Abstract
Cyclin E–Cdk2 is essential for S phase entry. To identify genes interacting with cyclin E, we carried out a genetic screen using a hypomorphic mutation of Drosophila cyclin E (DmcycEJP), which gives rise to adults with a rough eye phenotype. Amongst the dominant suppressors of DmcycEJP, we identified brahma (brm) and moira (mor), which encode conserved core components of the Drosophila Brm complex that is highly related to the SWI–SNF ATP-dependent chromatin remodeling complex. Mutations in genes encoding other Brm complex components, including snr1 (BAP45), osa and deficiencies that remove BAP60 and BAP111 can also suppress the DmcycEJP eye phenotype. We show that Brm complex mutants suppress the DmcycEJP phenotype by increasing S phases without affecting DmcycE protein levels and that DmcycE physically interacts with Brm and Snr1 in vivo. These data suggest that the Brm complex inhibits S phase entry by acting downstream of DmcycE protein accumulation. The Brm complex also physically interacts weakly with Drosophila retinoblastoma (Rbf1), but no genetic interactions were detected, suggesting that the Brm complex and Rbf1 act largely independently to mediate G1 arrest.
Keywords: Brahma/cell cycle/cyclin E/retinoblastoma/S phase
Introduction
The coordination of cell proliferation and differentiation is fundamental for development of multicellular organisms. The G1 to S phase transition is a critical point in the cell cycle where a cell makes the decision to proliferate or differentiate. Entry into S phase is driven by G1 cyclin–Cdk protein kinases (reviewed by Ekholm and Reed, 2000). In multicellular organisms, there are three classes of G1 cyclins, cyclin D, E and A, that are rate limiting and essential for the G1 to S phase progression. Cyclin E and cyclin A form a complex with Cdk2, while cyclin D forms a complex with Cdk4 or Cdk6. The activity of these cyclin complexes is regulated by the binding of the p21CIP1 and p16INK4a families of inhibitor proteins (reviewed by Sherr and Roberts, 1995). The only essential target of the cyclin D–Cdk4(6) protein kinase is the retinoblastoma (Rb) tumor suppressor protein (reviewed by Dyson, 1998; Harbour and Dean, 2000). Rb in its unphosphorylated form binds to the E2F/DP transcription factor, forming an inactive complex at S phase gene promoters. Phosphorylation of Rb by cyclin D–Cdk4(6) is important to inactivate Rb and allow S phase gene transcription. Cyclin E–Cdk2 and cyclin A–Cdk2 are also required for further phosphorylation and complete inactivation of Rb. However, cyclin E–Cdk2 and cyclin A–Cdk2 have other essential roles in promoting entry into S phase, possibly by phosphorylating proteins involved in the initiation of DNA replication (reviewed by Ekholm and Reed, 2000).
The regulation of entry into S phase is similar between Drosophila and mammalian cells. Fly homologs of many of the essential mammalian proteins have been identified and characterized, including cyclin E, cyclin D, cyclin A, Cdk2, Cdk4(6), p21 (Dacapo), E2F, DP and Rb (reviewed by Edgar and Lehner, 1996). In Drosophila, cyclin E (DmcycE) appears to be the most important cyclin in the G1 to S phase transition. DmcycE is expressed in proliferating cells and is down-regulated as cells exit into G1 phase (Richardson et al., 1993, 1995). Furthermore, mutant and overexpression studies have shown that Drosophila DmcycE is both sufficient and rate limiting for the G1 to S phase transition (Knoblich et al., 1994; Richardson et al., 1995).
Two E2F (E2F1 and E2F2), one DP and two Rb (Rbf1 and Rbf2) homologs are present in flies (reviewed by Dyson, 1998; Harbour and Dean, 2000). As in mammalian cells, E2F/DP regulates the S phase genes, such as PCNA and RNR2, and induces cells into S phase. Mutant and ectopic expression analyses have shown that Rbf1 abrogates E2F1/DP function during Drosophila development (Du et al., 1996a; Du and Dyson, 1999; Du, 2000). In Drosophila, DmcycE–Cdk2 is also able to phosphorylate Rbf1, thereby leading to inactivation of Rbf1 and entry into S phase. However, in many tissues in the embryo, except nervous system cells, DmcycE transcription is also regulated by E2F1/DP (Duronio and O’Farrell, 1995; Sauer et al., 1995; Jones et al., 2000). Thus, in many tissues, DmcycE and E2F1/DP work in a positive feedback loop to induce entry into S phase by abrogating Rbf1. In addition, other positive and negative cell cycle regulators are likely to play an important role in potentiating the G1 to S phase transition, including growth factors and cell–cell communication signaling pathways.
In order to identify novel G1/S phase regulators in Drosophila, we have taken advantage of a hypomorphic DmcycE mutation (DmcycEJP), which exhibits defects in both eye and wing development. The DmcycEJP mutant displays a rough eye phenotype due to a reduction in S phases during eye development and exhibits wing notching and shortening of the L5 wing vein (Secombe et al., 1998). We carried out a genetic modifier screen of X-ray and ethyl methanesulfonate (EMS) mutagenized flies to isolate dominant suppressors and enhancers of the DmcycEJP rough eye phenotype (to be reported in detail elsewhere). By genetic analysis, we have identified two of the suppressors to be the brahma (brm) and moira (mor) genes, which are members of the SWI–SNF group of chromatin remodeling, general transcriptional regulatory genes (Tamkun et al., 1992; Crosby et al., 1999; reviewed by Tamkun, 1995). brm and mor alleles were isolated originally as dominant suppressors of a Polycomb (Pc) mutant that resulted in aberrant expression of homeotic genes (Kennison and Tamkun, 1988). Due to their positive regulatory affect on homeotic gene expression, brm and mor have been classed in the trithorax group (trx-G) of genes (Kennison and Tamkun, 1992), generally thought to act as global transcriptional activators. Both Brm and Mor are components of the fly counterpart of the yeast SWI–SNF complex that utilizes the energy of ATP hydrolysis to remodel chromatin, thereby overcoming the repressive effects of chromatin structure on transcription. The SWI–SNF complex, originally identified in yeast, is a large (∼2 MDa) multisubunit complex composed of 8–11 stably associated proteins (reviewed in Kingston and Narlikar, 1999; Peterson and Workman, 2000). Several of the core subunits are highly conserved among metazoan SWI–SNF counterparts, known as the Brm complex in Drosophila (Dingwall et al., 1995; Papoulas et al., 1998) and the hBrm and Brg1 complexes in mammals (Wang et al., 1996). The biochemical properties of the purified yeast and mammalian SWI–SNF complexes have been examined in detail (Peterson and Workman, 2000); however, the biological roles of the complex in metazoan development are not well understood. The Drosophila Brm complex has been purified and shown to contain homologs of several yeast SWI–SNF proteins, including Brm (SWI2/SNF2), BAP155 (Mor/SWI3), BAP45 (Snr1/SNF5) and BAP60 (SWP73/RSC6), as well as novel proteins BAP111 (a HMG-like protein), BAP74 (Hsp70 cognate 4), BAP55 (actin-related protein) and BAP47 (actin) (Papoulas et al., 1998). A potential SWI1 homolog was not identified among the purified Brm complex components; although the trx-G gene osa/eyelid encodes a protein with limited homology to SWI1, osa mutants strongly interact genetically with Brm complex genes and Osa may be a component of some Brm complexes (Collins et al., 1999). Other purified Brm complexes appear to contain Osa, as well as several additional unidentified proteins, but not BAP74 (Kal et al., 2000). Therefore, similarly to mammalian SWI–SNF complex counterparts (hBrm and Brg1 complexes; Wang et al., 1996), the composition of the Brm complex may be heterogeneous, varying in different developmental contexts. In addition to brm, mor and osa, specific mutations have been described in the Brm complex gene snr1, which shows genetic interactions with brm (Dingwall et al., 1995; Triesman et al., 1997; Vazquez et al., 1999), and Hsc70-4 (Mollaaghababa et al., 2001).
Several recent studies have provided strong connections between metazoan SWI–SNF complexes and regulation of the cell cycle. In yeast, the SWI–SNF complex is not essential for viability, and whole genome analyses of swi/snf mutants have shown roles in activation and repression of transcription (Holstege et al., 1998; Sudarsanam and Winston, 2000; Sudarsanam et al., 2000). A screen for modifiers of E2F1/DP function in Drosophila identified new alleles of brm and mor as enhancers of the rough eye phenotype associated with ectopic expression of E2F1 and DP in the developing Drosophila eye imaginal disc (Staehling-Hampton et al., 1999). In support of this, mammalian homologs of Brm and Mor (hBrm/Brg1 and BAF55, respectively) have been recently reported to be present in cyclin E complexes and to be phosphorylated by cyclin E–Cdk2 (Shanahan et al., 1999). Significantly, human homologs of Brm (hBrm and Brg1) inhibit entry into S phase and achieve this at least in part by cooperation with the tumor suppressor, Rb (Dunaief et al., 1994; Muchardt et al., 1998; Reyes et al., 1998; Shanahan et al., 1999). Furthermore, Rb can bind to Brg1 and hBrm (Dunaief et al., 1994; Strober et al., 1996; Trouche et al., 1997), and the ability of Rb to induce G1 arrest has been shown to depend upon hBrm and Brg1 (Strobeck et al., 2000a,b; Zhang et al., 2000). However, the precise mechanism by which the mammalian Brm complexes cooperate with Rb to achieve G1 arrest is unclear. The recent identification of two Rb–Brg1 (hBrm) complexes, one of which also includes a histone deacetylase (Hdac), has revealed more complexity (Zhang et al., 2000). Hdac is required for gene repression by removing the acetyl groups from histones and binds to and cooperates with Rb in repression of E2F-dependent gene transcription (reviewed by Harbour and Dean, 2000; Zhang and Dean, 2001). The Hdac–Rb–Brg1(hBrm) complex appears to be important to repress cyclin E transcription, while the Rb–Brg1(hBrm) complex is involved in repression of the cyclin A and cdc2 genes (Zhang et al., 2000). The Hdac-associated co-repressor protein Sin3a has also been detected in hBrm and one of two different Brg1 complexes (Sif et al., 2001), although the exact biological role it plays in the specific function of these complexes is not clear. The importance of the Brm complex in cell cycle regulation is reinforced by the identification of truncating mutations of the hSNF5/INI1/SMARCB1 gene, a human homolog of snr1, in pediatric rhabdosarcomas and other tumors (Versteege et al., 1998; Sevenet et al., 1999).
