitive to the microbial metabolite, lactacystin (Fenteany et al., 1995); it is also markedly inhibited by the peptide aldehyde, MG132 (Palombella et al., 1994). Therefore, these two compounds and two serine protease inhibitors, phenylmethylsulfonyl fluoride (PMSF) and tosyl-lysine chloromethyl ketone (TLCK), were used to characterize peptide hydrolysis by PA200–proteasome complexes. According to the data in Table II, activated cleavage of LY-MCA was inhibited >90% by MG132 and lactacystin, whereas PMSF and TLCK had no inhibitory effect. Similar results were obtained with the T-site substrate, LRR-MCA, and the PGPH substrate, LLE-βNA (Table II). These results coupled with the absence of any obvious peptidase motifs among PA200 homologs demonstrate that PA200 is not a latent peptidase, rather it is a bona fide proteasome activator.
Table II. Effects of protease inhibitors on peptide hydrolysis by PA200–proteasome complexes.
| Inhibitor | Concentration (µM) | % inhibition |
|
|
|
|
|
|---|---|---|---|---|---|---|---|
| LY-MCA |
LRR-MCA |
LLE-βNA |
|||||
| Proteasome | Proteasome + PA200 | Proteasome | Proteasome + PA200 | Proteasome | Proteasome + PA200 | ||
| Lactacystin | 50 | 96 | 99 | 84 | 85 | 34 | 29 |
| MG132 | 50 | 94 | 99 | 74 | 75 | 85 | 86 |
| PMSF | 100 | –3 | 1 | –35 | –34 | –41 | –102 |
| TLCK | 100 | 1 | 13 | 0 | –1 | 26 | –17 |
The entries presented above are means from at least three experiments using a single preparation of PA200 that increased proteasomal hydrolysis of LY, LRR and LLE by 3-, 2- and 6-fold, respectively.
Association of PA200 with the 26S proteasome
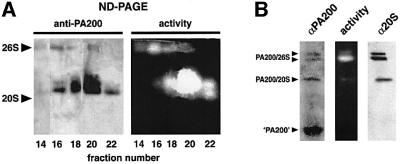
Using dilutions of purified PA200 and 20S proteasome as standards, the amount of each component in testis extract was estimated by western blotting. For each 10 µg of crude extract, we measured (within a factor of two) 100 ng of proteasome and 16 ng of PA200. These values permit us to calculate that there is one PA200 molecule for every two proteasomes. Assuming a uniform distribution of the two components among the different cell types in testis, there are more than enough 20S proteasomes to bind all available PA200 molecules. However, some early experiments with rabbit reticulocyte lysate provided evidence that PA200 also associates with the 26S proteasome. In these studies, DEAE-enriched proteasome fractions were sedimented on glycerol gradients and individual gradient fractions were then analyzed on native gels. Fluorogenic peptide overlays and western blotting with anti-PA200 revealed co-localization of PA200 and the rabbit 26S proteasome (see Figure 6A). Furthermore, western blot analysis of HeLa cell extracts separated on native gels shows clear staining of 26S proteasomes by anti-PA200 (Figure 6B). Since anti-PA200 does not react with 20S proteasomes (Figure 2C) or the 19S RC (data not shown), it appears that, in addition to binding 20S proteasomes, PA200 molecules associate with the 26S proteasome. Pre sumably they bind one end of the 20S proteasome while the 19S RC binds the other.
Fig. 6. Association of PA200 with the 26S proteasome. (A) Fractions obtained by DEAE chromatography of rabbit reticulocyte lysate enriched in 20S and 26S proteasomes were pooled and sedimented at 113 000 g for 22 h on a 25–45% glycerol gradient. The gradient was collected by bottom puncture, and samples of fractions 14, 16, 18, 20 and 22 were resolved on a 4.5% native polyacrylamide gels; the gels were overlaid with LLVY-MCA to detect 26S and 20S proteasomes (activity, right) prior to transfer to a nitrocellulose membrane for western blotting with anti-PA200 (left). Note that small amounts of anti-PA200 immunoreactivity coincides with 26S proteasome activity in fractions 14–20. (B) HeLa cells were extracted in 0.5% Triton X-100 and samples were analyzed in multiple lanes of a native acrylamide gel. Some lanes were overlaid with LLVY-MCA to localize 26S and 20S proteasomes (activity), while other lanes were transferred to nitrocellulose for western blotting with anti-PA200 (αPA200) or antibodies to 20S proteasome subunits (α20S). Note that as in (A), anti-PA200 immunoreactivity coincides with two forms of the 26S proteasome as well as with a 20S species. The component denoted ‘PA200’ (bottom left) represents the 60 kDa immunoreactive species.
