Abstract
We recently described an erythroid ε-globin gene repressor activity, which we named DRED (direct repeat erythroid-definitive). We show that DRED binds with high affinity to DR1 sites in the human embryonic (ε-) and fetal (γ-) globin gene promoters, but the adult β-globin promoter has no DR1 element. DRED is a 540 kDa complex; sequence determination showed that it contains the nuclear orphan receptors TR2 and TR4. TR2 and TR4 form a heterodimer that binds to the ε and γ promoter DR1 sites. One mutation in a DR1 site causes elevated γ-globin transcription in human HPFH (hereditary persistence of fetal hemoglobin) syndrome, and we show that this mutation reduces TR2/TR4 binding in vitro. The two receptor mRNAs are expressed at all stages of murine and human erythropoiesis; their forced transgenic expression reduces endogenous embryonic εy-globin transcription. These data suggest that TR2/TR4 forms the core of a larger DRED complex that represses embryonic and fetal globin transcription in definitive erythroid cells, and therefore that inhibition of its activity might be an attractive intervention point for treating sickle cell anemia.
Keywords: erythroid/HPFH/nuclear orphan receptor/repressor/sickle cell anemia
Introduction
Changes in the program of expression of each of the five human β-type globin genes during embryonic development are a consequence of the influences of both local and distant transcriptional control elements. Transcript abundance from the globin genes is determined by the locus control region (LCR), a dispersed group of DNase I hypersensitive sites lying 5′ to the five human genes (Forrester et al., 1987; Grosveld et al., 1987). In contrast, temporal control of individual globin genes during erythropoiesis appears to be largely a property of more local regulatory influences: even individual human β-type globin genes are generally expressed at their proper developmental times (albeit at quite low levels) in transgenic mice in the absence of the LCR (Magram et al., 1985; Townes et al., 1985; Raich et al., 1990; Dillon and Grosveld, 1991).
In order to investigate one aspect of how globin gene temporal specificity might be established, we showed that clustered mutations in the ε-globin promoter, incorporated into a 150 kb human β-globin yeast artificial chromosome (YAC), induced this exclusively embryonic gene to continued high-level expression into adulthood (Tanimoto et al., 2000). This mutation was originally intended to generate a high affinity binding site for an erythroid transcriptional activator, EKLF, since EKLF had been shown to be mandatory for adult β-globin transcriptional activation (Perkins et al., 1996; Wijgerde et al., 1996). To our surprise, we found that this mutational effect acted independently of EKLF (by breeding the mutant β-globin YAC transgene into an EKLF-null mutant background). We therefore investigated the possibility that continued adult erythroid expression of ε-globin in the mutant YAC might have been due to inadvertent mutation of a previously unrecognized ε-globin repressor binding site. We discovered that definitive erythroid cells indeed harbored a novel DNA binding activity that could represent this purported repressor, which we named DRED (direct repeat erythroid-definitive), and we showed that DRED bound to the ε-globin DR1 (direct repeat) promoter elements. We further demonstrated that DRED was not COUP-TF2, a nuclear receptor expressed exclusively in embryonic erythroid cells (Filipe et al., 1999).
Of numerous genetic lesions shown to affect human β-globin transcription, one specific class of mutations cause continued synthesis of γ-globin from the fetal stage into adulthood. These mutations are collectively termed HPFH (hereditary persistence of fetal hemoglobin) syndromes, and lead to elevated synthesis (up to 30%) of γ-globin in definitive erythroid cells which normally have only very low levels (usually <1%) of tetrameric hemoglobin F (HbF, α2γ2). HPFH mutations include both small and large deletions in the β-globin locus, as well as point mutations in the two γ-globin gene promoters (Stamatoyannopoulos and Neinhuis, 1994). Of particular interest here, one non-deletion HPFH mutant allele (referred to as the Greek –117 Aγ HPFH) was found to lie near the CCAAT box of the Aγ-globin gene; importantly, this mutation alters a DR1 sequence element in the Aγ-globin gene promoter (Gelinas et al., 1985).
Here we report the purification of DRED binding activity, as well as its cloning and initial characterization. The DR1-specific binding activity of DRED is composed of two nuclear orphan receptors that are expressed in both embryonic and adult erythroid cells. These two proteins have been shown to repress transcription collaboratively in other physiological settings, and we show that they do so here as well, implying that these orphan receptors are likely candidates to be a part of the DRED ε- and γ-globin gene repressor complex.
Results
DRED binding site specificity and purification
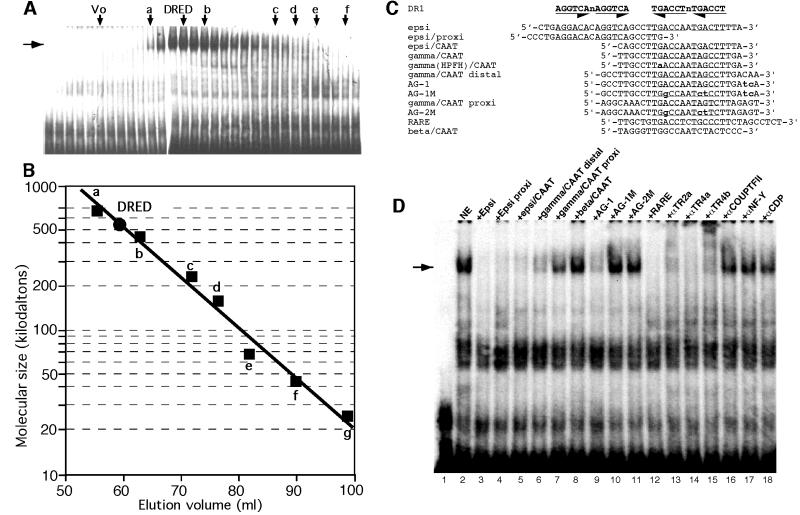
After testing several strategies for purification based on the recovery of DRED electrophoretic gel mobility shift assay (EMSA) activity, we finally adopted differential precipitation, gel exclusion and ion exchange chromatography steps followed by a final DNA sequence affinity column (see Materials and methods). Nuclear extracts from mouse erythroleukemia (MEL) cells were tested for their degree of purification and percentage of EMSA activity recovered after individual fractionation steps. We first selected DRED activity by ammonium sulfate precipitation followed by gel filtration. The peak of DRED binding activity eluted from the gel exclusion column (Figure 1A) as a species of ∼540 kDa (Figure 1B), suggesting either that DRED was huge or that it was composed of a relatively stable admixture of multiple components.