Here, we have investigated the role of the Brm complex in the G1 to S phase transition in flies. We isolated brm and mor alleles in a screen for dominant suppressors of a DmcycE hypomorphic allele. We show that other Brm complex genes also interact genetically with DmcycE. Although the most obvious manner in which the Brm complex mediates negative regulation of S phase is via transcriptional regulation of DmcycE or E2F1/DP target genes, neither we nor others have observed any transcriptional effects. Consistent with this, we find that Brm and Snr1 physically interact with DmcycE in vivo. In addition we show that Rbf1 and Brm complex proteins weakly associate in vivo; however, we have observed no genetic interactions between brm or mor and rbf1. This suggests that the Brm complex and Rbf1 function largely independently in negatively regulating the G1 to S phase transition.
Results
brm and mor alleles suppress the DmcycEJP rough eye phenotype by increasing S phases
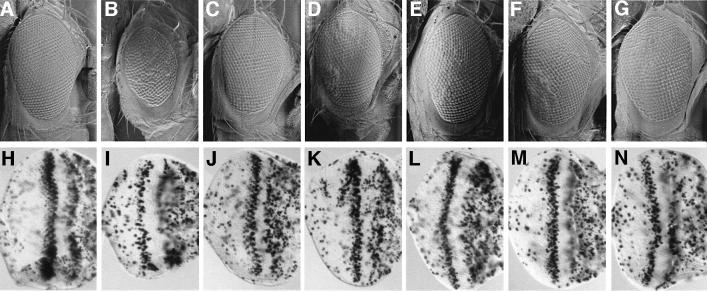
Based on deficiency mapping and complementation tests, two alleles of brahma (an X-ray-induced allele, 25S14 and an EMS-induced allele, E6S8) and one allele of moira (an X-ray-induced allele, 35S1) were isolated in a DmcycEJP modifier screen (to be published elsewhere). These alleles, as well as previously isolated alleles of brahma (brm2) and moira (mor1), suppressed the DmcycEJP rough eye phenotype (Figure 1C–F compared with B; data not shown). In addition, when Brm function was reduced in a DmcycEJP background by using the brm dominant-negative transgene, brmK804R (Papoulas et al., 1998), suppression of the DmcycEJP rough eye phenotype was also observed (Figure 1G). To test whether the suppression of the DmcycEJP rough eye phenotype by brm and mor was due to an increase in S phases, we carried out bromodeoxyuridine (BrdU) labeling of DmcycEJP; brm/+ and DmcycEJP; mor/+ eye imaginal discs from third instar larvae. Halving the dosage of brm or mor, using either alleles obtained in our genetic screen or previously isolated alleles, as well as the brm dominant-negative transgene, resulted in a dramatic increase in the number of S phases relative to DmcycEJP eye discs (Figure 1J–N compared with I). No significant effects on adult eye phenotype or entry into S phase were observed with mutant alleles of brm or mor alone (results not shown), indicating that brm and mor alleles only increase S phases in the sensitized DmcycEJP background. These results show that brm and mor interact genetically with DmcycE and are consistent with a role for Brm and Mor in negatively regulating entry into S phase.
Fig. 1. brm and mor dominantly suppress the DmcycEJP rough eye phenotype by increasing S phases. (A–G) Scanning electron micrographs of adult eyes. (A) Wild-type (w1118); (B) DmcycEJP; (C) DmcycEJP; brm25S14/+; (D) DmcycEJP; brm2/+; (E) DmcycEJP; mor35S1/+; (F) DmcycEJP; mor1/+; and (G) brmK804R; DmcycEJP. (H–N) Third instar larval eye imaginal discs labelled with BrdU. (H) Wild-type (w1118); (I) DmcycEJP; (J) DmcycEJP; brm25S14/+; (K) DmcycEJP; brm2/+; (L) DmcycEJP; mor35S1/+; (M) DmcycEJP; mor1/+; and (N) brmK804R; DmcycEJP. Adult eyes and larval imaginal discs are orientated anterior to the right in this and all subsequent figures.
Other Brm complex genes interact genetically with cyclin E
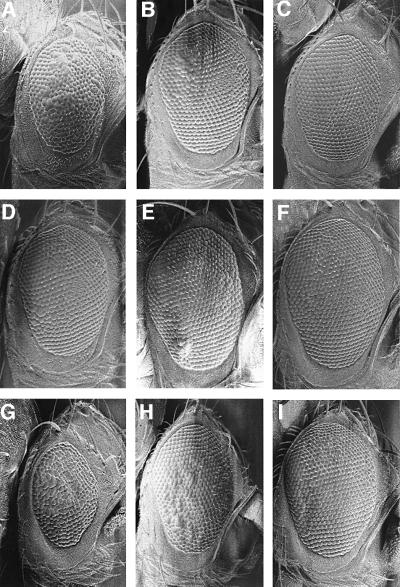
Since Brm and Mor are conserved core components of the fly SWI–SNF chromatin remodeling complex (Brm complex), we wished to test genes encoding other proteins present in this complex for a genetic interaction with DmcycE. The Drosophila Brm complex has been shown to consist of Brm, Mor/BAP155, BAP111 (HMG domain protein), BAP74 (Hsp70 cognate Hsc4), BAP60 (SWP73/RSC6 homolog), BAP55 (actin-related protein), BAP47 (actin 5C or 42A) and Snr1–BAP45 (Papoulas et al., 1998). The Osa/Eyelid protein, related to yeast SWI1, has also been shown to exist in some, but not all, Brm complexes (Collins et al., 1999; Kal et al., 2000). Using loss-of-function alleles of Hsc70-4 and snr1, and deficiencies of BAP111 and BAP60, we examined whether halving the dosage of these genes modified the rough eye phenotype of DmcycEJP (Figure 2; Table I). BAP55 could not be tested since no deficiencies covering this gene exist and, due to the uncertainty of the identity of BAP47, deficiencies of Actin5C or Actin42A were not investigated. Our results showed that decreasing the dosage of snr1 using several different alleles resulted in moderate suppression of the DmcycEJP eye phenotype (Figure 2B and C compared with A). However, strong suppression was observed when a C-terminal deletion of snr1 that acts as a dominant-negative (snr1-cdel.3) was ectopically expressed in the eye using the GAL4/UAS binary system (Brand and Perrimon, 1993) in DmcycEJP flies heterozygous for snr1R3 (Figure 2H and I compared with G). Expression of snr1-cdel.3 via the Act5C-GAL4 driver, which is expressed ubiquitously at high levels, resulted in better suppression than the GawB[69B]-GAL4 driver, which is expressed at low levels in the eye imaginal disc (Figure 2G–I). Dominant suppression of the DmcycEJP eye phenotype was also observed with osa (Figure 2D and E compared with A) and with one of the two Hsc70-4 alleles tested (Table I). The difference in interaction of the two Hsc70-4 alleles with DmcycEJP may reflect different allele strengths, specific functions affected by one allele but not the other, or genetic background affects. However, Hsc70-4 may not be a core component of the Brm complex since it is not always present in purified Brm complexes, nor have genetic interactions been detected between brm and Hsc70-4 (Kal et al., 2000; Mollaaghababa et al., 2001). Strong suppression was also observed with two deficiencies that removed BAP111 (Figure 2F compared with A; Table I) and BAP60/RSC6 (Table I). We also tested two deficiencies that were identified as enhancers of the brm dominant-negative allele, brmK804R (Papoulas et al., 1998). Both of these deficiencies, E(brm)25D-26B and E(brm)64E1-65C, showed strong suppression of the DmcycEJP rough eye phenotype (Table I). Thus, all Drosophila Brm complex genes and brm-interacting genes tested interact genetically with DmcycE.
Fig. 2. Dominant suppression of DmcycEJP rough eye phenotype by SWI–SNF genes. Scanning electron micrographs of DmcycEJP adult eyes. (A–F) In the background of w/+; b DmcycEJP bw/DmcycEJP. (A) +; (B) snr101319/+; (C) snr1R3/+; (D) osa00090/+; (E) osaS3263b/+; (F) Df(BAP111)/+. (G–I) In the background of w; DmcycEJP, which is slightly more extreme than w/+; b DmcycEJP bw/DmcycEJP. (G) +; (H) P[w+; UASGALhsp70-snr1-cdel.3] snr1R3/P[w+; 69B-GAL4]. (I) P[w+; UASGALhsp70-snr1-cdel.3] snr1R3/P[w+; Act5C-GAL4].
