Abstract
To elucidate the basis of sequence-specific single-stranded (ss) DNA recognition by K homology (KH) domains, we have solved the solution structure of a complex between the KH3 domain of the transcriptional regulator heterogeneous nuclear ribonucleoprotein K (hnRNP K) and a 10mer ssDNA. We show that hnRNP K KH3 specifically recognizes a tetrad of sequence 5′d-TCCC. The complex is stabilized by a dense network of methyl-oxygen hydrogen bonds involving the methyl groups of three isoleucine residues and the O2 and N3 atoms of the two central cytosine bases. Comparison with the recently solved structure of a specific protein–ssDNA complex involving the KH3 and KH4 domains of the far upstream element (FUSE) binding protein FBP suggests that the amino acid located five residues N-terminal of the invariant GXXG motif, which is characteristic of all KH domains, plays a crucial role in discrimination of the first two bases of the tetrad.
Keywords: hnRNP K/KH domains/NMR/sequence-specific ssDNA recognition/solution structure
Introduction
The vast majority of transcription factors target double-stranded DNA. However, one class of DNA binding proteins, characterized by the presence of KH domains, has been shown to specifically bind and transactivate single-stranded cis elements within the c-myc promoter both in vivo and in vitro (Duncan et al., 1994; Michelotti et al., 1996a,b; Tomonaga and Levens, 1996). In particular, heterogeneous nuclear ribonucleoprotein K (hnRNP K) binds to the CT element (Michelotti et al., 1996a; Tomonaga and Levens, 1996) and the FUSE binding protein (FBP) binds to the far upstream element (FUSE) (Duncan et al., 1994; Michelotti et al., 1996b) located 100–150 and 1500 bp, respectively, upstream of the c-myc promoter. Single-stranded (ss) DNA within these cis elements is induced by torsion and flexural strain exerted on the DNA during the course of transcription (Duncan et al., 1994), and the recognition of single-stranded cis elements by transcriptional regulators provides a mechanism for the re-establishment of transcription after mitosis (Michelotti et al., 1997) and the tight control of oncogenes (He et al., 2000; Liu et al., 2001).
Recently, we solved the structure of a complex between the KH3 and KH4 domains of FBP (FBP3/4) and a ssDNA 29mer from FUSE (Braddock et al., 2002). In the FBP3/4–FUSE complex, KH3 and KH4 bind in a specific orientation and register to the ssDNA 29mer, which is preserved in complexes of the individual KH domains bound to shorter oligonucleotides, 9–10 bp in length, encompassing their respective target sites in the larger complex. Although the ssDNA binding site is located within a relatively narrow groove that generally favors pyrimidines over purines, the origin of further sequence-specific base recognition appears to be subtle. To explore the molecular basis of ssDNA sequence-specific recognition by KH domains, we have now solved the structure of the KH3 domain of hnRNP K bound to the same ssDNA 10mer used previously for the complex of FBP–KH4 with ssDNA.
hnRNP K is a 463-residue modular protein (Matunis et al., 1992; Bomsztyk et al., 1997; Ostareck-Lederer et al., 1998). Interactions with nucleic acids (ssDNA and RNA) are mediated by three KH domains (Tomonaga and Levens, 1995). hnRNP K KH1 (residues 32–112) and KH2 (residues 142–217) are located at the N-terminal end of the protein, separated by a 30-residue linker, a spacing that is essentially the same as that between the KH3 and KH4 domains of FBP (Braddock et al., 2002). hnRNP K KH3 is isolated from the other two KH domains and located at the C-terminal end of the protein (residues 389–459). The intervening 172-residue stretch between KH2 and KH3 is involved in protein–protein interactions with multiple partners that include other transcription factors (such as TATA binding protein and various zinc finger-containing transcriptional repressors), as well as proteins involved in diverse signal transduction pathways (such as a number of tyrosine and serine/threonine kinases, and the Vav proto-oncoprotein) (Bomsztyk et al., 1997). Among the latter is a class of insulin-sensitive pathways that signal nucleic acid-directed processes involving hnRNP K (Ostrowski et al., 2001).
