Abstract
The multisubunit TRAP/Mediator complex is a mammalian counterpart of the yeast Mediator that shows diverse coactivation functions. Genetic ablation of the murine TRAP100 component of this complex has revealed that it is not essential for cell viability per se. However, null mutant mice die at an early developmental stage with severe malformations, and cultured TRAP100-deficient cells exhibit attenuated functions of a wide variety of transcriptional activators on ectopic reporters. The TRAP100-deficient TRAP/Mediator complex also lacks TRAP95 and TRAP150β/SUR2, which together with TRAP100 may form a submodule, and contains a reduced amount of SRB10/CDK8. Nevertheless, the residual complex shows unaltered binding both to RNA polymerase II and, with the exception of the oncoprotein E1A, to various activators. These findings suggest that TRAP/Mediator is broadly involved in transcription and that a TRAP100-containing submodule plays a secondary role, beyond primary activator interactions and RNA polymerase recruitment by the TRAP complex, in magnifying effects of activators on the general transcriptional machinery.
Keywords: embryonic lethality/RNA polymerase II/TRAP100–TRAP95–SUR2 submodule/TRAP220/TRAP/Mediator
Introduction
TRAP/Mediator is a mammalian transcriptional regulatory complex that contains >25 polypeptides and is, in part, phylogenetically conserved from yeast to human (reviewed in Malik and Roeder, 2000; Myers and Kornberg, 2000; Rachez and Freedman, 2001). The TRAP complex was isolated originally as a thyroid hormone receptor (TR)-associated protein (TRAP) complex that mediates ligand-dependent TR function on naked DNA in vitro (Fondell et al., 1996). Subsequently identified complexes that have proved to be closely related or identical to the TRAP complex include SMCC (Gu et al., 1999; Ito et al., 1999), DRIP (Rachez et al., 1999), ARC (Näär et al., 1999), NAT (Sun et al., 1998), human/mouse Mediator (Jiang et al., 1998; Boyer et al., 1999), PC2 (Malik et al., 2000) and CRSP (Ryu et al., 1999) (reviewed in Malik and Roeder, 2000; Myers and Kornberg, 2000; Ito and Roeder, 2001). Apart from TR-dependent transcription, the TRAP complex (herein called TRAP/Mediator) and related complexes mediate the function of diverse activators that include vitamin D receptor (VDR) (Rachez et al., 1998; Yuan et al., 1998), tumor suppressor p53, herpes simplex activator VP16, synthetic activator AH (Gu et al., 1999; Ito et al., 1999), SREBP, NF-κB (Näär et al., 1999), SP1 (Ryu et al., 1999) and adenovirus oncoprotein E1A (Boyer et al., 1999).
TRAP-dependent activation is observed on naked DNA templates in systems reconstituted with highly purified general transcription factors, indicating a primary (but not necessarily exclusive) function at post-chromatin remodeling steps and in accordance with the apparent lack of intrinsic histone acetyltransferase activities (Fondell et al., 1999; Ito et al., 1999; Rachez et al., 1999). Consistent with the demonstration of a yeast Mediator–RNA polymerase holoenzyme complex (reviewed in Myer and Young, 1998) and a model in which activator-bound Mediator recruits RNA polymerase II, the human TRAP, SMCC and DRIP complexes have been found to interact with RNA polymerase II (Gu et al., 1999; Rachez et al., 1999; Chiba et al., 2000; C.-X.Yuan, S.Malik and R.G.Roeder, unpublished).
In yeast, genetic and biochemical studies have shown both general and gene-selective functions for distinct Mediator subunits, as well as a modularity within the complex (reviewed in Myers and Kornberg, 2000). In mammals, activator-selective functions have been inferred from demonstrated interactions of TRAP220 with TR and other nuclear receptors (in a ligand-dependent manner), TRAP170/RGR1 with glucocorticoid receptor, TRAP80 with VP16 and p53, and TRAP150β/SUR2 with E1A (reviewed in Malik and Roeder, 2000). In mice, genetic analyses have confirmed activator-selective functions of TRAP220 and further shown that TRAP220 is essential for embryonic development, but not for cell viability (Ito et al., 2000; Zhu et al., 2000), whereas SRB7 has been found to be essential for both very early embryonic development and (presumably) cell viability (Tudor et al., 1999). Other genetic analyses have indicated that TRAP240, TRAP230 and TRAP80 are essential for optimal development (TRAP240 and TRAP230) and cell viability (TRAP80) in Drosophila; that TRAP230 is involved in the Wnt and β-catenin pathway in Caenorhabditis elegans; and that mutations in TRAP230 correlate with human schizophrenia, dementia and hypothyroidism (Philibert et al., 2001; reviewed in Ito and Roeder, 2001; Rachez and Freedman, 2001).
TRAP100 is phylogenetically unique to higher organisms, being present in mammals and Drosophila, but evidently not in yeast or C.elegans. Although TRAP100 has six or seven of the LXXLL motifs that have been identified as nuclear receptor recognition motifs in various coactivators (reviewed in Glass and Rosenfeld, 2000), including TRAP220, it shows no receptor interactions in vitro (Yuan et al., 1998) and has only a very modest effect on nuclear receptor function in transfected cells (Rachez et al., 1998; Zhang and Fondell, 1999). To investigate the physiological function of TRAP100, as well as its mechanism of action, we have undertaken a mouse genetic approach to inactivate the Trap100 gene.
Results
Trap100–/– mice show embryonic lethality
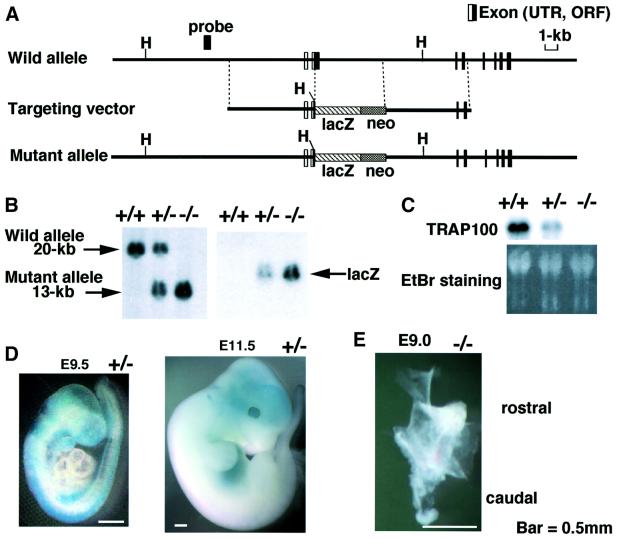
Embryonic stem (ES) cell clones with a disrupted Trap100 locus were used to obtain germline chimeras that in turn were used to generate heterozygous F1 mutant mice (Figure 1A). The loss of Trap100 expression was confirmed by Southern and northern blot analyses of the pooled embryos (Figure 1B and C). F1 Trap100+/– mice were apparently normal. An analysis of liveborn progeny obtained from crosses between F1 Trap100+/– mice revealed that the Trap100 mutation is recessive embryonic lethal (Table I). During E8.5–E10.5 stages of development, Trap100–/– embryos with a 129SvJ/C57BL6 hybrid background were detected at a somewhat lower frequency than expected (Table I), indicating lethality at an even earlier stage of development in a small subset of null embryos. Null embryos after E9.5 were apparently dead and degenerated.
