Abstract
The transcription factor TFIID is a large multiprotein complex, composed of the TATA box-binding protein (TBP) and 14 TBP-associated factors (TAFs), which plays a key role in the regulation of gene expression by RNA polymerase II. The three-dimensional structure of yeast (y) TFIID, determined at ∼3 nm resolution by electron microscopy and image analysis, resembles a molecular clamp formed by three major lobes connected by thin linking domains. The yTFIID is structurally similar to the human factor although the clamp appears more closed in the yeast complex, probably reflecting the conformational flexibility of the structure. Immunolabelling experiments showed that nine TAFs that contain the histone fold structural motif were located in three distinct substructures of TFIID. The distribution of these TAFs showed that the previously reported pair-wise interactions between histone fold domain (HFD)-containing TAFs are likely to occur in the native yTFIID complex. Most of the HFD-containing TAFs have been found in two distinct lobes, thus revealing an unexpected and novel molecular organization of TFIID.
Keywords: histone fold domains/immunolabelling/three-dimensional model/transcription factor IID/yTAFs
Introduction
In eukaryotes, transcription initiation of protein-coding genes by RNA polymerase II is a multistep process requiring the coordinated interaction of multiple proteins. A set of basal transcription factors, TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH, in addition to RNA polymerase II, assemble at promoters in vitro and form a pre-initiation complex (PIC) competent for specific and accurate transcription (Orphanides et al., 1996; Hampsey and Reinberg, 1999). The general transcription factor TFIID is a stable multisubunit complex which in yeast cells contains the TATA box-binding protein (TBP) and 14 TBP-associated factors (TAFs) (Sanders and Weil, 2000). The cloning of the Drosophila (d) and human (h) TAFs revealed the evolutionary conservation of TFIID since most if not all yeast TAFs have homologues in higher metazoans (Gangloff et al., 2001a). This observation led to a unified nomenclature which is summarized in Table I (Tora, 2002). TFIID is instrumental in core promoter recognition, and its essential role in activated transcription is thought to be mediated by direct and selective interactions between specific TAFs and activation domains (Sauer and Tjian, 1997; Bell and Tora, 1999). TFIID and, more generally, all TAF-containing complexes carry a histone acetyl transferase activity which was shown to be important for remodelling the chromatin structure (Mizzen et al., 1996; Grant et al., 1998; Brand et al., 1999b).
Table I. New RNA polymerase II TAF nomenclature including the correspondence with the S.cerevisiae TAFs and their known D.melanogaster and H.sapiens orthologues (Tora, 2002).
| New name | S.cerevisiae | D.melanogaster | H.sapiens |
|---|---|---|---|
| TAF1 | Taf145/130p | TAFII230 | TAFII250 |
| TAF2 | Taf150p or TSM1 | TAFII150 | TAFII150 |
| TAF3 | Taf47p | TAFII155 or BIP2 | TAFII140 |
| TAF4 | Taf48p or MPT1 | TAFII110 | TAFII130/135 |
| TAF5 | Taf90p | TAFII80 | TAFII100 |
| TAF6 | Taf60p | TAFII60 | TAFII80 |
| TAF7 | Taf67p | (AAF54162) | TAFII55 |
| TAF8 | Taf65p | Prodos | (BAB71460) |
| TAF9 | Taf17p | TAFII40 | TAFII32/31 |
| TAF10 | Taf25p | TAFII24 | TAFII30 |
| TAF11 | Taf40p | TAFII30β | TAFII28 |
| TAF12 | Taf61/68p | TAFII30α | TAFII20/15 |
| TAF13 | Taf19p or FUN81 | (AAF53875) | TAFII18 |
| TAF14 | Taf30p |
One of the most striking structural features of TFIID subunits is that several TAFs contain a stretch of amino acids with sequence homology to histones, comprising the histone fold domain (HFD). Initial sequence alignments showed obvious homologies with histone H3 (TAF9), H4 (TAF6) and H2B (TAF12) (Kokubo et al., 1994; Hoffmann et al., 1996). These homologies were confirmed by X-ray diffraction studies which revealed that dTAF9 and dTAF6 form a heterotetramer and interact through a characteristic histone fold which is reminiscent of the interactions found between histones H3 and H4 (Xie et al., 1996). Subsequent structural studies showed that hTAF11 and hTAF13 also contain an HFD that is used as an interaction interface to form heterodimers (Birck et al., 1998). Based on sequence alignment, specific heterodimerization of bacterial co-expressed TAFs and interactions in two-hybrid assays, a total of nine yeast TAFs were found to contain an HFD (Gangloff et al., 2001c). In addition to the initially described Drosophila homologues of TAF6–9 and TAF11–13 heterodimers, the following HFD-containing pairs were identified in yTFIID: TAF4–12 (Gangloff et al., 2000; Reese et al., 2000; Sanders and Weil, 2000), TAF3–10 and TAF8–10 (Gangloff et al., 2001c). These results indicate that dimerization of HFD-containing yTAFs is highly specific and that the HFD is a repeated structural motif essential for the organization of TFIID. It has been suggested that nucleosome-like octameric structures could be present in the TFIID architecture (Hoffmann et al., 1996; Gangloff et al., 2001b). However, the five histone-like pairs currently identified are too numerous to form a single octamer and insufficient to form two octamers, unless the stoichiometry of yTAFs differs from one per complex. The observations that TAF10 has two dimerization partners (TAF3 and TAF8) (Gangloff et al., 2001c) and that the dTAF6–9 pair was observed to form hetero tetramers at least under crystallization conditions (Xie et al., 1996) suggest that some yTAFs might be present in more than one copy in TFIID.
To elucidate the molecular organization of TFIID, a better understanding of the distribution of TAFs in native complexes is required. Low resolution structures of the hTFIID and of the TAF-containing complex, TFTC, were obtained recently by electron microscopy and digital image analysis (Andel et al., 1999; Brand et al., 1999a). The TFIID complex forms a molecular clamp and is composed of three or four lobes organized into a horseshoe-like structure. Immunolabelling experiments localized hTBP in the central density of hTFIID, facing the central cavity, and supporting the hypothesis that this groove is a major site for DNA binding (Andel et al., 1999; Nogales, 2000). Here we determine the three-dimensional (3-D) structure of the yTFIID complex at higher resolution and show its structural similarity to the human factor. To gain insights into the spatial organization of yTAFs in the native yTFIID, nine HFD-containing TAFs were immunolabelled and localized within the 3-D structure of yTFIID. The immunolabelling experiments demonstrated that all HFD-containing TAFs were found in two locations and that the previously described specific heterodimers are present in the native yTFIID.