The CT element consists of four imperfect direct repeats of sequence 5′d-CCCTCCCA (Tomonaga and Levens, 1995). The structure of the FBP3/4–FUSE complex showed that each KH domain recognizes a core sequence of 4–5 bases and that only 6–7 bases make contact with each KH domain (Braddock et al., 2002). FBP KH4 interacts with the sequence 5′d-TATTCCC (with the core recognition sequence in bold) both in the context of the whole FBP3/4–FUSE complex and a smaller complex comprising only KH4 and a ssDNA 10mer of sequence 5′d-ATATTCCCTC, which we termed M5′. Since M5′ contains the TCCC portion of the CT element, and since the number of bases that contact KH domains is small, we anticipated that hnRNP K KH3 would also bind M5′ sequence specifically but in a different register from FBP KH4, thereby permitting a detailed comparison of the factors involved in determining sequence-specific recognition of ssDNA by KH domains.
Results and discussion
Structure determination
The hnRNP K KH3 construct employed in the present study comprises the last 85 C-terminal residues of hnRNP K (residues 379–463, which correspond to residues 5–89 in the current numbering scheme). Upon titration of the M5′ ssDNA into hnRNP K KH3, the majority of 1H-15N cross-peaks observed in a 1H-15N correlation spectrum gradually shift from their free to bound positions as DNA is added; that is, their chemical shifts are weighted averages of the shifts in the free and bound states, characteristic of fast exchange on the chemical shift time scale. The difference in 1HN shifts between the free and bound states for these cross-peaks is ≤200 Hz at a spectrometer frequency of 600 MHz. Non-linear least-squares fitting of the titration data yields an equilibrium dissociation constant of ∼3 ± 1.5 µM, which is comparable to that for the binding of FBP KH4 to M5′. In the case of two residues, Gly32 and Ile49, the 1H-15N cross-peaks corresponding to the free state (at 9.02/112.6 and 8.76/123.6 p.p.m., respectively) decrease in intensity as DNA is added, and the cross-peaks for the bound state (at 10.3/117.8 and 10.09/117.4 p.p.m.) only appear after the addition of ∼0.8–0.9 equivalents of DNA; at a ratio slightly in excess of 1:1 DNA:protein, the bound 1H-15N cross-peaks for these two residues have essentially the same line-widths as the other cross-peaks. Thus, for Gly32 and Ile49, the free and bound 1H-15N cross-peaks are in slow exchange on the chemical shift time scale. At 600 MHz, the shift differences in the 1HN resonances for Gly32 and Ile49 are 766 and 804 Hz, respectively. One can therefore conclude that the dissociation rate constant for the hnRNP K KH3–ssDNA M5′ complex must be >1200 s–1 but <5000 s–1 (with the limits given by 2πΔν, where ν is the chemical shift difference in Hz).
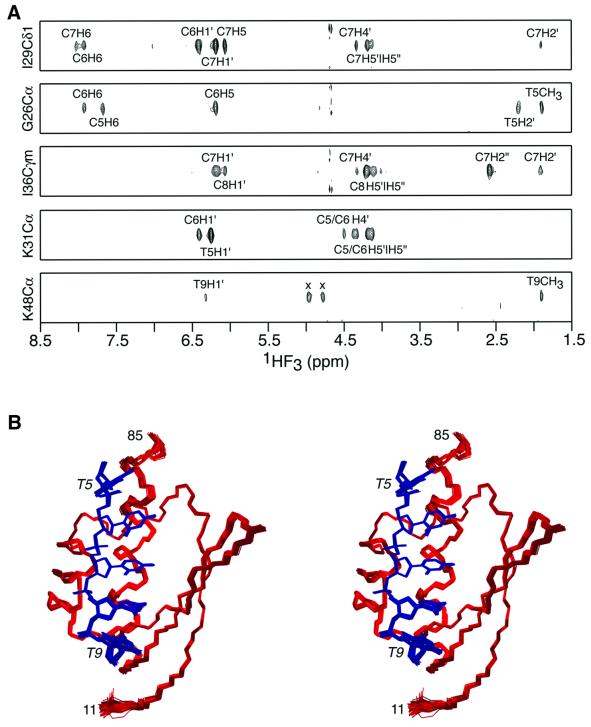
The structure of the hnRNP K KH3–ssDNA M5′ complex was solved by multidimensional NMR (Clore and Gronenborn, 1998a) on the basis of 1986 experimental NMR restraints, including 73 intermolecular NOE derived inter-proton distance restraints and 287 residual dipolar couplings measured in two different alignment media (pf1 phage and polyethylene glycol/hexanol). A summary of the structural statistics is provided in Table I. The quality of the experimental data is illustrated in Figure 1A by selected strips taken from a three-dimensional (3D) 13C-separated/12C-filtered NOE experiment recorded on a sample containing 15N/13C-labeled protein and unlabeled DNA, which permits one to specifically observe only NOEs between protein protons attached to 13C and DNA protons attached to 12C. A stereoview showing the superposition of the final ensemble of 125 simulated annealing structures is shown in Figure 1B.