Fig. 1. Disruption of the Trap100 gene. (A) The wild-type allele of the mouse Trap100 gene (above), the targeting vector with the LacZ and pGK-neo cassettes (middle) and the predicted mutant allele resulting from homologous recombination (below) are presented. H denotes the HindIII restriction site. (B) Southern blot analysis of offspring obtained by a heterozygous cross. Genomic DNAs from wild-type (+/+), heterozygous (+/–) and pooled homozygous (–/–) F2 embryos were digested with HindIII and hybridized with the 5′ external probe or lacZ. (C) Northern blot analysis of TRAP100 mRNA in pooled E9.0 embryos of the indicated genotypes. Ethidium bromide staining of total RNA is shown as a control. (D) Expression of TRAP100 protein during embryogenesis as visualized by whole-mount lacZ staining. (E) E9.0 Trap100–/– embryo, dissected together with the yolk sac.
Table I. Genotype of offspring of the TRAP100 knockouts by heterozygous crossing.
| Developmental stage | +/+ | +/– | –/– | (Empty deciduae) |
|---|---|---|---|---|
| E8.5 | 10 | 18 | 5 | (7) |
| E9.5 | 16 | 27 | 7 | (7) |
| E10.5 | 52 | 95 | 23a | (28) |
| E11.5 | 17 | 38 | 0 | (20) |
| At weaning (3 weeks old) | ||||
| 129SVJ/C57BL6J F2 hybrid | 99 | 207 | 0 | |
| Pure 129SvJ | 39 | 68 | 0 |
aDead and some degenerated.
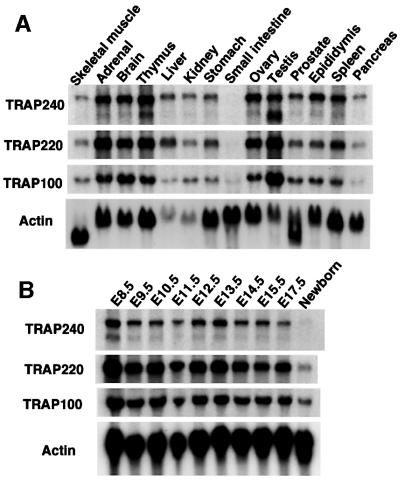
Spatiotemporal expression of TRAP100 in wild-type mice was exactly the same as that of TRAP220 and TRAP240 (Figure 2). Thus, northern blot analysis revealed ubiquitous and somewhat variable TRAP100 mRNA expression levels that were highest in testis, strong and sustained throughout embryogenesis, and attenuated immediately after birth (Figure 2), suggesting a critical role for TRAP100 in the progression and maintenance of embryonic development. To determine the TRAP100 expression pattern in embryos, β-galactosidase activity was measured in Trap100+/– embryos that contain lacZ fused to the translation start site of the Trap100 gene. The expression pattern coincided with the TRAP220 expression pattern (Ito et al., 2000), being strong throughout the primitive central nervous system (CNS), the hepatic primordium and the earliest limb bud stage (Figure 1D). However, these expression patterns contrast with that of the TRAP/Mediator subunit PAQ/ARC105/TIG-1, which exhibits strong and sustained expression specifically in the first and second pharyngeal arches and distal limbs throughout E10.5–E12.5 stages (Berti et al., 2001). These latter results suggest specialized roles for PAQ in the morphogenesis processes of organs derived from these pharyngeal arches, as further indicated by congenital malformations in the human DiGeorge syndrome which presumably is caused by PAQ haploinsufficiency (Berti et al., 2001).
Fig. 2. Expression of TRAP subunits in mouse tissues and embryos. Total RNAs isolated from mice and embryos were subjected to northern blot analysis with cDNA probes as indicated. Adult mouse tissues (A) and developing embryos (B) are shown.
Histopathology of Trap100–/– embryos
Since the live null embryos (F2 129SvJ and C57BL6 hybrid and backcrossed strain with C57BL6) at E9.0 were too tiny and poorly differentiated for further analysis (Figure 1E), we attempted to rescue the embryonic phenotype by backcrossing the mice to other strains. Three backcrossings to the CD1 outbred strain resulted in embryos that survived for an additional day and showed substantially better embryonic growth and differentiation.
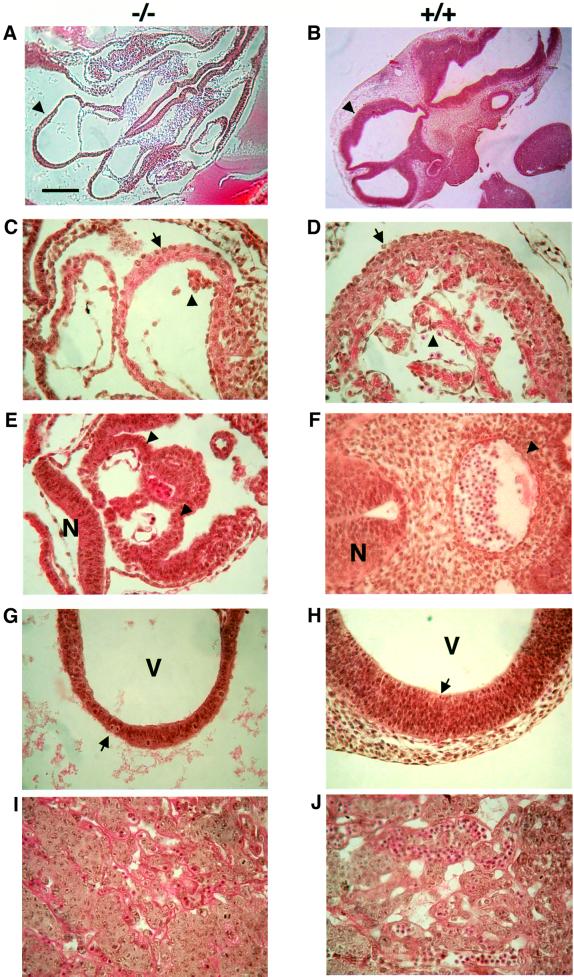
However, gross inspection and fine histological examinations of the live E10.0 Trap100–/– embryos revealed a wide variety of severe developmental abnormalities. Overall, null embryos appear severely hypoplastic compared with both the other genotyped siblings and Trap220–/– embryos (Figure 3A and B; Ito et al., 2000). More specifically, the Trap100–/– embryos are moderately to severely anemic, but the existence of some nucleated red cells indicates that yolk sac hematopoiesis is only partially blocked. Although weak heartbeats were observed in E10.0 null embryos, histological examination revealed cardiac hypoplasia, with both very thin heart walls and poor trabeculation, which is even more severe than that observed in E9.5 Trap220–/– embryos and that inevitably causes severe heart failure. However, some differentiated cardiomyocytes do exist, indicating a preserved differentiation potency of myoblasts (Figure 3C and D). The vascular system, including both great vessels and collateral small vessels, is also ill developed and narrow in the Trap100–/– embryo compared with the wild-type control, as typified in the aorta and labyrinthine placenta (Figure 3E, F, I and J). Also of note, the two dorsal aortas are not yet fused at this stage of the Trap100–/– embryo (Figure 3E and F).