Results
Three-dimensional model of yeast TFIID
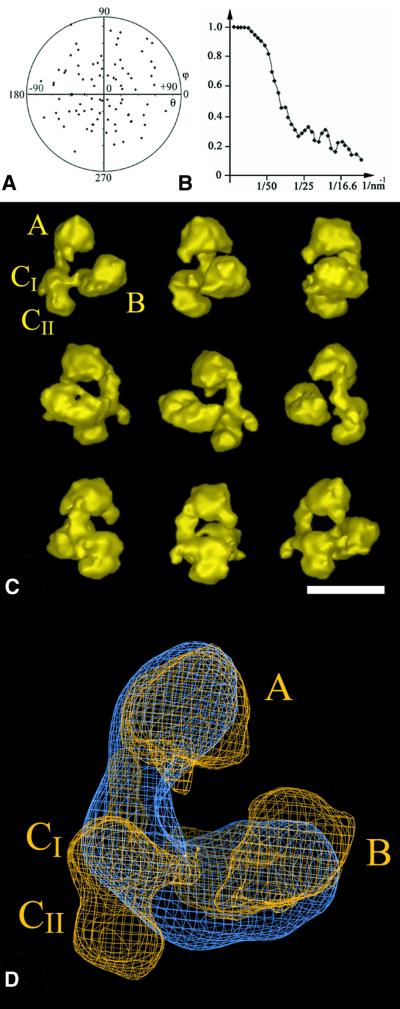
The yTFIID complex, N-terminally haemagglutinin (HA) tagged on TAF, was purified and functionally characterized as previously described (Sanders and Weil, 2000). This preparation of TFIID is capable of both binding TATA-DNA specifically and supporting specific RNA polymerase II transcription in vitro. Indeed, on a TBP molar basis, the transcriptional activity of this TFIID preparation was found to be 25–50 times higher than TBP alone for both basal and activated transcription in vitro (Sanders et al., 2002). When processed for electron microscopy, yTFIID appeared as a homogeneous dispersion of globular particles in which triangular-shaped molecules were frequently observed (Figure 1A). A total of 2904 pairs of 60°-tilted and untilted molecular images were recorded and the untilted views were analysed numerically to obtain characteristic noise-free views of the particle (Figure 1B–E). The triangular-shaped views (Figure 1B) represented 50–60% of all molecular images, suggesting that yTFIID was oriented preferentially on the supporting carbon film. When viewed through this orientation, the particle was composed of three distinct lobes, A, B and C, arranged to form an asymmetric triangle ∼25 nm high by 18 nm wide. Only 2-nm thick linking domains connected lobes A and B to lobe C in order to form a horseshoe-shaped particle opened between lobes A and B. Lobe A was oval shaped (∼12.5 × 9 nm in size), lobe B showed a triangular shape (11 × 11 nm), whereas lobe C had an elongated shape (12 × 6.5 nm) and was divided into two subdomains (CI and CII). The asymmetry of this molecular view was due to the orientation of the septum in lobe C as well as to the relative positions of the lobes: lobe B is closer to lobe C than is lobe A, and lobe A is collinear with the long axis of lobe C, whereas lobe B is positioned perpendicularly to the long axis of lobe C. The opening of the clamp between lobes A and B was less discriminating because slight variations in orientation resulted in overlapping of these lobes. These structural features preclude any in-plane confusion between the lobes. Moreover, the image analysis reproducibly identified a mirror-symmetric view resulting from the upside-down adsorption of the TFIID molecule onto the carbon foil (Figure 1C). Additional TFIID views (Figure 1D and E) were identified which correspond to different orientations of the yTFIID complex where the different lobes were superimposed.
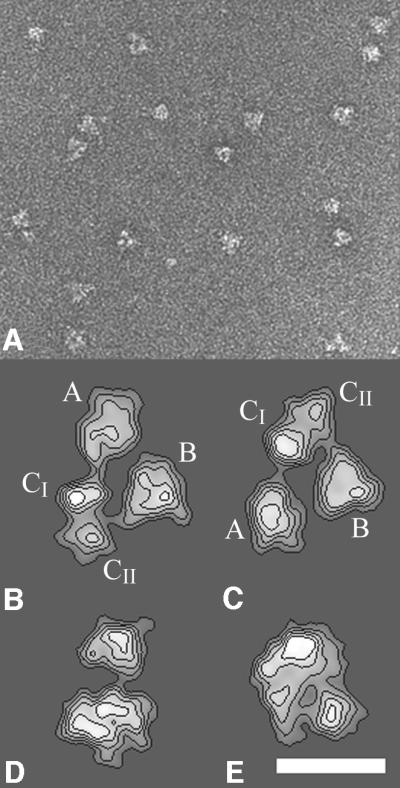
Fig. 1. Electron microscopy and image analysis of the yeast TFIID complex. (A) Electron micrograph of yTFIID molecules adsorbed on a carbon film and negatively stained with uranyl acetate showing the homogeneity in size of the complexes. (B–E) Gallery of the most representative yTFIID views obtained upon averaging aligned images clustered into homogeneous classes. The stain-excluding protein densities are outlined by contours of equal density. The two views in (B) and (C) are related by a mirror symmetry and correspond to two upside down orientations of yTFIID where the molecule interacts by two opposite sides with the carbon foil. The lobes are identified as A, B, CI and CII throughout the text. The bar represents 170 nm in (A) and 15 nm in (B–E).
The 60°-tilted images corresponding to the most abundant triangular views (Figure 1B) were used to reconstruct a preliminary 3-D model using the conical tilt series method (Radermacher et al., 1987). This model was then used to determine the viewing directions of the untilted molecular images. To increase the occurrence of rare orientations, which yield novel views of the molecule, the untilted data set was raised to 3475 images. A total of 93 different views of the yTFIID molecule were used, and the orientation plot shows that the viewing directions were distributed equally (Figure 2A). The resolution tests gave values of 32 or of 28 Å for the 0.5 Fourier shell correlation (FSC) and 3σ criteria, respectively (see Materials and methods and Figure 2B).
Fig. 2. 3-D model of the yTFIID complex. (A) Diagram showing the orientations of the 93 class averages used in the final reconstruction. Each class average is represented by a point in a (θ, φ) coordinate system. (B) Diagram representing the Fourier shell correlation coefficient versus the resolution between two independent reconstructions. (C) Surface representation of the 3-D reconstruction of yTFIID at a resolution of 2.8–3.2 nm. The model is turned around a vertical axis by increments of 40°. (D) Alignment and superimposition of the yTFIID model (yellow) with a previously determined hTFIID model (blue) (Brand et al., 1999a). The lobes are identified as A, B, CI and CII as indicated in the text. The bar represents 20 nm in (C) and 6 nm in (D).
The 3-D structure of the yTFIID complex appeared as a tri-lobed, horseshoe-shaped particle 27 × 22 × 16 nm in size (Figure 2C). The two large lobes A and B are connected to lobe CI by 2-nm thin and 4-nm long linkers so that the opening of the claw is between lobes A and B. The density threshold was set to delimit a volume of 1.2 MDa (assuming a protein density of 1.4 g/cm3) which corresponds to the apparent mass of the complex determined by gel filtration (see Sanders et al., 2002; Supplementary data available at The EMBO Journal Online). Lobe A is 13 × 8.5 × 8 nm in dimensions and is elongated in a direction normal to the plane formed by the three lobes. Lobe B is 12 × 8 × 8 nm in size and is formed by two distinct subdomains separated by a septum which was not detected in the preferential triangular view because of the superimposition of the subdomains in projection (central panel in Figure 2C). The central lobe C, 10 × 5 × 7.5 nm in size, is subdivided into two domains and a small protein density protrudes from domain CI. When turned 120° around a vertical axis, the relative positions of the lobes and the linkers form an almost closed ring of protein densities with a central hole of ∼4 nm in diameter (Figure 2C). The different parts of yTFIID seem to wrap around a thread contacting the groove between the two subdomains in lobe B, lobe CI, the A–C linker and lobe A, respectively, over a total distance of 16 nm.
