Abstract
Tn7 insertion into its specific target site, attTn7, is mediated by the proteins TnsA, TnsB, TnsC and TnsD. The double-strand breaks that separate Tn7 from the donor DNA require the Tns proteins, the transposon and an attTn7 target DNA, suggesting that a prerequisite for transposition is the formation of a nucleoprotein complex containing TnsABC+D, and these DNAs. Here, we identify a TnsABC+D transposon–attTn7 complex, and demonstrate that it is a transposition intermediate. We demonstrate that an interaction between TnsB, the transposase subunit that binds to the transposon ends, and TnsC, the target DNA-binding protein that controls the activity of the transposase, is essential for assembly of the TnsABC+D transposon–attTn7 complex. We also show that certain TnsB residues are required for recombination because they mediate a TnsB–TnsC interaction critical to formation of the TnsABC+D transpo son–attTn7 complex. We demonstrate that TnsA, the other transposase subunit, which also interacts with TnsC, greatly stabilizes the TnsABC+D transpo son–attTn7 complex. Thus multiple interactions between the transposase subunits, TnsA and TnsB, and the target-binding transposase activator, TnsC, control Tn7 transposition.
Keywords: DNA–protein complex/DNA transposition/enzyme activation/Tn7/transposase
Introduction
Diverse processes involving DNA, such as replication, transcription and recombination, are often dependent on the recognition of multiple DNA sites by DNA-binding proteins that through interactions with other proteins and/or with each other, assemble into higher-order nucleoprotein complexes (Echols, 1990; Grosschedl, 1995). In the recombination reaction transposition, a discrete DNA segment moves between non-homologous sites in a reaction that involves multiple DNA sites, and often multiple proteins. Here, we identify and characterize an elaborate higher-order nucleoprotein complex containing the bacterial transposon Tn7 and the target DNA. We demonstrate that this complex is a transposition intermediate that plays a central role in controlling Tn7 transposition.
A key event early in transposition is the pairing of the transposon ends, a reaction that identifies the DNA segment that forms the transposon, and permits coordinated DNA cleavage and joining reactions at the transposon termini (Haren et al., 1999). The Tn7 transposase is responsible for recognition and pairing of the transposon ends (R.Sarnovsky and N.Craig, in preparation), and similar transposase-mediated end pairing has been observed with bacteriophage Mu (Mizuuchi et al., 1992; Watson and Chaconas, 1996) and the Tn10 transposon (Sakai et al., 1995). However, beyond the end-pairing step, these transposition systems differ in the timing of the initial transposon end cleavage reaction, as well as the strategy for mediating formation of a transposon–target DNA complex (Haren et al., 1999).
A distinguishing feature of Tn7 transposition is the central role that the target DNA plays in regulating the Tn7 transposase (Peters and Craig, 2001b; Craig et al., 2002). It was observed previously that the Tn7 transposase does not initiate the cleavages at the transposon ends that separate the transposon from the flanking donor DNA unless several Tn7 target-binding proteins and a specific target DNA are also present (Bainton et al., 1993). We have hypothesized that the dependence of transposon end cleavage on target DNA reflects a requirement for the formation of a higher-order nucleoprotein complex containing the transposon, the target DNA and the Tns proteins (Bainton et al., 1993). Formation of this complex is proposed to activate the transposase to initiate the transposition process. In contrast, some other transposons, for example Tn10, interact with the target DNA at a later stage of the transposition process. Excision of Tn10 occurs independently of target recognition; the element excises from the donor site and subsequently recognizes and inserts into an appropriate target site (Sakai and Kleckner, 1997).
In the presence of the Tn7-encoded transposition proteins, TnsA, TnsB, TnsC and TnsD, Tn7 inserts into a specific site in the Escherichia coli chromosome called attTn7 (Bainton et al., 1993). The Tn7 target-selection protein TnsD binds to attTn7 and recruits TnsC (Bainton et al., 1993; Kuduvalli et al., 2001), an ATP-dependent DNA-binding protein (Gamas and Craig, 1992). When bound to TnsD–attTn7, TnsC becomes competent to activate the TnsAB transposase (Bainton et al., 1993; Stellwagen and Craig, 1997b; Lu and Craig, 2000). Thus, TnsC is central to the target-dependent regulation of Tn7 transposition: TnsC mediates activation of the TnsAB transposase, but is only able to do so when bound to TnsD–attTn7 DNA.
The Tn7 transposase is a heteromeric complex containing the TnsA and TnsB subunits. TnsB binds specifically to multiple sites within the left and right transposon ends (Arciszewska and Craig, 1991), and mediates end pairing (R.Sarnovsky and N.Craig, in preparation). Specific interaction of TnsA alone with the Tn7 ends has not been detected (Bainton et al., 1993), and we presume that it is recruited to the ends of Tn7 through interaction with TnsB. The TnsA and TnsB subunits catalyze distinct chemical reactions. The 3′ transposon ends are cut by TnsB, and the 5′ transposon ends are cut by TnsA (May and Craig, 1996; Sarnovsky et al., 1996). The resulting double-strand breaks (DSBs) at the transposon termini excise it from the donor site. TnsB then joins the exposed 3′ hydroxyl groups at the ends of the transposon to the target DNA, generating a simple insertion (Sarnovsky et al., 1996). Although TnsA and TnsB have distinct roles in the chemical steps of transposition, no transposon end cutting, or joining of the transposon end to the target DNA is detectable in the presence of either protein alone, and previous studies have suggested that the activities of TnsA and TnsB are interdependent (Sarnovsky et al., 1996; Biery et al., 2000b).
How do the Tn7 transposase subunits and the transposon ends interact with TnsC bound to TnsD–attTn7? After a transposon–attTn7 target DNA complex is formed, how does TnsC activate the TnsAB transposase? An interaction between TnsA and TnsC has been detected in solution (Stellwagen and Craig, 2001), and TnsA binding to a TnsC–TnsD–attTn7 complex has also been observed (Lu and Craig, 2000; Stellwagen and Craig, 2001). However, because TnsA does not exhibit any apparent intrinsic affinity for the transposon ends, TnsB, which specifically recognizes the Tn7 ends, must also be involved in promoting association of the transposon with the target DNA. Several scenarios can be imagined: TnsB may directly interact with TnsC–TnsD–attTn7, and/or TnsB may interact indirectly with TnsC–TnsD–attTn7 through TnsA.
Here, we have investigated interactions between the Tns proteins and substrate DNAs by using chemical cross-linking and gel electrophoresis. We demonstrate that TnsABC+D facilitate the assembly of a pre-recombination complex containing the Tn7 left (L) and right (R) ends, and attTn7 target DNA that we have named the (L,R)–Tgt complex. Moreover, we show that the TnsABC+D (L,R)–Tgt complex is relevant to transposition by demonstrating that it acts as an authentic reaction intermediate.
We also show that the TnsB and the TnsA transposase subunits each have critical roles in formation of the TnsABC+D (L,R)–Tgt complex. We have identified TnsB mutations that strongly decrease TnsABC+D transposition. Although these TnsB mutants can effectively recognize and pair the transposon ends, they are defective in their interaction with TnsC-bound target DNA, and hence are unable to form a transposon–target DNA complex. The finding that these transposition-defective mutants fail to form the TnsABC+D (L,R)–Tgt complex supports the view that this complex is critical to recombination. We also show TnsA greatly stabilizes the TnsABC+D (L,R)–Tgt complex, and suggest that this increased stability is key to activation of the TnsAB transposase.
Taken together, our findings suggest that an elaborate network of interactions between the TnsAB transposase, and the transposition regulator TnsC, are essential to the formation of a TnsABC+D nucleoprotein complex containing Tn7 and target DNA. Assembly of this higher-order nucleoprotein complex provides a mechanism for the control of the initiation of transposition by the presence of a particular target DNA.