Table I. Structural statisticsa.
| <SA> | ()r | |
|---|---|---|
| R.m.s. deviation from experimental restraintsa | ||
| Distances (Å) (1289) | 0.037 ± 0.002 | 0.033 |
| Torsion angles (°) (266) | 0.38 ± 0.04 | 0.27 |
| 13Cα/β chemical shifts (Hz) (144) | 1.1 ± 0.1 | 1.1 |
| Dipolar coupling R-factorsb (%) | ||
| PEG/hexanol | ||
| 1DNH (63) | 3.7 ± 0.1 | 2.8 |
| 1DNC′ (44) | 16.4 ± 0.4 | 15.2 |
| 2DHNC′ (40) | 14.5 ± 0.2 | 14.3 |
| Phage pf1 | ||
| 1DNH (56) | 8.9 ± 0.3 | 7.5 |
| 1DNC′ (42) | 22.0 ± 0.5 | 22.2 |
| 2DHNC′ (42) | 22.5 ± 0.4 | 25.2 |
| % residues in most favorable region of Ramachandran mapc | 92.8 ± 1.6 | 95.2 |
| Coordinate precision (Å)d | ||
| Protein backbone + DNA heavy atoms | 0.23 ± 0.05 | |
| Protein heavy atoms + DNA heavy atoms | 0.63 ± 0.05 |
a<SA> are the final 125 simulated annealing structures for the hnRPK KH3–ssDNA complex. ![]() is the average structure obtained by averaging the coordinates of the 125 simulated annealing structures best-fitted to each other (with respect to residues 11–85 of the protein and nucleotides 5–9 of the ssDNA). (
is the average structure obtained by averaging the coordinates of the 125 simulated annealing structures best-fitted to each other (with respect to residues 11–85 of the protein and nucleotides 5–9 of the ssDNA). (![]() )r is the corresponding restrained regularized mean structure derived from
)r is the corresponding restrained regularized mean structure derived from ![]() . The number of terms for the various restraints is given in parentheses. None of the structures exhibits interproton distance violations >0.5 Å or torsion angle violations >5°. There are 1235 structurally useful interproton distance restraints: 1094 interproton distances within the protein [315 intraresidue, and 282 sequential (|i – j|) = 1), 197 medium (1 < |i – j| ≤ 5) and 300 long (|i – j| > 5) range inter-residue restraints]; 68 within the DNA; and 73 intermolecular between protein and DNA. Fifty-four distance restraints for 27 backbone hydrogen bonds located in helices and sheets were added during the final stages of refinement using standard criteria (Clore and Gronenborn, 1998a). The torsion angle restraints are subdivided into 210 restraints for the protein and 55 for the DNA. The r.m.s. deviations from idealized covalent geometry are 0.004 Å for bonds, 0.7° for angles and 0.5° for improper torsions.
. The number of terms for the various restraints is given in parentheses. None of the structures exhibits interproton distance violations >0.5 Å or torsion angle violations >5°. There are 1235 structurally useful interproton distance restraints: 1094 interproton distances within the protein [315 intraresidue, and 282 sequential (|i – j|) = 1), 197 medium (1 < |i – j| ≤ 5) and 300 long (|i – j| > 5) range inter-residue restraints]; 68 within the DNA; and 73 intermolecular between protein and DNA. Fifty-four distance restraints for 27 backbone hydrogen bonds located in helices and sheets were added during the final stages of refinement using standard criteria (Clore and Gronenborn, 1998a). The torsion angle restraints are subdivided into 210 restraints for the protein and 55 for the DNA. The r.m.s. deviations from idealized covalent geometry are 0.004 Å for bonds, 0.7° for angles and 0.5° for improper torsions.