The previously analysed 3-D structure of hTFIID was superimposed on the envelope of yTFIID (Figure 2D). Three criteria were used to align the yeast structure on the human model: (i) the relative positions of the centres of the three lobes; (ii) the thickness of the connector domain; and (iii) the position of TBP (data not shown). Together, these criteria converge to a unique orientation and the lobes of the yTFIID model were labelled from A to C as proposed earlier for hTFIID (Andel et al., 1999). The overlay shows the conservation in size and in overall shape between the two complexes; however, yTFIID appears sharper due to the slightly better resolution. A major difference lies in the relative positions of lobes A and B which are closer to each other in yTFIID than in hTFIID by ∼3 nm. Interestingly, the CII domain of yTFIID appears to project out of the hTFIID envelope, whereas a similar density seems to be missing in yTFIID in the linking domain between lobe B and C. A better superposition would be achieved by a rotation of lobe C to fill in this missing density, which suggests a concerted movement of lobes B and C in order to close the clamp and exclude domain CII.
Immunolocalization of TAF10
In order to locate TAF10 within yTFIID, the endogenous complex was incubated with a 5-fold molar excess of a yTAF10-specific monoclonal antibody (mAb 38TA), whose epitope was mapped between amino acids 149 and 180. A total of 1023 images of putative immune complexes (IC) (n1 in Table II) were recorded, and a subset of 278 IC images (n2 in Table II) corresponding unambiguously to the characteristic yTFIID view shown in Figure 1B were selected and analysed further to reveal a specific antibody-binding site. Particular care was taken during the selection to recognize the discrimination features so as to avoid any in-plane misalignment or confusion with the mirror-symmetric view. For each IC, the labelled site was plotted onto one out of nine sectors subdividing the contour of the molecular view (Figure 3A and Table II). Unexpectedly, this analysis revealed two strong antibody-binding sites; one in lobe CII (51% of the antibodies bound to sector 7) and another in lobe A (23% of the antibodies bound to sector 3). The binding was considered to be specific when found above background by >3σ, the background values being estimated by the average binding frequency and the standard deviation of the seven other sectors. This analysis indicated that the two sites were specific since the average background value represented 4% of the bound antibodies and since this value was exceeded by 31.8 and 12.6σ for the sites in lobes CII and A, respectively (Figure 3A and Table II). The difference in labelling frequency between lobes CII and A was not analysed further since the accessibility of the epitopes might be different in the two lobes due to a different environment of TAF10 and since the binding of the antibody could interfere with the adsorption of the complex, thus favouring the observation of complexes labelled on one lobe versus the other. A noise-free view was produced for each labelled site by image averaging (left panel in Figure 3B and C for lobes CII and A, respectively) and the difference image between the labelled and the unlabelled views revealed a statistically significant density due to the TFIID-bound antibody (right panel in Figure 3B and C for lobes CII and A, respectively). Altogether, these observations show that TAF10 can reside in two different locations within yTFIID.
Table II. Labelling statistics.
| yTAF labelled | No. of images |
Lobe A |
Lobe B |
Lobe C |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n1 | n2 | Sector 1 | Sector 2 | Sector 3 | Sector 4 | Sector 5 | Sector 6 | Sector 7 | Sector 8 | Sector 9 | |
| TAF10 | 1023 | 278 | 1.6 | 0.8 | 12.6 | 0.8 | 0.4 | 1.2 | 31.8 | 0.4 | 0.4 |
| TAF8 | 908 | 599 | 1.1 | 5.9 | 0.3 | 1 | 0.5 | 1.5 | 8.1 | 6.7 | 0.6 |
| TAF3 | 723 | 486 | 0.7 | 4.9 | 0.3 | 1 | 9.1 | –1 | 0 | 1.7 | 0.7 |
| TAF6 | 1545 | 264 | 0.6 | 4.5 | 11 | 1.0 | 7.1 | 1.0 | 0.3 | 1.0 | 0.3 |
| TAF9 | 1575 | 784 | 0.6 | 3.6 | 0.9 | 0.3 | 2.2 | 3.4 | 1.5 | 0.6 | 1.0 |
| TAF11 | 532 | 157 | 0.5 | 1.0 | 13 | 4.9 | 4.5 | 0.9 | 1.4 | 0.1 | 0.9 |
| TAF13 | 477 | 136 | 0.4 | 1.6 | 3.4 | 0.6 | 3.2 | 0.1 | 1.6 | 1.0 | 1.0 |
| TAF4 | 420 | 148 | 1.8 | 0.9 | 0.9 | 0.3 | 5.1 | 0 | 3.9 | 0.9 | 0.6 |
| TAF12 | 576 | 142 | 0.6 | 0.8 | 1.0 | 0.6 | 12.4 | 1.0 | 12.0 | 1.3 | 0.9 |
For each antibody, used to detect the position of yTAFs, the number of recorded images of ICs (n1) and the number of selected tri-lobed views (n2) are indicated. The proportion of tri-lobed views varies from one experiment to the other since only unambiguous images were used and since the binding of the antibody may affect the orientation of the particle on the supporting film. Each lobe was divided into three sectors as shown in Figure 3A. The values reported for each sector represent deviations from the average binding frequency (in σ folds). The values indicated for TAF8 were obtained from the analysis of the upside-down view. Bold characters represent values >3σ.
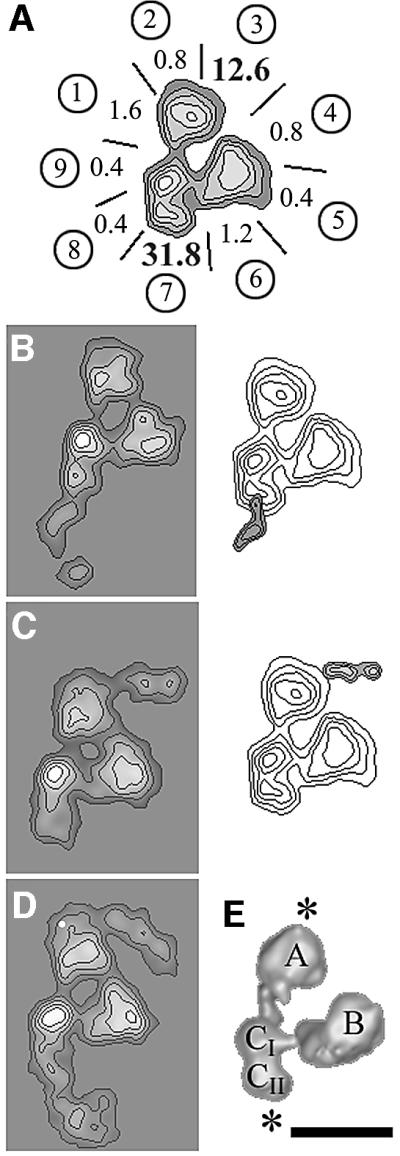
Fig. 3. Immunolabelling of TAF10 with a subunit-specific monoclonal antibody. (A) Mapping of the yTAF10-specific antibody-binding sites at the outer contour of the tri-lobed yTFIID view. Each lobe A, B and C is divided into three sectors labelled from 1 to 9 as in Table II. The values indicated for each sector represent deviations from the average binding frequency in sigma folds. Labelling in sectors 3 and 7 is significantly above background. (B) Average images of yTFIID molecules labelled in lobe C (left panel) and difference map with unlabelled yTFIID molecules (right panel). (C) Average images of yTFIID molecules labelled in lobe A (left panel) and difference image with unlabelled molecules (right panel). (D) Average image of yTFIID molecules simultaneously labelled in both lobes A and C. (E) Surface representation of the 3-D model of yTFIID overlaid with the proposed locations of TAF10 in lobes A and C. The bar represents 15 nm.