Results
The Tns proteins promote the formation of complexes containing Tn7 and attTn7 target DNA
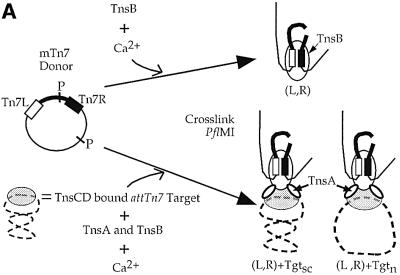
We assembled Tn7 higher-order nucleoprotein complexes by mixing the Tns proteins, a donor plasmid containing a miniTn7 element (mTn7) bounded by the left and right transposon ends and an attTn7 target DNA in buffer containing ATP and Ca2+ (Figure 1A). This method for complex assembly mimics the in vitro TnsABC+D transposition reaction, except Ca2+ is used in place of Mg2+ (Bainton et al., 1993). This divalent cation change was made because although transposition–integration systems including Tn7, Mu, Tn10 and HIV require Mg2+ for the transposon end cleavage and target joining reactions, Ca2+ can often promote the assembly of pre-recombination higher-order nucleoprotein complexes (Haren et al., 1999). We then added a protein cross-linker, glutaraldehyde, which is required to stabilize the complexes during their subsequent separation by gel electrophoresis. The reaction products were digested with a restriction enzyme (Figure 1A), subjected to gel electrophoresis, and visualized by Southern blotting using a probe that recognizes the mTn7 element (Figure 1B).
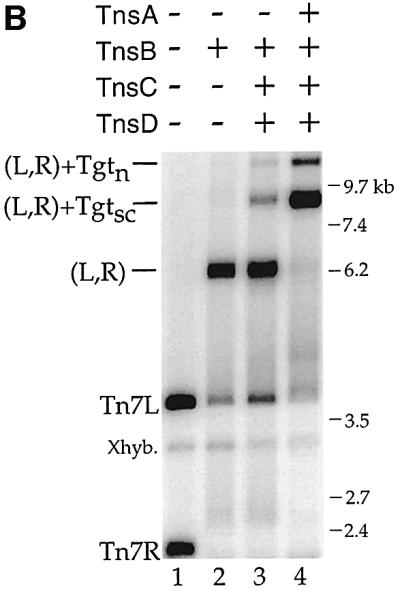
Fig. 1. (A) Schematic of Tn7 higher-order nucleoprotein complexes. (Left) The donor plasmid contains a mTn7 element (thick line) flanked by white and black rectangles representing the left (Tn7L) and right (Tn7R) transposon ends, and two PflMI restriction sites (P). (Top) Formation of a complex containing Tn7L and Tn7R (L,R) held together by TnsB represented as a white oval. (Bottom) TnsABC+D promote formation of complexes containing Tn7L, Tn7R and supercoiled (Tgtsc) or relaxed (Tgtn) target DNAs. The TnsC and TnsD proteins are depicted as a gray oval bound to the target DNA. (B) TnsB, TnsBC+D and TnsABC+D proteins mediate the formation of Tn7 nucleoprotein complexes. The mobilities of the DNAs, relative to the mobility of a DNA ladder (the ladder is not shown), are indicated on the right side. In lane 1, the mTn7-specific probe detects PflMI fragments containing Tn7L (3.7 kb) and Tn7R (2.2 kb). The cross-hybridization band (Xhyb) arises from binding of the mTn7-specific probe to the target DNA, which is present in excess to the donor DNA. Quantification of five independent cross-linking experiments indicated that TnsABC+D (L,R)–Tgtsc+n complex (lane 4) incorporated 4.9 ± 2.0-fold more of the total mTn7 donor DNA than the TnsBC+D (L,R)–Tgtsc+n complex (lane 3).
What combinations of Tns proteins form Tn7 nucleoprotein complexes? As described elsewhere (R.Sarnovsky and N.Craig, in preparation), TnsB alone can form a new species that migrates much more slowly than either of the individual L or R end-containing fragments (Figure 1B, lane 1 versus 2). This new band reflects the ability of TnsB to specifically bind to and pair the transposon ends, resulting in the formation of a (L,R) complex (Figure 1A).
In the presence of TnsABC+D, we observed two new bands with slower mobilities than the (L,R) paired ends complex (Figure 1B, lane 4). We found these new bands contain target DNA by evaluating a Southern blot of the cross-linked TnsABC+D reaction with a target-specific probe (Figure 2A, lane 9 and Supplementary figure 8, lane 11, available at The EMBO Journal Online). We suggest that the most abundant new band contains (L,R) and supercoiled target DNA, (L,R)–Tgtsc, and the other contains nicked target DNA, (L,R)–Tgtn (Figure 1A). In support of these designations, when both the donor and target plasmids were linearized with restriction enzymes, the two target containing bands were converted to a single species (data not shown). Thus, TnsABC+D, the proteins required for Tn7 insertion, promote the formation of a nucleoprotein complex that contains the DNA substrates involved in transposition.
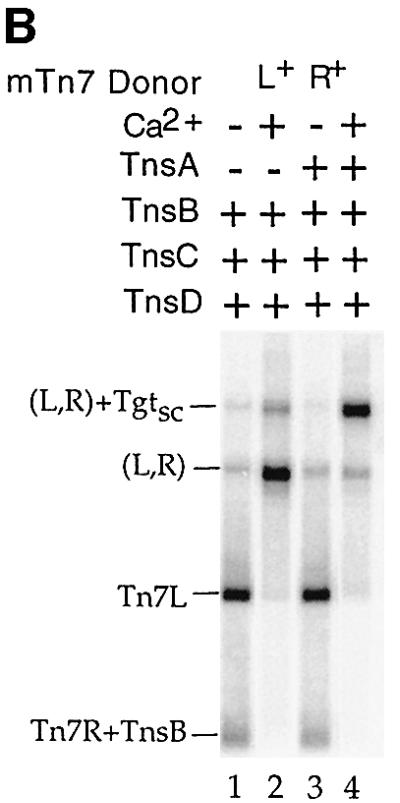
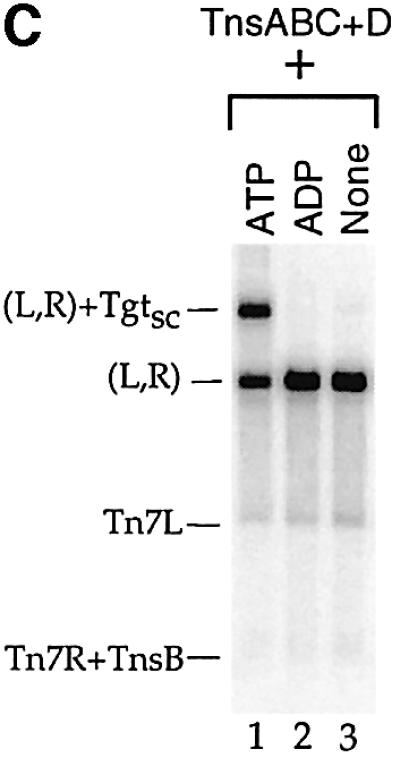
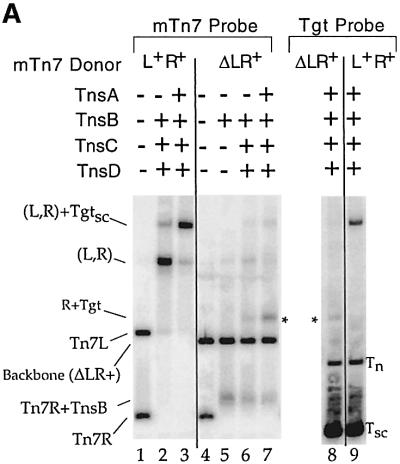
Fig. 2. (A) The TnsABC+D (L,R)–Tgt complex contains target DNA and requires a donor DNA with two Tn7 ends. Lanes 1–7 were probed with mTn7-specific DNA, and lanes 8 and 9 were probed with target-specific DNA. In lane 5, TnsB shifts the PflMI fragment containing Tn7R, labeled Tn7R+TnsB. In lane 7, just above the backbone fragment of the ΔLR+ plasmid, there is a new band labeled *. The mobility of this band is consistent with a complex containing a single Tn7R fragment and attTn7 target DNA. In lane 8, the target-specific probe hybridizes to the * band, as well as to the excess nicked (Tn) and supercoiled (Tsc) target DNAs. In lane 9, the target-specific probe recognizes the TnsABC+D (L,R)–Tgtsc complex. (B) Ca2+ stimulates formation of the TnsABC+D and TnsBC+D (L,R)–Tgtsc complexes. (C) ATP is required for TnsABC+D (L,R)–Tgtsc complex formation.