bThe dipolar coupling R-factor, which scales between 0 and 100%, is defined as the ratio of the r.m.s. deviation between observed and calculated values to the expected r.m.s. deviation if the vectors were randomly oriented, given by [2Da2(4 + 3η2)/5]1/2 where Da is the magnitude of the axial component of the alignment tensor and η the rhombicity (Clore and Garrett, 1999). The values of DaNH and η, derived from the distribution of normalized dipolar couplings as described (Clore et al., 1998a), are 15.5 and 0.37 Hz for the dipolar couplings measured in polyethylene glycol (PEG)/hexanol, and 8.8 and 0.58 Hz for the dipolar couplings measured in pf1 phage.
cCalculated with the program PROCHECK (Laskowski et al., 1993). The dihedral angle G-factors for φ/ψ, χ1/χ2, χ1 and χ3/χ4 are 0.03 ± 0.04, 0.46 ± 0.01, –0.19 ± 0.16 and –0.15 ± 0.18, respectively.
dThe precision of the coordinates is defined as the average atomic r.m.s. difference between the individual 125 simulated annealing structures and the corresponding mean coordinates obtained by best-fitting to residues 11–85 of the protein and 5–9 of the ssDNA 10mer. The values for the coordinate precision also refer to the same residues.
Fig. 1. Solution structure of the hnRNP K KH3–ssDNA complex. (A) Strips from a 3D 13C-separated/12C-filtered NOE experiment illustrating intermolecular NOEs between protein protons attached to 13C and DNA protons attached to 12C (along the F3 axis). (The two peaks in the lower panel marked by an x are residual diagonal peaks.) (B) Stereoview showing a superposition of the final 125 simulated annealing structures. The protein backbone (residues 11–85) is shown in red, and the DNA (nucleotides 5–9) in blue. Residues 1–10 and 86–90 of the protein are disordered in solution. Only the nucleotides that interact with the protein are shown. Although the whole ssDNA 10mer displays NOEs characteristic of right-handed DNA, the orientation of the nucleotides that do not contact the protein relative to the core complex (residues 11–85 and nucleotides 5–9) is poorly determined by the experimental data.
Residues 1–10 and 86–89 of the protein are disordered in solution [as evidenced by 15N-{1H}NOE values <0.5 and the absence of any non-sequential inter-residue 1H-1H NOEs]. Although the pattern of inter-nucleotide NOEs and 31P chemical shifts is characteristic of right-handed DNA, the long-range structure of nucleotides 1–4 and 10, which are not in contact with the protein, could not be determined from the NMR data. The structures of the KH3 domain (residues 11–85) and nucleotides 5–9, which directly contact KH3, are defined with high precision (Figure 1B). It should be noted that the high precision of nucleotides 5–9 is predominantly due to the large number of intermolecular NOE contacts (a total of 73) in conjunction with two other factors: (i) the high precision of the protein backbone and aliphatic interfacial side chain coordinates afforded by extensive NOE and protein backbone dipolar coupling data; and (ii) the utilization of a multidimensional torsion angle database potential of mean force to ensure that sampling during simulated annealing is biased towards conformations that are likely to be energetically possible, as defined by those conformations that are known to be physically realizable from high resolution crystal structures (Kuszewski et al., 1996; Clore and Kuszewski, 2002).
Overall structure of the hnRNP K KH3–ssDNA complex
An overall view of the hnRNP K KH3–ssDNA M5′ complex is shown in Figure 2A. The structure of hnRNP K KH3 is typical of a KH domain comprising three α-helices (residues 22–29, 34–43 and 67–84) packed on top of a three-stranded anti-parallel β-sheet (residues 14–20, 45–51 and 58–64) arranged in β1–α1–α2–β2– β3–α3 topology, with the invariant GXXG motif located in the loop connecting helices 1 and 2. The backbone atomic r.m.s. difference between the present structure and that of the unbound G26R mutant of hnRNP K KH3 solved previously (Baber et al., 1999) is ∼1 Å; the only significant differences are around the site of mutation (residues 25–33), which includes the GXXG motif, and in the variable loop region (residues 52–57). The conformation of the GXXG motif in the current structure conforms to that found in the X-ray structure of NOVA-2 KH3 (Lewis et al., 2000) and the NMR structure of FBP KH3/KH4 (Braddock et al., 2002): namely, positive φ angles at positions 1, 3 and 4, with the φ/ψ angles at positions 3 and 4 in a left-handed helical conformation. The back bone r.m.s. differences between hnRNP K KH3, and NOVA-2 KH3, FBP KH3 and FBP KH4, excluding the variable loop (residues 51–58), are 1.2, 1.0 and 1.4 Å, respectively, consistent with a sequence identity of ∼30% (the corresponding values for the free G26R mutant structure are 0.3–0.4 Å higher).