To investigate further whether a single yTFIID molecule can contain two TAF10 molecules, the yTFIID preparation was incubated with a 20-fold molar excess of mAb 38TA in order to identify double-labelled particles. Under these conditions, a significant proportion of complexes labelled by two antibodies were identified and the noise-free image clearly showed that both lobes A and CII can be labelled simultaneously at the same locations as were the singly labelled particles (Figure 3D). This experiment showed that a minimum of two copies of TAF10 are present in yTFIID complexes.
Immunoprecipitation experiments suggest the presence of multiple TAF10 molecules in yTFIID
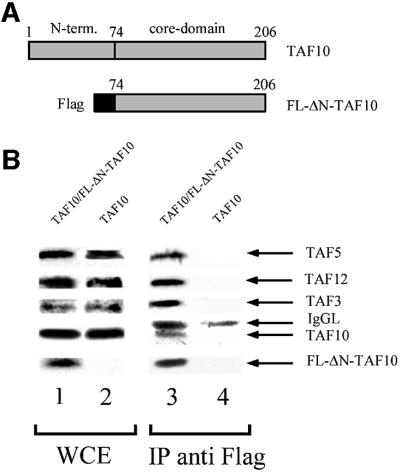
To confirm the immunolabelling results, we carried out an immunoprecipitation experiment with yeast cell extracts prepared from strains in which TAF10 was expressed either alone or together with its N-terminally truncated form (Flag-ΔN-TAF10) that contained a Flag tag on its N-terminal end (Figure 4A; Kirschner et al., 2002). The cell extracts prepared from these two strains were first tested for the presence of the different TAF10 derivatives by western blot analysis (Figure 4B, lanes 1 and 2), and then incubated with anti-Flag antibody-containing beads for immunoprecipitation. The antibody-bound complexes were then washed and eluted by competition with the epitope peptide. The eluates were tested for specific TAF content by western blot analysis. The truncated Flag-ΔN-TAF10 protein was able to co-immunoprecipitate the full-length TAF10 (Figure 4B, lane 3) together with TAF3, TAF12 and TAF5, whereas in the control immunoprecipitation, where the truncated Flag-ΔN-TAF10 protein was absent, no TAF10 protein was detected (Figure 4B, lane 4). This result shows that the truncated Flag-tagged TAF10 and the full-length TAF10 can be incorporated into the same TAF-containing complex, indicating that these complexes contain at least two copies of yTAF10.
Fig. 4. Immunoprecipitation of yTAF10-tagged yTFIID. (A) Schematic representation of the wild-type full-length yTAF10 and of the Flag-tagged core yTAF10 (FL-ΔN-TAF10). (B) Immunoblots of whole-cell extracts (WCE, lanes 1 and 2) and of complexes immunoprecipitated (IP anti Flag) with an anti-Flag antibody (lanes 3 and 4). WCE and IP containing both TAF10 and FL-ΔN-TAF10 were analysed in lanes 1 and 3, whereas the control WCE and IP were analysed in lanes 2 and 4. The upper panels were probed with the indicated yTAF-specific antibody, whereas the lowest panel was probed with the anti-Flag antibody. The blot reveals that the complexes immunoprecipitated with an anti-Flag antibody contain both TAF10 and Flag-tagged ΔN-TAF10, indicating that at least two TAF10 molecules can be incorporated into the complexes. IgG light chains (IgGL) leaking from the resin are also indicated in lanes 3 and 4.
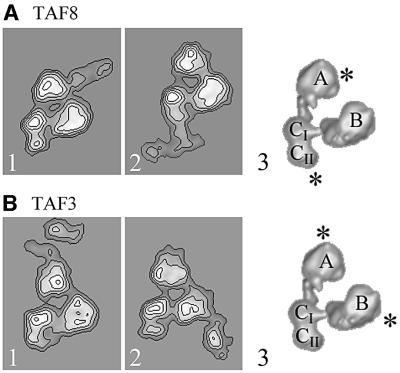
Immunolocalization of yTAF8 and yTAF3
We next mapped the HFD-containing heterodimerization partners of TAF10, TAF8 and TAF3 (Gangloff et al., 2001c) within yTFIID using cognate subunit-specific polyclonal antibodies. The analysis of yTFIID molecules incubated with either anti-TAF3 or anti-TAF8 IgGs revealed specific labelling of two distinct lobes for each antibody (Table II). In the case of TAF8, lobes A and CII were labelled and the antibody interaction site correlated almost exactly with the labelling pattern observed for TAF10 (Figure 5A). This co-localization is consistent with the hypothesis that TAF8 is a heterodimerization partner of TAF10 and indicates that yTFIID probably contains two copies of TAF8. The possibility that yTFIID molecules lie upside down, thereby producing mirror-symmetric views (see Figure 1B and C) and confounding our interpretation of the immunolocalization studies, was investigated. This image subpopulation was separated accurately from the direct views, and the labelling pattern was found to be identical for both molecular views (data not shown), indicating that the antibody-binding pattern is independent of the molecular orientation. The TAF3-specific antibodies bound specifically to lobes A and B (Figure 5B; Table II). In lobe A, TAF3 is found close to TAF10, whereas in lobe B the localization of TAF3 is clearly distinct from that of TAF10, which was not found in this lobe.
Fig. 5. Immunolabelling of the HFD-containing yTFIID subunits TAF3 and TAF8. (A) Average images of yTFIID molecules labelled with TAF8-specific antibodies in lobe A (panel 1) or in lobe C (panel 2). Surface representation of the 3-D model of yTFIID overlaid with the proposed locations of TAF8 in lobes A and C (panel 3). (B) Average images of yTFIID molecules labelled with TAF3-specific antibodies in lobe A (panel 1) or in lobe B (panel 2). Surface representation of the 3-D model of yTFIID overlaid with the proposed locations of TAF3 in lobes A and B.
Immunolocalization of the six other HFD-containing yTAFIIs
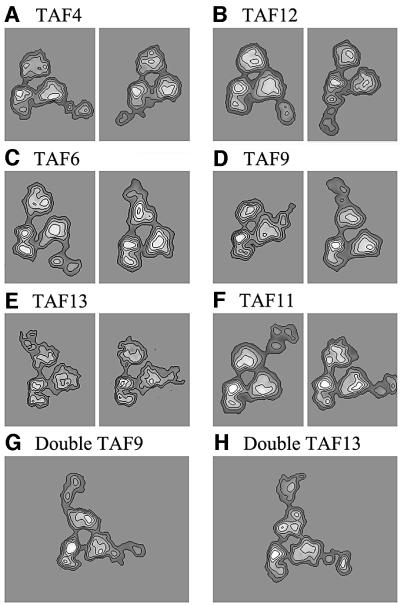
The distribution of the remaining six HFD-containing TAFs was probed using subunit-specific polyclonal antibodies. In the case of TAF12, lobes B and CII were labelled, indicating that TAF12 can be found in two distinct locations (Figure 6A and Table II). In the conditions tested, only two double-labelled yTFIID molecules were found in our data set, a number too low to allow us to calculate an average view, but indicated that at least some yTFIID complexes can contain two TAF12 molecules. The labelling pattern was similar with the TAF4-specific antibody (Figure 6B and Table II). These results indicate that TAF4 and TAF12 are present in two copies in yTFIID and that they co-localize, consistent with their property of forming heterodimers in solution. Importantly, the labelled sites are different from those labelled for TAF10 or TAF8, results excluding the possibility of any systematic errors in our subunit mapping analysis.