Table I. Interaction of DmcycEJP with Brm complex genes.
| Brm complex gene | Allele | Suppression of DmcycEJP |
|---|---|---|
| mor (moira) | 1 (hypomorph) | +++ |
| (SWI3 homolog) 89B1 | 35S1 (X-ray) | +++ |
| brm (brahma) | 2 (amorph) | ++ |
| (SWI2 homolog) 72A3 | 25S14 (X-ray) | ++ |
| E6S8 (EMS) | ++ | |
| Snr1 | 01319 (P allele) | ++ |
| (SNF5-related 1) 83A5-6 | R3 (lethal recessive) | ++ |
| Act5C-snr1-cdel.3, snr1R3 | +++ | |
| osa (eyelid) | 00090 (P allele) | ++ |
| (SWI1 homolog) 90C1-2 | krycheck | ++ |
| s3263b (P allele) | ++ | |
| Hsc70-4 (Hsp70 cognate 4) | 03550 (P allele) | + |
| (Bap74) 88E8-9 | L3929 (P allele) | – |
| BAP111 (HMG-like) | Df(1)lz-90b24 | +++ |
| 8C9-13 | Df(1)M38-c5 | +++ |
| BAP60 (RSC6/SWP73) 11D5-10 | Df(1)c246 | +++ |
| E(brm)25D-26B | Df(2L)cl-h3 | +++ |
| E(brm)64E1-65C | Df(3L)ZN47 | +++ |
brm and snr1 alleles suppress the DmcycEJP wing defects
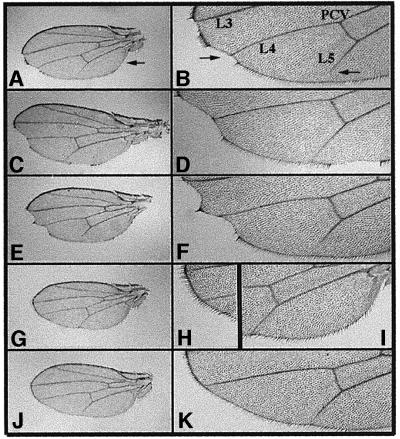
To explore further the genetic interaction between Brm complex genes and DmcycE, we examined another phenotype of DmcycEJP, that of wing notching and L5 wing vein truncation (Secombe et al., 1998; Table II; Figure 3). The control DmcycEJP flies show mild notching at the wing blade periphery at a penetrance of ∼96%, and only 50% of wings show a complete L5 vein (Table II; Figure 3A–F). Halving the dosage of brm strongly suppressed the wing blade notching and the truncated L5 wing vein phenotypes of DmcycEJP flies (Figure 3J and K). In addition, ectopic expression of the dominant-negative snr1-cdel.3 transgene using either Act5C-GAL4 or GawB[69B]-GAL4, in combination with reducing the dosage of the endogenous snr1 gene, resulted in the suppression of both the wing notching and the truncated wing vein phenotypes (Table II; Figure 3G–I; data not shown). Expression of the dominant-negative snr1 transgene via the stronger Act5C-GAL4 driver resulted in a greater suppression of both of these phenotypes (Table II). Thus, brm and snr1 show a dosage-sensitive suppression of DmcycEJP wing and eye phenotypes.
Table II. Suppression of DmcycEJP wing defects by snr1 and brm.
| Genotype | No. of wings examined | Notching |
% complete L5 | ||
|---|---|---|---|---|---|
| None | Mild | Severe | |||
| DmcycEJP; X-GAL4/TM3 | 192 | 4% | 23% | 73% | 50% |
| DmcycEJP; GawB[69B]-GAL4/UAS-snr1-cdel.3, snr1R3 | 72 | 55% | 28% | 17% | 87% |
| DmcycEJP; Act5C-GAL4/UAS-snr1-cdel.3, snr1R3 | 32 | 91% | 9% | 0% | 100% |
| DmcycEJP; brm2/TM3 | 56 | 82% | 18% | 0% | 100% |
Fig. 3. Suppression of DmcycEJP wing phenotypes by mutations in brm and by ectopic expression of a snr1 deletion transgene. Wings were dissected from flies homozygous for the DmcycEJP mutation on the second chromosome and either wild-type or heterozygous for various mutations and/or transgenes carried on the third chromosome. Wings shown in (A), (C), (E), (G) and (J) are at the same magnification, as are the magnified views of the same wings shown in (B), (D), (F), (H), (I) and (K). (A and B) Flies homozygous for DmcycEJP alone shown as a whole wing view (A) or at increased magnification (B). Note the notching at the posterior/distal wing margin, the missing hairs along the posterior/proximal wing blade and shortening of the fifth longitudinal vein (L5), indicated by arrows in (A) and (B). Also indicated are the positions of the L3 and L4 longitudinal veins and the posterior cross-vein (PCV). (C–K) DmcycEJP containing heterozygous mutations and/or transgenes on the third chromosome. (C and D) w; DmcycEJP; P[w+; Act5C-GAL4]/TM3. Note the phenotypes similar to those observed in (A) and (B). (E and F) w; DmcycEJP, P[w+; UASGALhsp70-snr1-cdel.3], snr1R3/TM3. (G–I) w; DmcycEJP, P[w+; UASGALhsp70-snr1-cdel.3], snr1R3/P[w+; Act5C-GAL4]. Note the suppression of both the wing margin and L5 defects in flies that ubiquitously overexpress the snr1-cdel.3 truncated transgene with a heterozygous snr1R3 mutation. Shown in (H) and (I) are magnified views of the wing margin (shown in G) between the L3 and L4 veins. (J and K) w; DmcycEJP; brm2/TM3. Note the suppression of the wing defects similar to those observed with overexpression of the snr1-cdel.3 truncation transgene shown above.
Interaction of DmcycEJP with trithorax and Polycomb group genes and global transcriptional regulators
To determine whether the genetic interaction observed between DmcycEJP and several Brm complex genes was specific or reflected a more global effect on transcription regulation, we also examined a number of trx-G and Pc-G genes for whether they could dominantly modify the DmcycEJP eye phenotype. As shown in Table III, most trx-G or Pc-G gene mutants did not show any strong dominant interactions with DmcycEJP, although subtle effects were observed with several.
Table III. Interaction of DmcycEJP with trx-G and Pc-G genes.
| Trithorax gene | Allele | Effect on DmcycEJP |
||
|---|---|---|---|---|
| Level of suppression | No effect | Level of enhancement | ||
| Mod(mdg4) modifier of Mdg4 93D7 | L3101 (P allele) | +++ | ||
| 03852 (P allele) | +++ | |||
| Trl (Trithorax-like) 70F1-2 | 13C (hypomorph) | ++ | ||
| R85 (hypomorph) | + | |||
| R67 (lethal recessive) | + | |||
| 62 (lethal recessive) | + | |||
| skd (skuld, S(Pc), S(Sevact)) 3-51 | 2 (hypomorph) | ++ | ||
| kto (kohtalo) 76B1-D5 | 1 (hypomorph) | + | ||
| dev/btl (devenir/breathless/FGFR) 70D2 | 1 (hypomorph) | + | ||
| 00208 (P allele) | + | |||
| Hth (Homothorax) 86C | 5E04 (lethal recessive) | + | ||
| trx (trithorax) 88B3 | 1 (hypomorph) | ++ (when homozygous) | ||
| E2 (amorph) | + | |||
| 00347 (semi-lethal, P allele) | No effect | |||
| urd (urdur) 87F12-15 | 2 (hypomorph) | + | ||
| lawC (leg arista wing complex) | EF520 (loss of function) | No effect | ||
| sls (sallimus, S(Pc)) 62C1-3 | 1 (recessive lethal) | + | ||
| ash1 (absent, small or homeotic1) 76B9 | B1 (hypomorph) | + | ||
| 22 (amorph) | + | |||
| ash2 (absent, small or homeotic2) 96A17 | 1 (amorph) | + | ||
| 18 (recessive lethal) | + | |||
| lid (little imaginal discs/E(ash1)) | 1 (lethal recessive) | + | ||
| (RBP2 homolog) 26A-B | 2 (lethal recessive) | + | ||
| kis (kismet) 21B7 | 1 (loss of function) | No effect | ||
| |
07812 (P allele) |
|
No effect |
|
| Polycomb gene |
|
|
|
|
| Pc (Polycomb) 78C9-78D | 7 (EMS allele) | ++ | ||
| 1 (amorph) | + | |||
| 4 (loss of function) | + | |||
| 2 (antimorph) | No effect | |||
| 6 (EMS allele) | No effect | |||
| Scr (Sex combs reduced) 84A5-B1 | 1 (loss of function) | + | ||
| Scm (Sex combs on midleg) 85E1-10 | D1 (loss of function) | No effect | ||
| ph-d (polyhomeotic distal) 2D1-5 | 503 (amorph) | No effect | ||
| Pcl (Polycomblike) 55B | 11 (amorph) | No effect | ||
| E(Pc) (Enhancer of Polycomb) 47F13-17 | 1 (lethal recessive) | + | ||
| Psc (Posterior sex combs) 49E1 | 1 (hypomorph) | No effect | ||
| Asx (Additional sex combs) 51A2 | 1 (gain of function) | No effect | ||
Among the trx-G genes tested that did affect the DmcycEJP phenotype, the most striking dominant suppression was observed with mod(Mdg4), which encodes a BTB/POZ domain transcription factor (Read et al., 2000). Moderate suppression was observed upon halving the dosage of skuld, which was isolated as a suppressor of Pc (Kennison and Tamkun, 1988). Although the product encoded by skuld is not known, its genetic map position (3–51) does not correlate with any known gene encoding a Brm complex subunit (Papoulas et al., 1998). Moderate suppression was also observed with one mutant allele of Trithorax-like (Trl13C), while other alleles showed slight suppression (Table III). However, it is likely that a homozygous viable mutant obtained as a dominant suppressor of DmcycEJP in our genetic screen (65S19) is an allele of Trl, since when crossed to known Trl alleles it gives a Trl abdominal transformation phenotype (data not shown). The Trl gene encodes the GAGA factor, which is a BTB/POZ domain transcription factor that regulates the expression of many genes in collaboration with the NURF chromatin remodeling complex (Farkas et al., 1994; Tsukiyama and Wu, 1995). A hypomorphic allele of trithorax (trx1) suppressed the DmcycEJP eye phenotype when homozygous, but stronger alleles of trithorax did not show dominant suppression. In contrast, some mutant alleles of other trx-G genes (sallimus, ash1, ash2 and lid) resulted in slight dominant enhancement of the DmcycEJP rough eye phenotype (Table III).
Many of the Pc-G genes are thought to function in opposition to trx-G genes in transcriptional regulation of specific targets. Of the Pc-G genes tested, very few showed any significant interactions with DmcycEJP (Table III). Thus, although some dominant genetic interactions were observed with trx-G or Pc-G genes and DmcycEJP, most of these were subtle compared with interactions observed with Brm complex genes. This suggests that while global transcriptional regulation may be loosely linked to the G1 to S phase transition, the Brm complex plays a more important role. The strong suppression of DmcycEJP observed with mod(Mdg4) also suggests that mod(Mdg4) may play an important role in DmcycE regulation and entry into S phase.