We also note that in the presence of TnsBC+D, a (L,R)–Tgtsc DNA species of the same mobility as that formed by TnsABC+D is observed (Figure 1B, lane 3). Hybridization with a target-specific probe confirmed that this complex contains target DNA (Supplementary figure 8, lane 12). Thus TnsB, which binds specifically to the ends of the transposon, can also interact with the TnsC–TnsD-bound target DNA in the absence of TnsA. Since the DNA substrates are likely the predominating determinants of complex mobility under these conditions, it is not surprising that the absence of TnsA has no affect on the mobility of this complex. The amount of the TnsBC+D complex is, however, much less than that formed in the presence of TnsABC+D.
DNA substrate and cofactor requirements for TnsABC+D (L,R)–Tgt complex formation
TnsABC+D transposition in vitro requires donor DNA containing a Tn7 element bounded by left and right ends (Gary et al., 1996), target DNA containing attTn7 and the cofactors ATP and Mg2+ (Bainton et al., 1993). Does formation of the TnsABC+D (L,R)–Tgt complex have the same substrate and cofactor requirements?
We examined the ability of a Tn7 element lacking the left end of Tn7 (ΔLR+) to promote complex formation. In contrast to the ability of the Tn7 with both L and R ends to form a donor DNA–target DNA complex (Figure 2A, lane 3), the ΔLR+ Tn7 element did not support significant TnsABC+D (L,R)–Tgt complex formation (Figure 2A, lane 7). Furthermore, no donor–target complexes were observed when ΔLR+ and ΔRL+ plasmids were mixed and incubated with the Tns proteins and target DNA (data not shown).
With regard to target DNA, we have found that DNA lacking attTn7 does not support formation of the TnsABC+D or the TnsBC+D (L,R)–Tgt complexes (data not shown), consistent with the previous finding that TnsC binds stably to DNA only in the presence of TnsD–attTn7 (Bainton et al., 1993).
Two cofactors are required for TnsABC+D in vitro transposition: Mg2+ and ATP (Bainton et al., 1993). The active sites of both TnsA and TnsB use Mg2+ to promote DNA-strand breakage and joining (Sarnovsky et al., 1996; Hickman et al., 2000). Either Mg2+ or Ca2+, when present at the same concentration as is used in the in vitro transposition reaction, greatly stimulate the formation of the (L,R) paired ends complex formed by TnsB (R.Sarnovsky and N.Craig, in preparation). Formation of the TnsABC+D (L,R)–Tgt and the TnsBC+D (L,R)–Tgt complexes were also greatly stimulated by the presence of 15 mM Ca2+ (Figure 2B, lanes 2 and 4).
Tn7 transposition also requires ATP because the ATP state of the transposition regulator TnsC determines its ability to stimulate the activity of the TnsAB transposase (Bainton et al., 1993; Stellwagen and Craig, 2001), and to bind to TnsD–attTn7 target DNA (Bainton et al., 1993). Formation of the TnsABC+D (L,R)–Tgt complex is also dependent on ATP (Figure 2C).
The dependence of the TnsABC+D and TnsBC+D (L,R)–Tgt complexes on the same DNA substrates and cofactors that are required for transposition supports the view that these complexes depend on the Tns interactions that underlie TnsABC+D transposition.
The TnsABC+D (L,R)–Tgt complex is competent for transposition in the presence of Mg2+
The DNA substrate and cofactor requirements for TnsABC+D (L,R)–Tgt complex formation suggest this complex is a transposition intermediate. To determine whether this Ca2+ assembled complex is competent for recombination in the presence of Mg2+, we puri fied an uncut (L,R)–Tgt complex from a TnsABC+D cross-linking reaction (see Materials and methods). We found that, without restriction digestion, agarose gel electrophoresis readily separated the cross-linked TnsABC+D (L,R)–Tgt complex from the faster mobility (L,R) paired ends complex and the unreacted DNA substrates. The agarose gel slice containing the (L,R)–Tgt complex was excised, incubated in the absence or presence of Mg2+, and the DNAs were isolated by electroelution, precipitated and then analyzed (Figure 3B). Incubation of the complex with Mg2+ resulted in formation of the Tn7 transposition product, the simple insert (SI) where the transposon has been excised from the donor DNA and joined to the target DNA (Figure 3B).
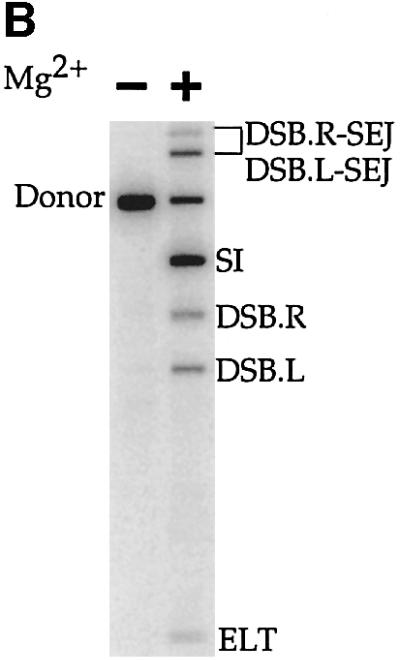
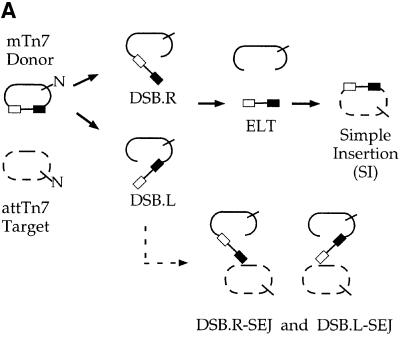
Fig. 3. The TnsABC+D (L,R)–Tgt complex is competent for transposition. (A) The products of TnsABC+D in vitro transposition. The donor plasmid (solid lines) contains a mTn7 element flanked by rectangles representing the transposon ends. The target plasmid (dashed lines) contains attTn7. Both plasmids contain an NdeI (N) restriction site, which is used to linearize the DNAs after transposition has occurred facilitating their separation by gel electrophoresis. A DSB at Tn7R or Tn7L results in DSB.R and DSB.L; an excised linear transposon (ELT) is the result of DSB at both ends; a simple insertion results from joining both transposon ends to the target DNA. DSB.R and DSB.L can be joined to the target DNA, resulting in single-end joining, i.e. DSB.R-SEJ and DSB.L-SEJ, respectively. (B) In situ transposition of the TnsABC+D (L,R)–Tgt complex in the presence of Mg2+. The products of the transposition reaction are labeled as described in (A).
We conclude that the TnsABC+D (L,R)–Tgt complex is a transposition intermediate that can promote the Tn7 cleavage and joining reactions when supplied with Mg2+. The ability of this complex to mediate recombination after cross-linking with glutaraldehyde is striking.
We also tested whether the cross-linked TnsBC+D (L,R)–Tgt complex could act as a transposition intermediate by adding Mg2+, or Mg2+ and TnsA, to an agarose gel slice containing the isolated complex. No recombination was observed in either case (data not shown). It may be that in the absence of TnsA, glutaraldehyde modification of the TnsBC+D proteins precludes interaction with TnsA, or that TnsA is unable to enter the gel slice in a productive way.