Fig. 6. Immunolabelling of six HFD-containing yTAFs. (A) Average images of yTFIID molecules labelled with TAF4-specific antibodies in lobe B (left panel) and in lobe C (right panel). (B) yTFIID molecules labelled with TAF12-specific antibodies in lobe B (left panel) and in lobe C (right panel). (C) yTFIID molecules labelled with TAF6-specific antibodies in lobe B (left panel) and in lobe A (right panel). (D) yTFIID molecules labelled with TAF9-specific antibodies in lobe B (left panel) and in lobe A (right panel). (E) yTFIID molecules labelled with TAF13-specific antibodies in lobe A (left panel) and in lobe B (right panel). (F) yTFIID molecules labelled with TAF11-specific antibodies in lobe A (left panel) and in lobe B (right panel). (G) yTFIID molecules simultaneously labelled in both lobes A and B by TAF9-specific antibodies. (H) yTFIID molecules simultaneously labelled in both lobes A and B by TAF13-specific antibodies.
The HFD-containing TAF6 and TAF9 subunits were reported to form specific heterodimers capable of forming higher ordered structures with the TAF4–12 pair (Selleck et al., 2001). We reasoned that if such structures form in vivo, then these four subunits should co-localize to the same lobe of TFIID. Immunomapping experiments located TAF6 in two distinct lobes: lobe A was labelled between sectors 2 and 3 and lobe B was labelled in sector 7 (Figure 6C). The antibodies directed against TAF9 bound to sector 2 of lobe A and to sector 6 of lobe B (Figure 6D). These results indicate that TAF6 and TAF9 co-localize and are thus likely to form the previously described histone-like dimers. Moreover, the TAF6–9 and TAF4–12 pairs both map in lobe B where they could form the predicted octameric structure. However, our results indicate that these two pairs can also be found separately; the TAF6–9 pair in lobe A and the TAF4–12 pair in lobe C. In order to demonstrate that two copies of TAF9 can be present in the same yTFIID molecules, the purified complex was incubated with a 20-fold molar excess of anti-TAF9 antibodies. In these conditions, 29 out of 127 specifically labelled yTFIID complexes bound two IgG molecules. Despite the inherent flexibility of the double-labelled molecules, an average image could be calculated from 19 molecules and this average image clearly showed the two antibody molecules bound simultaneously to their specific site (Figure 6E). This experiment indicates that TAF9, probably together with its HFD-containing partner TAF6, can be present in at least two copies in yTFIID.
Finally, we investigated the localization of TAF11 and TAF13, which were also reported to form specific heterodimers, within yTFIID. Again, both subunits were found to be co-localized in lobe A and lobe B (Figure 6F and G). In the case of TAF13, ∼7% of yTFIID particles were double labelled and showed, upon averaging, the same antibody-binding site as the single-labelled complexes (Figure 6H).
Discussion
Structural homology between human and yeast TFIID
The 3-D model of yTFIID presented in this report emphasizes its structural homology with the human factor that has been independently described by two groups (Andel et al., 1999; Brand et al., 1999a). The similar architecture of TFIID formed of three major lobes organized into a molecular clamp most probably reflects the extended subunit conservation among eukaryotes. It is thus reasonable to assume that the locations of the different TAFs in the yeast complex are liable to reflect the molecular organization of hTFIID. As a consequence of the high homogeneity of the yTFIID preparation used in this study, the resulting structure revealed greater detail regarding the shape of the three lobes comprising TFIID, the 20 Å thin linking domains between the lobes and the connections of both linkers to the central domain CI. Upon alignment, the major differences between the two models are the presence of domain CII in yTFIID and the more open conformation of the clamp in the human model. These differences probably reflect a conformational flexibility of the structure resulting in a concerted movement of lobes B and C in order to close the clamp and to exclude domain CII. A comparable conformational change involving a movement of the clamp was detected previously upon analysis of hTFIID (Brand et al., 1999a). Such structural rearrangements within TFIID may be of functional importance in the sequential assembly of the PIC nucleated by the binding of TFIID to the promoter.
Most of the described HFD-containing TAF heterodimers exist in native yTFIID
HFDs were first identified in human TAF6, TAF9 and TAF12 based on sequence homologies with histones H4, H3 and H2B, respectively, and on their ability to form specific heterodimers (Kokubo et al., 1994; Hoffmann et al., 1996). Two atomic structures of HFD–TAF heterodimers showed that the canonical histone fold consisting of two short α-helices flanking a long central α-helix is conserved in the TAFs and that their dimerization interface mimics the typical inter-histone contacts found in H2A–H2B and H3–H4 heterodimxers (Xie 1996; Birck et al., 1998). Subsequent sequence analysis, pair-wise interaction studies between overexpressed TAFs and yeast suppression genetics showed that putative HFDs are conserved in nine yeast TAFs and that this motif may determine specific pair-wise interactions resulting in five heterodimers (Birck et al., 1998; Reese et al., 2000; Gangloff et al., 2001c; reviewed in Gangloff et al., 2001b). It has been argued that interaction studies using fragments of the HFD–TAFs may not necessarily represent interactions that occur in vivo (Albright and Tjian, 2000). We thus investigated whether the distribution of HFD–TAFs within the native yTFIID complex is consistent with their abilities to form specific heterodimers in vitro.
Our immunolabelling data showed that the two partners of four heterodimers (namely the TAF4–12, TAF11–13, TAF8–10 and TAF6–9 pairs) were all found to map within the native complex in a fashion consistent with the formation of the predicted HFD pairs (Figure 7). Despite the intriguing observation that each HFD–TAF was found in two distinct lobes, the distribution of the partners of these four heterodimers coincided for both locations. The observation that most pair-wise interactions detected in vitro are preserved in the native structure is consistent with genetic experiments in yeast, which showed that temperature sensitive (Ts) mutations of some HFD– yTAFs can be suppressed by the overexpression of their heterodimerization partner (Michel et al., 1998; Reese et al., 2000; Gangloff et al., 2001c).
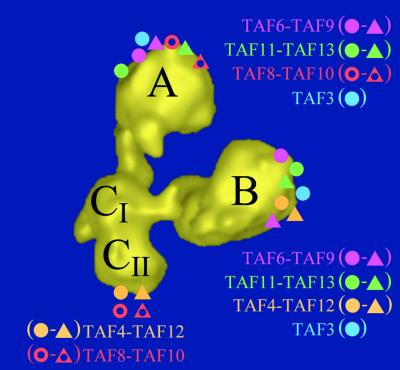
Fig. 7. Summary of the immunolabelling experiments. yTAF-specific antibody-binding sites schematically represented on the 3-D model of yTFIID. Each studied HFD-containing yTAF pair is shown in a different colour, and each partner of a pair is identified by a circle or a triangle as indicated. The size of the coloured areas does not match the size of the corresponding polypeptide or the precision of the labelling.