Intracellular localization of PA200
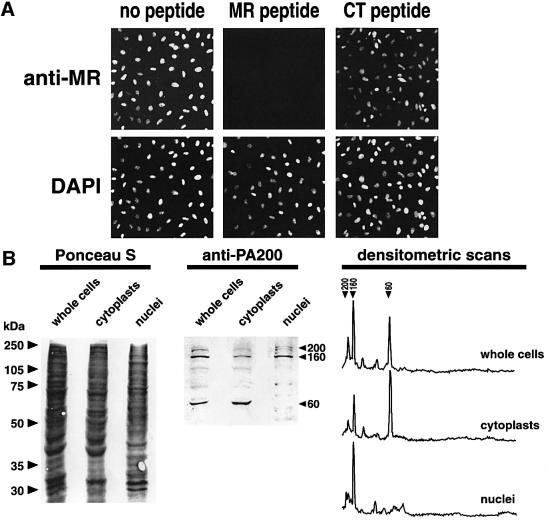
Anti-PA200 antibodies were used to localize the molecule within mammalian culture cells. HeLa cells were fixed in formaldehyde and stained with each antipeptide antibody (anti-NH, -MR or -CT) followed by FITC secondary antibody. Nuclei were brightly stained by both the DNA-specific compound DAPI (Figure 7A, lower panels) or by anti-PA200, and there was little cytoplasmic immunofluorescence. At mitosis PA200 is located throughout the cell with no evidence for association with condensed chromosomes (see Figure 8). Nuclear staining was abolished when the immunizing MR peptide was mixed with anti-MR prior to staining (see Figure 7A, central panel), whereas addition of CT peptide had no effect on nuclear staining by anti-MR (Figure 7A, upper right). Equivalent results were obtained for anti-NH in that NH-peptide eliminated nuclear staining, but MR peptide had no effect (data not shown). With anti-CT, nuclear staining was reduced, although not eliminated, by CT-peptide perhaps because anti-CT is such an avid antibody (see Figure 2A).
Fig. 7. Intracellular distribution of PA200. (A) Indirect immunofluorescent HeLa cell stain. Cells growing on coverslips were fixed in 3.7% formaldehyde and stained with 1:300 anti-MR antiserum (see Figure 2) followed by FITC-goat anti-rabbit IgG. Note the excellent correspondence between staining by the DNA specific dye DAPI and FITC-fluorescence (left panels). Nuclear fluorescence was abolished when anti-MR was pre-incubated with 20 µg MR peptide prior to staining (center) but was not affected by preincubation with 20 µg CT peptide (right). (B) Distribution of PA200 in cytoplasts and nuclei prepared by cytochalasin-B-mediated enucleation. HeLa cells were enucleated as described in Materials and methods, and the distribution of PA200 was determined by western blotting of cytoplast and karyoplast proteins separated on 10% SDS–PAGE gels. The blot shown in the central panel and densitometry of it (right) clearly demonstrate that the 60 kDa immunoreactive species is confined to the cytoplasm, and that the 200 and 160 kDa species are located within nuclei.
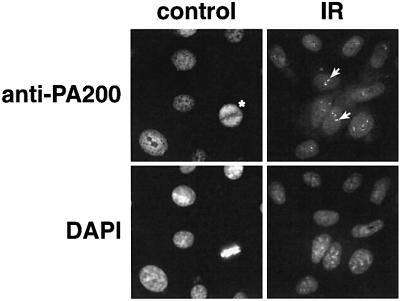
Fig. 8. Intracellular distribution of PA200 following exposure of HeLa cells to ionizing radiation (IR). HeLa cells growing on coverslips were exposed to 50 Gy from a 137Cs source and fixed at 2, 6 or 22 h post-irradiation, then stained with anti-PA200 and DAPI. Note that the relatively uniform, albeit finely punctate, distribution of PA200 in control cells is distinctly different from the focal aggregates or foci of PA200 (white arrows) seen in HeLa nuclei 22 h after irradiation. Note also the even distribution of PA200 in the metaphase cell (control, asterisk).
The intracellular distribution of PA200 was also examined by cell fractionation. In contrast to the immunofluorescence results, virtually all HeLa PA200 was present in the post-nuclear supernate (not shown). Since proteins can leak from nuclei during homogenization and centrifugation, we attribute the presence of PA200 in the cytosolic fraction to leakage. This interpretation is supported by experiments involving cytochalasin B-mediated enucleation. HeLa cells were grown on coverslips, exposed to cytochalasin B and the coverslips were centrifuged in an inverted orientation at 6800 g for 1 h. This resulted in the enucleation of 80% of the attached HeLa cells. Western blot analysis of SDS–PAGE samples from the resulting karyoplasts and cytoplasts revealed full-length PA200 to be present largely in nuclei (Figure 7B). In fact, densitometry showed that <20% of full-length PA200 molecules remained in the cytoplasts, and these could be accounted for by whole cells left on the coverslips (Figure 7B). The immunofluorescence results combined with those obtained by cytochalasin B-mediated enucleation clearly show the 160 and 200 kDa forms of PA200 to be nuclear.