We also examined whether other global transcriptional regulators dominantly interacted with DmcycEJP. Hdacs are important in changing gene expression states (reviewed by Kouzarides, 1999). Sin3a interacts biochemically and genetically with Hdacs and is thought to tether Hdacs to transcription repressor proteins such as Mad–Max and Rb, which are important in the G1 to S phase transition. In Xenopus oocytes, a SWI2 family member, Mi-2, forms a complex that includes the Hdac Rpd3, the deacetylase-associated protein Sin3a and the Rb-associated protein RpAp46/48 (Wade et al., 1998). Among these genes, specific mutations exist in flies for rpd3, one of the four known Hdacs that act to enhance gene silencing at heterochromatic regions (de Rubertis et al., 1996), and for sin3a (Pennetta and Pauli, 1998). No effect on the DmcycEJP eye phenotype was observed when we halved the dosage of rpd3 (data not shown), suggesting that Rpd3 is not rate limiting for DmcycE function or is redundant with other Hdacs. Surprisingly, mild enhancement of the DmcycEJP eye phenotype was observed when the dosage of sin3a was decreased using homozygous viable P element alleles (not shown). This enhancement was increased when the dosage of sin3a was decreased further using a deficiency of sin3a. However, since this deficiency also removes other genes including ISWI (nurf-140/chrac), another SWI2 family nucleosome remodeling gene (Deuring et al., 2000), it is possible that this enhancement is due to halving the dosage of ISWI or to other genes as well as sin3a. This genetic interaction was opposite to what was expected, given that Sin3a is required for Hdac-mediated repression. The mechanism by which this occurs requires further investigation.
brm and mor do not function to suppress DmcycEJP by increasing DmcycE levels
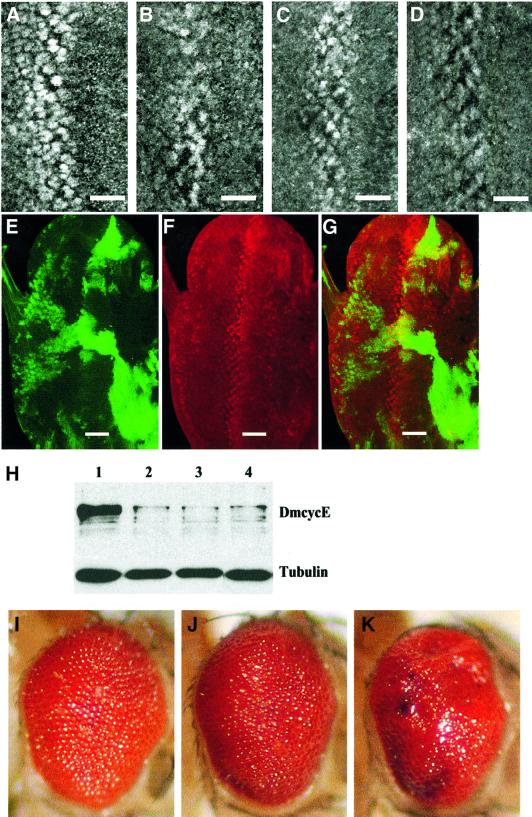
The DmcycEJP mutation is a hypomorph that exhibits decreased DmcycE protein levels (Secombe et al., 1998). To explore whether brm and mor suppression of the DmcycEJP eye phenotype was due to effects on DmcycE expression, we examined DmcycE protein levels in eye discs by immunostaining and western blot analyses (Figure 4). DmcycE antibody staining of eye imaginal discs from DmcycEJP; brm/+ and DmcycEJP; mor/+ larvae revealed that there was no significant increase in DmcycE protein levels relative to DmcycEJP alone (compare Figure 4B with C and D; data not shown). To explore this further, we used the eyeless-FLP; UAS-GFP, Tb-GAL4, FRT, Tb-GAL80 system (Lee and Luo, 1999) to generate clones of cells overexpressing the dominant-negative brm transgene (UAS-brmK804R) within otherwise wild-type eye imaginal discs (Figure 4E–G). In this system, clones expressing UAS-brmK804R are marked by green fluorescent protein (GFP) (Figure 4E). If Brm acts to repress DmcycE transcription, we would expect that in UAS-brmK804R-expressing clones DmcycE should be ectopically expressed. By DmcycE antibody staining, no ectopic expression of DmcycE was observed in UAS-brmK804R-expressing clones (Figure 4F and G). Furthermore, we examined DmcycE protein levels in DmcycEJP; brm/+ and DmcycEJP; mor/+ third instar larval eye imaginal discs by western analysis (Figure 4H). No significant increase in DmcycE protein level was observed when brm or mor dosage was halved relative to DmcycEJP eye discs (Figure 4H). Thus, the Brm complex appears to function downstream of DmcycE protein accumulation to inhibit S phase entry. Consistent with these data, brm and mor mutants dominantly enhanced the rough eye phenotype produced by the ectopic expression of DmcycE (Figure 4I–K). In these flies, DmcycE is produced independently of its normal transcriptional regulation using the GMR-GAL4 driver, and apoptosis is inhibited by GMR-driven expression of the caspase inhibitor p35 (GMR-p35), leading to an overgrown and rough eye phenotype (Figure 4I). Only genes that act downstream of DmcycE transcription are expected to show modification of this rough eye phenotype. Halving the dosage of either brm or mor enhanced this rough eye phenotype and increased the number of S phase cells (Figure 4J and K; data not shown). Thus, genetically, the Brm complex functions downstream of DmcycE transcription. Furthermore, the study of Staehling-Hampton et al. (1999) showed that the E2F target genes, rnr2 and dhfr, were also not affected by brm or mor mutants. Taken together, these results suggest that the Brm complex has a role, independent of DmcycE protein accumulation and E2F-dependent gene transcription, in mediating negative regulation of S phase.
Fig. 4. The Brm complex does not affect DmcycE protein levels and functions genetically downstream of DmcycE transcription. DmcycE antibody staining of larval eye imaginal discs from (A) wild-type; (B) DmcycEJP; (C) DmcycEJP; brm25S14/+; and (D) DmcycEJP; mor1/+. (E–G) DmcycE antibody staining and GFP fluorescence from an ey-FLP, UAS-GFP; Tb-GAL4 FRT(82B) GAL80/FRT(82B) UAS-brmK804R eye disc. (E) GFP (green) marks the clones expressing UAS-brmK804R. (F) DmcycE antibody staining. (G) Merge. Note that in clones expressing UAS-brmK804R distant from the normal band of cyclin E staining, cyclin E is not expressed ectopically. The bar indicates the position of the morphogenetic furrow. (H) Western analysis of DmcycE protein (upper panel) or tubulin (lower panel) in eye imaginal discs from wild-type (lane 1); DmcycEJP (lane 2); DmcycEJP; brm25S14/+ (lane 3); and DmcycEJP; mor1/+ (lane 4). Since DmcycEJP affects the eye imaginal disc but not the antennal disc, the antennal disc was removed from the eye disc before protein was prepared. Quantitation of band intensities from the DmcycE immunoblot normalized to tubulin showed that the level of DmcycE in DmcycEJP eye discs was not increased by halving the dosage of brm or mor. (I–K) Adult eyes from (I) GMR-GAL4, UAS-DmcycE/+; GMR-p35/+; (J) GMR-GAL4, UAS-DmcycE/+; GMR-p35/brm25S14; and (K) GMR-GAL4, UAS-DmcycE/+; GMR-p35/mor35S1.
DmcycE physically associates with the Brm complex in vivo
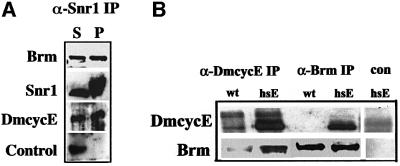
The genetic interactions we have observed with DmcycE and the Brm complex genes is consistent with the observation in mammalian cells that cyclin E–Cdk2 can form a complex with the Brahma homolog Brg1 and the Moira homolog BAP155 (Shanahan et al., 1999). To confirm that a biochemical interaction was also occurring between these proteins in Drosophila, we initially examined embryonic extracts, which have higher levels of DmcycE protein than larval tissues (Figure 5A). Embryonic extract immunoprecipitated with an anti-Snr1 antibody co-precipitated Brm and DmcycE, but not a control nuclear transcription factor. Comparison of the precipitated versus supernatant protein fractions revealed that a significant portion of DmcycE in the embryonic extract was co-precipitated efficiently with Snr1, along with Brm. We have also observed co-precipitation of Brm complex with the Cdk2 protein (data not shown; C.Zraly and A.Dingwall, in preparation). Co-precipitation of Cdk2 and DmcycE is expected, since Cdk2 is the sole catalytic partner of DmcycE in Drosophila (Sauer et al., 1995).
Fig. 5. Brm and Snr1 form a complex with cyclin E. (A) Snr1 and Brm form a complex with DmcycE in embryos. Native wild-type embryo extracts (500 µg) were incubated with affinity-purified Snr1 rabbit antibodies and precipitated with protein G–Sepharose beads. The presence of Brm, Snr1, DmcycE and a control nuclear protein was examined in the supernatant (S) and in the pelleted material eluted from the Sepharose beads (P) by immunoblotting. The supernatant tracks represent one-tenth of the immunoprecipitated tracks. (B) Brm forms a complex with DmcycE in larval brains/discs. Larval extracts were prepared from w1118 or a line transgenic for hsp70-DmcycE and incubated with an anti-DmcycE (8B10) or anti-Brm antibodies and precipitated with protein A–Sepharose beads. Pelleted proteins were examined for the presence of Brm (lower panel) and DmcycE (upper panel) by immunoblotting. The control immunoprecipitation was carried out using DmcycE pre-immune serum.