Unexpectedly, the distribution of the TAF10-specific antibodies did not overlap entirely with the labelling pattern of its two putative heterodimerization partners, TAF8 and TAF3. Instead, the two locations of TAF10 correlated exactly with those of TAF8, strongly suggesting that this HFD–TAF pair is present both in lobe A and in lobe C. In contrast, TAF3 co-localized with TAF10 only in lobe A and not in lobe B (Figure 7). A possible explanation for these results is that TAF3 may have another, as yet undiscovered, partner. This hypothesis is supported by a systematic analysis of the TAF content of a collection of yTFIID complexes harbouring different Ts mutants of TAF10 which showed that mutations in the TAF10 HFD can result in the loss of TAF8, but not in the loss of TAF3 (Kirschner et al., 2002). Alternatively, the following explanations can be put forward to reconcile the strong genetic and the selective physico-chemical interaction found between the HFDs of TAF10 and TAF3 with our observations. On one hand, it is conceivable that TAF10 interacts with TAF3 in lobe A and with TAF8 in lobe C. This would, however, imply that both TAF8 and TAF3 have another partner in lobes A and B, respectively. On the other hand, the stoichiometry of TAF10 could be of 4:1 so that two TAF8–10 pairs would be present in lobes A and C, respectively, and two TAF3–10 heterodimers would be present in lobes A and B, respectively. This possibility would require that the epitope recognized by the TAF10-specific monoclonal antibody would not be accessible when TAF10 interacts with TAF3 in lobe B.
All HFD-containing TAFs are found in two locations
The immunolabelling experiments showed that all HFD–yTAFs are located in two distinct sites, suggesting that they may all be present in two copies in yTFIID (Figure 7). Our experiments were validated carefully for antibody-binding specificity and for unambiguous orientation of the TFIID views in order to avoid any bias. As a first control, this approach revealed a single binding site in the case of TBP and TAF1 (data not shown), indicating that the 3σ criterion, used here to validate the antibody-binding specificity, is suitable to detect single binding sites. Secondly, the possibility of confusing the preferential tri-lobed yTFIID view with its upside-down view was considered, but the image analysis protocol clearly discriminated between these two molecular orientations. Moreover, a consistent labelling pattern was observed for both views (data not shown), demonstrating that the same sites were labelled regardless of the orientation of the TFIID molecule. Thirdly, a significant amount of double-labelled complexes was observed for three different antibodies, showing that a single yTFIID molecule can host two identical subunits. Finally, the binding pattern was different from one antibody to the other, discounting the possibility of a systematic confusion of the lobes. Altogether, these control experiments validate the finding that all HFD-containing yTAFs can be located in two distinct positions in yTFIID. However, because of incomplete antibody labelling, our results cannot be interpreted directly as stoichiometry data. The TAF copy number would be underestimated when an epitope(s) is masked or when two identical subunits are too close to each other to be resolved. Alternatively, the stoichiometry would be overestimated when a subunit is detected in two distinct but incompletely occupied locations.
Independent lines of evidence showed for two histone-like TAFs that their copy number is >1. The co-immunopurification of full-length TAF10 with its truncated FLAG-tagged version suggests that two copies of this subunit are present in yTFIID, assuming that SAGA and TFIID have the same TAF10 content. Earlier experiments used a similar approach to demonstrate that two copies of the human homologue of yTAF10 can be incorporated into hTFIID (Hoffmann et al., 1996). The extensive data obtained for yTAF10, which showed that two statistically significant binding sites are correlated with double-labelled complexes and co-immunopurification, may be extrapolated to the other HFD–TAFs to propose that all these subunits are present in two copies in the native yTFIID structure.
Higher ordered HFD-containing yTAFs structures in native TFIID
Our immunolabelling data show that HFD–yTAFs are present in all three lobes of yTFIID which thus form three distinct substructures, each containing a unique combination of HFD–yTAFs. Three heterodimers and TAF3 were detected in lobes A and B, whereas lobe CII harbours two heterodimers (Figure 7). The question of having higher ordered or even nucleosome-like structures in yTFIID was raised since high molecular weight assemblies of recombinant HFD–TAF heterodimers were described previously. However, the predicted subcomplexes have never been detected in vivo and it was questioned whether the HFD–TAFs assemble into nucleosome-like structures or whether these motifs could merely constitute protein– protein interaction interfaces in the endogenous TFIID (Albright and Tjian, 2000).
The Drosophila TAF9 and TAF6 form a (H3–H4)2-like heterotetramer (Xie et al., 1996) and the human homologues were proposed to associate into an octamer with multiple copies of TAF12 (Hoffmann et al., 1996). Using purified recombinant yTAFs, Tan and colleagues were able to form an octamer-like structure in vitro consisting of two copies each of TAF6–9 and TAF4–12 heterodimers (Selleck et al., 2001). Support for the idea that the latter structure would occur in vivo comes from the observation that Ts mutants of TAF6 or TAF9 can be suppressed not only by the overexpression of their cognate heterodimerization partners but also by overexpression of TAF12 (Michel et al., 1998). Our results reveal that this set of yTAFs co-localizes in lobe B, although our experiments failed to resolve whether two copies of these yTAFs are present in lobe B as would be required to form an octamer. It can thus not be concluded whether this set of yTAFs assembles into a bone fide nucleosome structure. Two observations, however, weaken a model where these four yTAFs would form an octamer in lobe B. First, our data show that the TAF4–12 and the TAF6–9 heterodimer interaction is not unique since each pair is found independently of the other in a distinct lobe and thus in a distinct substructure. As a consequence, the direct heterodimer interactions required to form a nucleosome-like particle would have a degenerated or regulated specificity. Secondly, additional HFD–yTAFs are found in lobe B such as TAF3 and the TAF11–13 pair, indicating that this substructure of yTFIID contains more HFD– yTAFs than those forming the putative nucleosome- like structure. We thus favour the hypothesis that the HFD–yTAFs heterodimers are recruited onto yTFIID through more complex interactions than those occurring in the nucleosome.
Yeast suppressor genetics have described another network of interactions between TAF3, 10, 11, 13 and 6, suggesting that these subunits interact in a second substructure (Gangloff et al., 2001b). Consistent with these results, these HFD–yTAFs were found to co-localize in lobe A, but again this combination is not unique since these polypeptides can be found independently of one another in a different lobe. The intriguing observation that TAF6 can be present in two distinct assemblies contradicts a simple model where lobes A and B would each contain a stable octameric structure, but is entirely consistent with the HFD–yTAF distribution revealed by our immunolabelling experiments.
Taken together, our results do not contradict the existing HFD–TAF pair-wise interaction data but favour a more complicated organization of these TAFs within TFIID. The original proposal that the HFD–TAFs form a compact nucleosome-like structure (Hoffmann et al., 1996) has to be revisited in the light of the three-lobed structure of TFIID and the finding that each substructure contains a unique combination of histone-like TAFs. In favour of a nucleosome-like structure was the observation that the complex induces negative supercoiling upon interaction with DNA (Oelgeschlager et al., 1996). Since all three lobes of TFIID were found to contain HFD–TAFs, it is possible that two, or even all three, subdomains of TFIID could form a dedicated structure modifying the topology of DNA and stably anchoring TFIID to the promoter. Further cryo-electron microscopy observations using promoter DNA-bound TFIID will reveal how and where the different lobes of TFIID contact core promoters.