Fig. 1. DNA binding characteristics and specificity of DRED. (A) MEL cell nuclear extracts were separated by filtration on a Superdex 200 column (Materials and methods) relative to known molecular mass standards: a, thyroglobulin (669 kDa); b, ferritin (440 kDa); c, catalase (232 kDa); d, aldolase (158 kDa); e, albumin (67 kDa); f, ovalbumin (43 kDa); and g, chymotrypsinogen A (25 kDa). The peak of DRED EMSA activity (horizontal arrow) elutes coincident with a predicted molecular mass of 540 kDa (B). (C) Oligonucleotides used in EMSA and DRED purification. Unless otherwise stated, each of the oligonucleotides referred to in the text were blunt ended, double-stranded DNAs. The top line shows the position of two inverted DR1 consensus sequence elements aligned above the two DR1 elements in the ε-globin gene promoter (epsi), while the remaining oligonucleotides are described in the text. The Greek –117 Aγ HPFH point mutation [gamma(HPFH)/CAAT] is indicated in lower case bold, as are all of the oligonucleotides that differ from parental (wild type) sequences. Genuine or postulated (test) DR1 elements are underlined. (D) EMSA was performed as described in Materials and methods using radiolabeled ‘epsi’ bearing two DR1 elements (Tanimoto et al., 2000) as the probe. MEL cell nuclear extract (5 µg) was incubated with a 200-fold molar excess of indicated competing oligonucleotides before the addition of radiolabeled probe.
When crude nuclear extracts from MEL cells were tested for DRED binding specificity using the relevant region where we originally detected DRED binding activity (the ‘epsi’ oligonucleotide; Figure 1C) as a radiolabeled probe, we found that both individual DR1 elements in the ε-globin promoter (‘epsi/proxi’ and ‘epsi/CAAT’; Figure 1C) competed effectively for DRED complex formation (Figure 1D, lanes 4 and 5), as did the ‘gamma/CAAT distal’ and consensus retinoic acid response element (RARE) oligonucleotides (Figure 1D, lanes 6 and 12). In contrast, a comparable segment of the adult β-globin gene promoter that lacks a DR1 element (β/CAAT), or a segment of the γ-globin promoter that contains a duplicated CAAT element lying closer to the gene (Figure 1C, γ/CAAT proxi) were far less effective competitors for DRED binding.
To examine more closely the details of sequence requirements for DRED binding, we found that an oligonucleotide containing a two-base mutation far away from the DR1 CAAT box of the Aγ gene (Figure 1C, AG-1) was an effective competitor (Figure 1D, lane 9), while mutation of an additional 3 bp within the DR1 element (AG-1M) abrogated competition (Figure 1D, lane 10). Since the mutation in AG-1M lies within the DR1 element, but does not affect the partially overlapping ‘CCTTG’ motif, DRED must be a different protein than the one proposed to bind and repress transcription from the multiple γ-globin CCTTG promoter elements (Lee et al., 2000). In summary, these data confirmed our earlier suspicion that the DNA binding component of DRED was probably a member of the extended family of nuclear receptors (Tanimoto et al., 2000).
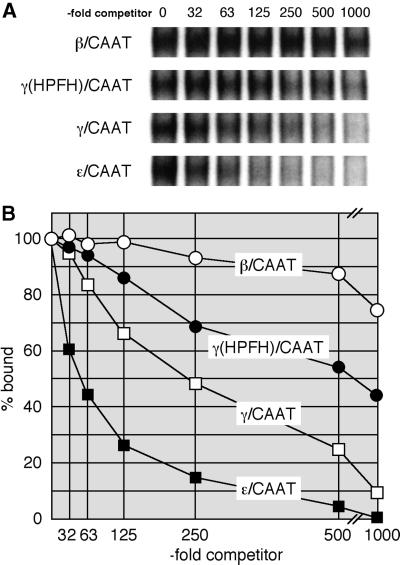
To examine DRED affinity for the DR1 element in greater detail, we performed competitive equilibrium EMSA experiments. To do so, a constant concentration of MEL cell nuclear extract was added to an aliquot of radiolabeled probe (Epsi) (Tanimoto et al., 2000) plus varying quantities of different competitor oligonucleotides (see Materials and methods). After electrophoresis, the amount of DRED complex in each reaction was quantified on a phosphorimager. The data depicted in Figure 2 show that the ε-globin CAAT probe containing one DR1 element competes most effectively, and that the same protein–DNA complex is strongly inhibited by a γ-globin DR1 oligonucleotide (γ/CAAT; Figure 1C), while the analogous segment of the adult β-globin gene promoter (β/CAAT), which contains no DR1 element, is an ineffective competitor. An oligonucleotide bearing the –117 Aγ HPFH point mutation [γ(HPFH)/CAAT; Figure 1C] is a less effective competitor (∼3-fold) than the wild-type γ/CAAT DR1 oligonucleotide. Thus, a single DR1 element in the ε-globin gene promoter binds to DRED with a higher affinity than does the γ-globin DR1 element, but the –117 HPFH point mutation in the DR1 sequence that leads to elevated γ-globin transcription in vivo binds to DRED with 3-fold lower affinity than the wild-type γ-globin DR1 sequence.
Fig. 2. Competitive EMSA analysis of DRED binding to CAAT boxes from the β-like globin gene promoters. (A) Equilibrium competition EMSA was performed as described in Materials and methods using radiolabeled ‘epsi’ as probe. A 32-, 63-, 125-, 250-, 500- or 1000-fold molar excess of competing oligonucleotide [β/CAAT, γ(HPFH)/CAAT, γ/CAAT or ε/CAAT] was incubated with MEL cell nuclear extract before the addition of the radiolabeled probe. (B) The DRED EMSA bands shown in (A) were quantified on a phosphorimager and the relative amount of complex remaining at each competitor concentration was plotted relative to the same sample with no added competitor (set at 100%). The values represent the average of two independent experiments.
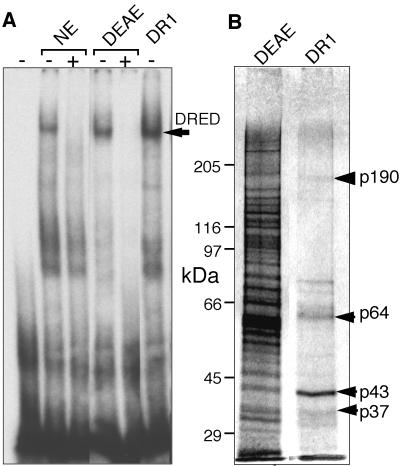
In executing the final phase of the overall purification strategy that we finally adopted, the peak of binding activity from a DEAE column was twice applied to, and eluted from, an ε-globin DR1 sequence-specific affinity column (Figure 3A). The 0.4 M eluate from the sequence-specific affinity column containing peak DRED activity revealed four major protein species (Figure 3B, p37 to p190) as well as a number of other bands that were either less abundant or did not correlate well with DRED EMSA activity eluted from the final column.
Fig. 3. DRED purification. The DRED complex was purified as described in Materials and methods. (A) EMSA was performed by incubating crude nuclear extract (NE; 2 µg of protein), the pooled DEAE Sepharose fraction (DEAE, 0.2 µg, 10 µl) or the peak fraction from the DNA sequence affinity column (DR1, 10 µl) with radiolabeled ‘epsi’ probe with (+) or without (–) preincubation with excess unlabeled probe. The arrow indicates the mobility of the DRED complex. (B) Proteins recovered in the peaks of the DEAE Sepharose and DR1 sequence affinity fractions (40 µl each) were separated by SDS–PAGE and silver-staining.