Mutations in the C-terminal domain of TnsB decrease TnsABC+D transposition
The ability of the TnsBC+D proteins to form a complex containing the transposon and the target DNAs reveals that TnsB can interact directly with the TnsC–TnsD–attTn7 target DNA complex, an interaction that has not been previously observed. We suspected that the TnsB C-terminus could be important for this interaction because of the properties of a recombination-defective TnsB mutant, TnsB amino acids (aa) 1–662, which lacks the 40 C-terminal amino acids present in wild-type TnsB aa1–702 (R.Sarnovsky and N.Craig, in preparation). While TnsB aa1–662 can recognize the ends of Tn7 and promote end pairing, TnsABC+D transposition with TnsB aa1–662 is profoundly defective (data not shown).
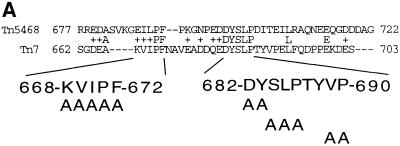
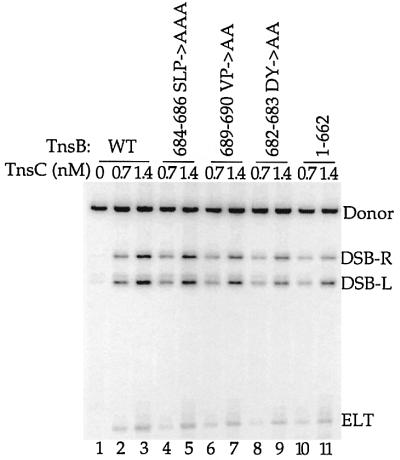
We hypothesized that the C-terminal 40 amino acids of TnsB might be required for the interaction of TnsB, and thus the transposon ends, with TnsC–TnsD–attTn7 DNA. Thus, we utilized an alignment of Tn7 TnsB with TnsB from Tn5468, a Tn7-like transposon isolated from Thiobacillus ferroxidans (Oppon et al., 1998), as a guideline for site-directed mutagenesis of the C-terminus of TnsB (Figure 4A). We found that alanine substitutions across two conserved regions, Tn7 TnsB aa682–683 DY→AA and aa684–686 SLP→AAA, dramatically reduced TnsABC+D transposition in vivo. Using a quantitative λ hop assay (see Materials and methods), we found that the transposition frequency in vivo promoted by these mutants was decreased by at least 6 × 104-fold from that of TnsABWTC+D. We also found that another alanine substitution mutant, TnsB aa689–690 VP→AA, substantially decreased TnsABC+D transposition in vivo by 6 × 103-fold. As examined by western blotting, these mutations had no effect on the TnsB half-life (data not shown).
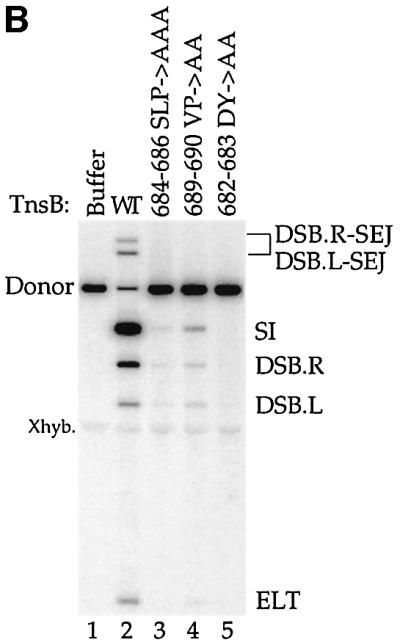
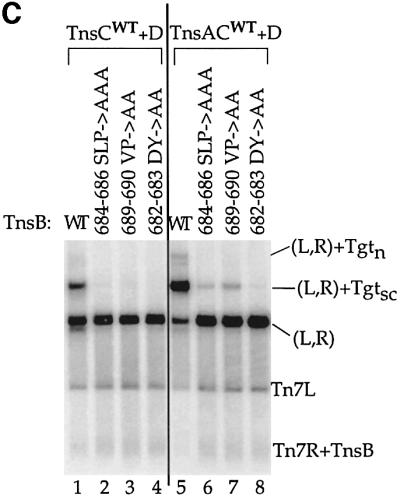
Fig. 4. Mutagenesis of the C-terminus of TnsB strongly decreases transposition and interaction with the target DNA. (A) Alignment of the amino acid sequences from the C-terminus of Tn7 TnsB with Tn5468 TnsB. (B) TnsB alanine mutants are defective for TnsABC+D transposition in vitro. The reaction products are labeled as outlined in Figure 3A. Total recombination (mTn7-specific signal – unreacted donor/mTn7-specific signal) was quantified and the TnsB aa689–690 VP→AA mutant promoted ≥2.5-fold more recombination than the other TnsB alanine mutants. (C) TnsB alanine mutants are defective for TnsABC+D and TnsBC+D (L,R)–Tgt complex formation.
In contrast to the defects in recombination observed with the above TnsB mutants, we found that alanine substitutions in another conserved region, TnsB aa668–672 KVIPF→AAAAA had no effect on TnsABC+D transposition in vivo.
To probe the properties of the transposition-defective alanine substitution mutants, we purified these proteins and analyzed their activities in vitro. Consistent with the profound transposition defects observed in vivo, TnsABC+D transposition in vitro with these mutants was sharply reduced (Figure 4B, lanes 2, 3, 4 and 5). As was also true in vivo, the residual level of transposition promoted by TnsB aa689–690 VP→AA was greater than the amount of recombination displayed by the TnsB aa682–683 DY→AA and aa684–686 SLP→AAA mutants (Figure 4B, compare lane 4 with lanes 3 and 5). Thus, the TnsB alanine mutants display the same defects in TnsABC+D transposition in vivo and in vitro.
The C-terminal domain of TnsB is essential for interaction with TnsC–TnsD–attTn7
To determine whether the transposition-defective TnsB alanine mutants are defective in nucleoprotein complex assembly, we examined their ability to pair the transposon ends and to form TnsABC+D (L,R)–Tgt complexes. The transposition-defective TnsB alanine mutants efficiently formed the (L,R) paired ends complex (Figure 4C, lanes 6, 7 and 8), revealing that these mutations do not affect TnsB-mediated end recognition and pairing. In striking contrast, the transposition-defective TnsB alanine mutants display drastically reduced formation of the TnsABC+D (L,R)–Tgt complex (Figure 4C, lanes 6, 7 and 8). Of the three transposition-defective mutants, TnsB aa689–690 VP→AA, which displays the highest level of recombination both in vivo and in vitro, displays the highest residual level of complex formation (Figure 4C, lane 7). This correlation between the amount of transposition and the amount of donor–target complex is consistent with the view that TnsABC+D (L,R)–Tgt complex formation plays an important role in TnsABC+D transposition.
We also found the transposition-defective TnsB alanine mutants did not form the TnsBC+D (L,R)–Tgt complex (Figure 4C, lanes 2, 3 and 4). Thus, formation of the TnsABC+D and TnsBC+D complexes depends upon the same residues within the C-terminus of TnsB.
The C-terminal domain of TnsB mediates interactions between TnsB and TnsC
In the experiments described above, TnsB interacted with TnsC+TnsD-bound attTn7 target DNA. Thus, it is unclear which of the target-binding proteins are required to form a complex between TnsB bound to the transposon ends and attTn7. It is known, however, that TnsC is the activator of the transposase because transposition can occur in the presence of only TnsABC when a TnsC gain-of-function mutant, TnsCA225V, is used (Stellwagen and Craig, 1997b). TnsCA225V activates the TnsAB transposase to promote simple insertions into target DNA with relatively little insertion site selectivity (Stellwagen and Craig, 1997b; Biery et al., 2000b). We found that the TnsB alanine mutants, which are defective for TnsABC+D transposition, are also defective for TnsABCA225V transposition (Figure 5A).