Materials and methods
Yeast strains, cell transformation and yeast TFIID purification
The yeast strain and the protocol used to purify yTFIID containing the HA-tagged TAF1 subunit were described previously (Sanders and Weil, 2000). Strains used for the TAF10 interaction study were: DKY106 [MATa ura3-Δ1 his3-Δ200,leu2, Δtrp1::(ERE)3-URA3 taf25::KANMX2 YEP90HIS-yTAF25] and DKY107 [MATa ura3-Δ1 his3-Δ200,leu2, Δtrp1::(ERE)3-URA3 taf25::KANMX2 YEP90HIS-yTAF25 + YEP10FL TRP-ΔN-yTAF25]. These strains were generated from the strain PL3 (2n) (Pierrat et al., 1992). The YEP90HIS-yTAF25 plasmid was generated by cloning the full-length yTAF10 cDNA into the XhoI–BamHI sites of the YEP90HIS vector (Pierrat et al., 1992). The YEP10FLTRP-ΔN-yTAF25 plasmid was constructed as described in Kirschner et al. (2002). Saccharomyces cerevisiae strains were propagated according to standard procedures on appropriate selective medium without histidine and/or tryptophan. Yeast cell transformation was carried out as described by Gietz et al. (1992).
Electron microscopy and image processing
The yTFIID fractions were diluted to 20 µg/ml in 20 mM Tris–HCl pH 7.4, 150 mM NaCl and 20% glycerol, and 5 µl of this preparation were placed on a 10 nm thick carbon film treated by a glow discharge in air. A mild glutaraldehyde fixation at a final concentration of 0.01% for 10 s was performed before adsorption to stabilize the complex. After 2 min adsorption, the grid was negatively stained with a 2% (w/v) uranyl acetate solution. The images were formed on a Philips CM120 Transmission Electron Microscope operating at 100 kV with a LaB6 cathode. Areas covered with individual molecules were recorded at a magnification of 45 000× on SO163 photographic plates or at 35 000× on a Pelletier cooled slow scan CCD camera (Model 794, Gatan, Pleasanton).
The micrographs were digitized at 18 µm raster size, resulting in a pixel spacing of 0.4 nm on the object, and processed using the IMAGIC software package (van Heel et al., 1996) (Image Science Software, Berlin, Germany). For the 3-D model of yTFIID, the 60°-tilted and the untilted molecular images, 128 × 128 pixels in size, were extracted from the original micrographs and analysed as described previously (Schultz et al., 2000). Briefly, the untilted TFIID images were aligned iteratively and analysed by multivariate statistical methods (van Heel and Frank, 1981). A preliminary 3-D model was calculated by weighted back-projection from a conical tilt series consisting of the tilted views of similarly oriented TFIID molecules (Radermacher et al., 1986). This model was refined by aligning the untilted data set against reprojections of the 3-D densities along 105 equally spaced directions. The newly aligned images were clustered into 100 classes and the angular assignment of each class average was performed using sinogram correlation functions against reprojections of the model (van Heel, 1988). The resolution of the final reconstruction was estimated from the FSC obtained by comparing two independent reconstructions generated by randomly splitting the data set in half. The resolution was determined according to two criteria: the 0.5 cut-off FSC curve (0.5 FSC criterion) and the intersection point of the 3σ curve with the FSC curve (3σ criterion) (van Heel, 1987).
Immunoelectron microscopy
Specific polyclonal rabbit antibodies recognizing yTFIID components were generated and affinity purified as described (Sanders et al., 1999; Sanders and Weil, 2000). To generate the anti-yTAF10 mouse mAb 38TA-2H2, the full-length recombinant yTAF10 was overexpressed in Escherichia coli, purified and used to immunize mice as described (Brou et al., 1993). The epitope recognized by this antibody was mapped between amino acids 149 and 180 in a non-conserved region of TAF10, putatively organized into a loop structure.
For immunoelectron microscopy, a 3- to 5- fold molar excess of antibodies was incubated for 30 min at 20°C with purified yTFIID at a final protein concentration of 20 µg/ml. The labelled complexes were identified by a stain-excluding domain protruding out of the yTFIID molecule whose size and shape were consistent with those of an IgG molecule. Images of such complexes were aligned against references obtained upon reprojecting the 3-D model of the unlabelled yTFIID along 25 equally spaced directions. The aligned images were partitioned into classes corresponding to characteristic views of the particles, and only the most abundant tri-lobed molecular views in which the discriminating features were recognized were analysed further for labelling specificity and location. In order to determine more precisely the antibody-binding site, a ring-shaped mask surrounding the outer shape of the particle was generated and the density variations, due to the binding of the antibody, were analysed using a multivariate statistical approach. The images were then partitioned according to this peripheral density variation. Generally, a class of unlabelled particles was detected as well as several classes of labelled particles differing in antibody orientation or with slight variations in the position of the binding site. A density difference image was calculated between the unlabelled and the labelled class averages in order to locate the tip of the antiboby when superimposed on the yTFIID particle.
Whole-cell extract (WCE) preparation and western blot analysis
Cultures were grown to midlog phase, harvested by centrifugation for 5 min at 3400 r.p.m., washed in cold phosphate-buffered saline (PBS), re-centrifuged and the pellets were frozen at –80°C and thawed prior to use. One volume of buffer B [40 mM Tris–HCl pH 7.8, 350 mM NaCl, 0.1% Tween, 10% glycerol, 1× protease inhibitor cocktail (PIC: 2.5 mg/ml leupeptin, pepstatin, chymostatin, antipain and aprotinin)] was added to the pellet and cells were broken by addition of acid-washed glass beads (0.5 mm diameter from Sigma). Tubes were vortexed vigorously six times for 20 s interspaced with 1 min cooling on ice. WCEs were clarified by centrifugation for 30 min at 4°C and the protein concentration was determined before storage of the protein extracts at –80°C. WCEs (15 µl) were boiled in SDS sample buffer, loaded on an 11% SDS– polyacrylamide gel and analysed by SDS–PAGE. After transfer to a nitrocellulose membrane, the blots were probed with subunit-specific antibodies and then revealed with peroxidase-conjugated goat anti-mouse secondary antibodies (Jackson Immuno Research). Chemiluminescence detection was performed according to the manufacturer’s instructions (Amersham).
Immunoprecipitation analysis
Immunoprecipitation was carried out using anti-Flag M2–agarose beads (Sigma) equilibrated in buffer B. WCEs (500 µg) were mixed with 60 µl of M2–beads each, and incubated for 12 h at 4°C by rotation. Beads were washed three times with 10 vols of buffer B, and twice with 10 vols of buffer C (40 mM Tris–HCl pH 7.9, 150 mM NaCl, 0.1% Tween, 10% glycerol, 1× CPI). Bound protein complexes were eluted with 50 µl of buffer C, containing 2.5 mg/ml of the Flag peptide for 2 h at 4°C. From these eluates, equal volumes (15 µl) were analysed by western blotting.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank M.Schatz (Image Science Software, Berlin, Germany) for helpful advice with the IMAGIC software and for customizing the package. This work was supported by the Institut National de la Santé et de la Recherche Médicale, the Centre National pour la Recherche Scientifique, the Hôpital Universitaire de Strasbourg (HUS), the Association pour la Recherche sur le Cancer, the NIH (grant GM 52461 to P.A.W.) and the Human Frontier Science Program (grant no. RG 196/98 to L.T and P.A.W., and grant no. RG 336/99 to P.S.).