As the DmcycEJP mutant phenotype result from decreased S phases during larval imaginal disc development (Secombe et al., 1998), we examined whether DmcycE was stably associated with the Brm complex in larval tissues. When anti-DmcycE was used for the immunoprecipitation, Brm was readily detectable by immunoblotting (Figure 5B). However, in Brm immunoprecipitates, DmcycE was not detected (Figure 5B), probably because most cells in these extracts are arrested in G1 and do not contain DmcycE. To increase the level of DmcycE, we ectopically expressed DmcycE in larvae by using the hsp70-DmcycE transgene (Richardson et al., 1995). This resulted in a dramatic increase in DmcycE and in the ability of DmcycE and Brm to be co-immunoprecipitated (Figure 5B, hsE tracks). Thus, Brm and DmcycE physically associate in both larval and embryonic tissues.
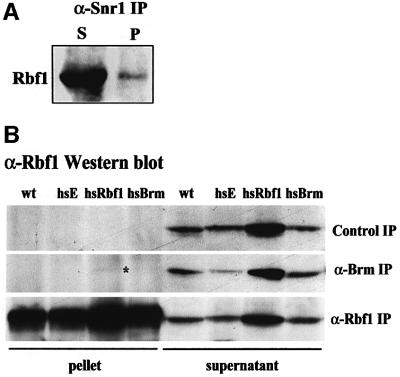
Rbf1 physically associates with the Brm complex in vivo
In mammalian cells, Brm and Brg1 can bind to Rb (Dunaief et al., 1994; Strober et al., 1996; Trouche et al., 1997). To determine whether Drosophila Rbf1 could also form stable associations with the Brm complex in vivo, we carried out co-immunoprecipitation experiments from both embryo and larval tissue extracts. As shown above in Figure 5, both Brm and DmcycE are present in anti-Snr1 immunoprecipitates from wild-type embryonic extracts. Similarly, we observed co-precipitation of Rbf1 with Snr1, although only a small portion of total Rbf1 was associated with Snr1 [Figure 6A, compare supernatant (S) with pellet (P) lanes]. We next examined associations in larval tissues where the relative amount of Brm complex and Rbf1 is much reduced compared with embryos. In anti-Brm immunoprecipitates, Rbf1 was not detectable in wild-type larval extracts, but could be weakly detected when Rbf1 levels were increased after heat shock induction of an hsp70-rbf1 transgene in larvae (Figure 6B, compare supernatant with pellet lanes). Conversely, in anti-Rbf1 immunoprecipitates, low levels of Brm were detected by immunoblotting in heat-shocked hsp70-rbf1 larval extracts (results not shown). In contrast, E2F1 was readily detected in Rbf1 immunoprecipitates (data not shown). These results show that Rbf1 physically associates with Brm complex proteins in Drosophila, albeit at relatively low levels compared with DmcycE and Brm or Snr1.
Fig. 6. Brm and Snr1 physically interact weakly with Rbf1. (A) Snr1 and Rbf1 physically interact weakly in embryos. Embryo extracts were incubated with affinity-purified anti-Snr1 antibodies and precipitated with protein G–Sepharose beads. The presence or Rbf1 was examined in the supernatant (S) and in the pelleted material eluted from the beads (P) by immunoblotting. (B) Rbf1 and Brm weakly physically interact in larval brains/discs. Extracts prepared from w1118 or heat-shocked hsp70-DmcycE (hsE), hsp70-rbf1 (hsRbf1) or hsp70-GAL4, UAS-brmK804R (hsBrm) larvae were immunoprecipitated with anti-Rbf1 or anti-Brm antibodies. Pelleted proteins and the supernatants were examined for the presence of Rbf1 by immunoblotting. Rbf1 was detected weakly in Brm immunoprecipitates in the hsRbf1 track (*). The control immunoprecipitation was carried out using protein A–Sepharose beads alone. (A and B) The supernatant tracks represent one-tenth of the immunoprecipitated tracks. Quantitation of band intensities showed that only a small fraction of total Rbf1 is co-immunoprecipitated with Snr1 or with Brm.
brm or mor do not interact genetically with rbf1
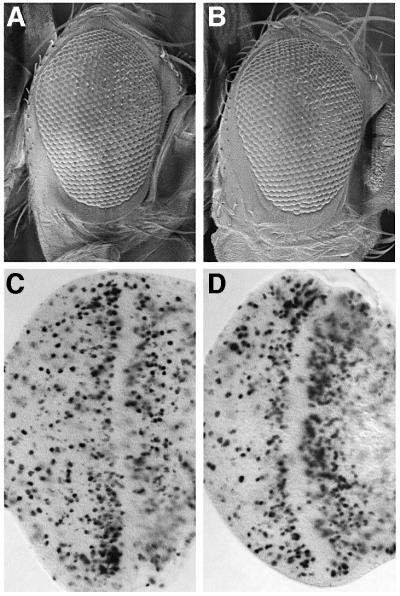
In mammalian cells, co-transfection of Brm and Rb cooperates to mediate G1 arrest (reviewed by Muchardt and Yaniv, 2001) and it is possible that a similar cooperation occurs in Drosophila. To obtain evidence for a functional interaction between the Brm complex and Rbf1, we first looked for genetic interactions between rbf1 and brm or mor alleles. Using the rbf111 null allele, we examined transheterozygous combinations of rbf111 and brm or mor mutants for adult phenotypes, including eye or bristle patterning effects. In this assay, we did not observe any significant specific defects (data not shown). To compromise the function of rbf1 further, we made use of a female sterile allele of rbf1, rbf1120a (Du, 2000; Bosco et al., 2001). Transheterozygous rbf111/rbf1120a females are viable but sterile due to defects in the endoreplication cycles of the follicle cells that surround the egg chamber (Bosco et al., 2001). During oogenesis, there are two phases of endoreplication in the follicle cells (Royzman et al., 1999). Until stage 10A, genomic endoreplication occurs asynchronously, while at stage 10B these cycles switch to synchronous amplification of discrete foci including the chorion genes. In rbf1 female sterile mutants, there are defects both in the switch from endoreplication to chorion gene amplification and in over-replication of the chorion gene foci (Bosco et al., 2001). We examined follicle cell S phases by BrdU labeling of rbf111/rbf1120a females, also heterozygous for brm, mor or both. Under these circumstances, we failed to observe any significant enhancement of the replication defects of rbf111/rbf1120a follicle cells (data not shown). Furthermore, we investigated whether halving the dosage of brm or mor could enhance the S phase defect of rbf111/rbf1120a in eye imaginal discs. rbf111/rbf1120a eye discs have a severe disruption in the post-morphogenetic furrow S phase band, but have excessive S phases in the posterior region (Figure 7C compared with Figure 1H). Despite this disruption, the adult eye phenotype of rbf111/rbf1120a flies is only mildly disorganized (Figure 7A compared with Figure 1A). Halving the dosage of brm, mor or both did not significantly increase the number of S phases in the posterior region of the eye disc, nor was the rbf111/rbf1120a adult eye phenotype affected (Figure 7B and D; data not shown). In addition, we examined if we could detect interactions between rbf1 and brm by using the rough eye phenotype generated by overexpression of the brm dominant-negative allele in the developing eye (GMR-GAL4; UAS-brmK804R; data not shown). Reducing or increasing the dosage of rbf1 in this background did not significantly affect the GMR-GAL4; UAS-brmK804R eye phenotype or affect S phases (data not shown).
Fig. 7. brm or mor do not interact genetically with rbf1. Scanning electron micrographs of adult eyes from (A) rbf1120a/rbf111 and (B) rbf1120a/rbf111; brm25S14, +/+, mor35S1. (C and D) BrdU labeling of eye imaginal discs from (C) rbf1120a/rbf111 and (D) rbf1120a/rbf111; brm25S14, +/+, mor35S1.
Since we only observed a physical interaction between Brm and Rbf1 in larval tissues when Rbf1 was overexpressed, we wished to determine if under these conditions a genetic interaction between Brm and Rbf1 could be observed. Overexpression of rbf1 in the eye, using four copies of a GMR-rbf1 transgene, results in a mild rough eye phenotype, while two copies of GMR-rbf have no significant effect (Du et al., 1996a). We wished to determine whether we could enhance this phenotype by co-expression of Brm complex components. Flies overexpressing brm or mor are not available; however, UAS-osa transgenic flies have been described (Collins et al., 1999). We observed that overexpression of wild-type osa via the GMR driver resulted in a mild rough eye phenotype (data not shown). However, co-expression of rbf1 and osa did not result in an enhanced rough eye phenotype or in a decrease in S phase cells in eye imaginal discs relative to GMR-osa or GMR-rbf1 alone (results not shown). In summary, using several different genetic assays, we were unable to detect any significant functional interactions between rbf1 and Brm complex genes.
Discussion
In this study, we have presented evidence that brm, mor and snr1 interact genetically with DmcycE. This interaction is specific to the Brm complex genes, rather than being a general feature of the Trithorax group (trx-G) of global transcriptional activators, since very few trx-G genes other than Brm complex genes strongly suppressed the DmcycEJP eye phenotype. This is consistent with the finding that Ash1 and Ash2, at least, are present in high molecular weight complexes distinct from the Brm complex (Papoulas et al., 1998). We also showed that Brm and Snr1 form complexes with DmcycE in embryo and larval brain/disc extracts. Although we have not examined all components of the Brm complex, the dominant genetic interactions that we have observed between DmcycE, and mor and osa alleles and deficiencies removing BAP60 and BAP111 suggest that these gene products may also be present in a complex with DmcycE, along with Brm and Snr1.
The DmcycEJP suppression we observed occurred by reducing the dosage of Brm complex genes, as well as by reducing Brm complex function with the dominant-negative brmK804R transgene. The brmK804R mutation abolishes Brm function by blocking ATP binding, but does not affect the assembly or stability of the complex (Elfring et al., 1998). This suggests that an ATP-dependent function is required for the Brm complex to negatively regulate S phase entry.