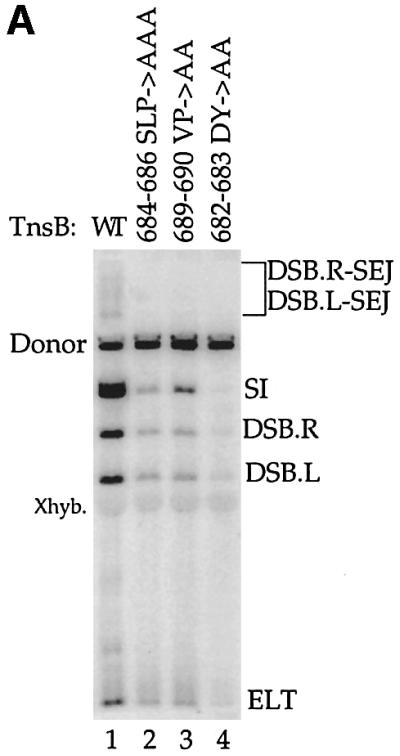
Fig. 5. TnsB and TnsCA225V can mediate formation of a (L,R)–Tgt complex, and this reaction depends on residues within the C-terminus of TnsB. (A) TnsB alanine mutants strongly decrease in vitro TnsABCA225V transposition. (B) TnsB alanine mutants are defective for interaction with TnsACA225V- and TnsCA225V-bound target DNA.
We also examined formation of transposon–target complexes in the presence of TnsCA225V. In the presence of TnsABWTCA225V, a transposon containing complex with mobility slower than the (L,R) paired ends complex was observed (Figure 5B, lane 5). We have shown that the migration of this complex and the TnsABC+D (L,R)–Tgt complex are the same (Supplementary figure 8, lanes 4 and 5), and we also demonstrate that this complex contains target DNA by hybridization with a target-specific probe (Supplementary figure 8, lane 10). Thus, TnsCA225V is able to bind to target DNA in the absence of TnsD and interact with the TnsAB transposase to form a (L,R)–Tgt complex.
TnsB and TnsCA225V can also promote formation of a (L,R)–Tgt complex (Figure 5B, lane 1 and Supplementary figure 8, lanes 3 and 9). Therefore, an interaction between TnsB and TnsCA225V, in the absence of both TnsA and TnsD, is sufficient to mediate formation of a complex containing the transposon ends and the target DNA. However, we note that the amount of (L,R)–Tgt complex formed in the presence of TnsABCA225V is much greater than the amount of complex formed in the presence of TnsBCA225V. Thus, as was observed with TnsBC+D, the presence of TnsA increases the amount of (L,R)–Tgt complex.
The transposition-defective TnsB alanine mutants are also defective in formation of TnsBCA225V and TnsABCA225V (L,R)–Tgt complexes (Figure 5B, lanes 2–4 and 6–8). An attractive idea is that the mutated residues within the C–terminus of TnsB make molecular contacts with TnsC.
One other possible interpretation is that the TnsB mutations do not affect TnsB–TnsC interaction; rather they disrupt a direct interaction between TnsB and the target DNA, i.e. an interaction that is TnsC independent. We have tested this model by utilizing relaxed in vitro conditions, i.e. high glycerol and Mn2+, to promote TnsAB-only catalyzed transposon-end breakage and intramolecular joining (Biery et al., 2000a). We have found that using these relaxed conditions, the TnsB mutants, in collaboration with TnsA, are able to mediate efficient end cleavage and intramolecular joining. Thus, the TnsB mutants are not defective for interaction with an intramolecular DNA target (Supplementary figure 9, even lanes).
Taken together, these experiments show that interaction between TnsB and TnsC mediates the formation of (L,R)–Tgt complexes in which the transposon ends are bound by TnsB and the target DNA is bound by TnsC. Furthermore, TnsA appears to assist in the formation of the (L,R)–Tgt complex.
TnsA increases the stability of a (L,R)–Tgt complex
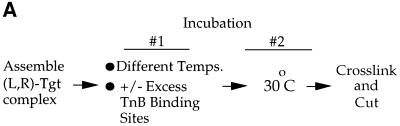
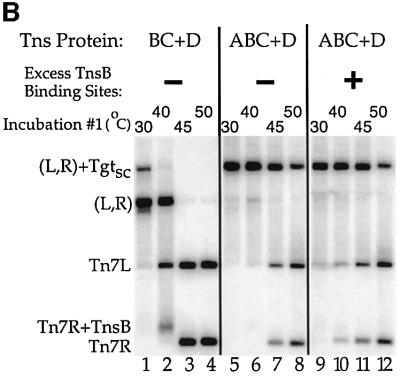
As described above, TnsBC+D can promote formation of the (L,R)–Tgt complex. However, transposition requires TnsA as well as TnsBC+D. To better understand the effect of TnsA on (L,R)–Tgt complex assembly, we examined the stability of the TnsBC+D and TnsABC+D nucleoprotein complexes by applying a temperature challenge before cross-linking (Figure 6A). As a control, a large excess of TnsB-binding site competitor DNA was added to some of these reactions such that if the (L,R)–Tgt complex were to disassociate at the elevated temperatures, the competitor DNA would block reassociation of TnsB with the Tn7 ends and thus would also block reformation of the (L,R)–Tgt complex.
Fig. 6. The TnsABC+D (L,R)–Tgt complex is more stable than the TnsBC+D (L,R)–Tgt complex. (A) Design of the thermal challenge experiment. (L,R)–Tgt complexes were assembled with the TnsBC+D and TnsABC+D proteins at 30°C, then challenged for 10 min at various temperatures as indicated below. In some reactions, a 120-fold molar excess of TnsB-binding site competitor DNA (see Materials and methods) was added at the beginning of the temperature challenge. The reactions were placed at 30°C for 10 min before cross-linking and subsequent analysis. (B) The presence of TnsA stabilizes the TnsABC+D (L,R)–Tgt complex to temperature challenge.
We found that 90% of the TnsABC+D (L,R)–Tgt complex was still present after incubation at 40°C (Figure 6B, lane 5 versus 6). Moreover, little reduction in the amount of (L,R)–Tgt complex was observed even in the presence of excess TnsB-binding sites, indicating that TnsABC+D (L,R)–Tgt reassociation does not mask thermal instability (Figure 6B, lanes 5–8 versus 9–12). By contrast, the TnsBC+D (L,R)–Tgt complex was completely disassociated by the 40°C challenge (Figure 6B, lane 1 versus 2). Thus, TnsA plays a key role in the stability of the TnsABC+D (L,R)–Tgt complex.
TnsC can also activate the TnsAB transposase via an interaction that does not depend on the C-terminal domain of TnsB
As demonstrated above, formation of a critical transposition intermediate, the TnsABC+D (L,R)–Tgt complex, depends upon the interaction of the C-terminal domain of TnsB with TnsC. Can TnsC activation of the TnsAB transposase occur in the absence of the C-terminal TnsB–TnsC interaction?
Under our standard in vitro conditions for TnsABC+D recombination, no transposon end cleavage is observed without TnsD and attTn7. However, by modifying the reaction conditions, i.e. omitting TnsD, target DNA and ATP, and increasing the glycerol concentration, TnsC will activate TnsAB-catalyzed cleavage of the transposon ends (Figure 7, lanes 1–3) (Stellwagen and Craig, 2001). Using these relaxed in vitro conditions, we tested the ability of TnsC to activate TnsA and TnsB1–662, or TnsA and the TnsB alanine mutants to catalyze cleavage of the transposon ends (Figure 7, lanes 4–11). Interestingly, TnsC activates the transposon end cleavage activity of the TnsAB transposase without regard for the changes to the C-terminus of TnsB. Moreover, in a related assay that also utilizes relaxed in vitro reaction conditions, i.e. Mn2+ and glycerol, to promote activation of the TnsAB transposase in the absence of the other Tns proteins (Biery et al., 2000a), we found that TnsC also stimulated recombination without regard for the changes to the C-terminus of TnsB (Supplementary figure 9, lanes 3, 5, 7 and 9).