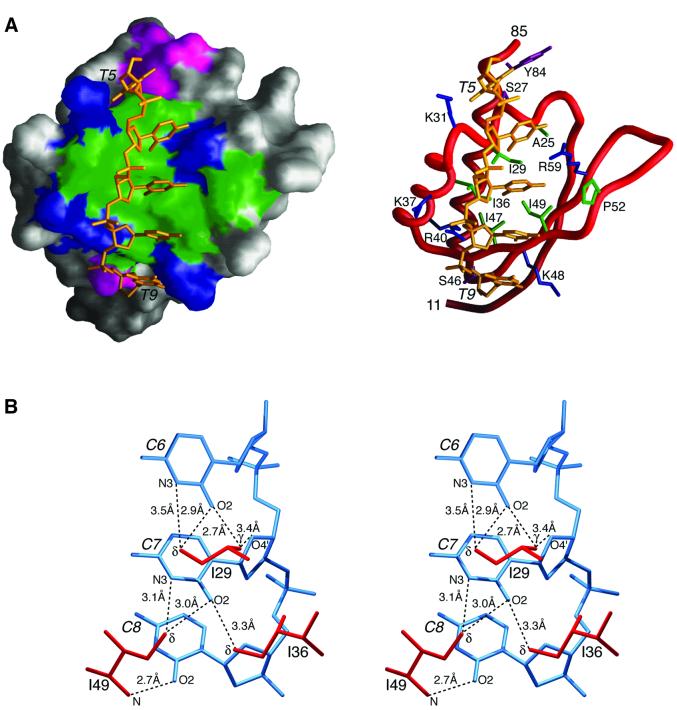
Fig. 2. ssDNA binding by hnRNP K KH3. (A) Overall complex. The protein is displayed as a molecular surface (left) and as a backbone tube (right); hydrophobic, uncharged hydrophilic, positively charged and negatively charged residues located in the ssDNA binding site are depicted in green, magenta, blue and red, respectively; the ssDNA heavy atoms are in gold. (B) Detailed stereoview showing the hydrogen-bonding interactions of the methyl groups of Ile29, Ile36 and Ile49 with the O2 and N3 atoms of the cytosine bases. Nucleotide numbering is in italics.
Nucleotides 5–9 (TCCCT) of the M5′ ssDNA contact the protein and are located in a narrow hydrophobic groove lined by four clusters of positively charged residues (Lys31, Lys37/Arg40, Lys48 and Arg59) and capped top (Tyr84 and Ser27) and bottom (Ser46) by a hydrophilic residue(s) (Figure 2A). The ssDNA has a right-handed helical twist, but is more extended and underwound relative to right-handed double-stranded DNA. The binding site on hnRNP K KH3 defined by the present structure is fully consistent with a previous chemical shift mapping study using the longer ssDNA sequence 5′d-TTCCCCTCCCCATTT (Baber et al., 2000). A summary of the protein–ssDNA contacts is provided in Figure 3B (left).