Nanoelectrospray mass spectrophotometric sequence analysis of peptides generated from all four major bands recovered from the SDS–polyacrylamide gel (Figure 3B) revealed co-purification of several well characterized nucleic acid binding proteins: the CDP (CCAAT displacement protein) homeoprotein in the p190 band, the leader-binding protein-1a (LBP-1a) and orphan nuclear receptors TR2 and TR4 in p64, and the upstream stimulatory factor-1 (USF-1) in p43. In particular, detection of unique signature peptides for murine (m)TR2 (S203PLAATPT FVTDSETAR219; DDBJ/EMBL/GenBank accession No. U28265) and mTR4 (I35QIVTAVDASGSSK48; DDBJ/EMBL/GenBank accession No. U11688) nuclear orphan receptors were of particular interest, since we had already concluded that the DRED DNA binding activity might contain nuclear receptor family members. We therefore focused on whether TR2 and/or TR4 might comprise the DNA binding component of DRED.
DRED is partially composed of a heterodimer of the nuclear orphan receptors TR2 and TR4
In order to determine whether or not the EMSA band that we identified as DRED contained TR2 and/or TR4, we performed further EMSA studies on MEL cell nuclear extracts, by including in the reactions antibodies that might be expected to compete with, or block the interactions with, possible constituents of the DRED repressor. The addition of anti-TR2 (αTR2a) or anti-TR4 antibodies (αTR4a and αTR4b) to the EMSA reactions ablated the entire DRED band (Figure 1D, lanes 13–15). Importantly, addition of any of these three antibodies affected the whole complex, allowing us to conclude that TR2 and TR4 both participate in DRED DNA binding. Inclusion of an antibody to the nuclear orphan receptor COUP-TF2, which is expressed exclusively in primitive erythroid lineage cells (Filipe et al., 1999), did not affect the DRED EMSA band (Figure 1D, lane 16; Tanimoto et al., 2000), nor did inclusion of antibodies recognizing the CAAT-box factor CDP (Barberis et al., 1987; Mantovani et al., 1988, 1989; Superti-Furga et al., 1988) or USF-1 (Gregor et al., 1990; data not shown), which co-purified with TR2 and TR4 (Figure 1D). The inclusion of antibodies against another CAAT factor (NF-Y) or antibodies that recognize proteins known to associate with transcription factors as part of chromatin co-repressor complexes (e.g. mSin3; Ayer et al., 1995) also failed to produce a reaction (data not shown).
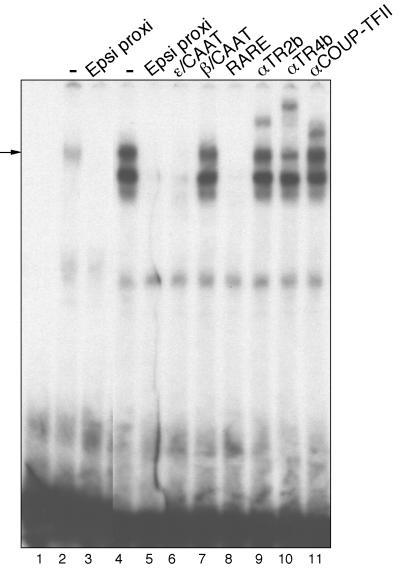
In contrast to the single EMSA product observed in nuclear extracts prepared from definitive erythroid cells, several complexes from primitive erythroid cells appeared to bind specifically to the ε-globin DR1 elements: the supershift experiments show that these factors include both DRED and COUP-TF2 (Figure 4, lanes 9–11). Thus, embryonic erythroid cells contain multiple protein species that can bind to the DR1 element (Filipe et al., 1999), including at least two different factors that are not present in definitive erythroid cells.
Fig. 4. Embryonic erythroid cells contain multiple DR1 binding proteins. MEL (lanes 2 and 3) or K562 (lanes 4–11) nuclear extracts (5 µg of protein each) were incubated with the indicated competing oligonucleotides (Figure 1C) or antibodies before the addition of radio labeled ‘epsi proxi’ probe. See the text and Figure 1D legend for details.
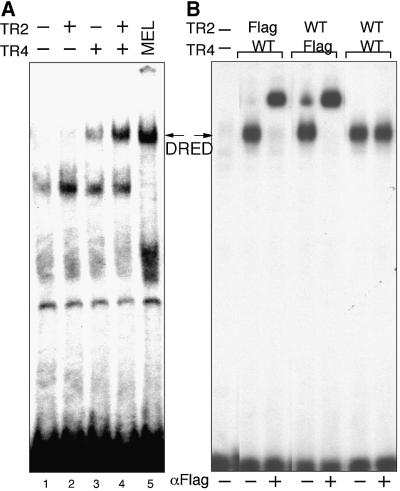
We then tested TR2 and TR4, both individually and together, for binding specificity after transfection into non-erythroid cells. Upon transfection of the wild-type receptors, nuclear extracts were prepared and examined for ‘epsi/proxi’ (Figure 1C) binding activity. When TR2 alone is transfected into quail fibroblast (QT6) cells, no new EMSA bands appear (Figure 5A, lane 2), while transfection of TR4 alone yields a weak EMSA product (Figure 5A, lane 3) that co-migrates with authentic MEL cell-derived DRED binding activity (Figure 5A, lane 5, arrow). This complex is more intense (suggesting either higher affinity or more abundant binding) when both TR2 and TR4 are co-transfected into QT6 cells (Figure 5A, lane 4).
Fig. 5. Reconstitution of DRED binding by expression of TR2 and TR4 in tissue culture cells. (A) TR2 or TR4 expression plasmids were transiently transfected into QT6 quail fibroblasts. Two days after transfection, nuclear extracts were prepared and subjected to EMSA using the ‘epsi proxi’ probe. MEL cell nuclear extract was used as a control. (B) Amino/Flag-epitope-tagged or wild-type TR2 or TR4 expression plasmids were transiently transfected into 293T cells. Two days after transfection, nuclear extracts were prepared and subjected to EMSA with the ‘epsi proxi’ probe. Nuclear extracts were preincubated with anti-Flag monoclonal antibody (+) or not (–), before the addition of the radiolabeled ‘epsi proxi’ probe. The arrows indicate the migration position of authentic DRED.
To confirm that TR2 and TR4 are able to bind to the ε-globin promoter DR1 elements as a heterodimer, we prepared N-terminal Flag epitope-tagged versions of both TR2 and TR4, and repeated the transfections into the human embryonic kidney (293T) cell line. As shown in this experiment, the only EMSA product formed with nuclear extracts from TR2 plus TR4-transfected cells is a heterodimer, regardless of which molecule contains the Flag epitope (Figure 5B). Since inclusion of the anti-Flag antibody supershifts the entire EMSA band (i.e. there is no remaining antibody-unreactive EMSA product) when either the TR2 or TR4 molecules are epitope tagged, we conclude that DRED is composed of a heterodimer of TR2 and TR4 orphan nuclear receptors.