Formation of PA200 intranuclear foci after γ-irradiation
According to sequences deposited in the DDBJ/EMBL/GenBank database, the budding yeast homolog of PA200 is encoded by two adjacent genes. One of these is BLM3, so named because its mutation results in bleomycin sensitivity (Moore, 1991), and the other is YFL006. However, yPA200 is actually encoded by a single gene that includes BLM3 and YFL006 (see Supplementary figure S1). Nonetheless, mutation of yPA200 results in bleomycin sensitivity, and because bleomycin causes DNA double strand breaks (DSBs), we suspected that PA200 might be involved in DNA repair. The intranuclear distribution of many DNA repair factors changes upon DNA damage. For example, Rad52p (Lisby et al., 2001) and 53BPI (Schultz et al., 2000; Anderson et al., 2001) form discrete intranuclear foci after DNA damage. Given our suspicion that PA200 is involved in DNA repair, we examined its intranuclear distribution after irradiating HeLa cells with γ-rays (50 Gy), UV (254 nm, 34J/m2) or after treating the cells with 5 mM H2O2 for 30 min. The micrographs in Figure 8 clearly reveal the formation of PA200 nuclear foci in virtually all cells at 22 h after exposure of HeLa to ionizing radiation; the same pattern was seen at 6 h, but in only 20% of the irradiated cells. In contrast, PA200 remained dispersed within HeLa nuclei after UV irradiation or H2O2 treatment (not shown). The redistribution of PA200 within HeLa nuclei following exposure of the cells to ionizing radiation provides further support for the protein’s involvement in DNA repair, presumably of DSBs.
Discussion
PA200 is a proteasome activator
The experiments presented above constitute the initial characterization of PA200, a novel nuclear protein, and they provide substantial evidence that PA200 is a proteasome activator. The protein was originally identified as a component of a slow-migrating, activated form of the 20S proteasome in rabbit reticulocyte lysate (Hoffman et al., 1992). Antibodies to PA200 specifically stain 20S–PA200 complexes separated on native gels (Figure 2C); they also stain rabbit reticulocyte and HeLa 26S proteasomes (Figure 6A and B). Recent large-scale proteomics screens identified yeast PA200 bound to the yeast proteasome (Gavin et al., 2002; Ho et al., 2002), indicating that association of PA200 with proteasomes is conserved among diverse organisms. More convincing evidence that PA200 is a proteasome activator is provided by the fact that purified bovine testis PA200 forms a complex with the 20S proteasome (Figure 5), and in so doing, it stimulates fluorogenic peptide hydrolysis as much as 10-fold (Table I). Use of proteasome inhibitors eliminated the possibility that PA200 is, itself, a peptidase (Table II).
Comparison of PA200 to other proteasome activators
Three other cellular components, the 19S RC, PA28α/β and PA28γ, are also capable of stimulating proteasomal hydrolysis of fluorogenic peptides. Comparison of proteasome activation by PA200 to that seen with the other activators reveals that it is more like PA28α/β and PA28γ than the 19S RC. The latter is the only known activator capable of stimulating proteolysis as well as peptide hydrolysis. The RC is also unique in its requirement for ATP to stimulate peptide bond hydrolysis by the proteasome (Hoffman and Rechsteiner, 1994). PA200, PA28α/β and PA28γ bind and activate the proteasome in the absence of nucleotides (Rechsteiner, 1998). Also unlike the 19S RC, they do not enhance degradation of intact proteins.
The proteasome contains three active β-subunits, CT, T and PGPH, based respectively on their preferences for hydrophobic, basic or acidic residues as the carbonyl donor to the peptide bond being cleaved. The RC, PA28s and PA200 activate these subunits in distinct ways. Rabbit reticulocyte RC stimulates ∼3-fold the hydrolysis of fluorogenic peptides diagnostic for each of the catalytic subunits (Hoffman and Rechsteiner, 1994). Likewise, PA28α/β stimulates hydrolysis of the three diagnostic peptides, but to a much greater extent, i.e. 10- to 20-fold (Dubiel et al., 1992; Ma et al., 1992). In contrast, PA28γ stimulates hydrolysis of LRR-MCA, a trypsin-like substrate, by an order of magnitude while inhibiting the hydrolysis of peptides cleaved at the CT site (Li et al., 2001). A unique pattern of proteasome activation is seen with PA200. It stimulates proteasomal hydrolysis of the PGPH substrates, LLE-βNA and IETD-MCA, almost three times more than the hydrolysis of fluorogenic peptides diagnostic for the CT and T active sites (Table I).
Mechanism for proteasome activation
There are two mechanisms, not mutually exclusive, by which activators can stimulate proteasome peptide hydrolysis. They can open pores to the central catalytic chamber or they can alter the conformation of the active β-subunits thereby speeding catalysis. Crystallography of REGα– proteasome complexes (Whitby et al., 2000) shows that REGα (PA28α) binds the ends of the proteasome and opens a channel into the central chamber. Since binding of REGα activates all three catalytic subunits comparably, there is no need to invoke conformational changes in the catalytic β-subunits. On the other hand, REGγ only activates the T subunit (Realini et al., 1997), and we have presented evidence that REGγ suppresses hydrolysis at the CT and PGPH sites (Li et al., 2001). PA200 also binds the ends of the proteasome (J.Ortega and A.Steven, personal communication). The preferential hydrolysis of peptides with acidic residues at P1 could be due to PA200 generating a channel into the proteasome that facilitates entry of negatively charged peptides or PA200 could induce activating conformational changes preferentially in the PGPH catalytic subunit. Recently we used a combinatorial library of fluorogenic tetrapeptides to characterize REGα/β– and REGγ–proteasome complexes (Harris et al., 2001). Similar analyses of PA200–proteasome complexes should permit us to determine the relative contributions of selective entry or allosteric mechanisms to activation by PA200.