Fig. 3. Histopathology of Trap100–/– embryos. The E10.0 Trap100–/– embryos (A, C, E, G and I) and Trap100+/+ littermates (B, D, F, H and J) are presented. (A and B) Overview of embryos in horizontal sections. Arrows indicate telencephala. (C and D) Hearts. Arrows indicate ventricular walls, and arrowheads trabeculation. (E and F) Transverse sections at the level of the dorsal aorta. Arrowheads indicate the aortic wall, and N indicates neural tubes. Note also the divided dorsal aortas and the open neural tube in the Trap100–/– embryo. (G and H) Telencephala. Arrows indicate mitoses within the neuroepithelial layers. V indicates ventricles. Note the irregular mitotic events in the Trap100–/– neural tube. (I and J) Labyrinthine placentae. Note the poorly developed vasculature in the Trap100–/– placenta. Scale bar (A) = 300 µm; (B) = 600 µm; (C–E) = 75 µm; (F) = 150 µm; (G–J) = 75 µm.
Apart from cardiovascular defects, poor and abnormal development of the CNS is also apparent. Compared with the Trap100+/+ neural tube, the Trap100–/– neural tube is quite thin (even thinner than the Trap220–/– neural tube; Ito et al., 2000) and, in contrast to Trap220–/– neural tube, is open (Figure 3A, B and E–H). Further, although several mitoses are observed in the Trap100–/– neuroepithelial cell layer, they arise randomly in all layers (Figure 3G). This contrasts with the normal organized mitosis that takes place in the innermost layer of the neural tube, where cells normally (and in the Trap220–/– neural tube as well) migrate toward the outer layer of the neural tube and differentiate into mature neurons (Figure 3H).
Finally, although the small size of the Trap100–/– placenta is compatible with the small size of the Trap100–/– embryo, poor vascular development in the labyrinthine placenta, as well as the accompanying anemia, probably restrict a proper nutritional supply to the embryo. However, the aforementioned abnormal development in other organs probably represents primary defects since the nutritional supply is provided mainly by the yolk sac up to the E9.5 stage. Thus, embryonic death most probably reflects a combination of factors that include anemia, heart failure and poor nutritional supply due to the poor placental vasculature, as well as an intrinsic impairment of cell growth (see below).
Widely impaired transcriptional activation in Trap100 mutant cells
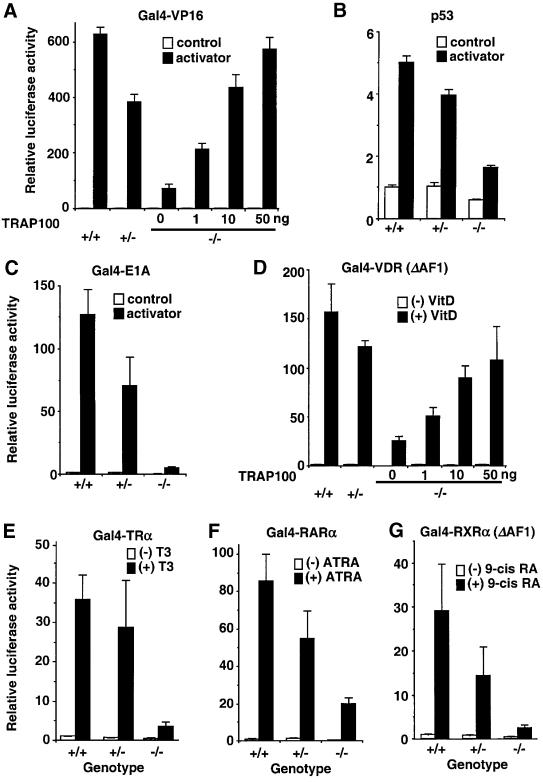
It has been shown that TRAP220 is a potent nuclear receptor coactivator in vitro and in vivo (reviewed in Ito and Roeder, 2001), whereas ectopically expressed TRAP100 showed only a modest positive effect on receptor functions in transfection assays (Rachez et al., 1998; Zhang and Fondell, 1999). In order to check the role of TRAP100 in a more physiological setting in living cells, we attempted to culture primary cells from the mutant embryos. Initial attempts to isolate mouse primary embryonic fibroblasts (MEFs) were unsuccessful; the cells do not proliferate well under conventional culture conditions, which implies a defective autonomous cell growth program in Trap100–/– cells. However, cells could be cultured successfully under the special conditions normally used for ES cells, as described in Materials and methods. Fibroblastoid cells of all three genotypes were recovered from E9.0 sibling embryos (F2 129SvJ and C57BL6 hybrid) and cultured in the same manner, and cells with a similar morphology were chosen for transient transfection assays using a luciferase reporter. The basal (activator-independent) level of transcription from an ectopic reporter containing Gal4-binding sites was ∼50% lower in Trap100–/– cells than in Trap100+/– or Trap100+/+ cells (Figure 4; figure 1 of the Supplementary data available at The EMBO Journal Online). Activator-dependent transcription from ectopic reporters was more severely attenuated in TRAP100-deficient cells, in a gene dose-dependent manner and by all ectopic activators tested; these included Gal4-fused nuclear receptors TRα, RARα, VDR and RXRα, as well as a variety of other activators such as p53 and Gal4-fused VP16 and E1A. Among these activators, the function of Gal4-E1A was most severely affected in Trap100–/– cells (Figure 4C). However, consistent with the viability of Trap100–/– cells, significant residual activation by all activators was apparent in Trap100–/– cells. Further, the levels of basal transcription and the levels of activator-dependent transcription by the representative activators Gal4-VP16 and Gal4-VDR(ΔAF1) were both restored to near normal levels in Trap100–/– cells by expression of ectopic human TRAP100 (Figure 4A and D; Supplementary figure 1). These results demonstrate a broad suppression of basal and activated transcription in Trap100–/– cells. This in turn indicates either severely but incompletely attenuated functions of TRAP100-deficient TRAP/Mediator complex or, less likely, the existence of alternative TRAP/Mediator complex-independent pathway(s) for these diverse activator functions.
Fig. 4. Defective activator-driven transcriptional activation in TRAP100-deficient embryonic cells. (A) Gal4-VP16-driven transcription. The luciferase reporter with Gal4-binding sites upstream of the SV40 promoter was used in all experiments except that for p53 (B). (B) p53-driven transcription from the luciferase reporter with the natural mdm2 promoter. (C) Gal4-E1A-driven transcription. (D) Gal4-VDR-driven transcription. (E) Gal4-TRα-driven transcription. (F) Gal4-RARα-driven transcription. (G) Gal4-RXRα-driven transcription. Values (means ± SD of a representative experiment performed in quadruplicate) are plotted as the fold increase against the value of Trap100+/+ cells without an activator or ligand. Reporter expression levels were normalized to the Renilla luciferase (control) expression levels, which were comparable for wild-type and TRAP100-deficient cells.
Given possible limitations of assays using overexpressed activators and transiently introduced artificial reporters, we next tested the effect of TRAP100 deficiency on expression of an endogenous gene, namely metallothionein II (MT-II). In mammalian cells, MT-II expression is induced by both glucocorticoids and heavy metals (including cadmium and zinc ions) through, respectively, the glucocorticoid- and heavy metal- response elements of the MT-II promoter (Richards et al., 1984). Therefore, we examined MT-II gene expression in response to dexamethasone, Cd2+ and Zn2+ in Trap100+/+ and Trap100–/– cells. While Trap100+/+ cells showed 4.2-, 12.0- and 11.1-fold increases in MT-II expression in response to dexamethasone, Cd2+ and Zn2+, respectively, Trap100–/– cells exhibited attenuated increases of only 2.4-, 4.3- and 3.2-fold in response to these agents (Figure 5). These results indicate that the glucocorticoid receptor and heavy metal induction pathways for endogenous MT-II are both significantly impaired (2- to 4-fold) in Trap100–/– cells, and further support the above conclusions from the ectopic activator and gene reporter assays that TRAP100 deficiency leads to generalized effects on transcription.