TR2 and TR4 are expressed in both primitive and definitive murine and human erythroid cells
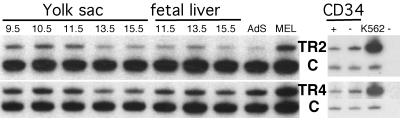
We next examined embryonic and adult blood cells for the presence and relative abundance of both TR2 and TR4 mRNAs. For RNA analysis, primers corresponding to unique sequences within either cDNA were generated (Materials and methods). RNA was isolated from murine embryos throughout gestation from yolk sacs and fetal livers as the source of primitive and definitive embryonic erythroid cells, respectively, or anemic adult spleens (AdS) as the source of definitive adult erythroid cells. The level of HPRT mRNA was used as the internal co-amplification control. Figure 6 shows that TR2 mRNA is present in yolk sac erythrocytes at 9.5 days post-coitum (d.p.c.), and at this stage of erythroid development the mouse embryo contains only primitive, nucleated erythrocytes. The abundance of TR2 mRNA in erythroid cells diminished modestly during embryonic development, and is relatively constant even during adult erythropoiesis (in anemic adult spleen cells; Figure 6, upper left panel). TR4 mRNA levels during murine gestation remained essentially constant (Figure 6, lower left panel). Thus, both mRNAs were expressed in both primitive and definitive murine erythroid lineage cells. We also tested human CD34+ and CD34– adult bone marrow cells for TR2 and TR4 mRNA accumulation (using an S14 co-amplification control; right panels), and found that both TR2 and TR4 were modestly more abundant (<50%) in the CD34– fraction, but both were significantly more abundant in K562 (primitive erythroid) cultured cells. We conclude that TR2 and TR4 are expressed in both primitive and definitive murine and human erythroid lineage cells, but clearly during murine development the abundance of TR2 and TR4 mRNA does not vary significantly between primitive and definitive erythropoiesis.
Fig. 6. TR2 and TR4 are both expressed throughout murine and human hematopoiesis. Total RNA was prepared from mouse tissues or from cell lines, and used for cDNA synthesis (see Materials and methods). TR2 (top panels) and TR4 (bottom panels) transcript abundances were normalized to the expression levels of co-amplified mouse HPRT mRNA (C, left panels) or human ribosomal protein S14 mRNA (C, right panels). RT–PCR was performed with primers corresponding to TR2 (top panels) or TR4 (bottom panels). cDNAs used for the reactions corresponded to the cell types shown above each lane. For the human erythroid cells, total RNA was prepared from fractionated human bone marrow cells (CD34+ or CD34–) and was used for cDNA synthesis. RNAs recovered from the murine definitive erythroid cell line MEL or the human primitive erythroid cell line K562 were used as the positive controls.
Forced expression of TR2 and TR4 in myeloerythroid lineages reduces endogenous murine εy gene transcription
Finally, we addressed the possible functional consequences of TR2/TR4-forced expression on globin gene transcription in vivo. To do so, we linked the Flag-tagged TR2 and TR4 cDNAs to an erythroid expression vector, placing them under GATA1-HRD (GATA-1 gene hematopoietic regulatory domain) transcriptional control. The 6.1 kb GATA1-HRD cassette consists of 3.9 kb of 5′ DNA flanking the GATA-1 gene erythroid first exon (exon IE), the first intron and the second exon, including the GATA-1 translational start site. This segment of the GATA-1 gene has been shown to be necessary and sufficient for generating high level expression of linked cDNAs in primitive and definitive erythroid cells (Onodera et al., 1997). We have previously employed this vector to investigate the specific requirements of erythroid cells for GATA-1 (as compared with GATA-2 or GATA-3; Takahashi et al., 2000), and the role of small Maf proteins in megakaryopoiesis and platelet generation (Motohashi et al., 2000).
During the abbreviated developmental process of erythropoiesis in the mouse, the βH1 gene is expressed most abundantly at ∼10.5 d.p.c. in the yolk sac, where it comprises ∼40% of total β-type mRNA, while εy transcription comprises the remaining 60% (Whitelaw et al., 1990). εy contains two promoter DR1 sites, while βH1 has one, and thus we assumed that any effect of increased TR2/TR4 levels would be most sensitively reflected in altered εy transcription. Since the ratios of adult α-globin to GATA-1 transcript levels do not vary significantly in the yolk sac between 9.5 and 11.0 d.p.c. (Whitelaw et al., 1990), we normalized the expression of εy and βH1 at 10.5 d.p.c. to α-globin mRNA levels, and the abundance of transgenic TR2 and TR4 mRNAs separately to GATA-1 abundance. We then examined the consequences of expressing variously elevated levels of transgenic TR2 and TR4 mRNAs on the endogenous murine embryonic β-type globin genes.
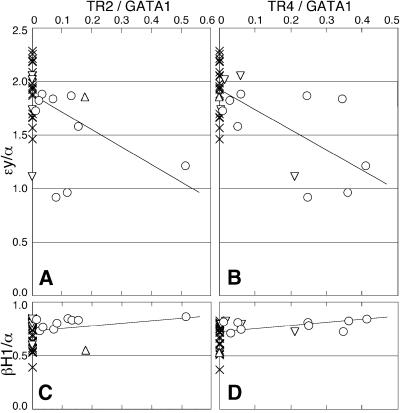
The GATA1-HRD.TR2 and GATA1-HRD.TR4 DNAs were co-injected into fertilized eggs; the injected embryos were implanted into pseudopregnant foster mothers and allowed to develop to 10.5 d.p.c. The yolk sacs from injected embryos were then analyzed for approximate TR2/TR4 transgene copy number using PCR, and then for TR2 and/or TR4 transgene-derived mRNA levels as well as endogenous murine GATA-1, adult α-, embryonic εy- and embryonic βH1-globin mRNA accumulation, by RT–PCR. Transgene-specific TR2 or TR4 transcripts were detected by PCR using a GATA1 exon IE sense primer, which was common to both transgenes, and individual antisense TR2 or TR4 cDNA primers. The abundance of transgene-derived TR2 and TR4 transcripts was normalized to endogenous GATA-1 RT cDNA co-amplified by duplex PCR, and expression of εy- or βH1-globins was normalized to co-amplified α-globin. The normalized data (Figure 7) show that εy abundance diminishes markedly with increasing TR2 or TR4 transgene-derived transcript accumulation, while βH1 abundance does not vary significantly, or might even increase slightly (the result anticipated if εY and βH1 are in ‘competition’ for LCR activity at this stage) (Choi and Engel, 1988; Wijgerde et al., 1995). The correlation between TR2 or TR4 expression and diminished εy abundance is statistically significant (P < 0.5 and < 0.1%, respectively, by Pearson’s correlation coefficient test). These data demonstrate that TR2 and TR4, when forcibly expressed in myeloerythroid lineage cells, repress endogenous εy transcription, and thus strongly support the working hypothesis that DRED is an erythroid repressor of DR1-dependent β-type globin transcription, and also that the TR2/TR4 heterodimer is the DNA recognition component of DRED.