Multiple forms of PA200
PA28γ is predominantly nuclear, PA28α/β is predominantly cytoplasmic and the RC is distributed between the two compartments (Brooks et al., 2000). Like PA28γ, the 200 and 160 kDa forms of PA200 are present within HeLa nuclei, whereas the 60 kDa immunoreactive band is cytoplasmic (see Figure 7B). Although both the 200 and the 160 kDa species react with three antipeptide antibodies, the 60 kDa protein only reacts with anti-MR after SDS–PAGE. Because the 60 kDa species is so prominent on western blots (Figure 7B), we were surprised to find that anti-MR just stains HeLa nuclei (Figure 7A). Presumably the 60 kDa epitope is either masked in the cytoplasm or destroyed during fixation prior to fluorescence microscopy. Alternatively, it is possible that cytochalasin B causes the 60 kDa protein to relocate to the cytoplasm. Because all three bands are present in HeLa cells lysed directly in hot SDS–PAGE sample buffer, it is unlikely that the 160 and 60 kDa immunoreactive forms arise by proteolysis during sample preparation. They could be naturally processed forms of PA200, or they could arise by alternative splicing. In this regard, it should be noted that while most PML isoforms are nuclear, some splice variants are present in the cytoplasm (Salomoni and Pandolfi, 2002).
The multiple molecular weight forms seen with PA200 have not been reported for PA28α/β or PA28γ. Post-translational modification also distinguishes PA200 from the other two proteasome activators. Although it was reported some years ago that PA28α/β had to be phosphorylated to activate the proteasome (Li et al., 1996), we have not observed modifications of either PA28α/β or PA28γ, and to our knowledge, neither have other investigators. The 2D PAGE gels presented in Figure 2B provide evidence that PA200 is phosphorylated, since treatment with alkaline phosphatase shifted the isoelectric point of the most acidic form of PA200 to a value which matches the predicted pI of human PA200. Surprisingly, several other isoelectric variants were unaffected by alkaline phosphatase treatment, raising the possibility that PA200 is subject to several post-translational modifications or that some phosphate groups are refractory to alkaline phosphatase.
Oligomerization of PA200
PA28α/β and PA28γ are heptamers (Zhang et al., 1999; Li et al., 2001) and the RC is an even higher order multimer (Gorbea et al., 2000). As a single polypeptide chain capable of activating the proteasome, PA200 is unique among the four known activators. In fact, PA200 appears to exist in a monomer–dimer equilibrium. This is evident in Supplementary figure S5, where the distribution of PA200 on the glycerol gradient is not symmetric; rather there is a shoulder of faster sedimenting molecules. Moreover, the western blot in Figure 5 clearly shows two electrophoretic forms of PA200 following native gel electrophoresis. Interestingly the faster-migrating form, assumed to be monomeric PA200, appears to preferentially bind the proteasome. Conceivably, as has been proposed for Rad23p and Ddi1p (Bertolaet et al., 2001), the activity of PA200 is controlled by its oligomerization state.
Evidence that PA200 plays a role in DNA repair
There are several strong indications that yPA200 functions in DNA repair. BLM3 was originally discovered in a screen for sensitivity to the DNA damaging agent bleomycin (Moore, 1991). Transcript profiles of yeast exposed to the DNA damaging agent methyl methanesulfonate revealed ∼5-fold increases in the message for yPA200 (Jelinsky et al., 2000). Consistent with our finding that PA200 is a proteasome activator, recent large-scale proteomic screens have identified the BLM3-YFL006 gene product bound to the yeast 20S proteasome (Gavin et al., 2002; Ho et al., 2002). More interesting, the BLM3-YFL006 protein was also found complexed with the chromatin component Sir4p (Ho et al., 2002). Upon DNA damage, Sir4p has been shown to leave telomeres and relocate at DSBs, where it binds yKu70, a well-established component of DSB repair (Martin et al., 1999; Mills et al., 1999; Doherty and Jackson, 2001). Several properties of mammalian PA200 also implicate the protein in DNA repair. Both the PA200 message and the full-length protein are abundant in testis, where DSBs occur during meiotic recombination. Furthermore, PA200 is a nuclear phosphoprotein, and like a number of DNA repair factors (Schultz et al., 2000; Tashiro et al., 2000; Anderson et al., 2001; Bischof et al., 2001; Lisby et al., 2001; Mirzoeva and Petrini, 2001), it forms intranuclear foci following γ-irradiation (see Figure 8). These observations indicate that mammalian PA200 also functions in the repair of DSBs.