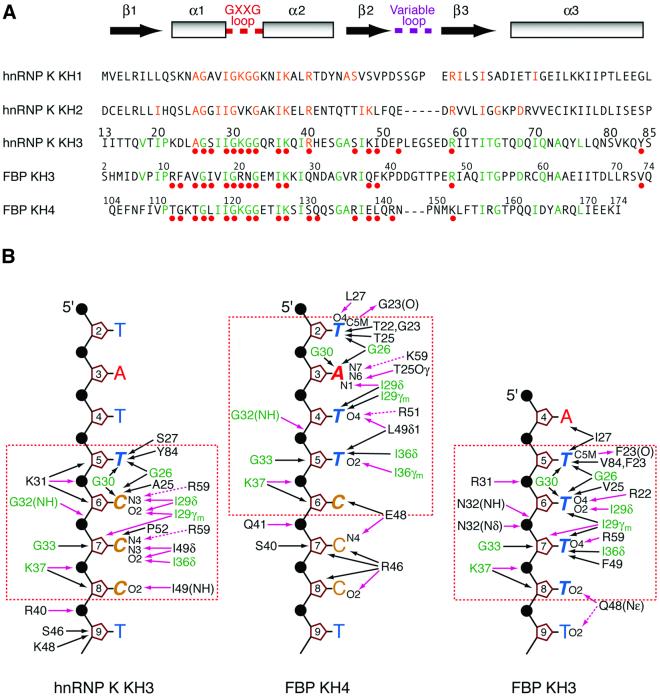
Fig. 3. Comparison of the interaction of hnRNP K KH3, FBP KH3 and FBP KH4 with ssDNA. (A) Structure-based sequence alignment of the KH domains of hnRNP K and the KH3 and KH4 domains of FBP. The location of the secondary structure elements is shown at the top of the figure. The loop connecting strands β2 and β3 is variable in length (five residues in hnRNP K KH1, six residues in hnRNP K KH3 and FBP KH3, seven residues in hnRNP K KH2, and nine residues in FBP KH4). Residues that contact the DNA in the complexes of hnRNP K KH3, FBP KH3 and FBP KH4 with ssDNA are indicated by the solid red circles. Residues in the KH3 and KH4 domains of FBP that are identical in the KH domain of hnRNP K KH3 are colored green; residues in the KH1 and KH2 domains of hnRNP that are identical in the KH3 domain of hnRNP K are colored orange. The numbering scheme for hnRNP K KH3 is shown above the hnRNP K KH3 sequence; the numbering of FBP KH3 and KH4 used by Braddock et al. (2002) is indicated in smaller numerals above their respective sequences. For the structurally aligned sequences shown in the figure, the overall percentage sequence identities of hnRNP K KH1, hnRNP K KH2, FBP KH3 and FBP KH4 to hnRNP K KH3 are 23, 24, 26 and 32%, respectively. (B) Summary of interactions between hnRNP K KH3, FBP KH4 and FBP KH3 and ssDNA. The protein residue and ssDNA nucleotide numbering employed is that of the hnRNP K KH3–ssDNA complex. Residues at the protein–ssDNA interface that are common to all three complexes are shown in green. Hydrogen bonds and salt bridges are represented by purple arrows; the dashed arrows indicate potential electrostatic interactions, either direct or water mediated. The core recognition sequences are enclosed by the dashed orange box. The ssDNA 10mers used for the complexes with hnRNP K KH3 and FBP KH4 are identical. The core recognition elements for hnRNP K KH3 and FBP KH3 are the tetrads 5′d-TCCC and 5′d-TTTT, respectively; the core recognition element for FBP KH4 is a pentad of sequence 5′d-TATTC. The first two nucleotides of each recognition sequence form a vice that grips helix 1 in all three complexes, and the nature of the amino acid at positions 25 (numbering scheme of hnRNP K KH3) is critical for determining the register of ssDNA binding: Ala for the KH domains of hnRNPK, Thr for FBP KH4 and Val for FBP KH3. The intermolecular contacts observed in the FBP KH3 and KH4 complexes were analyzed from the coordinates of Braddock et al. (2002) (accession code 1J4W).
The protein–ssDNA interface
Base contacts only involve the tetrad TCCC. The first two nucleotides of the tetrad (TC) clasp helix 1, with the bases of T5 and C6 located on the top and bottom sides of the helix, respectively, in the view shown in Figure 2A. The Cα atom of Gly26 is partially intercalated between the bases of T5 and C6, which readily accounts for the observation that the G26R mutant no longer binds ssDNA (Baber et al., 1999), since the longer Arg side chain would introduce severe steric clash. Interactions with the bases of C6 and C7 are characterized by an extensive network of hydrogen bonds (heavy atom donor/acceptor distances ≤3.5 Å) involving the methyl groups of Ile29, Ile36 and Ile49 with the O2 and N3 atoms of the C bases (Figure 2B): the δ-methyl of Ile29 is hydrogen bonded to the N3 and O2 atoms of C6; the γ-methyl of Ile29 to the O2 and sugar O4′ atoms of C6; the δ-methyl of Ile36 to the O2 atom of C7; and the δ-methyl of Ile49 to the O2 and N3 atoms of C7. Although weak in nature, CH---O hydrogen bonds have been observed in both proteins (Derewenda et al., 1995) and protein–duplex DNA complexes (Mandel-Gutfreund et al., 1998). In the latter, the methyl group of T and the H5 atom of C have been observed to act as donors and side-chain oxygen atoms of the protein as acceptors (Mandel-Gutfreund et al., 1998). In this instance, however, the donor methyl groups originate from the protein. These CH---O hydrogen-bonding interactions are supplemented by a hydrogen bond from the backbone amide of Ile49 to the O2 atoms of C8, and putative hydrogen bonds (distances between heavy atoms <3.5 Å in 85% of the simulated annealing structures) from the guanidino group of Arg59 to the N3 atom of C3 and possibly via a water molecule to the N4 amide group of C7 as well. In addition to interactions with the bases, there are four electrostatic interactions with phosphate groups involving the backbone amide of Gly32 (whose 1H resonance at 10.3 p.p.m. is downfield shifted by 1.28 p.p.m. relative to that in the free protein) and the side chains of Lys31, Lys37 and Arg40, as well as a rather limited number of contacts with the deoxyribonucleotide sugars (Figure 3B).