Fig. 7. TR2 and TR4 erythroid-restricted expression represses εy-globin transcription. Transgenic founder embryos co-injected with TR2 and TR4 cDNAs transcriptionally directed by GATA1-HRD (see text) were analyzed for accumulation of transgenic TR2 or TR4 mRNAs, as well as for εy- and βH1-globin mRNAs in E10.5 yolk sacs by semi-quantitative RT–PCR. TR2 and TR4 mRNA levels were normalized to GATA-1 mRNA abundance, and εy- and βH1-globin mRNA levels were normalized to endogenous α-globin mRNA abundance (see text). Symbols: crosses, embryos expressing no transgene (n = 25); open circles, embryos expressing both TR2 and TR4 (n = 9); triangles, embryos expressing only TR2 (n = 1); inverted triangles, embryos expressing only TR4 (n = 4). Each of the lines was drawn by the least squares method. Analyzed by Pearson’s correlation coefficient test, the correlation between TR2 or TR4 expression and diminished εy abundance was statistically significant (A and B), while there was no significant correlation between expression of TR2 or TR4 and βH1 (C and D).
Discussion
We showed here that DRED is (initially) a high molecular weight complex of ∼540 kDa, and that this complex binds with high affinity to both the human ε- and γ-globin DR1 promoter elements. We also showed, through equilibrium EMSA competition experiments, that DRED binds to the Greek HPFH point mutation with 3-fold lower affinity than it does to the wild-type Aγ gene promoter DR1 site. When we purified this DR1 binding activity from MEL cells, we found that among the transcription factors recovered from the most highly purified fraction were two whose proteolytic products corresponded uniquely to the TR2 and TR4 orphan nuclear receptors. Antibodies that recognize either TR2 or TR4 ablated (or supershifted) the entire DRED EMSA band, demonstrating that DRED is composed, at least in part, of a TR2/TR4 heterodimer. This conclusion was underscored by cell-based transfection of Flag-tagged TR2 and TR4 orphan receptors. Mutant oligonucleotide competition experiments demonstrated that the TR2/TR4 binding site is restricted to sequences uniquely within the DR1 element. We also showed that both TR2 and TR4 are expressed in both primitive and definitive murine and human erythroid cells, and that forced expression of TR2 and TR4 in erythroid cells led to repression of endogenous murine embryonic εy-globin gene transcription. Thus, these biochemical and genetic data, taken together, constitute strong preliminary evidence that DRED is a bona fide human ε- and γ-globin gene repressor, and that the TR2/TR4 heterodimer provides the scaffold upon which the repressor complex with DNA is assembled.
Testicular receptors (TR) TR2 and TR4 were originally isolated because of their high nucleic acid sequence homology to the conserved DNA binding domain of steroid hormone receptors (Chang and Kokontis, 1988; Chang et al., 1994). Both nuclear receptors can form homodimers to enable one mode of DR element recognition, but they prefer to heterodimerize (Lee et al., 1998). Importantly, TR2 acting with an unknown partner molecule has been shown to repress erythropoietin gene transcription mediated through its hypoxia-inducible enhancer (Lee et al., 1996), and the TR2/TR4 heterodimer has been implicated in repression of the retinoic acid receptor β gene through a RARE sequence (Lee et al., 1998). Although not previously known to regulate genes in the blood compartment, expression of the two receptors has been documented in hematopoietic cells (Chang and Kokontis, 1988; Chang et al., 1994).
Assuming that changes in the abundance and activity of the cellular transcription factor milieu are responsible for the differential activity of globin genes during development, then changes in the activity of multiple factors will contribute to the final transcriptional output. In searching for factors that are expressed in a uniquely stage-specific manner in erythroid cells, only a few candidates have been conjectured to exist. The fact that an alteration in the activity of individual transcription factors can dramatically effect the equilibrium of γ- compared with β-globin transcription is without question: depleted adult β-globin synthesis in germline-targeted EKLF mutant mice is accompanied by induced levels of fetal γ-globin transcription (Perkins et al., 1996; Tanimoto et al., 2000), even though EKLF is normally present in both fetal and adult erythroid cells (Southwood et al., 1996).
We showed here that TR2 and TR4 are expressed at all stages of erythropoiesis. If the model proposed here for ε- and γ-globin repression by DRED is correct, then the present conundrum is that the DRED repressor (as well as the EKLF activator) exerts its effect in a developmental stage-specific fashion even though TR2 and TR4 are expressed throughout erythropoiesis. A number of possibilities could resolve the apparent inconsistency between the stage-independent expression and stage-dependent transcriptional effects of these proteins, including alterations in their relative abundance (Kulessa et al., 1995), changes in the activity of the factors by post-translational modification (e.g. Zhang and Bieker, 1998; Hung et al., 2001) or differential cytoplasmic versus nuclear retention at different developmental stages (e.g. Itoh et al., 1999).
We were unable to demonstrate directly that the TR2/TR4 heterodimer regulates either ε- or γ-globin transcription by transient transfection into tissue cultured cells. We did not detect significant changes in the transcriptional activity of either promoter, whether or not either or both of the TR2 and TR4 expression plasmids were included, after transient transfection into either erythroid or non-erythroid cells (see, for example, Figure 5) (W.Lin and O.Tanabe, unpublished observations). The possible reasons for this failure are many and complex, e.g. TR2 and/or TR4 are not limiting in formation of the 540 kDa DRED complex (Figure 1A and B), or the TR2/TR4 heterodimer participates in a chromatin remodeling step that is not readily detectable in transient transfection assays. Therefore, rather than pursue this fruitless line of investigation, we initiated gain-of-function studies in transgenic mice. Given the current evidence that: (i) there are no other proteins that bind specifically to the DR1 element in definitive erythroid cells (Figures 1D, 2 and 4) (Tanimoto et al., 2000), (ii) the DR1 element is the specific in vivo target of repression at the adult stage (Figure 2) (Tanimoto et al., 2000), (iii) the TR2/TR4 heterodimer has been shown to repress the transcription of other cellular genes (Chinpaisal et al., 1997; Lee et al., 1998) and (iv) forced expression of TR2 and TR4 in primitive erythroid cells suppresses transcription of the human ε-globin gene ortholog (murine εy) in vivo (Figure 7), we are reasonably confident that the hypothesis regarding the involvement of TR2/TR4 in adult stage repression of embryonic/fetal globin gene transcription will be supported after more extensive in vivo studies.
DRED as a molecular target for sickle cell disease?
Sickle cell anemia was the first inherited human disorder to be elucidated at the molecular level: the characteristic sickled appearance of red blood cells recovered from homozygous individuals was shown to result from a single amino acid substitution in the adult β-globin polypeptide chain (Glu6Val; Ingram, 1957). HbF is known to inhibit sickle polymer formation caused by HbS (α2βS2) (Sunshine et al., 1978; Behe and Englander, 1979) and is also known to be clinically beneficial in reducing sickle cell disease (SCD)-related morbidity (Powars et al., 1984). Therefore, virtually all current efforts focus on treatment strategies that will elevate the level of γ-globin relative to βS-globin in adult erythroid cells.