Fig. 7. Mutation of the C-terminus of TnsB does not significantly affect TnsC-stimulated TnsAB cleavage. The reaction products are labeled as outlined in Figure 3A.
These findings are notable because, as we showed above, under our ‘standard’ in vitro conditions the C-terminal residues of TnsB are required for the TnsB–TnsC interaction that underlies (L,R)–Tgt complex formation. One explanation for this TnsC-dependent activation of recombination is that the alternative conditions promote interactions between TnsA and TnsC and/or between TnsA and TnsB that occur under our standard conditions only when the C-terminal TnsB–TnsC interaction is also present. Alternatively, the relaxed in vitro conditions may promote a TnsB–TnsC interaction other than the C-terminal one described above.
Discussion
TnsABC+D promote the formation of a transposition intermediate containing the Tn7 ends and attTn7 target DNA
Previous observations revealed that the initiation of Tn7 insertion into attTn7 requires TnsABC+D, a transposon with both ends, the cofactors Mg2+ and ATP, and attTn7 target DNA (Bainton et al., 1993). Thus, it was proposed that the initial cleavage events at the Tn7 ends are dependent upon the TnsABC+D-mediated formation of a nucleoprotein complex containing the transposon ends and the attTn7 target DNA. (Bainton et al., 1993; May and Craig, 1996; Sarnovsky et al., 1996). Here, we have shown that the TnsABC+D proteins do indeed mediate formation of a nucleoprotein complex that depends upon the same DNA substrates as the TnsABC+D transposition reaction. Formation of this complex requires Ca2+, a condition under which recombination does not occur. However, when isolated transposon–attTn7 target complex is incubated with Mg2+, transposition does occur, yielding recombinant DNA species that are the same as the products of a standard TnsABC+D transposition reaction. Thus, the TnsABC+D (L,R)–Tgt complex is an authentic transposition intermediate.
Previous studies suggest a role for each Tns protein in formation of the transposon–attTn7 target complex. The TnsA and TnsB proteins collaborate to form the transposase, which executes the chemistry of transposition (Sarnovsky et al., 1996). The attTn7 target DNA is bound by TnsD, which then recruits TnsC, the protein that mediates activation of the TnsAB transposase (Stellwagen and Craig, 1997b). Footprinting studies of TnsCD bound to attTn7 suggest that TnsC forms an extended ‘platform’ on the target DNA around the point of Tn7 insertion (Kuduvalli et al., 2001). Thus an attractive model is that the target DNA and the transposon DNA are brought into close proximity through interactions that occur between the TnsA, TnsB and TnsC proteins.
A TnsB–TnsC interaction is necessary for association of the ends of Tn7 with attTn7 target DNA
Which Tns protein–protein interactions underlie formation of the transposon–attTn7 target complex? We used a TnsC mutant, TnsCA225V, which activates transposition in the absence of TnsD, to show that the TnsAB transposase bound to the ends of Tn7 can form a complex with TnsCA225V bound to a target DNA. Moreover, a (L,R)–Tgt complex is also formed in the presence of only TnsB, the subunit of the transposase that binds specifically to the ends of Tn7, and TnsCA225V. Thus, an interaction between TnsB and TnsC is sufficient to form a nucleoprotein complex containing the DNA substrates for Tn7 transposition.
Does a TnsB–TnsC interaction also underlie formation of the TnsABC+D (L,R)–Tgt complex? We suspected that the C-terminus of TnsB was important for mediating a TnsB–TnsC interaction because a 40 amino acid C-terminal deletion, TnsB aa1–662, has been shown to be defective for TnsABC+D transposition, but not for transposon end pairing (R.Sarnovsky and N.Craig, in preparation). Thus, we mutated residues within this region to alanine, and found that some substitutions dramatically decreased TnsABC+D and TnsABCA225V transposition. As was seen with TnsB aa1–662, these TnsB C-terminal alanine mutations did not affect the ability of TnsB to pair the transposon ends. However, TnsABC+D (L,R)–Tgt complex formation was dramatically reduced with the transposition-defective TnsB alanine substitution mutants. These TnsB mutations adversely affected formation of all (L,R)–Tgt complexes, whether the target DNA was bound by TnsC–TnsD or TnsCA225V, and regardless of the presence or absence of TnsA.
Thus, interaction of TnsB with TnsC requires residues within the C-terminus of TnsB, and this interaction is necessary for association of the Tn7 ends with attTn7 DNA. Moreover, there is a strong correlation between the ability of the TnsB mutants to execute TnsABC+D transposition, and their ability to form the TnsABC+D (L,R)–Tgt complex, supporting the importance of (L,R)–Tgt complex formation in Tn7 transposition.
TnsA stabilizes the TnsABC+D (L,R)–Tgt complex
As discussed above, a TnsB–TnsC interaction is critical to TnsABC+D transposition because it is required for TnsABC+D (L,R)–Tgt complex formation. Interactions between TnsA and TnsC are also important for transposition. A previous study showed that mutations within a specific region of TnsA abolish TnsABC+D transposition (Lu and Craig, 2000). Similar to the TnsB alanine mutants, these TnsA mutants are blocked for interaction with TnsC bound to TnsD–attTn7. Thus, both subunits of the Tn7 transposase must apparently interact with TnsC for efficient TnsABC+D transposition.
A TnsB–TnsC interaction is sufficient to mediate association of the transposon and target DNA, but when TnsA is also part of this complex it becomes a transposition intermediate. What effect does TnsA have on the (L,R)–Tgt complex? We have shown here that (L,R)–Tgt complexes formed in the presence of TnsA are much more stable than TnsBC+D or TnsBCA225V (L,R)–Tgt complexes. We speculate that the TnsA–TnsC interaction described above is required for this effect of TnsA on (L,R)–Tgt complex stability.
Thus, our studies have revealed that both transposase subunits, TnsA and TnsB, interact with the transposase regulator TnsC, and each subunit plays a different critical role in the formation of a transposition-competent (L,R)–Tgt complex.
Evidence for a TnsA–TnsB interaction suggests an interaction network between TnsA, TnsB and TnsC
The work described above has revealed that both transposase subunits interact with TnsC, which in turn interacts with the target DNA. Other observations suggest TnsA–TnsB interactions are also important to transposition. For example, wild-type TnsAB alone can promote Tn7 end cleavage and intramolecular joining in the presence of Mn2+ and a high concentration of glycerol (Biery et al., 2000a). In addition, certain mutations in TnsA and TnsB allow recombination in the absence of the other Tns proteins (Lu and Craig, 2000).
We have shown that when the in vitro reaction conditions are relaxed by the addition of high glycerol and Mg2+, or high glycerol and Mn2+, the TnsAB transposase can be activated by TnsC to promote recombination, even when the TnsB C-terminal alanine mutants are used. An attractive hypothesis to account for the activation of these TnsB mutants is that, under these particular conditions, a TnsA–TnsB interaction recruits TnsA to the transposon ends, and a TnsA–TnsC interaction activates the TnsAB transposase. Alternatively, these relaxed conditions may promote an interaction between TnsB and TnsC that is independent of the C-terminal residues of TnsB.
Thus, multiple interactions between the TnsABC proteins, TnsA–TnsC, TnsB–TnsC and TnsA–TnsB, form an interaction network that is built around the Tn7 ends and the target DNA.