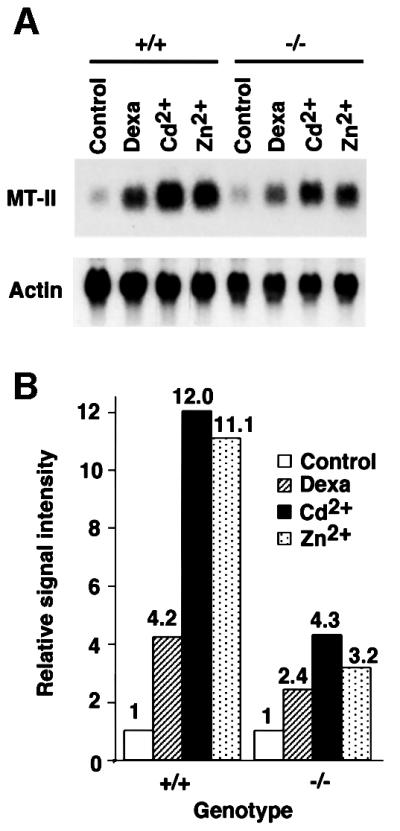
Fig. 5. Expression of the endogenous metallothionein-II gene in embryonic cells. (A) Embryonic cells were incubated for 10 h in the absence or presence of dexamethasone (10–6 M), CdCl2 (10–5 M) or ZnCl2 (10–4 M), and total RNAs were used for northern blot analysis with cDNA probes as indicated. (B) Relative induction of the metallothionein-II gene based on quantification of the data in (A).
Trap220 and Trap100 dose dependency in embryonic development and housekeeping gene expression
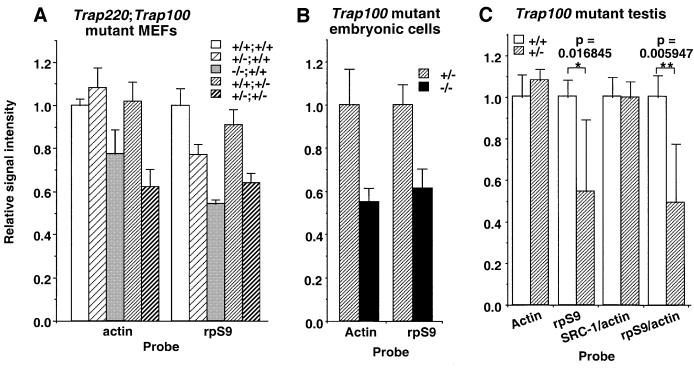
Motivated by the finding of Trap100 dose-dependent defects in transient transfection assays of embryonic cells, and in light of dose-dependent effects of Trap220 in both animals and cultured cells (Ito et al., 2000), we investigated the possibility that a deficiency in one TRAP gene enhances the phenotypic effects of a deficiency in the other TRAP gene. For this purpose, we checked endogenous gene expression patterns in single and double mutant cells. The Trap220+/+Trap100+/+, Trap220+/– Trap100+/+, Trap220–/–Trap100+/+, Trap220+/+Trap100+/– and Trap220+/–Trap100+/– MEFs were isolated from corresponding E10.0 embryos, and total RNAs extracted from passage 2 of these cells were tested by northern blot. Among the tested genes, the levels of some housekeeping genes such as actin and ribosomal protein S9 (rpS9) were decreased in a gene dose-dependent manner by the loss of either both Trap220 alleles or one Trap220 and one Trap100 allele (Figure 6A). Similarly, an analysis of embryonic cells, derived from E9.0 embryos and specially cultured as described above, showed a more pronounced decrease in actin and rpS9 gene expression in Trap220+/+ Trap100–/– cells relative to Trap220+/+Trap100+/– cells (Figure 6B).
Fig. 6. Gene dosage-dependent impairment of transcription of housekeeping genes in embryonic cells and testis. Relative signal intensities of quantitated RNA blot analyses are indicated. Results of five or six samples of 129SvJ/C57BL6 hybrid MEFs (A), embryonic cells (B) or 12-week-old testes (C) are shown. Values are expressed as the relative change ± SD from the mean expression level of each mRNA in Trap100+/+ MEFs (A), Trap100+/– cells (B) or Trap100+/+ testes (C). The levels of rRNAs were exactly the same (data not shown).
Both Trap220 and Trap100 expression levels are highest in testes in adult mice, and we previously reported attenuated actin expression but up-regulated ribosomal protein (e.g. rpS9) gene expression in testes of Trap220+/– mice (Ito et al., 2000). To check if levels of expression of these genes are altered similarly in Trap100+/– mice, F1 testes were analyzed by northern blot. Contrary to expectations from the Trap220+/– results, actin expression in heterozygotes was indistinguishable from wild-type controls, whereas rpS9 expression was lower than the controls (Figure 6C). Together with the above result of attenuated rpS9 expression in Trap220+/– MEFs, these results suggest that up-regulation of ribosomal proteins is likely to be a specialized event in adult Trap220+/– testis and further indicate tissue- or cell-specific roles for TRAP220 in special circumstances in contrast to a more general role for TRAP100. These results, taken together, indicate that all four Trap220 and Trap100 alleles are necessary for fully normal development and gene expression both in vivo and in cultured cells, and that any additional gene deletion results in phenotypic deterioration relative to the alteration caused by another mutation.
The TRAP100-deficient TRAP complex also lacks the TRAP150β/SUR2 and TRAP95 subunits and most of the SRB10/CDK8 subunit
In view of the early lethality of the very tiny Trap100–/– embryos, the stable culture of Trap100–/– cells is indispensable for investigating the mechanisms underlying the Trap100 mutant phenotypes. Although all trials of long-term culture of Trap100+/+ and Trap100+/– embryonic cells failed, all three trials of Trap100–/– embryonic cells in the same media allowed culture for a prolonged period of time. Some colonies of cuboid-shaped cells that presumably were of immature fibroblastic lineage emerged and became predominant after passage 25. These Trap100–/– cells, along with MEFs derived from an E9.0 p53 null embryo as a wild-type control, were then used in the subsequent studies.
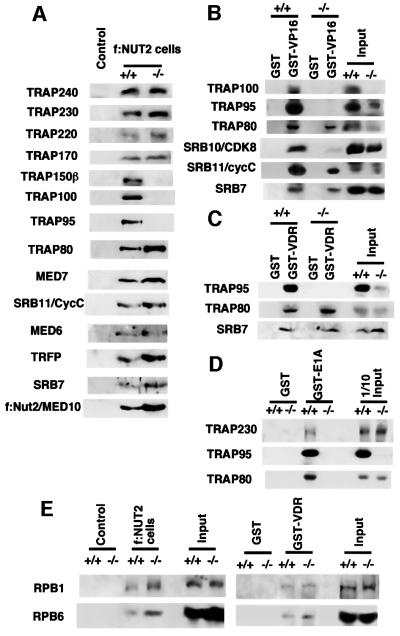
In order to isolate and characterize the TRAP/Mediator complex from these cells, they were stably transfected with a FLAG-tagged NUT2/MED10 expression vector containing the hygromycin resistance gene. Corresponding complexes were isolated from derived nuclear extracts by immunoprecipitation with M2–agarose. These preparations then were checked for the existence of individual subunits of the complete TRAP/Mediator by immunoblot analysis. Among the 14 subunits tested, TRAP150β/SUR2, TRAP100 and TRAP95 were selectively missing in the mutant TRAP/Mediator complex (Figure 7A). Since SRB10/CDK8 is not readily detected in M2–agarose immunoprecipitates from control FLAG-NUT2 cell lines (Malik et al., 2000), the presence of SRB10 within complexes from Trap100–/– cells was assessed by binding of complexes within nuclear extracts to a GST–VP16 column (see below). Indeed, SRB10 was greatly lowered in isolates from Trap100–/– cells relative to those from Trap100+/+ cells (Figure 7B), although the fact that the GST–VP16 matrix probably purifies a mixture of complexes could complicate interpretation of the data. It is notable that the protein levels of TRAP95 and SUR2 were significantly reduced in Trap100–/– cells compared with Trap100+/+ cells, although levels of other subunits (including SRB10) were comparable between these cells (Figure 7B; Supplementary figure 2). However, a northern blot analysis of these cells disclosed that the mRNA levels of TRAP95 and SUR2, like those of other TRAP/Mediator subunits, are comparable between these cells (Supplementary figure 2). These results suggest that the free subunits are destabilized and subject to an enhanced turnover in mutant cells. They also indicate the existence of a stable TRAP100-deficient mutant TRAP complex that, coincidentally, lacks TRAP150β/SUR2, TRAP95 and most of SRB10/CDK8. They further suggest that these latter subunits, together with TRAP100, may comprise a structural unit (submodule) whose association with the TRAP core complex is dependent upon TRAP100.