We would suggest that DRED may be an unusually attractive target for possible pharmacological intervention in SCD, or possibly even in β-thalassemia. TR2 and TR4 are nuclear orphan receptors that both contain a putative ligand binding domain. If these are genuine receptors and are regulated by some currently unidentified ligand (or perhaps even different ligands), and these in vivo modulators can be identified, it would then be a straightforward task, through conventional pharmacological studies, to search for antagonists that could confer stronger responses at lower doses with fewer toxic side effects than we encounter with current therapies. These discoveries may in turn lead to the identification of novel synthetic agents capable of inducing HbF with few physiological side effects in the CNS or male germline, where both of these receptors are abundant. Even if identification of bona fide TR2/TR4 ligand(s) is not imminent, one can envision inactivation approaches based on potential masking of the DNA binding domain or disruption of the heterodimer interface. Other components of the half megadalton DRED complex, once identified, should provide an even greater number of targets for intervention, which could be exploited in order to interfere with DRED repressor activity.
Materials and methods
EMSA
Nuclear extracts preparation, binding reactions and electrophoresis were performed as described previously (Tanimoto et al., 2000).
Oligonucleotides
The oligonucleotides used as EMSA probes and competitors (Tanimoto et al., 2000) are shown in Figure 1C. Competitor DNAs containing various sequences were incubated with MEL cell nuclear extracts (5 µg) for 15 min at 4°C, and then labeled probe (Figure 1C, epsi) was added and incubated for an additional 30 min at 4°C. The –117 Aγ HPFH transition mutation (G to A) is shown in bold in the γ(HPFH)/CAAT oligonucleotide (Figure 1C).
The following oligonucleotides were used as primers for cloning the entire open reading frames (ORFs) of murine TR2 and TR4 cDNAs (DDBJ/EMBL/GenBank accession Nos U28265 and U11688): TR2, 5′-GGCtctagaCCATTCGATCATGGCGACCATA-3′ (T2-5′) and 5′-CCG ctcgagTCAAAGGCTGTGACCAATTATTTG-3′ (T2-3′); TR4, 5′-GGC tctagaATCTCCAGGGATGACCAGCCC-3′ (T4-5′) and 5′-CCGctcgag TTGGTTGTGCACTATAGACTGGCT-3′ (T4-3′). XbaI or XhoI sites (lower case letters) were appended to the 5′-end of each primer for efficient cloning. Initiation or termination codons are underlined.
For semi-quantitative RT–PCR analysis of human and murine TR2 and TR4 expression in various hematopoietic tissues throughout development, the following primers were used: mTR2, 5′-GATTCTTTAA AAGAAGCATCCG-3′ and 3′-CGAAGTTGTCTTTTTTAGATGTA-5′ (223 bp amplicon); hTR2, 5′-CTGATCTGTCTGCACAACACCTG-3′ and 3′-GAAGCATCGGGTAATTGACGTTGA-5′ (210 bp amplicon); mTR4 and hTR4, 5′-TCAGTTGTGAAGGTTGCAAAGG-3′ and 3′-GCCCTCTTTGGTTCGTTAACA-5′ (218 bp amplicon). Murine HPRT (Suwabe et al., 1998) or human S14 (Leonard et al., 1993) oligonucleotide primers were used as internal amplification controls.
For quantitative RT–PCR analysis of yolk sacs from 10.5 d.p.c. transgenic founder embryos, the following oligonucleotides were used as primers: GATA1, 5′-CGTCATACCACTAAGGTGGCTGAAT-3′ (GATA1-IE) and 5′-GTGGAATCTGATGGTGAGGACA-3′ (GATA1- R1; 157 bp amplicon); transgene-derived TR2, GATA1-IE and 5′-AAACAACTGGTTGACACCTGC-3′ (T2-R1; 336 bp amplicon); transgene-derived TR4, GATA1-IE and 5′-AGCCAGGATCACCTT CCCAG-3′ (T4-R1; 324 bp amplicon); εy-globin, 5′-ACCCTCAT CAATGGCCTGTGGA-3′ (mEy-F1) and 5′-CATGGGCTTTGACC CTTGGG-3′ (mEy-R2; 166 bp amplicon); βH1-globin, 5′-ATCA TGGGAAACCCCCGGA-3′ (mBH1-F1) and 5′-GGGTGAATTCCTT GGCAAAATGAGT-3′ (mBH1-R1; 211 bp amplicon); α-globin, 5′-GCTGCCTGGGGGAAGATTGG-3′ (mAlph-F1) and 5′-GGGTGAAA TCGGCAGGGTGG-3′ (mAlph-R1; 322 bp amplicon). All primer sets for RT–PCR were designed to span introns in order to amplify only cDNA sequences.
The ‘epsi’ oligonucleotide (Figure 1C) was also used for construction of the sequence-specific DNA affinity column; in this case, a ‘GATC’ sequence was appended to the 5′-end of both the sense and antisense strands for efficient ligation prior to CNBr/Sepharose crosslinking.
Antibodies
Antibodies were purchased from or donated by Santa Cruz Biotechnology, and are designated sc-9087 (TR2a), sc-8617 (TR2b), sc-9086 (TR4a), sc-8620 (TR4b), sc-7714x (CP-1/NF-Y), sc-6578x (COUP-TFII), sc-6327x (CDP), sc-8983x (USF-1), sc-994x (mSin3A) and sc-768x (mSin3B).
DRED purification
DRED was purified from nuclear extracts of MEL cells by following EMSA activity. Nuclear extracts were prepared from 2 × 1010 cells grown in 20 l of medium (see below). DRED activity was precipitated from the extracts with ammonium sulfate (25–40% saturation), redissolved in buffer A (20 mM Tris–HCl pH 7.6, 1 mM EDTA, 2 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, 0.5 mM PMSF, 10 µg/ml TPCK, 2 µg/ml leupeptin, 2 µg/ml aprotinin, 1 µg/ml pepstatin A, 0.1 mM benzamidine) plus 100 mM NaCl, and then applied to a Superdex 200 gel filtration column (1.6 × 60 cm; Amersham Pharmacia). The single peak of DRED activity was pooled, diluted in buffer A, and then applied to a DEAE Sepharose FF column (1.6 × 10 cm; Amersham Pharmacia). The peak activity eluted from the DEAE column at 170 mM NaCl (using a linear 40–500 mM NaCl gradient) in buffer A. After pooling and dilution of the active fractions, and the addition of NP-40 to 0.01% (final) and poly(dI–dC) to 7 µg/ml (final), the DEAE peak was applied to a multimerized ‘epsi’ sequence-specific DNA affinity column (0.7 × 1.3 cm) (Kadonaga, 1996). DRED activity was eluted as a single peak at 0.4 M NaCl (using a stepwise 0.1 M increasing NaCl gradient in buffer A plus 0.1% NP-40). The active fractions were pooled, applied and eluted once again to the same affinity column.
Protein sequence analysis
Proteins recovered from the most active fractions from the DNA affinity column were precipitated with 20% trichloroacetic acid at 0°C for 2 h. The precipitate was washed with acetone and then applied to a 10% SDS–polyacrylamide gel (Invitrogen). After electrophoresis, the protein bands were visualized with Colloidal Blue Stain (Invitrogen). The bands were excised and subjected to sequence analyses at the Harvard Microchemistry Facility by microcapillary reverse-phase, high-performance liquid chromatography nano-electrospray tandem mass spectrometry on a Finnigan LCQ DECA quadrupole ion trap mass spectrometer.