Target DNA control of Tn7 transposition
The work presented here has provided molecular insight into how attTn7 target DNA controls the initiation of transposition. The nature of the target DNA also plays an important role in two additional aspects of Tn7 transposition. Transposition can occur by another pathway that utilizes TnsABC+E. TnsE is an alternative activator of the TnsABC transposition machinery, which ignores attTn7, and instead directs Tn7 insertion preferentially into DNA that is undergoing the lagging strand DNA synthesis that occurs in recipient cells during conjugation (Wolkow et al., 1996; Peters and Craig, 2001a). An attractive hypothesis is that TnsABC+E, by analogy to TnsABC+D, mediates the formation of a nucleoprotein complex containing the transposon and a structure found on lagging strand DNA.
Tn7 transposition is directed towards particular target DNAs in the presence of TnsD or TnsE. However, Tn7 displays target or transposition immunity, i.e. Tn7 insertion is decreased into DNAs that already contain Tn7 (Craig et al., 2002). As is true in directing Tn7 towards certain DNAs such as attTn7, the interaction of TnsC with a prospective target DNA also plays a central role in target immunity. Immune DNAs are poor targets for Tn7 insertion in vitro because TnsC is actively displaced from immune DNA (Stellwagen and Craig, 1997a). This displacement then precludes the interaction of TnsAB and the transposon ends with the immune target.
Thus, Tn7 is attracted to particular target DNA due to the formation of a nucleoprotein complex containing the ends of the transposon and that target DNA, as in the case of TnsABC+D-mediated insertion into attTn7. Alternatively, Tn7 will ignore DNA that already contains a copy of Tn7 because the displacement of TnsC prevents the assembly of a complex containing the ends of the transposon and the immune target.
Comparison of Tn7 and bacteriophage Mu
It is of interest to compare Tn7 transposition with transposition of bacteriophage Mu (Craig et al., 2002). Mu transposition is mediated by the MuA transposase, which resembles TnsB in that it binds to the ends of the transposon and mediates DNA breakage and joining at the 3′ ends of the transposon, and MuB, an ATP-utilizing protein that, like TnsC, interacts with the target DNA and activates the transposase. A significant difference between the Mu and Tn7 systems is that the Tn7 transposase contains two essential subunits, TnsA that mediates breakage at the 5′ ends of the transposon, and TnsB that cleaves the transposon 3′ ends (Sarnovsky et al., 1996). Additionally, unlike MuB, TnsC is unable to interact with a target DNA and activate transposition in the absence of another protein, TnsD and its cognate target site, attTn7 (Bainton et al., 1993).
We have established here that a critical interaction in Tn7 transposition occurs between the transposase subunit TnsB, and the ATP-utilizing target-binding protein TnsC. Previous work has established that interaction between TnsA and TnsC is also important to transposition (Lu and Craig, 2000). Thus, in contrast to a single type of transposase activator interaction, as occurs with Mu, two distinct Tn7 transposase subunits must interact with the Tn7 transposase activator TnsC. The activation of both Tn7 transposase subunits is important because each one mediates one of the strand scissions necessary for excision of Tn7 from the donor DNA. By contrast, in the Mu replicative transposition reaction, MuA promotes only 3′ end nicks so that coordination between two cleavage reactions at each transposon end is not necessary. Interactions between TnsA and TnsB likely add an additional layer of complexity to the control of Tn7 transposase activity.
There are also significant differences in Tn7 and Mu transposase activation with regard to the necessity for an appropriate target DNA bound by a transposase activator, TnsC or MuB, respectively. The Mu system is flexible; MuA-catalyzed end breakage and joining can occur even in the absence of MuB (Craigie and Mizuuchi, 1987; Surette et al., 1987; Mizuuchi et al., 1992). However, pre-recombination complexes containing the MuA-bound transposon ends and MuB-bound target DNA have been observed (Naigamwalla and Chaconas, 1997), and Mu transposition is most efficient when these complexes are formed (Baker et al., 1991; Mizuuchi et al., 1992; Naigamwalla and Chaconas, 1997).
In contrast, as this work has shown, Tn7 transposition is strictly dependent on the capture of the target DNA by TnsC and apposition of the transposon ends and target DNA prior to cleavage at the transposon ends. Moreover, although the transposon ends bound by TnsB can form a complex with TnsC and the target DNA, another protein, TnsA, is still required for transposition.
Thus, while the Mu and Tn7 systems share some features, i.e. a protein that binds specifically to the transposon ends and an ATP-utilizing protein involved in target selection, the Tn7 transposase is subject to more elaborate regulation. This rigid control of transposase activity arises from the use of a target-selecting protein to recruit TnsC to particular DNA sites, and the need for coordination between two transposase subunits.
Further dissection of the protein–protein and protein–DNA interactions that mediate each of these systems will continue to provide important insights into the assembly and regulation of nucleoprotein complexes that will be relevant to other complex nucleic acid–protein transactions.
Materials and methods
Plasmids
The L+R+ donor plasmid pEMΔ contains a 1.6 kb miniTn7 element (Bainton et al., 1993), and the target plasmid pRM2 contains attTn7 (–342 to +165) (McKown et al., 1988). The ΔLR+ plasmid, pZS833, is a pEMΔ derivative constructed by deletion of Tn7L (166 bp) and 50 bp of flanking DNA.
pZS564 was constructed by insertion of the BglII–NciI fragment of Tn7 encoding TnsA and TnsB into the NdeI and EcoRI sites of the pSC101 derivative pTA106 (Peters and Craig, 2001a). The cloned fragment includes the TnsAB promoter as well as 2.5 of the four Tn7R binding sites (Waddell and Craig, 1988). The TnsB alanine derivatives of pZS564, pZS793 (684–686 SLP→AAA), pZS794 (682–683 DY→AA), and pZS795 (689–690 VP→AA), were constructed by PCR-based site-directed mutagenesis. pZS568 was constructed by ligation of the Tn7 HpaI fragment encoding the TnsC and TnsD genes to the XmnI–AvaI fragment of pACYC184 in the same orientation as the tetracyclin resistance gene.
pZS785 (684–686 SLP→AAA), pZS786 (682–683 DY→AA), and pZS787 (689–690 VP→AA), were constructed by PCR-based site-directed mutagenesis of pRS513 (R.Sarnovsky and N.Craig, in preparation), a pCYB1 (New England Biolabs) based vector, for expression of a TnsB–intein–chitin-binding domain (intein–CBD) fusion protein.
The TnsB binding site competitor DNA, used in Figure 6B, is pZS832, which contains the Tn7 ends and flanking DNA from pEMΔ, but is not recognized by the mTn7 probe used in this experiment (R.Sarnovsky and N.Craig, in preparation).
Tns proteins
TnsA, TnsC and His-tagged TnsD were purified as described (Gamas and Craig, 1992; May and Craig, 1996; Sharpe and Craig, 1998). The TnsB proteins were expressed from pRS513, pZS785, pZS786 and pZS787, and purified to ≥95% homogeneity as intein–CBD fusion proteins, as described elsewhere (R.Sarnovsky and N.Craig, in preparation). After purification on a chitin column, TnsB was separated from the intein–CBD tag by intein-mediated self-cleavage (New England Biolabs).
Transposition in vitro
The standard TnsABC+D transposition reaction (100 µl) contains 3.3 nM target DNA, 0.21 nM donor DNA, 25.7 mM HEPES (pH 7.5), 1.95 mM Tris (pH 7.5), 2.1 mM dithiothreitol (DTT), 2.01 mM ATP, 19.25 mM NaCl, 16.5 mM KCl, 0.12 mM CHAPS, 1.2% glycerol, 10 µg tRNA, 909 nM bovine serum albumin, 0.062 mM EDTA, 0.12 mM MgCl and 15 mM MgAc. The following Tns proteins were added: 12 nM TnsA, 6.2 nM TnsB, 4.8 nM TnsC and 3.7 nM TnsD. In the first stage of the reaction, TnsC, TnsD and the target DNA were incubated for 25 min at 30°C. Next, mTn7 donor DNA, TnsA, TnsB and MgAc were added. The TnsC and TnsD proteins are pre-assembled on attTn7 because Mg2+ strongly inhibits binding of these proteins to attTn7 (Bainton et al., 1993). The reaction was incubated at 30°C for 20 min, stopped with phenol/chloroform, the DNA precipitated, then digested with NdeI and analyzed by 0.6% agarose gel electrophoresis in 0.5× TBE buffer. The DNAs were Southern transferred to a GeneScreen Plus membrane (NEN Research Products), and the blot was hybridized with a mTn7-specific probe (Sarnovsky et al., 1996). Following phosphorimaging, the reaction products were quantified using ImageQuant software (Molecular Dynamics).