Fig. 7. Subunit composition of the TRAP100-deficient TRAP complex and its interactions with activators and RNA polymerase II. (A) Immunoblot analysis of FLAG-NUT2-containing TRAP complexes from the Trap100+/+ and Trap100–/– cells immunopurified on M2–agarose. Nuclear extract from untagged Trap100+/+ cells was used as a control. The FLAG-NUT2 expression level was estimated to be ∼10% of the endogenous level. (B and C) Immunoblot analysis of TRAP complexes from the Trap100+/+ and Trap100–/– nuclear extracts bound to GST–VDR AF2 (B) or GST–VP16 (C). (D) Immunoblot analysis of the binding to GST–E1A of the FLAG-NUT2 TRAP complex immunopurified (M2–agarose) from Trap100+/+ and Trap100–/– cells. (E) Immunoblot analysis of RPB1 and RPB6 subunits of RNA polymerase II co-purified with the TRAP complex from Trap100+/+ and Trap100–/– cells on M2–agarose (left panel) or on GST–VDR AF2 (right panel). The amount of the co-purified TRAP complex in each lane was normalized to an equal amount of TRAP80 detected by immunoblot (data not shown).
TRAP100-deficient TRAP/Mediator complex can bind to activators VP16 and VDR but not to E1A
Next we asked if the mutant TRAP complex can associate with activators and the RNA polymerase II core complex as effectively as the wild-type TRAP complex. To test the association with activators, GST–VP16, GST–VDR(AF2) and GST–E1A(CR3) fusion proteins (immobilized on glutathione–Sepharose) were used to pull-down the TRAP complex from nuclear extracts or from an M2-purified preparation, and the eluted complexes were checked by immunoblot analysis. Indeed, in nuclear extracts, the mutant TRAP complex bound GST–VP16 and GST–VDR (but not GST) as avidly as did the wild-type TRAP complex (Figure 7B and C), consistent with previous data indicating that VP16 and VDR interact, respectively, with TRAP80 and TRAP220 (Yuan et al., 1998; Ito et al., 1999), and with current data indicating the preservation of these two TRAPs in the mutant complex (above). This view was confirmed by the observation that the TRAP complex was not pulled-down by GST–VDR(AF2) from the nuclear extract of Trap220–/– MEFs, and that the bound TRAP100-deficient complex had the same subunit composition as the unbound complex (data not shown).
In contrast, the purified mutant TRAP complex did not bind to immobilized GST–E1A (Figure 7D), consistent with previous data indicating that E1A interacts with the TRAP complex through a subunit (TRAP150β/hSUR2) (Boyer et al., 1999) that is missing in the mutant TRAP complex (above). These results are consistent with the observation that, of the activators tested, E1A showed the most profound loss of activity in Trap100–/– cells. However, Trap100–/– cells still showed a residual 5-fold activation by ectopic Gal4-E1A, which may indicate either persistent binding of a small amount of TRAP150β/SUR2 to the mutant TRAP complex (especially in the presence of the activator) or an alternative pathway that either is independent of the TRAP complex or acts through another TRAP subunit. The former possibility may be more probable, as the SUR2 deficiency in ES cells completely abolishes Gal4-E1A function (Stevens et al., 2002). Taken together, these results indicate that the mutant TRAP complex can bind efficiently to at least some activators, but not to E1A.
TRAP100-deficient TRAP/Mediator complex can bind to RNA polymerase II
Next, to check an association with RNA polymerase II, the affinity-purified FLAG-NUT2 TRAP complexes and the nuclear extract TRAP/Mediator complexes that were bound to GST–VDR(AF2) were analyzed (after elution) by immunoblot analysis. Both the RPB1 and RPB6 subunits of RNA polymerase II were detected in both wild-type and mutant TRAP/Mediator preparations (Figure 7E). More interestingly, although comparable levels of RNA polymerase II were detected in both wild-type and mutant TRAP complexes that were bound (from nuclear extracts) to VDR, the level of RNA polymerase II associated with the TRAP complexes that were affinity purified (on M2–agarose) via FLAG-NUT2 appeared to be slightly higher (1.5-fold) for the mutant complex than for the control preparation. In view of the proposal that the TRAP complex associated with VDR (and presumably with other activators) binds more RNA polymerase II than the free complex (Chiba et al., 2000), the latter result might indicate a greater association of activators with the TRAP complex in mutant cells relative to wild-type cells. Alternatively, the RNA polymerase II might have an intrinsically higher affinity for the TRAP100-deficient complex. In any case, these results clearly show, overall, that the mutant TRAP complex is sufficiently intact to bind RNA polymerase II normally, and they further suggest that it can bind activators and RNA polymerase II simultaneously.
Discussion
TRAP100 is required for embryogenesis
Two previous genetic studies of the murine TRAP/Mediator have shown that TRAP220 and SRB7 are both essential for embryogenesis but with quite different degrees of phenotypic severity. Trap220–/– embryos are viable up to E11.0, and the differentiation of primitive organs expected at this stage is apparent but incomplete (Ito et al., 2000; Zhu et al., 2000). In contrast, Srb7–/– embryos are viable only up to the blastocyst stage, through which maternal SRB7 remains (Tudor et al., 1999). In the present study, the phenotypic severity of the Trap100–/– embryos, which actually lack a submodule consisting of TRAP150β, TRAP100 and TRAP95 (discussed below), is intermediate between that of the Trap220 and Srb7 mutations; and the Trap100 and Trap220 double mutations augment the phenotypic severity. These results, in toto, indicate that TRAP/Mediator is essential for cell viability per se, but that a given mutant phenotype is variable and dependent upon the subunit composition of the residual TRAP/Mediator.
Trap100–/– embryos suffer severe deterioration in the growth and differentiation of organs throughout the entire embryo, although the differentiation program is preserved in various types of committed cells such as neuronal cells, cardiomyocytes, nucleated erythrocytes and endothelia. An impaired autonomous cell growth program seems to underlie these abnormalities.