A role for the Brm complex downstream of DmcycE
The genetic interactions with DmcycE or E2F1/DP and Brm complex genes initially were thought to be due most probably to effects on DmcycE transcription or E2F/DP-dependent transcription, given the role of the Brm complex in transcriptional regulation (Tamkun, 1995). Surprisingly, the results of this study suggest that the Brm complex functions downstream of DmcycE transcription and protein accumulation. (i) No significant effect on DmcycE protein levels in DmcycEJP eye discs was observed when the dosage of brm or mor was halved. (ii) The rough eye phenotype due to overexpression of DmcycE from the GMR driver was enhanced by halving the dosage of brm and mor, indicating that Brm and Mor act to inhibit S phase entry downstream of DmcycE transcription. (iii) DmcycE forms a complex with Brm and Snr1. Taken together, these data provide strong evidence that the Brm complex does not inhibit the G1 to S phase transition by acting to down-regulate DmcycE transcription.
It is also likely that the Brm complex does not act to down-regulate E2F1/DP-dependent gene transcription, since no effect was observed for at least two E2F1/DP targets in brm mutants (Staehling-Hampton et al., 1999). Thus, mutations in Brm complex genes suppress the DmcycEJP mutant phenotypes by allowing progression into S phase without increasing either DmcycE protein levels or the expression of E2F1/DP-dependent genes. This suggests that one function of the Drosophila Brm complex is to restrict entry into S phase by inhibiting DmcycE–Cdk2 activity or by acting downstream of DmcycE–Cdk2 function. A function for Brm downstream of DmcycE–Cdk2 is consistent with reports that mammalian cyclin E can bind to and phosphorylate components of the Brm complex and thereby inactivate it (Shanahan et al., 1999). Thus the Brm complex may be acting as a curb to S phase entry that needs to be overcome by phosphorylation and inactivation by cyclin E–Cdk2.
Brm and Rbf1
Consistent with studies in cultured mammalian cells (Dunaief et al., 1994; Trouche et al., 1997; Zhang et al., 2000), we observed that the Rbf1 protein was present in complexes with Brm or Snr1 in larval and embryonic extracts. However, in embryos, only a small portion of total cellular Rbf1 was present in Snr1 immunoprecipitates, in contrast to a significant fraction of the cellular DmcycE, suggesting that most Brm complexes do not contain Rbf1. Our observation that Drosophila Rbf1 and Brm form a complex in vivo is consistent with studies in mammalian cells showing that hBrm and/or Brg1 can bind to and cooperate with Rb in transcriptional repression, and that hBrm and Brg1 are required for Rb-induced G1 arrest (reviewed by Muchardt and Yaniv, 2001; Zhang and Dean, 2001). However, in Drosophila, we were unable to obtain clear evidence for cooperation of brm or mor with rbf1 in S phase entry. It is possible that the phenotypes we were examining were not sensitive enough for S phase effects to be observed. However, the lack of a strong effect of Brm complex mutants on the rbf1 mutant S phase phenotype, when strong genetic interactions were observed with Brm complex genes and DmcycE, suggests that Rbf1 and Brm primarily function independently in negatively regulating S phase entry. Therefore, the suppression of the S phase defect of DmcycEJP by Brm complex mutants may not involve rbf1. Independent roles for Brm and Rb are also likely in mammalian cells since Rb knockout mice have a different mutant phenotype from that of Brg1 or Brm knockouts (reviewed by Muchardt and Yaniv, 2001).
In mammalian cells, Rb can form a complex containing both Brg1 and Hdac1, which is required to repress DmcycE transcription (Dahiya et al., 2000; Zhang et al., 2000) and may also have a role at replication origins (Lai et al., 2001). However, reducing the dose of the Drosophila Hdac gene, rpd3, did not suppress the DmcycEJP rough eye phenotype. It is possible that no interaction was observed for rpd3 and DmcycE, because there are a least three other Hdacs in flies that may perform overlapping functions with rpd3. However, mutations in sin3a, which encodes a Hdac-interacting protein, enhanced the DmcycEJP rough eye phenotype, suggesting that Sin3a functions in opposition to Brm in regulating DmcycE or S phase entry. Further studies using specific mutations in other Drosophila Hdacs, and Hdac-interacting proteins are required to analyze further their role in the G1 to S phase transition.
How does the Brm complex mediate negative regulation of the G1 to S phase transition?
Our results, along with those of Staehling-Hampton et al. (1999), suggest that the Brm complex is playing a role independent of DmcycE transcription and E2F/DP-dependent transcription in negatively regulating the G1 to S phase transition. One way in which this may occur is by transcriptional regulation of other critical G1/S phase genes. For example, there is evidence that in Drosophila, the Brm complex is important in negatively regulating Armadillo-dTCF target genes in the Wingless signaling pathway (Collins and Treisman, 2000). Although as yet there have been no studies showing directly that G1/S phase-inducing genes are targets of the Wingless signaling pathway in Drosophila, this is possible based on studies in mammalian cells (reviewed by Nollet et al., 1999). Furthermore, the Wingless pathway clearly has a role in cell proliferation in some Drosophila tissues (e.g. Neumann and Cohen, 1996). Whether this is the mechanism by which the Brm complex mediates negative regulation of cell cycle entry requires further investigation.
Another way in which the Brm complex may function is by restricting or regulating access to chromosomal origins of replication. Several studies have shown that ATP-dependent chromatin remodeling is important for modulating the initiation of chromosomal DNA replication (Hu et al., 1999; Li, 1999; Lipford and Bell, 2001). Our data are consistent with the view that the Brm complex may play a role in this process, possibly functioning to restrict entry into S phase by acting directly to remodel nucleosomes at replication origins. In this scenario, DmcycE–Cdk2 may then act to phosphorylate and inactivate the Brm complex, allowing assembly or function of the pre-replication complex and replication origin firing. Indeed, cyclin E–Cdk2 has been shown recently to be recruited by the Cdc6 pre-replication complex protein to replication origins at the G1 to S phase transition (Furstenthal et al., 2001).
Intriguingly, recent studies have shown that the E2F/DP complex also acts directly at replication origins. In the amplification of the chorion gene clusters during the ovarian follicle cell endoreplicative cycles, it has been shown that E2F1/DP is important in localizing the origin of replication complex specifically to the chorion gene origins and activating replication, and that Rbf1 is important in limiting DNA replication (Austin et al., 1999; Royzman et al., 1999; Bosco et al., 2001). This mechanism is not limited to these specialized cycles, since transcription-independent roles for E2F1 in inducing S phase have also been documented in the eye imaginal disc (Du, 2000). Taken together, these studies suggest that the E2F1/DP–Rbf1 complex plays a non-transcriptional role in S phase by acting directly at DNA replication origins (Bosco et al., 2001). In mammalian cells, a similar non-transcriptional role for Rb in DNA replication inhibition has been demonstrated (Knudsen et al., 1998), possibly through its functional association with the pre-replication complex protein Mcm7 (Sterner et al., 1998) and its localization to replication foci (Kennedy et al., 2000).
Given the data for a role for Rb–E2F/DP directly at replication origins and the evidence that chromatin remodeling is important in replication initiation, it is possible that Brm and Rbf1 may both have a role at replication origins to prevent premature origin firing in G1. However, the failure to detect a genetic interaction between brm complex genes and rbf1 suggests that they also have other important roles, independent of each other, in the G1 to S phase transition.
In summary, our results have shown that mutations in genes encoding components of the Brm chromatin remodeling complex can dominantly suppress a DmcycE hypomorphic allele by increasing the number of S phase cells without affecting cyclin E protein levels. Consistent with this view, DmcycE physically interacts with Brm and Snr1. Although a complex was also observed between the Brm complex and Rbf1, no genetic interactions were detected between Brm complex genes and rbf1, suggesting that Rbf1 and Brm function largely independently in negatively regulating the G1 to S phase transition. Taken together, these data suggest that the Brm complex negatively regulates entry into S phase, possibly in partial collaboration with Rbf1, and that this negative regulation can be abrogated by the action of cyclin E at the G1 to S phase transition.
Materials and methods
Fly strains and genetic manipulations
To examine genetic interactions between DmcycEJP and trx-G or Pc-G genes, stocks were generated that contained DmcycEJP (either heterozygous over CyO or homozygous) together with the test allele over a balancer chromosome. A stock of DmcycEJP isogenic for the second and third chromosomes was used for all crosses. Marked second chromosome DmcycEJP stocks (dp cl b DmcycEJP cn or b DmcycEJP cn bw) were used to generate recombinants of DmcycEJP and second chromosome genes. These stocks were outcrossed to DmcycEJP, and at least 50 progeny heterozygous for the test allele were scored for modification of the DmcycEJP rough eye phenotype. Mutant alleles of Brm complex, trx-G, Pc-G and Hdac complex genes (as listed in Table I) were tested for interaction with DmcycEJP at 25°C. Deficiencies used to examine the effect of halving the dosage of several Brm complex genes and the Brm-interacting genes identified by Papoulas et al. (1998) were: E(brmK804R)25D-26B, Df(2L)cl-h3; E(brmK804R)64E1-65C, Df(3L)ZN47; BAP111, Df(1)lz-90b24; Df(1)M38-c5; and BAP60, Df(1)C246. Flies showing a modified eye phenotype were analyzed further by scanning electron microscopy as previously described (Secombe et al., 1998). The GMR-GAL4, UAS-DmcycE; GMR-p35 stock was generated by recombination of second chromosome lines of GMR-GAL4 and UAS-DmcycE (containing a genomic DmcycE transgene under control of UASGAL4, obtained from C.Lehner) followed by crosses to obtain the stock with GMR-p35 (third chromosome). Flies co-expressing rbf1 and osa via the GMR driver were generated by first making a recombinant of GMR-GAL4 with UAS-osa (second chromosome) and the crossing to GMR-rbf1 flies. To analyze genetic interaction with the brm dominant-negative allele, a GMR-GAL4; UAS-brmK804R stock was generated, and crosses were carried out with GMR-rbf1 or rbf111 flies. To generate brmK804R-overexpressing clones in the eye, ey-FLP UAS-GFP; +; Tb-GAL4 FRT(82B) Tb-GAL80 flies were crossed to FRT(82B), UAS-brmK804R flies and larval progeny eye imaginal discs were dissected. Sources for fly stocks were: brmK804R transgenic flies, J.Tamkun; UAS-osa and ey-FLP, UAS-GFP; Tb-GAL4 FRT(82B), Tb-GAL80 flies, J.Treisman; lid alleles, A.Shearn; and Trl alleles, T.Greenberg. osa alleles isolated as enhancers of GMR-E2F1, DP were obtained from N.Dyson. All other fly stocks were obtained from the Bloomington stock center.