Possible mechanisms by which PA200 facilitates DNA repair
Because yPA200 binds both the proteasome and Sir4p, it is tempting to hypothesize that it serves as an adapter between the proteasome and chromatin proteins involved in DNA repair. As a component of the 26S proteasome (Figure 6), it could recruit the large ATP-dependent protease to sites of DNA damage. Once there, the 26S proteasome could degrade chromatin proteins in order to expose DNA to the requisite repair enzymes. In fact, yeast histone H2B is depleted in the immediate vicinity of DSBs (R.Schroff and M.Lichten, personal communication). On the other hand, there are several examples of an antagonistic relationship between DNA repair and the Ub–proteasome pathway. For example, the 26S proteasome and its subcomponent 19S RC have been reported to negatively affect nucleotide excision repair, perhaps by limiting the accumulation of Rad4 (Lommel et al., 2000; Gilette et al., 2001). Similarly, the Ub ligase components CUL4A or hHYD are reported to inhibit the repair of UV or X-ray lesions, respectively (X.Chen et al., 2001; Honda et al., 2002). The Ub carrier protein, Rad6, and several other Ub pathway components (Hofmann and Pickart, 1999; Ulrich and Jentsch, 2000; Torres-Ramos et al., 2002) also play important roles in repairing UV damage, but their exact function in the repair process is not well understood (Ulrich, 2002). Rad23p provides an example of a DNA repair component that inhibits proteolysis, for it suppresses elongation of Ub chains (Ortolan et al., 2000; L.Chen et al., 2001). Since chains containing four Ub molecules are required for efficient proteolysis by the 26S proteasome (Thrower et al., 2000), Rad23p presumably inhibits Ub-dependent proteolysis. PA200 does not promote degradation of intact proteins (Table I), so it too could antagonize protein degradation. For example, following DNA damage PA200 might compete more effectively with the 19S RC for free 20S proteasomes. This could reduce the level of 26S proteasomes and stabilize chromatin proteins during DNA repair. Clearly, a central question for future experiments is whether PA200 enhances or suppresses proteolysis following DNA damage.
In conclusion, we have shown that PA200 is a nuclear phosphoprotein that activates the proteasome. We have also presented evidence that PA200 plays a role in DNA repair. In mammalian cells the evidence is circumstantial, e.g. nuclear location, enrichment in testis and redistribution upon γ-irradiation. In yeast, the evidence is virtually compelling. Mutation of yPA200 confers sensitivity to bleomycin, and methyl methanesulfonate induces the message for yPA200. Moreover, yPA200 binds both the proteasome and SIR4p, which is part of a protein complex known to relocate to DSBs and bind yKu70. Although there is substantial evidence that PA200 contributes to efficient DNA repair, exactly how the proteasome activator functions in the process remains to be determined.
Materials and methods
Materials
Frozen bovine testis was obtained from Pel-Freez Biologicals (Rogers, AR). TSK-DEAE-650 M was from Supelco (Bellefonte, PA). Q Sepharose, Superdex 200 and Mono Q were obtained from Amersham-Pharmacia. DMEM and trypsin were purchased from Gibco-BRL. Penicillin, streptomycin, cytochalasin B, Ub, PMSF, TCLK, lactacystin and the fluorogenic peptide substrates LLVY-MCA, LY-MCA and LLE-βNA were obtained from Sigma. MG132 was from Calbiochem. LRR-MCA and IETD-MCA were from Peptides International, Inc. (Louisville, KY). LSTR-MCA was purchased from Peninsula Laboratories, Inc. (Belmont, CA). The Complete protease inhibitor cocktail was purchased from Roche Molecular Biochemicals (Mannheim, Germany). Imject maleimide activated mariculture keyhole limpet hemocyanin (mcKLH) was obtained from Pierce. Peroxidase-conjugated goat anti-rabbit IgG used as secondary antibody was obtained from Cappel (Aurora, OH). Calf intestine alkaline phosphatase was purchased from Roche Molecular Biochemicals. Protein silver stain reagents were obtained from Bio-Rad. Western blot chemiluminescence reagent and 125I were purchased from NEN Life Science Products, Inc. (Boston, MA).
Cell culture, cell fractionation and cytochalasin-mediated enucleation
The HeLa derivative, D98/AH2, was grown in DMEM containing 10% fetal calf serum and penicillin–streptomycin. For fractionation, cells were harvested by trypsinization, washed with phosphate-buffered saline (PBS) and suspended at 4 × 107 cells/ml in 0.25 M sucrose, 10 mM Tris pH 7.5, 1 mM Na EDTA. The cells were disrupted by 400 strokes in a tight-fitting Dounce homogenizer. Phase-contrast microscopy confirmed that >95% of the cells were broken prior to collecting the nuclear fraction by centrifuging in an Eppendorf microfuge for 10 min at 14 000 r.p.m. For enucleation, D98/AH2 cells were grown on 18 mm circular coverslips and enucleated by the method of Prescott et al. (1972) as described previously (Rechsteiner and Catanzarite, 1974).
Purification of PA200 from bovine testis
Frozen bovine testicles were shattered with a hammer and chisel, the albuginea was removed, and the remaining tissue was cut into small pieces. Diced tissue (125 g) was homogenized in a Waring blender in 300 ml of 0.25% Triton X-100, 10 mM Tris pH 7.0, 1 mM DTT, to which were added six tablets of Complete protease inhibitor cocktail (Roche). The homogenate was centrifuged at 100 000 g for 60 min, and the supernate was applied to a 100 ml DEAE 650-M column (Supelco) equilibrated in 10 mM Tris pH 7.0, 25 mM KCl, 10 mM NaCl, 5.5 mM MgCl2, 0.1 mM EDTA, 1 mM DTT and 10% glycerol. The column was developed at 1.5 ml/min with a linear gradient from 35 to 300 mM KCl over 10 column volumes in TSD/10% glycerol. See Supplementary figures S2–S5 for further description of the purification protocol.