Intriguingly, and in contrast to the situation in Trap220–/– embryos, the spatiotemporally organized alignment and proliferation of Trap100–/– cells are impaired in some organs, as evidenced by the random cell division of neuroepithelial layer cells, the open neural tube and the divided aortas in E10.0 embryos. These abnormalities may indicate, for example, impairments in the secretion of humoral organizers, the expression of cognate receptors on the cells or homeotic gene expression programs in the embryos. Further, these observations may represent functional distinctions between TRAP220 and TRAP100 in embryonic development.
Specificity of TRAP220 versus TRAP100 in transcriptional activation
While TRAP/Mediator is generally required for transcription, as evidenced by complete loss of basal and activator-dependent transcription in nuclear extracts lacking the complex (Mittler et al., 2001; Baek et al., 2002), cells with mutated TRAP/Mediator subunits retain transcriptional abilities that vary with the nature of the mutated subunit and the residual complex. In the most severe case, an Srb7 deletion appears to result in both cell lethality and a global decrease in transcription.
In contrast, TRAP220- and TRAP100-deficient cells are viable, although cells and animals with these deficiencies behave quite differently with respect to transcription. For example, while an attenuated transactivation function in transient transfection assays is limited (thus far) to nuclear receptors such as TRα, VDR, PPARγ and androgen receptor in Trap220–/– cells (Ito et al., 2000; unpublished data), Trap100–/– cells exhibit much broader defects in activator functions (this study). The broad block to transcription in Trap100–/– cells is evidenced further by decreased expression of several endogenous genes, including the constitutively expressed actin and rpS9 genes, and the glucocorticoid- and heavy metal ion-induced MT-II gene. Nevertheless, there are variations in the severity of the transcriptional block, which is most severe for Gal4-E1A. This may reflect the loss of both a general function of the TRAP100 module and a direct interaction site for E1A (below).
As further evidence of selective functions of TRAP100 and TRAP220, Trap220+/– mice exhibit mild dwarfism and pituitary hypothyroidism (Ito et al., 2000), whereas Trap100+/– mice do not. Further, while expression of rpS9 is up-regulated in the Trap220+/– testis (Ito et al., 2000), it is attenuated in the Trap100+/– testis. In addition, the haploinsufficiency of the TRAP/Mediator subunit PAQ/ARC105 in DiGeorge syndrome indicates a specialized role for PAQ in development of organs derived from first and second branchial arches (Berti et al., 2001). These differences may indicate more general roles of TRAP100, in contrast to more specialized roles of TRAP220 or PAQ, in transcriptional coactivation.
Novel mammalian TRAP submodules: TRAP150β/SUR2, TRAP100 and TRAP95
The successful long-term culture of Trap100–/– cells has enabled biochemical analyses and mechanistic insights into the function of TRAP100. The residual TRAP/Mediator complex in these cells lacks not only TRAP100, but also TRAP150β/SUR2 and TRAP95. This observation suggests that TRAP150β/SUR2, TRAP100 and TRAP95 constitute a structural submodule whose functional significance is evidenced by the transcription defects in Trap100–/– cells and by the phenotypic alterations of Trap100–/– cells in culture and in the embryo. The more severe phenotypes of Trap100–/– embryos and cells relative to the Trap220–/– phenotypes may reflect the loss of a greater number of subunits from TRAP/Mediator in Trap100–/– cells and consequent loss of both general and gene-specific functions. Indeed, the TRAP/Mediator in Trap220–/– cells seems to lack only the TRAP220 subunit (S.Malik, C.-X.Yuan, M.Ito and R.G.Roeder, unpublished).
The TRAP100-deficient TRAP/Mediator complex described here is reminiscent of the biochemically isolated mammalian core Mediator subcomplexes PC2 (Malik et al., 2000) and B-Med (Mittler et al., 2001). Although PC2 contains considerably fewer components than the TRAP100-deficient TRAP/Mediator complex, the presence of substoichiometric amounts of TRAP100 (Malik et al., 2000), and possibly of TRAP95 and SUR2 as well (S.Malik and R.G.Roeder, unpublished), provides further support for the existence of a TRAP100 submodule. This notion is also supported by the absence of TRAP95 in the B-Med complex (Mittler et al., 2001), although detailed information on the subunit composition of this subcomplex currently is unavailable. Consistent with these observations, the Mediator complex from cells lacking TRAP150β/SUR2 appears to be partially defective for TRAP100 and TRAP95 as well (Stevens et al., 2002).
Among the submodule components, TRAP150β/SUR2 is preserved from C.elegans to human, while TRAP100 and TRAP95 are unique to higher organisms, indicating that SUR2 apparently is incorporated into the C.elegans complex without the help of a molecule that corresponds to TRAP100. Whereas gene-specific functions currently are unknown for TRAP95 and TRAP100, other studies have indicated that the TRAP150β/SUR2 subunit mediates transcriptional activation by the adenoviral oncoprotein E1A through direct interaction (Boyer et al., 1999; Stevens et al., 2002). In support of this proposal, the mutant TRAP complex that lacks TRAP150β/SUR2 does not bind to E1A, and activation by Gal4-E1A is more severely attenuated than activation by other activators in Trap100–/– cells.
Our observation that TRAP/Mediator from Trap100–/– cells contains a significantly decreased amount of SRB10/CDK8, but a normal amount of its interacting regulatory partner SRB11/cyclin C, further suggests a role for the TRAP150β–TRAP100–TRAP95 submodule in stabilizing interactions of SRB10 with SRB11 within the TRAP complex. Although the SRB10–SRB11 pair has the potential to regulate TRAP/Mediator-dependent transcription negatively in yeast and human (Hengartner et al., 1998; Gu et al., 1999; Akoulitchev et al., 2000), attenuated transcription rather than enhanced transcription was observed in the current situation where TRAP100 deficiency results in a reduced association of SRB10. Although SRB10 could also have a positive role in transcription (Vincent et al., 2001), the current results most probably reflect a conditional negative function for SRB10 that is normally suppressed by other components (possibly including the TRAP100 module) of TRAP/Mediator. Given that TRAP95, TRAP100 and SUR2 have no yeast counterparts, the current results also suggest metazoan-specific mechanisms for recruitment and stabilization of the phylogenetically conserved SRB10–SRB11 complex within the Mediator.
Mechanism of attenuated transcription in TRAP100 deficiency
TRAP100 deficiency in cells results in a general decrease in transcription that is correlated with the loss of several subunits, which apparently form a submodule, from TRAP/Mediator. This could reflect general effects on either basal or activated transcription. Studies in yeast have provided insights into the role of specific Mediator subunits, within distinct submodules, in these processes. Thus, the Gal11 submodule (comprised of Gal11, Hrs1, Sin4 and Med2) has been implicated in the function of specific activators, through specific subunit interactions, but apparently plays no major role in RNA polymerase interactions or basal transcription (Lee et al., 1999; Myers et al., 1999; Dotson et al., 2000). Whereas these observations are consistent with indications that mammalian activators act through interactions with specific TRAP/Mediator subunits (Malik and Roeder, 2000), they contrast with the present observation of a general transcription deficiency in Trap100–/– cells.