The UAS-snr1-cdel.3 deletion transgene was constructed by first placing a 108 amino acid C-terminal deletion of the Snr1 open reading frame under the control of the GAL4(UAS) and minimal hsp70 promoter in pUAST (Brand and Perrimon, 1993). An insertion on the third chromosome was obtained following introduction by P-element transformation (C.Zraly and A.Dingwall, unpublished). The UAS-snr1-cdel.3, snr1R3 strain used in these studies (w; P[w+; UASGALhsp70-snr1-cdel.3], snr1R3/TM6B) was generated by recombining the UAS-snr1-cdel.3 transgene with the snr1R3 mutation (Dingwall et al., 1995). To induce expression of the transgene in a homozygous DmcycEJP background, w; DmcycEJP/CyO; UASGALhsp70-snr1-cdel.3, snr1R3/TM3 flies were crossed to either w; DmcycEJP/CyO; GawB[69B]-GAL4 or w; DmcycEJP/CyO; Act5C-GAL4/TM3 flies. Progeny of the genotype w; cycEJP/cycEJP; P[w+, UASGALhsp70-snr1-cdel.3], snr1R3/P[w+, Act5C-GAL4] and progeny homozygous for DmcycEJP and heterozygous for either UAS-snr1-cdel.3, snr1R3 or the GAL4 insertion were scored for suppression of the eye and wing defects. No obvious differences in suppression frequency or extent were correlated with the parental source of the GAL4 protein. Wings were dissected and mounted in DPX mountant (Fluka) for microscopic examination and photography.
Preparation of native extracts and immunoprecipitation assays
Native protein extracts from Oregon R embryos were prepared as described in Dingwall et al. (1995). For each immunoprecipitation experiment, ∼500 µg of native protein extract from Oregon R embryos was pre-cleared with protein G–Sepharose beads (Pharmacia), and then incubated with Snr1 affinity-purified antibody (Dingwall et al., 1995). Protein complexes were precipitated using protein G–Sepharose beads, and bound and unbound proteins were fractionated on a 10% SDS–polyacrylamide gel and analyzed by western blotting.
Protein extracts were prepared from dissected third larval instar head tissues following heat shock induction of w1118, hsp70-DmcycE or hsp70-rbf1 larvae. Equilibrated protein A–Sepharose CL-4B was incubated with anti-DmcycE monoclonal antibody (8B10), anti-Rbf1 monoclonal (gift of W.Du and N.Dyson; Du et al., 1996a) or rabbit polyclonal anti-Brm antiserum (gift of C.Muchardt and M.Yaniv) for 6 h at 4°C. By western analysis of immunoprecipitated pellets and supernatants, we have shown that the anti-Brm and anti-DmcycE monoclonal (8B10) antibodies can immunoprecipitate their respective proteins efficiently and specifically (data not shown). Protein A–Sepharose-cleared protein extract was added to the antibody-bound Sepharose and incubation continued overnight. After extensive washing of the Sepharose beads, bound proteins were fractionated on a 7.5% SDS–polyacrylamide gel and analyzed by western blotting. Other antibodies used for immunoprecipitation–western analysis included anti-DmcycE polyclonal antibody raised in rats (Crack et al., 2002), and anti-dE2F1 (Du et al., 1996b) and control antiserum directed against a nuclear transcription factor that does not physically interact with Snr1 (Zraly et al., 2002).
Comparison of DmcycE protein levels in DmcycEJP; brm/+ and DmcycEJP; mor/+ eye discs relative to DmcycEJP and wild-type was performed by dissecting 20 pairs of eye discs from each sample away from the antennal disc portion. The dissected tissue was homogenized in protein sample buffer before loading onto a 10% SDS–polyacrylamide gel. Western blot analyses were performed using the anti-DmcycE 8B10 monoclonal antibody followed by detection with a horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (Jackson Immuno chemicals) and enhanced chemiluminescence (ECL) detection. As a loading control, the blots were also incubated with an anti-β-tubulin antibody (E7; Developmental Studies Hybridoma Bank). The DmcycE and tubulin detection signals were measured by scanning densitometry of X-ray films to quantify the relative level of DmcycE protein in each sample.
Immunohistochemistry and BrdU labeling
DmcycE antibody staining of larval eye imaginal discs was carried out as described previously (Secombe et al., 1998) using the anti-DmcycE antibody raised in rats, followed by detection using indirect immunofluorescence with anti-rat biotin and streptavidin–rhodamine. BrdU labeling of eye discs and ovaries was carried out as described previously (Secombe et al., 1998; Royzman et al., 1999).
Acknowledgments
Acknowledgements
We gratefully acknowledge Nick Dyson for sharing unpublished information and supplying fly stocks, Christian Murchardt and John Tamkun for Drosophila Brm antisera, Wei Du for the Rbf1 antibody and rbf1 mutants, J.Treisman, J.Tamkun, A.Shearn, C.Lehner and T.Greenberg for other fly strains used in this study, Michelle Coombe for technical help, and Leonie Quinn and Anabel Herr for comments on the manuscript. This work was supported by an Australian Research Council Grant (A09601106), the Australian Research Council Special Investigator Award to R.S. and H.R. (A09703208), the Australian Research Council funding to the Centre for the Molecular Genetics of Development and by the Peter MacCallum Cancer Research Institute. H.R. is a Wellcome Senior Fellow in Medical Research. A.D. is a Basil O’Connor Research Scholar of the March of Dimes Birth Defects Foundation.
References
- Austin R., Orr-Weaver,T.L. and Bell,S.P. (1999) Drosophila Orc specifically binds to Ace3, an origin of DNA replication control element. Genes Dev., 13, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Bosco G., Du,W. and Orr-Weaver,T.L. (2001) DNA replication control through interaction of E2F-RB and the origin recognition complex. Nature Cell Biol., 3, 289–295. [DOI] [PubMed] [Google Scholar]
- Collins R.T. and Treisman,J.E. (2000) Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev., 14, 3140–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R.T., Furukawa,T., Tanese,N. and Treisman,J.E. (1999) Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J., 18, 7029–7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crack D., Coombe,M., Brumby,A., Secombe,J., Saint,R. and Richardson,H. (2002) Analysis of Drosophila cyclin E proteins during development: identification of an inhibitory zone within the morphogenetic furrow of the eye imaginal disc that blocks the function of cyclin EI but not cyclin EII. Dev. Biol., 241, 157–171. [DOI] [PubMed] [Google Scholar]
- Crosby M.A., Miller,C., Alon,T., Watson,K.L., Verrijzer,C.P., Goldman-Levi,R. and Zak,N.B. (1999) The trithorax group gene moira encodes a Brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol., 19, 1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya A., Gavin,M.R., Luo,R.X. and Dean,D.C. (2000) Role of the LXCXE binding site in Rb function. Mol. Cell. Biol., 20, 6799–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rubertis F., Kadosh,D., Henchoz,S., Pauli,D., Reuter,G., Struhl,K. and Spierer,P. (1996) The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature, 384, 589–591. [DOI] [PubMed] [Google Scholar]
- Deuring R. et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Dingwall A.K., Beek,S.J., McCallum,C.M., Tamkun,J.W., Kalpana,G.V., Goff,S.P. and Scott,M.P. (1995) The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell, 6, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W. (2000) Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain. Development, 127, 367–379. [DOI] [PubMed] [Google Scholar]
- Du W. and Dyson,N. (1999) The role of Rbf in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J., 18, 916–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Vidal,M., Xie,J.-E. and Dyson,N. (1996a) Rbf, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev., 10, 1206–1218. [DOI] [PubMed] [Google Scholar]
- Du W., Xie,J.E. and Dyson,N. (1996b) Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J., 15, 3684–3692. [PMC free article] [PubMed] [Google Scholar]
- Dunaief J.L., Strober,B.E., Guha,S., Khavari,P.A., Alin,K., Luban,J., Begemann,M., Crabtree,G.R. and Goff,S.P. (1994) The retinoblastoma protein and Brg1 form a complex and cooperate to induce cell cycle arrest. Cell, 79, 119–130. [DOI] [PubMed] [Google Scholar]
- Duronio R.J. and O’Farrell,P.H. (1995) Developmental control of the G1 to S transition in Drosophila: cyclin E is a limiting downstream target of E2F. Genes Dev., 9, 1456–1468. [DOI] [PubMed] [Google Scholar]
- Dyson N.(1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Edgar B.A. and Lehner,C.F. (1996) Developmental control of cell cycle regulators: a fly’s perspective. Science, 274, 1646–1652. [DOI] [PubMed] [Google Scholar]
- Ekholm S.V. and Reed,S.I. (2000) Regulation of G1 cyclin dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol., 12, 676–684. [DOI] [PubMed] [Google Scholar]
- Elfring L.K. et al. (1998) Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics, 148, 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas G., Gausz,J., Galloni,M., Reuter,G., Gyurkovics,H. and Karch,F. (1994) The Trithorax-like gene encodes the Drosophila GAGA factor. Nature, 371, 806–808. [DOI] [PubMed] [Google Scholar]
- Furstenthal L., Kaiser,B.K., Swanson,C. and Jackson,P.K. (2001) Cyclin E uses Cdc6 as a chromatin-associated receptor required for DNA replication. J. Cell Biol., 152, 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour J.W. and Dean,D.C. (2000) Chromatin remodeling and Rb activity. Curr. Opin. Cell Biol., 12, 685–689. [DOI] [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Hu Y.-H., Hao,Z.-L. and Li,R. (1999) Chromatin remodeling and activation of chromosomal DNA replication by an acidic transcriptional activation domain from BRCA1. Genes Dev., 13, 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Richardson,H. and Saint,R. (2000) Tissue-specific regulation of cyclin E transcription during Drosophila melanogaster embryo genesis. Development, 127, 4619–4630. [DOI] [PubMed] [Google Scholar]