Cloning murine TR2 and TR4 cDNAs and expression plasmid construction
The entire ORFs of the murine TR2 and TR4 cDNAs were amplified from anemic mouse spleen cDNA by PCR using high-fidelity Deep Vent DNA polymerase (New England Biolabs) and the T2-5′ and T2-3′ or T4-5′ and T4-3′ primer pairs (see above). Amplified cDNAs were cloned into pBluescript plasmid and verified by complete sequencing to be free of PCR-introduced mutations. Mammalian expression vectors for TR2 and TR4 cDNAs were constructed by ligating the cDNAs into the XbaI site of pEF-BOS (Mizushima and Nagata, 1990), thereby directing cDNA transcription from the elongation factor 1α promoter. For expression of Flag-tagged TR2 and TR4, the sequence encoding the Flag epitope was introduced at the 5′-end of both ORFs by PCR. The tagged cDNAs were then recloned into pEF-BOS. For the transgenic experiments, Flag-tagged TR2 and TR4 cDNAs were ligated to the KpnI–NotI fragment from IE3.9int-LacZ (GATA1-HRD; Onodera et al., 1997) and a simian virus 40 polyadenylation signal. The transgenic constructs were separated from vector DNA by restriction enzyme digestion, agarose gel electrophoresis and electroelution. Equimolar TR2 and TR4 constructs were co-injected into mouse oocytes.
Semi-quantitative RT–PCR analysis of TR2 and TR4 gene expression
Total RNA was extracted from yolk sac, fetal liver or anemic adult spleens using Isogene (Nippon Gene). First strand cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen) and random primer with 2 µg of total RNA in a 20 µl reaction volume. The amount of cDNA used for amplification was normalized to the expression level of co-amplified mouse HPRT or human S14 mRNA samples, respectively. PCR was performed in a 20 µl volume of 20 mM Tris–HCl pH 8.4, 50 mM KCl, 2.5 mM MgCl2, 200 µM each dNTP, 0.5 U AmpliTaq Gold polymerase (Perkin Elmer) and 0.1 µCi of [α-32P]dCTP at 94°C for 20 s, 55°C for 30 s, and 72°C for 45 s after initial incubation at 96°C for 10 min to activate the polymerase. Ten microlitres of each PCR reaction product were electrophoresed on 8% polyacrylamide gels, dried, and subjected to autoradiography and phosphorimager analysis.
Cell culture and transient transfection
The MEL (745A), QT6 and 293T cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. For transfection, 2 × 106 QT6 or 293T cells were plated in a 10-cm dish the day before transfection. Expression plasmid (20 µg) was mixed with 40 µl of lipofectamine 2000 reagent (Invitrogen) and then added to each dish according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were harvested and nuclear extracts were prepared.
Analysis of transgenic founder embryos
RNAs were extracted from the yolk sacs collected from transgenic founder embryos at E10.5, and first-strand cDNA was synthesized with Superscript II using 1 µg of total RNA in a 20 µl reaction volume. Duplex PCR was performed using 1 µl of cDNA in a 10 µl reaction volume containing 3.75 pmol of each gene-specific primer, 0.25 U of Taq DNA polymerase (Invitrogen), and 0.5 µCi of [α-32P]dCTP with temperature cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min. Cycle numbers were 24 for co-amplification of transgene-derived TR2 or TR4 with endogenous GATA1 cDNAs, 12 for co-amplification of εy- and α-globin cDNAs, and 13 for βH1- and α-globin cDNAs. Each PCR reaction was electrophoresed on 8% polyacrylamide gels, dried and subjected to phosphorimager analysis for quantitation of specific amplicons. The ratios of co-amplified cDNAs of transgene-derived TR2 or TR4 to endogenous GATA1, εy- or βH1- to α-globin were calculated as shown in Figure 7.
Acknowledgments
Acknowledgements
We thank K.-C.Lim and I.M.Klotz (Northwestern University) for thoughtful discussions of this manuscript, and W.Song and K.Lane for excellent technical assistance. We are very grateful to M.Baron and P.Frenette (Mount Sinai Medical School) for their generous gift of sorted human bone marrow cells and R.Hardison (Penn. State) for advice on erythroid cell transfections and helpful discussions. The expertise of the Harvard Microchemistry Facility, especially the initial guidance provided by W.Lane, is also gratefully acknowledged. MEL cells used for DRED purification were provided by the National Cell Culture Center. Salary support was provided by the JSPS and Cooley’s Anemia Foundation (K.T.) and the NIH (CA79447; A.D.C.). Infrastructure support for the Keck Biophysics Facility was provided by the Robert H. Lurie Comprehensive Cancer Center (P30 CA60553) and research support was from NIH (R01 HL24415; J.D.E.).
References
- Ayer D.E., Lawrence,Q.A. and Eisenman,R.N. (1995) Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell, 80, 767–776. [DOI] [PubMed] [Google Scholar]
- Barberis A., Superti-Furga,G. and Busslinger,M. (1987) Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone promoter. Cell, 50, 347–359. [DOI] [PubMed] [Google Scholar]
- Behe M.J. and Englander,S.W. (1979) Mixed gelation theory. Kinetics, equilibrium and gel incorporation in sickle hemoglobin mixtures. J. Mol. Biol., 133, 137–160. [DOI] [PubMed] [Google Scholar]
- Chang C. and Kokontis,J. (1988) Identification of a new member of the steroid receptor super-family by cloning and sequence analysis. Biochem. Biophys. Res. Commun., 155, 971–977. [DOI] [PubMed] [Google Scholar]
- Chang C., Da Silva,S.L., Ideta,R., Lee,Y., Yeh,S. and Burbach,J.P. (1994) Human and rat TR4 orphan receptors specify a subclass of the steroid receptor superfamily. Proc. Natl Acad. Sci. USA, 91, 6040–6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinpaisal C., Chang,L., Hu,X., Lee,C.H., Wen,W.N. and Wei,L.N. (1997) The orphan nuclear receptor TR2 suppresses a DR4 hormone response element of the mouse CRABP-I gene promoter. Biochemistry, 36, 14088–14095. [DOI] [PubMed] [Google Scholar]
- Choi O.-R. and Engel,J.D. (1988) Developmental regulation of β-globin gene switching. Cell, 55, 17–26. [DOI] [PubMed] [Google Scholar]
- Dillon N. and Grosveld,F. (1991) Human γ-globin genes silenced independently of other genes in the β-globin locus. Nature, 350, 252–254. [DOI] [PubMed] [Google Scholar]
- Filipe A., Li,Q., Deveaux,S., Godin,I., Romeo,P.H., Stamatoyannopoulos,G. and Mignotte,V. (1999) Regulation of embryonic/fetal globin genes by nuclear hormone receptors: a novel perspective on hemoglobin switching. EMBO J., 18, 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W.C., Takegawa,S., Papayannopoulou,T., Stammatoyannopoulos, G. and Groudine,M. (1987) Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res., 15, 10159–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas R., Endlich,B., Pfeiffer,C., Yagi,M. and Stamatoyannopoulos,G. (1985) G to A substitution in the distal CCAAT box of the A γ-globin gene in Greek hereditary persistence of fetal haemoglobin. Nature, 313, 323–325. [DOI] [PubMed] [Google Scholar]
- Gregor P.D., Sawadogo,M. and Roeder,R.G. (1990) The adenovirus major late transcription factor USF is a member of the helix– loop–helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev., 4, 1730–1740. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft,G.B., Greaves,D.R. and Kollias,G. (1987) Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell, 51, 975–985. [DOI] [PubMed] [Google Scholar]
- Hung H.L., Kim,A.Y., Hong,W., Rakowski,C. and Blobel,G.A. (2001) Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J. Biol. Chem., 276, 10715–10721. [DOI] [PubMed] [Google Scholar]
- Ingram V.M. (1957) Gene mutation in human hemoglobin: the chemical difference between normal and sickle cell hemoglobin. Nature, 180, 326–328. [DOI] [PubMed] [Google Scholar]
- Itoh K., Wakabayashi,N., Katoh,Y., Ishii,T., Igarashi,K., Engel,J.D. and Yamamoto,M. (1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev., 13, 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J.T. (1996) Purification of transcription factor AP-1 from HeLa cells. In Marshak,D.R., Kadonaga,J.T., Burgess,R.R., Knuth,M.W., Brennan,W.A. and Lin,S.-H. (eds), Strategies for Protein Purification and Characterization. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 127–203.