Generation of antibodies
Three peptides, LFNSITSFYHPSNNG, STSSYSQVRNKAQ and TKLP KKRKRDPGSVG, corresponding to three distinct regions of the PA200 protein (see Figure 1), were used for antibody production. A cysteine was added at the end of each peptide sequence for conjugation to the protein carrier, mcKLH, following instructions from Pierce. Peptide-coupled KLH was sent to Harlan Bioproducts for Science, Inc. (Indianapolis, IN) for antibody production.
Indirect immunofluorescent staining
HeLa cells were grown to confluency on fibronectin-coated coverslips, washed in PBS, fixed [15 min in 3.7% formaldehyde/universal buffer (UB) (50 mM Tris–HCl pH 7.6, 150 mM NaCl, 0.1% azide)], permeabilized (3 min in 0.2% Triton X-100/UB), blocked (1 h in 5% normal goat serum, 2% BSA, 0.2% gelatin, 0.02% azide pH 7.4), incubated with primary antibody (18 h at 4°C; 1:600 dilution PA200 sera in UB), washed in UB, incubated with secondary antibody and DAPI [1 h at 37°C; 1:300 AlexaFluor488-anti-rabbit and 0.3 µM DAPI (Molecular Probes, Eugene, OR)], washed in UB and mounted with ProLong (Molecular Probes). The cells were visualized with a Zeiss Axiophot microscope, photographed using a digital camera (Princeton Instruments, Trenton, NJ) and OpenLab software (Improvision, Boston, MA), and Adobe Photoshop for graphic presentation. For peptide blocking experiments, 20 µg peptide was incubated with antibody dilutions (15 min at 37°C) and then applied immediately to cells.
Organ survey
Organs from an adult male mouse were excised, minced, washed in PBS and extracted on ice using four strokes of a motor driven Teflon homogenizer rotating at 50 r.p.m. in 10 mM Tris pH 7.5, 1 mM DTT buffer containing 0.25% Triton X-100 and one Complete protease inhibitor tablet per 50 ml of buffer. Extracts were clarified by centrifugation for 20 min in an Eppendorf centifuge at 14 000 r.p.m. and protein concentration was determined using Coomassie Protein Assay Reagent (Pierce Chemical Company). Aliquots of 100 µg total protein from each extract were analyzed on SDS–10% PA mini gels using a Mini-Protean gel apparatus (Bio-Rad).
Electrophoresis
SDS–10% polyacrylamide gels were prepared and run according to the method of Laemmli (1970). The method of O’Farrell (1975) was used for 2D PAGE. A fraction of bovine testis PA200, purified through Mono Q, was incubated in the presence or absence of 30 U of alkaline phosphatase at 37°C for 40 min prior to isoelectric focusing. Native gel electrophoresis and substrate overlays were performed as described previously (Hoffman et al., 1992).
Western blotting and immunodetection
Following electrophoresis, proteins were transferred onto nitrocellulose membranes using the Bio-Rad Trans-Blot apparatus. To transfer high molecular weight proteins such as PA200 a modified Tris/glycine buffer containing 1% methanol was used. Transfers were stained with Ponceau S and blocked with 500 mM Tris pH 7.5, 4.5% NaCl and 10% dry milk prior to incubating with a mixture of each anti-peptide antibody at 1:1000 in blocking buffer containing 5% dry milk. Peroxidase conjugated goat anti-rabbit IgG was used as secondary antibody. Subsequent immunodetection was achieved by chemiluminescence detected on Kodak X-Omat AR film.
Proteasome and proteasome activator assays
Throughout purification of PA200 endogenous proteasomal activity was assayed by adding 5 µl aliquots from each fraction to 100 µl of 100 µM LLVY-MCA in 20 mM Tris pH 7.8, 5 mM MgCl2, 10 mM KCl, 1 mM DTT. After incubating at 37°C for 20 min, reactions were quenched by addition of 200 µl ETOH, and fluorescence was measured using a Perkin Elmer LS5 spectrofluorimeter at an excitation wavelength of 380 nm and an emission wavelength of 440 nm with MCA peptide substrates and at an excitation of 335 nm and emission of 410 nm for LLE-βNA. Proteasome activation was assayed by adding 5 µl of the various fractions in a 100 µl reaction mix containing 200 ng of purified bovine red blood cell (RBC) proteasome and 100 µM LLVY-MCA. After 20 min incubation the reactions were quenched with 200 µl ETOH. Purified PA200 binding and activation of the proteasome was assayed by incubating increasing concentrations of PA200 (145 ng to 435 ng) with 200 ng of purified bovine RBC proteasome for 30 min at room temperature. An 8-fold molar ratio of PA200 to 20S proteasome was saturating for activation. After incubation duplicated samples were run on a native polyacryamide gel electrophoresis for 100 min at 120 V (4°C). Gels containing duplicated samples were divided with one half being used to perform substrate overlay for activity detection while the other half was processed for immunodetection with anti-PA200 to locate PA200.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Carlos Gorbea for his help in assembling the proposed sequence for human PA200 and for assistance in preparing figures as well as for reading the manuscript. We greatly appreciate the help of Ray Warters with the irradiation experiments. We thank Tim Formosa, Christopher Hill, Geoff Goellner and Xiaolin Gao for helpful comments on the manuscript. We also are indebted to Robert Schroff, Michael Lichten, Joaquin Ortega and Alasdair Steven for allowing us to cite their unpublished findings. These studies were supported in part by a University of Utah Seed Grant.