References
- Albright S.R. and Tjian,R. (2000) TAFs revisited: more data reveal new twists and confirm old ideas. Gene, 242, 1–13. [DOI] [PubMed] [Google Scholar]
- Andel F., Ladurner,A.G., Inouye,C., Tjian,R. and Nogales,E. (1999) Three-dimensional structure of the human TFIID–IIA–IIB complex. Science, 286, 2153–2156. [DOI] [PubMed] [Google Scholar]
- Bell B. and Tora,L. (1999) Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res., 246, 11–19. [DOI] [PubMed] [Google Scholar]
- Birck C., Poch,O., Romier,C., Ruff,M., Mengus,G., Lavigne,A.C., Davidson,I. and Moras,D. (1998) Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionarily conserved motifs also found in the SPT3 family. Cell, 94, 239–249. [DOI] [PubMed] [Google Scholar]
- Brand M., Leurent,C., Mallouh,V., Tora,L. and Schultz,P. (1999a) Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science, 286, 2151–2153. [DOI] [PubMed] [Google Scholar]
- Brand M., Yamamoto,K., Staub,A. and Tora,L. (1999b) Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem., 274, 18285–18289. [DOI] [PubMed] [Google Scholar]
- Brou C., Chaudhary,S., Davidson,I., Lutz,Y., Wu,J., Egly,J.M., Tora,L. and Chambon,P. (1993) Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J., 12, 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff Y.G., Werten,S., Romier,C., Carre,L., Poch,O., Moras,D. and Davidson,I. (2000) The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol. Cell. Biol., 20, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff Y.G. et al. (2001a) The TFIID components human TAF(II)140 and Drosophila BIP2 (TAF(II)155) are novel metazoan homologues of yeast TAF(II)47 containing a histone fold and a PHD finger. Mol. Cell. Biol., 21, 5109–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff Y.G., Romier,C., Thuault,S., Werten,S. and Davidson,I. (2001b) The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci., 26, 250–257. [DOI] [PubMed] [Google Scholar]
- Gangloff Y.G., Sanders,S.L., Romier,C., Kirschner,D.B., Weil,P.A., Tora,L. and Davidson,I. (2001c) Histone folds mediate selective heterodimerisation of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 with SAGA component ySPT7. Mol. Cell. Biol., 21, 1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St. Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P.A., Schieltz,D., Pray-Grant,M.G., Steger,D.J., Reese,J.C., Yates,J.R. and Workman,J.L. (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell, 94, 45–53. [DOI] [PubMed] [Google Scholar]
- Hampsey M. and Reinberg,D. (1999) RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Genet. Dev., 9, 132–139. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Chiang,C.M., Oelgeschlager,T., Xie,X., Burley,S.K., Nakatani,Y. and Roeder,R.G. (1996) A histone octamer-like structure within TFIID. Nature, 380, 356–359. [DOI] [PubMed] [Google Scholar]
- Kirschner D.B., vom Bauer,E., Thibault,C., Sanders,S.L., Gangloff,Y.-G., Davidson,I., Weil,A.P. and Tora,L. (2002) Distinct mutations in yeast TAFII25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns. Mol. Cell. Biol., 22, 3178–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo T., Gong,D.W., Wootton,J.C., Horikoshi,M., Roeder,R.G. and Nakatani,Y. (1994) Molecular cloning of Drosophila TFIID subunits. Nature, 367, 484–487. [DOI] [PubMed] [Google Scholar]
- Michel B., Komarnitsky,P. and Buratowski,S. (1998) Histone-like TAFs are essential for transcription in vivo. Mol. Cell, 2, 663–673. [DOI] [PubMed] [Google Scholar]
- Mizzen C.A. et al. (1996) The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell, 87, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Nogales E. (2000) Recent structural insights into transcription preinitiation complexes. J. Cell Sci., 113, 4391–4397. [DOI] [PubMed] [Google Scholar]
- Oelgeschlager T., Chiang,C.M. and Roeder,R.G. (1996) Topology and reorganization of a human TFIID–promoter complex. Nature, 382, 735–738. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- Pierrat B., Heery,D.M., Lemoine,Y. and Losson,R. (1992) Functional analysis of the human estrogen receptor using a phenotypic transactivation assay in yeast. Gene, 119, 237–245. [DOI] [PubMed] [Google Scholar]
- Radermacher M., Wagenknecht,T., Verschoor,A. and Frank,J. (1986) A new 3-D reconstruction scheme applied to the 50S ribosomal subunit of E.coli. J. Microsc., 141, RP1–RP2. [DOI] [PubMed] [Google Scholar]
- Radermacher M., Wagenknecht,T., Verschoor,A. and Frank,J. (1987) Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc., 146, 113–136. [DOI] [PubMed] [Google Scholar]
- Reese J.C., Zhang,Z. and Kurpad,H. (2000) Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J. Biol. Chem., 275, 17391–17398. [DOI] [PubMed] [Google Scholar]
- Sanders S.L. and Weil,P.A. (2000) Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem., 275, 13895–13900. [DOI] [PubMed] [Google Scholar]
- Sanders S.L., Klebanow,E.R. and Weil,P.A. (1999) TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem., 274, 18847–18850. [DOI] [PubMed] [Google Scholar]
- Sanders S.L., Garbett,KA and Weil,P.A. (2002) Molecular characterization of Saccharomyces cerevisae TFIID. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer F. and Tjian,R. (1997) Mechanisms of transcriptional activation: differences and similarities between yeast, Drosophila and man. Curr. Opin. Genet. Dev., 7, 176–181. [DOI] [PubMed] [Google Scholar]
- Schultz P., Fribourg,S., Poterszman,A., Mallouh,V., Moras,D. and Egly,J.M. (2000) Molecular structure of human TFIIH. Cell, 102, 599–607. [DOI] [PubMed] [Google Scholar]
- Selleck W., Howley,R., Fang,Q., Podolny,V., Fried,M.G., Buratowski,S. and Tan,S. (2001) A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat. Struct. Biol., 8, 695–700. [DOI] [PubMed] [Google Scholar]
- Tora L. (2002) A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev., 16, 673–675. [DOI] [PubMed] [Google Scholar]
- van Heel M. (1987) Similarity measures between images. Ultramicroscopy, 21, 95–100. [Google Scholar]
- van Heel M. (1988) Angular reconstitution: a-posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy, 21, 111–124. [DOI] [PubMed] [Google Scholar]
- van Heel M. and Frank,J. (1981) Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy, 6, 187–194. [DOI] [PubMed] [Google Scholar]
- van Heel M., Harauz,G. and Orlova,E.V. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol., 116, 17–24. [DOI] [PubMed] [Google Scholar]
- Xie X., Kokubo,T., Cohen,S.L., Mirza,U.A., Hoffmann,A., Chait,B.T., Roeder,R.G., Nakatani,Y. and Burley,S.K. (1996) Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature, 380, 316–322. [DOI] [PubMed] [Google Scholar]