- Kulessa H., Frampton,J. and Graf,T. (1995) GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts and erythroblasts. Genes Dev., 9, 1250–1262. [DOI] [PubMed] [Google Scholar]
- Lee C.H., Chinpaisal,C. and Wei,L.N. (1998) A novel nuclear receptor heterodimerization pathway mediated by orphan receptors TR2 and TR4. J. Biol. Chem., 273, 25209–25215. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Young,W.J., Shih,C.Y. and Chang,C. (1996) Suppression of the human erythropoietin gene expression by the TR2 orphan receptor, a member of the steroid receptor superfamily. J. Biol. Chem., 271, 10405–10412. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Ngo,H., Kim,D. and Chung,J.H. (2000) Erythroid Kruppel-like factor is recruited to the CACCC box in the β-globin promoter but not to the CACCC box in the γ-globin promoter: the role of the neighboring promoter elements. Proc. Natl Acad. Sci. USA, 97, 2468–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard M., Brice,M., Engel,J.D. and Papayannopoulou,T. (1993) Dynamics of GATA transcription factor expression during erythroid differentiation. Blood, 82, 1071–1079. [PubMed] [Google Scholar]
- Magram J., Chada,K. and Costantini,F. (1985) Developmental regulation of a cloned adult β-globin gene in transgenic mice. Nature, 315, 338–340. [DOI] [PubMed] [Google Scholar]
- Mantovani R. et al. (1988) An erythroid specific nuclear factor binding to the proximal CACCC box of the β-globin gene promoter. Nucleic Acids Res., 16, 4299–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R., Superti-Furga,G., Gilman,J. and Ottolengi,S. (1989) The deletion of the distal CCAAT box region of the Aγ-globin gene in black HPFH abolishes the binding of the erythroid specific protein NF-E3 and of the CCAAT displacement protein. Nucleic Acids Res., 17, 6681–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima S. and Nagata,S. (1990) pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res., 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H., Katsuoka,F., Shavit,J.A., Engel,J.D. and Yamamoto,M. (2000) Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small maf proteins. Cell, 103, 865–875. [DOI] [PubMed] [Google Scholar]
- Onodera K. et al. (1997) GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc. Natl Acad. Sci. USA, 94, 4487–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A.C., Gaensler,K.M. and Orkin,S.H. (1996) Silencing of human fetal globin expression is impaired in the absence of the adult β-globin gene activator protein EKLF. Proc. Natl Acad. Sci. USA, 93, 12267–12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powars D.R., Weiss,J.N., Chan,L.S. and Schroeder,W.A. (1984) Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood, 63, 921–926. [PubMed] [Google Scholar]
- Raich N., Enver,T., Nakamoto,B., Josephson,B., Papayannopoulou,T. and Stamatoyannopoulos,G. (1990) Autonomous developmental control of human embryonic globin gene switching in transgenic mice. Science, 250, 1147–1149. [DOI] [PubMed] [Google Scholar]
- Southwood C.M., Downs,K.M. and Bieker,J.J. (1996) Erythroid Kruppel-like factor exhibits an early and sequentially localized pattern of expression during mammalian erythroid ontogeny. Dev. Dyn., 206, 248–259. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos G. and Neinhuis,A.W. (1994) Hemoglobin switching. In Stamatoyannopoulos,G., Nienhuis,A.W., Majerus,P. and Varmus,H. (eds), The Molecular Basis of Blood Diseases, 2nd edn. W.B. Saunders, PA, pp. 107–155.
- Sunshine H.R., Hofrichter,J. and Eaton,W.A. (1978) Requirement for therapeutic inhibition of sickle haemoglobin gelation. Nature, 275, 238–240. [DOI] [PubMed] [Google Scholar]
- Superti-Furga G., Barberis,A., Schaffner,G. and Busslinger,M. (1988) The –117 mutation in Greek HPFH affects the binding of three nuclear factors to the CCAAT region of the γ-globin gene. EMBO J., 7, 3099–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwabe N., Takahashi,S., Nakano,T. and Yamamoto,M. (1998) GATA-1 regulates growth and differentiation of definitive erythroid lineage cells during in vitro ES cell differentiation. Blood, 92, 4108–4118. [PubMed] [Google Scholar]
- Takahashi S. et al. (2000) GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood, 96, 910–916. [PubMed] [Google Scholar]
- Tanimoto K., Liu,Q., Grosveld,F., Bungert,J. and Engel,J.D. (2000) Context-dependent EKLF responsiveness defines the developmental specificity of the human ε-globin gene in erythroid cells of YAC transgenic mice. Genes Dev., 14, 2778–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townes T.M., Lingrel,J.B., Chen,H.Y., Brinster,R.L. and Palmiter,R.D. (1985) Erythroid-specific expression of human β-globin genes in transgenic mice. EMBO J., 4, 1715–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E., Tsai,S.-F., Hogben,P. and Orkin,S.H. (1990) Regulated expression of globin chains and the erythroid transcription factor GATA-1 during erythropoiesis in the developing mouse. Mol. Cell. Biol., 10, 6596–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijgerde M., Grosveld,F. and Fraser,P. (1995) Transcriptional complex stability and chromatin dynamics in vivo. Nature, 377, 209–213. [DOI] [PubMed] [Google Scholar]
- Wijgerde M., Gribnau,J., Trimborn,T., Nuez,B., Philipsen,S., Grosveld,F. and Fraser,P. (1996) The role of EKLF in human β-globin gene competition. Genes Dev., 10, 2894–2902. [DOI] [PubMed] [Google Scholar]
- Zhang W. and Bieker,J.J. (1998) Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl Acad. Sci. USA, 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]