References
- Anderson L., Henderson,C. and Adachi,Y. (2001) Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol., 21, 1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W., Walz,J., Zhül,F. and Seemüller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Bertolaet B.L., Clarke,D.J., Wolff,M., Watson,M.H., Henze,M., Divita,G. and Reed,S.I. (2001) UBA domains mediate protein–protein interactions between two DNA damage-inducible proteins. J. Mol. Biol., 313, 955–963. [DOI] [PubMed] [Google Scholar]
- Bischof O., Kim,S.-H., Irving,J., Beresten,S., Ellis,N.A. and Campisi,J. (2001) Regulation and localization of the bloom syndrome protein in reponse to DNA damage. J. Cell Biol., 153, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M., Ditzel,L., Groll,M., Hartmann,C. and Huber,R. (1999) The proteasome. Annu. Rev. Biophys. Biomol. Struct., 28, 295–317. [DOI] [PubMed] [Google Scholar]
- Brooks P. et al. (2000) Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J., 346, 155–161. [PMC free article] [PubMed] [Google Scholar]
- Chen L., Shinde,U., Ortolan,T.G. and Madura,K. (2001) Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO rep., 2, 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang,Y., Douglas,L. and Zhou,P. (2001) UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem., 276, 48175–48182. [DOI] [PubMed] [Google Scholar]
- Doherty A.J. and Jackson,S.P. (2001) DNA repair: how Ku makes ends meet. Curr. Biol., 11, R920–R924. [DOI] [PubMed] [Google Scholar]
- Dubiel W., Pratt,G., Ferrell,K. and Rechsteiner,M. (1992) Purification of an 11S regulator of the multicatalytic protease. J. Biol. Chem., 267, 22369–22377. [PubMed] [Google Scholar]
- Fenteany G., Standaert,R.F., Lane,W.S., Choi,S., Corey,E.J. and Schreiber,S.L. (1995) Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lacta cystin. Science, 268, 726–731. [DOI] [PubMed] [Google Scholar]
- Gavin A.-C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Gilette T.G., Huang,W., Russell,S.J., Reed,S.H., Johnston,S.A. and Friedberg,E.C. (2001) The 19S complex of the proteasome regulates nucleotide excision repair in yeast. Genes Dev., 15, 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbea C., Taillandier,D. and Rechsteiner,M. (2000) Mapping subunit contacts in the regulatory complex of the 26S proteasome. J. Biol. Chem., 275, 875–882. [DOI] [PubMed] [Google Scholar]
- Groll M., Bajorek,M., Kohler,A., Moroder,L., Rubin,D.M., Huber,R., Glickman,M.H. and Finley,D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol., 7, 999–1001. [DOI] [PubMed] [Google Scholar]
- Grziwa A., Baumeister,W., Dahlmann,B. and Kopp,F. (1991) Localization of subunits in proteasomes from Thermoplasma acidophilum by immunoelectron microscopy. FEBS Lett., 290, 186–190. [DOI] [PubMed] [Google Scholar]
- Harris J.L., Alper,P.B., Li,J., Rechsteiner,M. and Backes,B.J. (2001) Substrate specificity of the human proteasome. Chem. Biol., 8, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Hoffman L. and Rechsteiner,M. (1994) Activation of the multicatalytic protease. The 11S regulator and 20S ATPase complexes contain distinct 30-kilodalton subunits. J. Biol. Chem., 269, 16890–16895. [PubMed] [Google Scholar]
- Hoffman L., Pratt,G. and Rechsteiner,M. (1992) Multiple forms of the 20S multicatalytic and the 26S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J. Biol. Chem., 267, 22362–22368. [PubMed] [Google Scholar]
- Hofmann R.M. and Pickart,C.M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell, 96, 645–653. [DOI] [PubMed] [Google Scholar]
- Honda Y., Tojo,M., Matsuzaki,K., Anan,T., Matsumoto,M., Ando,M., Saya,H. and Nakao,M. (2002) Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J. Biol. Chem., 277, 3599–3605. [DOI] [PubMed] [Google Scholar]
- Hough R., Pratt,G. and Rechsteiner,M. (1986) Ubiquitin–lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. J. Biol. Chem., 261, 2400–2408. [PubMed] [Google Scholar]
- Jelinsky S.A., Estep,P., Church,G.M. and Samson,L.D. (2000) Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol., 20, 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B. and Lim,D.S. (2000) The many substrates and functions of ATM. Nat. Rev. Mol. Cell. Biol., 1, 179–186. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Li J., Gao,X., Ortega,J., Nazif,T., Joss,L., Bogyo,M., Steven,A. and Rechsteiner,M. (2001) Lysine 188 substitutions convert the pattern of proteasome activation by REGγ to that of REGs α and β. EMBO J., 20, 3359–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Lerea,K.M. and Etlinger,J.D. (1996) Phosphorylation of the proteasome activator PA28 is required for proteasome activation. Biochem. Biophys. Res. Commun., 225, 855–860. [DOI] [PubMed] [Google Scholar]