Nucleoprotein complex formation
Nucleoprotein complex formation mimics the TnsABC+D in vitro transposition reaction with a few changes. TnsA, TnsB, donor DNA and 15 mM CaAc were added to preassembled TnsCD-bound target DNA unless otherwise noted. The mixture was incubated at 30°C for 10 min, followed by a 10 min incubation with glutaraldehyde (Sigma) at 0.004%. The cross-linker was quenched by adding Tris pH 8.0 to 25 mM and lysine to 3 mM for 10 min at 22°C. Forty-eight units of PflMI (New England Biolabs) were added to the reaction as well as NaCl to 75 mM, Tris to 28.5 mM, MgCl2 to 0.7 mM, and DTT to 2.17 mM. After 1 h digestion at 22°C, 0.5× reaction was separated by 0.6% agarose gel electrophoresis in 1× TAE buffer, and visualized by blotting as described above. Uncut TnsABC+D complexes were gel purified using the same conditions.
Transposition in vivo
The λ hop transposition assay was used to determine transposition frequency in vivo as described elsewhere (McKown et al., 1988). TnsA and TnsB wild-type, or the various TnsB alanine mutants examined were expressed in trans from pZS564 or a pZS564 derivative, along with TnsCD from pZS568.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr Prasad Kuduvalli and other members of the Craig laboratory for advice and comments on the manuscript. This work was supported by grant P01 CA16519-28 (N.L.C) from the National Institutes of Health. N.L.C. is an investigator with the Howard Hughes Medical Institute.
References
- Arciszewska L.K. and Craig,N.L. (1991) Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic Acids Res., 19, 5021–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainton R.J., Kubo,K.M., Feng,J.-N. and Craig,N.L. (1993) Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell, 72, 931–943. [DOI] [PubMed] [Google Scholar]
- Baker T.A., Mizuuchi,M. and Mizuuchi,K. (1991) MuB protein allosterically activates strand transfer by the transposase of phage Mu. Cell, 65, 1003–1013. [DOI] [PubMed] [Google Scholar]
- Biery M.C., Lopata,M. and Craig,N.L. (2000a) A minimal system for Tn7 transposition: the transposon-encoded proteins TnsA and TnsB can execute DNA breakage and joining reactions that generate circularized Tn7 species. J. Mol. Biol., 297, 25–37. [DOI] [PubMed] [Google Scholar]
- Biery M.C., Steward,F., Stellwagen,A.E., Raleigh,E.A. and Craig,N.L. (2000b) A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res., 28, 1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N., Craigie,R., Gellert,M. and Lambowitz,A. (2002) Tn7. In Mobile DNA II. ASM Press, Washington, DC, pp. 423–456.
- Craigie R. and Mizuuchi,K. (1987) Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell, 51, 493–501. [DOI] [PubMed] [Google Scholar]
- Echols H. (1990) Nucleoprotein structures initiating DNA replication, transcription and site-specific recombination. J. Biol. Chem., 265, 14697–14700. [PubMed] [Google Scholar]
- Gamas P. and Craig,N.L. (1992) Purification and characterization of TnsC, a Tn7 transposition protein that binds ATP and DNA. Nucleic Acids Res., 20, 2525–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary P.A., Biery,M.C., Bainton,R.J. and Craig,N.L. (1996) Multiple DNA processing reactions underlie Tn7 transposition. J. Mol. Biol., 257, 301–316. [DOI] [PubMed] [Google Scholar]
- Grosschedl R. (1995) Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr. Opin. Cell Biol., 7, 362–370. [DOI] [PubMed] [Google Scholar]
- Haren L., Ton-Hoang,B. and Chandler,M. (1999) Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol., 53, 245–281. [DOI] [PubMed] [Google Scholar]
- Hickman A.B., Li,L., Mathew,S.V., May,E.W., Craig,N.L. and Dyda,F. (2000) Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell, 5, 1025–1034. [DOI] [PubMed] [Google Scholar]
- Kuduvalli P., Rao,J. and Craig,N.L. (2001) Target DNA structure plays a critical role in Tn7 transposition. EMBO J., 20, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F. and Craig,N.L. (2000) Isolation and characterization of Tn7 transposase gain-of-function mutants: a model for transposase activation. EMBO J., 19, 3446–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May E.W. and Craig,N.L. (1996) Switching from cut-and-paste to replicative Tn7 transposition. Science, 272, 401–404. [DOI] [PubMed] [Google Scholar]
- McKown R.L., Orle,K.A., Chen,T. and Craig,N.L. (1988) Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J. Bacteriol., 170, 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi M., Baker,T.A. and Mizuuchi,K. (1992) Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell, 70, 303–311. [DOI] [PubMed] [Google Scholar]
- Naigamwalla D.Z. and Chaconas,G. (1997) A new set of Mu DNA transposition intermediates: alternate pathways of target capture preceding strand transfer. EMBO J., 16, 5227–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppon J.C., Sarnovsky,R.J., Craig,N.L. and Rawlings,D.E. (1998) A Tn7-like transposon is present in the glmUS region of the obligately hemoautolithotrophic bacterium Thiobacillus ferrooxidans. J. Bacteriol., 180, 3007–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.E. and Craig,N.L. (2001a) Tn7 recognizes target structures associated with DNA replication using the DNA binding protein TnsE. Genes Dev., 15, 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.E. and Craig,N.L. (2001b) Tn7: smarter than we thought. Nat. Rev., 2, 806–814. [DOI] [PubMed] [Google Scholar]
- Sakai J. and Kleckner,N. (1997) The Tn10 Synaptic complex can capture a target DNA only after transposon excision. Cell, 89, 205–214. [DOI] [PubMed] [Google Scholar]
- Sakai J., Chalmers,R.M. and Kleckner,N. (1995) Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J., 14, 4374–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnovsky R., May,E.W. and Craig,N.L. (1996) The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J., 15, 6348–6361. [PMC free article] [PubMed] [Google Scholar]
- Sharpe P. and Craig,N.L. (1998) Host proteins can stimulate Tn7 transposition: a novel role for the ribosomal protein L29 and the acyl carrier protein. EMBO J., 17, 5822–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A. and Craig,N.L. (1997a) Avoiding self: two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J., 16, 6823–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A. and Craig,N.L. (1997b) Gain-of-function mutations in TnsC, an ATP-dependent transposition protein which activates the bacterial transposon Tn7. Genetics, 145, 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A.E. and Craig,N.L. (2001) Analysis of gain of function mutants of an ATP-dependent regulator of Tn7 transposition. J. Mol. Biol., 305, 633–642. [DOI] [PubMed] [Google Scholar]
- Surette M.G., Buch,S.J. and Chaconas,G. (1987) Transpososomes: stable protein–DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell, 49, 253–262. [DOI] [PubMed] [Google Scholar]
- Waddell C.S. and Craig,N.L. (1988) Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev., 2, 137–149. [DOI] [PubMed] [Google Scholar]
- Watson M.A. and Chaconas,G. (1996) Three-site synapsis during Mu DNA transposition: a critical intermediate preceding engagement of the active site. Cell, 85, 435–445. [DOI] [PubMed] [Google Scholar]
- Wolkow C.A., DeBoy,R.T. and Craig,N.L. (1996) Conjugating plasmids are preferred targets for Tn7. Genes Dev., 10, 2145–2157. [DOI] [PubMed] [Google Scholar]