- Kal A.J., Mahmoudi,T., Zak,N.B. and Verrijzer,C.P. (2000) The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev., 14, 1058–1071. [PMC free article] [PubMed] [Google Scholar]
- Kennedy B.K., Barbie,D.A., Classon,M., Dyson,N. and Harlow,E. (2000) Nuclear organization of DNA replication in primary mammalian cells. Genes Dev., 14, 2855–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J.A. and Tamkun,J.W. (1988) Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc. Natl Acad. Sci. USA, 85, 8136–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J.A. and Tamkun,J.W. (1992) Trans-regulation of homeotic genes in Drosophila. New Biol., 4, 91–96. [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Knoblich J., Sauer,K., Jones,L., Richardson,H.E., Saint,R.B. and Lehner,C.F. (1994) Cyclin E controls progression through S phase and its downregulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell, 77, 107–120. [DOI] [PubMed] [Google Scholar]
- Knudsen E.S., Buckmaster,C., Chen,T.T., Feramisco,J.R. and Wang,J.Y. (1998) Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev., 12, 2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (1999) Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev., 9, 40–48. [DOI] [PubMed] [Google Scholar]
- Lai A. et al. (2001) Rbp1 recruits the mSin3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol., 21, 2918–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. and Luo,L. (1999) Mosaic analysis with a repressible neurotechnique cell marker for studies of gene function in neuronal morphogenesis. Neuron, 22, 451–461. [DOI] [PubMed] [Google Scholar]
- Li R. (1999) Stimulation of DNA replication in Saccharyomyces cerevisiae by a glutamine- and proline-rich transcriptional activation domain. J. Biol. Chem., 274, 30310–30314. [DOI] [PubMed] [Google Scholar]
- Lipford J.R. and Bell,S.P. (2001) Nucleosome positioned by ORC facilitates the initiation of DNA replication. Mol. Cell, 7, 21–30. [DOI] [PubMed] [Google Scholar]
- Mollaaghababa R., Sipos,L., Tiong,S.Y., Papoulas,O., Armstrong,J.A., Tamkun,J.W. and Bender,W. (2001) Mutations in Drosophila heat shock cognate 4 are enhancers of Polycomb. Proc. Natl Acad. Sci. USA, 98, 3958–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C. and Yaniv,M. (2001) The SWI/SNF complex remodels the cell cycle. Oncogene, 20, 3067–3075. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Bourachot,B., Reyes,J.C. and Yaniv,M. (1998) Ras transformation is associated with decreased expression of the brm/SNF2α ATPase from the mammalian SWI–SNF complex. EMBO J., 17, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C.J. and Cohen,S.M. (1996) Distinct mitogenic and cell fate specification functions of Wingless in different regions of the wing. Development, 122, 1781–1789. [DOI] [PubMed] [Google Scholar]
- Nollet F., Berx,G. and van Roy,F. (1999) The role of the E-cadherin/catenin adhesion complex in the development and progression of cancer. Mol. Cell. Biol. Res. Commun., 2, 77–85. [DOI] [PubMed] [Google Scholar]
- Papoulas O., Beek,S.J., Moseley,S.L., McCallum,C.M., Sarte,M., Shearn,A. and Tamkun,J.W. (1998) The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development, 125, 3955–3966. [DOI] [PubMed] [Google Scholar]
- Pennetta G. and Pauli,D. (1998) The Drosophila Sin3 gene encodes a widely distributed transcription factor essential for embryonic viability. Dev. Genes Evol., 208, 531–536. [DOI] [PubMed] [Google Scholar]
- Peterson C.L. and Workman,J,L. (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev., 10, 187–192. [DOI] [PubMed] [Google Scholar]
- Read D., Butte,M.J., Dernburg,A.F., Frasch,M. and Kornberg.T.B. (2000) Functional studies of the BTB domain in the Drosophila GAGA and Mod(mdg4) proteins. Nucleic Acids Res., 28, 3864–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.C., Barra,J., Muchardt,C., Camus,A., Babinet,C. and Yaniv,M. (1998) Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J., 17, 6979–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H., O’Keefe,L.V., Reed,S.I. and Saint,R. (1993) A Drosophila G1-specific cyclin E homolog exhibits different modes of expression during embryogenesis. Development, 119, 673–690. [DOI] [PubMed] [Google Scholar]
- Richardson H., O’Keefe,L.V., Marty,T. and Saint,R. (1995) Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development, 121, 3371–3379. [DOI] [PubMed] [Google Scholar]
- Royzman .I., Austin,R.J., Bosco,G., Bell,S.P. and Orr-Weaver,T.L. (1999) ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev., 13, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K., Knoblich,J.A., Richardson,H. and Lehner,C.F. (1995) Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev., 9, 1327–1339. [DOI] [PubMed] [Google Scholar]
- Secombe J., Pispa,J. Saint,R. and Richardson,H. (1998) Analysis of a Drosophila cyclin E hypomorphic mutation suggests a novel role for cyclin E in cell proliferation. Genetics, 149, 1867–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenet N., Lellouch-Tubiana,A., Schofield,D., Hoang-Xuan,K., Gessler,M., Birnbaum,D., Jeanpierre,C., Jouvet,A. and Delattre,O. (1999) Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype–phenotype correlations. Hum. Mol. Genet., 8, 2359–2368. [DOI] [PubMed] [Google Scholar]
- Shanahan F., Seghezzi,W., Parry,D., Mahony,D. and Lees,E. (1999) Cyclin E associates with BAF155 and Brg1, components of the mammalian SWI–SNF complex, and alters the ability of Brg1 to induce growth arrest. Mol. Cell. Biol., 19, 1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. and Roberts,J.M. (1995) Inhibitor of mammalian G1-cyclin-dependent kinases. Genes Dev., 9, 1149–1163. [DOI] [PubMed] [Google Scholar]
- Sif S., Saurin,A.J., Imbalzano,A.N. and Kingston,R.E. (2001) Purification and characterisation of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev., 15, 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehling-Hampton K., Ciampa,P.J., Brook,A. and Dyson,N. (1999) A genetic screen for modifiers of E2F in Drosophila melanogaster.Genetics, 153, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner J.M., Dew-Knight,S., Musahl,C., Kornbluth,S. and Horowitz,J.M. (1998) Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with Mcm7. Mol. Cell. Biol., 18, 2748–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck M.W., Knudsen,K.E., Fribourg,A.F., DeCristofaro,M.F., Weissman,B.E., Imbalzano,A.N. and Knudsen,E.S. (2000a) BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl Acad. Sci. USA, 97, 7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck M.W., Fribourg,A.F., Puga,A. and Knudsen,E.S. (2000b) Restoration of retinoblastoma mediated signaling to Cdk2 results in cell cycle arrest. Oncogene, 19, 1857–1867. [DOI] [PubMed] [Google Scholar]
- Strober B.E., Dunaief,J.L., Guha,S. and Goff,S.P. (1996) Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol., 16, 1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P. and Winston,F. (2000) The Swi/Snf family—nucleosome remodeling complexes and transcriptional control. Trends Genet., 16, 345–351. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P., Iyer,V.R., Brown,P.O. and Winston,F. (2000) Whole-genome expression analysis of Snf/Swi mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J.W. (1995) The role of Brahma and related proteins in transcription and development. Curr. Opin. Genet. Dev., 5, 473–477. [DOI] [PubMed] [Google Scholar]
- Tamkun J.W., Deuring,R., Scott,M.P., Kissinger,M., Pattatucci,A.M., Kaufman,T.C. and Kennison,J.A. (1992) brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell, 68, 561–572. [DOI] [PubMed] [Google Scholar]
- Treisman J.E., Luk,A., Rubin,G.M. and Heberlein,U. (1997) eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev., 11, 1949–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D., Le Chalony,C., Muchardt,C., Yaniv,M. and Kouzarides,T. (1997) RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl Acad. Sci. USA, 94, 11268–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T. and Wu,C. (1995) Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell, 83, 1011–1020. [DOI] [PubMed] [Google Scholar]
- Vazquez M., Moore,L. and Kennison,J.A. (1999) The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development, 126, 733–742. [DOI] [PubMed] [Google Scholar]
- Versteege I., Sevenet,N., Lange,J., Rousseau-Merck,M.F., Ambros,P., Handgretinger,R., Aurias,A. and Delattre,O. (1998) Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature, 394, 203–206. [DOI] [PubMed] [Google Scholar]
- Wade P.A, Jones,P.L., Vermaak,D. and Wolffe,A.P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol., 8, 843–846. [DOI] [PubMed] [Google Scholar]
- Wang W. et al. (1996) Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. EMBO J., 15, 5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Zhang H.S. and Dean,D.C. (2001) Rb-mediated chromatin structure regulation and transcriptional repression. Oncogene, 20, 3134–3138. [DOI] [PubMed] [Google Scholar]
- Zhang H.S., Gavin,M., Dahiya,A., Postigo,A.A., Ma,D., Luo,R.X., Harbour,J.W. and Dean,D.C. (2000) Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC–Rb–hSWI/SNF and Rb–hSWI/SNF. Cell, 101, 79–89. [DOI] [PubMed] [Google Scholar]
- Zraly C.B., Feng,Y. and Dingwall,A.K. (2002) Genetic and molecular analysis of region 88E9;88F2 in Drosophila melanogaster, including the ear gene related to human factors involved in lineage-specific leukemias. Genetics, 160, 1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]