- Lisby M., Rothstein,R. and Mortensen,U.H. (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA, 98, 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel L., Chen,L., Madura,K. and Sweder,K. (2000) The 26S proteasome negatively regulates the level of overall genomic nucleotide excision repair. Nucleic Acids Res., 28, 4839–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Stock,D., Jap,B., Zwickl,P., Baumeister,W. and Huber,R. (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science, 268, 533–539. [DOI] [PubMed] [Google Scholar]
- Ma C.P., Slaughter,C.A. and De Martino,G.N. (1992) Identification, purification and characterization of a protein activator (PA28) of the 20S proteasome (macropain). J. Biol. Chem., 267, 10515–10523. [PubMed] [Google Scholar]
- Martin S.G., Laroche,T., Suka,N., Grunstein,M. and Gasser,S.M. (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell, 97, 621–633. [DOI] [PubMed] [Google Scholar]
- Mills K.D., Sinclair,D.A. and Guarente,L. (1999) MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell, 97, 609–620. [DOI] [PubMed] [Google Scholar]
- Mirzoeva O.K. and Petrini,J.H.J. (2001) DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol., 21, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C.W. (1991) Further characterizations of bleomycin-sensitive (blm) mutants of Saccharomyces cerevisiae with implications for a radiomimetic model. J. Bacteriol., 173, 3605–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell P.H. (1975) High-resolution two-dimensional electrophoresis of proteins. J. Biol. Chem., 250, 4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ortolan T.G., Tongaonkar,P., Lambertson,D., Chen,L., Schauber,C. and Madura,K. (2000) The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell Biol., 2, 601–608. [DOI] [PubMed] [Google Scholar]
- Palombella V.J., Rando,O.J., Goldberg,A.L. and Maniatis,T. (1994) The ubiquitin–proteasome pathway is required for processing the NF-κ B1 precursor protein and the activation of NF-κB. Cell, 78, 773–785. [DOI] [PubMed] [Google Scholar]
- Prescott D.M., Myerson,D. and Wallace,J. (1972) Enucleation of mammalian cells with cytochalasin B. Exp. Cell Res., 71, 480–485. [DOI] [PubMed] [Google Scholar]
- Realini C., Jensen,C., Zhang,Z., Johnston,S., Knowlton,R., Hill,C.P. and Rechsteiner,M. (1997) Characterization of recombinant REGα, REGβ and REGγ proteasome activators. J. Biol. Chem., 272, 25483–25492. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M. (1998) The 26S proteasome. In Peters,J.-M., Harris,J.R. and Finley,D. (eds), Ubiquitin and the Biology of the Cell. Plenum, New York, NY, pp. 147–189.
- Rechsteiner M.C. and Catanzarite,V. (1974) Pyridine nucleotide metabolism in enucleated culture cells. J. Cell Physiol., 84, 409. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M., Realini,C. and Ustrell,V. (2000) The proteasome activator 11S REG (PA28) and class I antigen presentation. Biochem. J., 345, 1–15. [PMC free article] [PubMed] [Google Scholar]
- Reits E.A., Benham,A.M., Plougaste,B., Neefjes,J. and Trowsdale,J. (1997) Dynamics of proteasome distribution in living cells. EMBO J., 16, 6087–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P. and Pandolfi,P.P. (2002) The role of PLM in tumor suppression. Cell, 108, 165–170. [DOI] [PubMed] [Google Scholar]
- Schultz L.B., Chehab,N.H., Malikzay,A. and Halazonetis,T.D. (2000) p53 binding protein1 (53BP1) is an early participant in the cellular reponse to DNA double-strand breaks. J. Cell Biol., 151, 1381–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I. et al. (2002) Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA, 99, 4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S., Walter,J., Shinohara,A., Kamada,N. and Cremer,T. (2000) Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol., 150, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman,L., Rechsteiner,M. and Pickart,C.M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J., 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ramos C.A., Prakash,S. and Prakash,L. (2002) Requirement of RAD5 and MMS2 for postreplication repair of UV-damage DNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 22, 2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H.D. (2002) Degradation or maintenance: actions of the ubiquitin system on eukaryotic chromatin. Eukaryot. Cell, 1, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H.D. and Jentsch,S. (2000) Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J., 19, 3388–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Whitby F.G., Masters,E.I., Kramer,L., Knowlton,J.R., Yao,Y., Wang,C.C. and Hill,C.P. (2000) Structural basis for the activation of 20S proteasomes by 11S regulators. Nature, 408, 115–120. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Krutchinsky,A., Endicott,S., Realini,C., Rechsteiner,M. and Standing,K.G. (1999) Proteasome activator 11S REG or PA28: Recombinant REGα/REGβ hetero-oligomers are heptamers. Biochemistry, 38, 5651–5658. [DOI] [PubMed] [Google Scholar]