Perhaps more relevant is the yeast Srb4 subcomplex comprised of Srb2, Srb4, Srb5, Srb6 and Med6 (Lee et al., 1999; reviewed in Myers and Kornberg, 2000). Srb4 and Srb6 appear to be generally required for all RNA synthesis (Holstege et al., 1998), and may act by antagonizing the effect of negative regulatory cofactors that restrict the intrinsic basal functions of the general transcription machinery (reviewed in Myers and Kornberg, 2000). In this regard, in vitro functions in basal transcription have been demonstrated for both yeast (Thompson et al., 1993; Kim et al., 1994; Lee et al., 1999) and human (Mittler et al., 2001; Baek et al., 2002) Mediator complexes. The Srb2 and Srb5 components of the Srb4 subcomplex have also been implicated in basal transcription in vitro (Thompson et al., 1993; Lee et al., 1999). However, in contrast to srb4 and srb6 deletions, which affect essentially all genes and result in lethality, srb2 and srb5 deletions result in a slow growth phenotype and affect expression of a smaller fraction of total yeast genes (Holstege et al., 1998). Significantly, yeast Mediator lacking Srb2 and Srb5 shows reduced levels of both basal and activated transcription, while maintaining normal activator and RNA polymerase II interactions; moreover, the artificial recruitment of the Srb2/Srb5-deficient Mediator (resulting from srb2 deletion) to the promoter fails to activate transcription (Lee et al., 1999).
Thus, from both functional and mechanistic standpoints, the human TRAP100–TRAP95–SUR2 submodule seems more similar to the yeast Srb2–Srb5 submodule. Genetic deletions of corresponding components (Trap100–/– versus srb2 and srb5) are non-lethal and have general effects on basal and activated transcription, and Mediator complexes lacking the submodules show persistent interactions both with RNA polymerase II and with a variety of activators. Hence, as proposed for yeast Mediator (Lee et al., 1999; Myers et al., 1999), human TRAP/Mediator may have functions beyond the simple recruitment of RNA polymerase II. It remains to be firmly established whether the TRAP100 submodule mainly facilitates TRAP/Mediator functions in basal transcription independently of activators or whether its function is linked mainly to activator– TRAP/Mediator interactions, although the present data (greater effect on activated transcription) favor the latter. In any case, it serves to amplify effects of activators mediated through interactions with specific subunits.
Materials and methods
Construction of the Trap100 targeting vector
To isolate the mouse Trap100 gene, a mouse 129SvJ genomic library (Stratagene) was screened with an oligonucleotide probe containing nucleotides –10 to 22 of the mouse TRAP100 cDNA (the first amino acid being nucleotides 1–3). Five overlapping clones contained a 34 kb genomic region that included exons encoding up to nucleotide 1056 of the murine TRAP100 cDNA. The 2.5 kb genomic fragment extending from nucleotide 4 in the first coding exon to the BamHI restriction site was replaced by lacZ in-frame and a PGK neo cassette (Figure 1A). This replacement deleted the entire Trap100 gene product. The targeting vector included a 6.6 kb upstream homologous region and a 6.2 kb downstream region.
Generation of the TRAP100 knockout mice
E14 ES cells were electroporated with the targeting vector (Ito et al., 2000), and 96 G418-resistant clones were screened by Southern blot analysis using the 5′ external probe. Out of seven clones that displayed evidence for homologous recombination, five clones were microinjected into blastocysts of C57BL/6J females and three independent ES clones generated germline chimeras. All animal experimentation was performed according to National Institutes of Health (NIH) guidelines in the Rockefeller University Laboratory Animal Research Center.
For genotyping, the genomic DNA was subjected to Southern blot analysis with a 5′ external probe or lacZ, or used for PCR analysis (primer sequences available on request).
Northern blot analysis
Total RNAs (10 µg) were used for an RNA blot analysis with cDNA probes of mouse TRAP100 and TRAP220, human TRAP240, specific mouse MT-II, human actin and mouse rpS9. Signal intensities were quantitated with a STORM840 image analyzer (Molecular Dynamics) (Ito et al., 2000).
Whole-mount lacZ staining and histology
Whole-mount lacZ staining and histological analysis of embryos were performed as described (Ito et al., 2000).
Preparation of embryonic cells
MEFs isolated from E10.0 embryos were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). The E9.0 Trap100–/–, Trap100+/– and Trap100+/+ embryonic cells were prepared in DMEM with 15% heat-inactivated FBS, 103 U/ml leukemia inhibitory factor (LIF), 1× non-essential amino acids, 1× nucleosides, 2 mM glutamine, 10–4 M β-mercaptoethanol and 100 U/ml penicillin/streptomycin, and, for the initial 2 weeks, cultured on mitomycin C-treated feeder cells seeded on gelatinized dishes.
For long-term culture, E9.0 MEFs derived from p53–/– mice (Jackson Laboratories) were used as Trap100+/+ embryonic cells. The Trap100–/– primary embryonic cells (above) were cultured in the above media until immature transformed cell colonies appeared and became predominant.
Transfection
For luciferase assays, embryonic cells were transiently transfected with various activator and reporter constructs and, for nuclear receptors, in the absence or presence (10–6 M) of corresponding ligands. Activator expression vectors driven by the cytomegalovirus (CMV) promoter (pCDM8, Invitrogen) were used at levels of 1 ng (Gal4-E1A and Gal4-VP16) or 25 ng [p53, Gal4-TRα, Gal4-VDR(ΔAF1), Gal4-RARα and Gal4-RXRα(ΔAF1)]. Reporter plasmids (100 ng) consisted of an SV40 promoter–luciferase reporter (pGL3-Promoter, Promega) with five Gal4-binding sites and an mdm2 promoter–luciferase reporter (Ito et al., 2000). The Renilla luciferase reporter pRL-SV40 (Promega) was used as a control (Ito et al., 2000).
For stable transfection of FLAG-tagged NUT2, an expression vector containing a hygromycin resistance gene, prepared by replacement of the neomycin resistance gene in the published expression vector (Malik et al., 2000) with the hygromycin resistance gene (S.Malik, unpublished data), was introduced into embryonic cells with lipofectamine (Gibco-BRL), and drug-resistant clones were selected.
Isolation and analysis of TRAP complexes
TRAP complexes were immunopurified from nuclear extracts (2 mg protein) prepared from embryonic cells expressing FLAG-tagged NUT2 by binding to M2–agarose in buffer BC180 with 0.5% NP-40 (Malik et al., 2000). Complexes were eluted with FLAG peptide and used for immunoblot analyses of subunit compositions. Nuclear extract from untagged cells was used as a control. Nuclear extracts (2 mg) from un tagged embryonic cells were also used to isolate complexes by incubation with 20 µg of immobilized GST–VP16, GST–VDR(AF2) or GST alone in buffer BC180 with 0.5% NP-40 [and 10–7 M 1α,25(OH)2D3 for GST–VDR] for 6 h. Bound complexes were eluted with 0.2% sarkosyl and used for immunoblot analyses. For binding of purified complexes, FLAG-tagged (NUT2) TRAP complex prepared from 20 mg of nuclear extract was incubated with 20 µg of immobilized GST–E1A (CR3) or GST alone in buffer BC180 with 0.5% NP-40 for 6 h. After washing with the same buffer, bound complexes were eluted with 0.2% sarkosyl and used for immunoblot analyses.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank S.Malik for the FLAG-NUT2 expression vector and anti-MED6, -MED7 and -NUT2 antibodies, H.J.Baek for the Gal4-E1A expression vector and anti-TRAP150β antibody, S.Yamamura for anti-TRAP95 antibody, Z.Wang for anti-RPB6 antibody, C.Yang and the Transgenic Facility of the Rockefeller University for help with ES cell manipulation and blastocyst injection, and members of the Roeder laboratory, particularly K.Ge, H.Kato, S.Malik, S.Yamamura, D.Zhang and J.Zhang, for useful discussions. This work was supported by a Human Frontier Science Program (HFSP) long-term fellowship to M.I. and grants from the NIH to R.G.R. and R.B.D.
References
- Akoulitchev S., Chuikov,S. and Reinberg,D. (2000) TFIIH is negatively regulated by cdk8-containing Mediator complexes. Nature, 407, 102–106. [DOI] [PubMed] [Google Scholar]
- Baek H.J., Malik,S., Qin,J. and Roeder,R.G. (2002) Requirement of TRAP/Mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAFIIs. Mol. Cell. Biol., 22, 2842–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti L. et al. (2001) Isolation and characterization of a novel gene from the DiGeorge chromosomal region that encodes for a Mediator subunit. Genomics, 74, 320–332. [DOI] [PubMed] [Google Scholar]
- Boyer T.G., Martin,M.E., Lees,E., Ricciardi,R.P. and Berk,A.J. (1999) Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature, 399, 276–279. [DOI] [PubMed] [Google Scholar]
- Chiba N., Suldan,Z., Freedman,L.P. and Parvin,J.D. (2000) Binding of liganded vitamin D receptor to the vitamin D receptor interacting protein coactivator complex induces interaction with RNA polymerase II holoenzyme. J. Biol. Chem., 275, 10719–10722. [DOI] [PubMed] [Google Scholar]
- Dotson M.R., Yuan,C.X., Roeder,R.G., Myers,L.C., Gustafsson,C.M., Jiang,Y.W., Li,Y., Kornberg,R.D. and Asturias,F.J. (2000) Structural organization of yeast and mammalian Mediator complexes. Proc. Natl Acad. Sci. USA, 97, 14307–14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell J.D., Ge,H. and Roeder,R.G. (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA, 93, 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell J.D., Guermah,M., Malik,S. and Roeder,R.G. (1999) Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc. Natl Acad. Sci. USA, 96, 1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C.K. and Rosenfeld,M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- Gu W., Malik,S., Ito,M., Yuan,C.-X., Fondell,J.D., Zhang,X., Martinez,E., Qin,J. and Roeder,R.G. (1999) A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell, 3, 97–108. [DOI] [PubMed] [Google Scholar]
- Hengartner C.J., Myer,V.E., Liao,S.-M., Wilson,C.J., Koh,S.S. and Young,R.A. (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell, 2, 43–53. [DOI] [PubMed] [Google Scholar]
- Holstege F.C.P., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Ito M. and Roeder,R.G. (2001) The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol. Metab., 12, 127–134. [DOI] [PubMed] [Google Scholar]
- Ito M. et al. (1999) Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell, 3, 361–370. [DOI] [PubMed] [Google Scholar]
- Ito M., Yuan,C.-X., Okano,H.J., Darnell,R.B. and Roeder,R.G. (2000) Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell, 5, 683–693. [DOI] [PubMed] [Google Scholar]
- Jiang Y.-W., Veschambre,P., Erdjument-Bromage,H., Tempst,P., Conaway,J.W., Conaway,R.C. and Kornberg,R.D. (1998) Mammalian Mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl Acad. Sci. USA, 95, 8538–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-J., Björklund,S., Li,Y., Sayre,M.H. and Kornberg,R.D. (1994) A multiprotein Mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell, 77, 599–608. [DOI] [PubMed] [Google Scholar]
- Lee Y.C., Park,J.M., Min,S., Han,S.J. and Kim,Y.-J. (1999) An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol., 19, 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S. and Roeder,R.G. (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci., 25, 277–283. [DOI] [PubMed] [Google Scholar]
- Malik S., Gu,W., Wu,W., Qin,J. and Roeder,R.G. (2000) The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell, 5, 753–760. [DOI] [PubMed] [Google Scholar]
- Mittler G., Kremmer,E., Timmers,H.T.M. and Meisterernst,M. (2001) Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO rep., 2, 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer V.E. and Young,R.A. (1998) RNA polymerase II holoenzymes and subcomplexes. J. Biol. Chem., 273, 27757–27760. [DOI] [PubMed] [Google Scholar]
- Myers L.C. and Kornberg,R.D. (2000) Mediator of transcriptional regulation. Annu. Rev. Biochem., 69, 729–749. [DOI] [PubMed] [Google Scholar]
- Myers L.C., Gustafsson,C.M., Hayashibara,K.C., Brown,P.O. and Kornberg,R.D. (1999) Mediator protein mutations that selectively abolish activated transcription. Proc. Natl Acad. Sci. USA, 96, 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näär A.M. Beaurang,P.A., Zhou,S., Abraham,S., Solomon,W. and Tjian,R. (1999) Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature, 398, 828–832. [DOI] [PubMed] [Google Scholar]
- Philibert R.A., Sandhu,H.K., Hutton,A.M., Wang,Z., Arndt,S., Andreasen,N.C., Crowe,R. and Wassink,T.H. (2001) Population-based association analyses of the HOPA12bp polymorphism for schizophrenia and hypothyroidism. Am. J. Med. Genet., 105, 130–134. [PubMed] [Google Scholar]
- Rachez C. and Freedman,L.P. (2001) Mediator complexes and transcription. Curr. Opin. Cell Biol., 13, 274–280. [DOI] [PubMed] [Google Scholar]
- Rachez C., Suldan,Z., Ward,J., Chang,C.P., Burakov,D., Erdjument-Bromage,H., Tempst,P. and Freedman,L.P. (1998) A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev., 12, 1787–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C., Lemon,B.D., Suldan,Z., Bromleigh,V., Gamble,M., Näär,A.M., Erdjument-Bromage,H., Tempst,P. and Freedman,L.P. (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature, 398, 824–828. [DOI] [PubMed] [Google Scholar]
- Richards R.I., Heguy,A. and Karin,M. (1984) Structural and functional analysis of the human metallothionein-IA gene: differential induction by metal ions and glucocorticoids. Cell, 37, 263–272. [DOI] [PubMed] [Google Scholar]
- Ryu S., Zhou,S., Ladurner,A.G. and Tjian,R. (1999) The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature, 397, 446–450. [DOI] [PubMed] [Google Scholar]
- Stevens J.L., Cantin,G.T., Wang,G., Shevchenko,A., Shevchenko,A. and Berk,A.J. (2002) Transcription control by E1A and MAP kinase pathway via Sur2 Mediator subunit. Science, 296, 755–758. [DOI] [PubMed] [Google Scholar]
- Sun X., Zhang,Y., Cho,H., Rickert,P., Lees,E., Lane,W. and Reinberg,D. (1998) NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell, 2, 213–222. [DOI] [PubMed] [Google Scholar]
- Thompson C.M., Koleske,A.J., Chao,D.M. and Young,R.A. (1993) A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell, 73, 1361–1375. [DOI] [PubMed] [Google Scholar]
- Tudor M., Murray,P.J., Onufryk,C., Jaenisch,R. and Young,R.A. (1999) Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev., 13, 2365–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent O., Kuchin,S., Hong,S.-P., Townley,R., Vyas,V.K. and Carlson,M. (2001) Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 5790–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C.-X., Ito,M., Fondell,J.D., Fu,Z.-Y. and Roeder,R.G. (1998) The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl Acad. Sci. USA, 95, 7939–7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. and Fondell,J.D. (1999) Identification of mouse TRAP100: a transcriptional coregulatory factor for thyroid hormone and vitamin D receptors. Mol. Endocrinol., 13, 1130–1140. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qi,C., Jia,Y., Nye,J.S, Rao,M.S. and Reddy,J.K. (2000) Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J. Biol. Chem., 275, 14779–14782. [DOI] [PubMed] [Google Scholar]