The KH domains of hnRNP K provide a molecular scaffold for interaction with both ssDNA and RNA (Bomsztyk et al., 1997). Although hnRNP K binds to ssDNA slightly more tightly than to RNA (Tomonaga and Levens, 1995), interactions with RNA have also been shown to be functionally important. In particular, binding of hnRNPK to the CU-rich differentiation control element DICE in the 3′-untranslated region of 15-lipoxygenase (LOX) mRNA mediates translation silencing of LOX in erythroid precursors (Ostareck et al., 1997). Recent SELEX experiments have shown that the optimal consensus RNA sequence motif is single-stranded 5′-UC3-4(U/A)(A/U) (Thisted et al., 2001), which is in effect the RNA equivalent of the ssDNA CT element. Thus, hnRNPK displays essentially the same base specificity for ssDNA and RNA. The conformation of ssRNA bound to hnRNP K KH3 is probably rather similar to that of ssDNA since the central three C nucleotides of the bound TCCCT ssDNA sequence adopt an RNA-like 3′-endo sugar pucker conformation. (The outer two Ts, as well as the other nucleotides in the M5′ ssDNA 10mer sequence exhibit predominantly 2′-endo sugar puckers.) Some changes in the details of binding are likely, however, as a consequence of the replacement of the H2” proton in the deoxyribose of DNA by the 2′-hydroxyl group in the ribose of RNA. Indeed, intermolecular NOEs to the H2” proton of C6 and C7 protons are observed from the γ-methyl groups of Ile29 and Ile36, respectively, and small coordinate shifts in an analogous RNA complex could readily accommodate a number of CH–O hydrogen bonds involving these methyl groups and the O2′ oxygen atoms of the ribose sugars.
Comparison of the hnRNP K KH3, FBP KH3 and FBP KH4 complexes with ssDNA
The KH4 domain of FBP binds specifically to the M5′ ssDNA 10mer (Braddock et al., 2002), but in a different register to hnRNP K KH3. The core recognition sequence in the FBP KH4–ssDNA complex is a pentad of sequence TATTC comprising nucleotides 2–6 of the M5′ ssDNA (Braddock et al., 2002), with nucleotides 2, 3, 4 and 5/6 occupying approximately the same location as the tetrad TCCC at nucleotide positions 5, 6, 7 and 8, respectively, in the hnRPK KH3 complex. The core recognition sequence for FBP KH3 is the TTTT tetrad (Braddock et al., 2002), which is positioned similarly to the TCCC tetrad in the hnRPK KH3–ssDNA complex. What is the basis of sequence-specific ssDNA recognition in these three complexes? A structure-based sequence alignment of hnRNP K KH3 with FBP KH3 and KH4 is shown in Figure 3A, and a comparison of the protein–DNA contacts in the three complexes is presented in Figure 3B.
Seventeen residues of hnRNP K KH3, 16 of FBP KH3 and 18 of FBP KH4 contact the ssDNA (Figure 3A). The sites overlap extensively with 14 residues in common. Moreover, 40–50% of the residues that interact with ssDNA are identical in the three complexes (Figure 3A). Nevertheless, it is clear that the three proteins recognize three different ssDNA sites, and that the conformation of the ssDNA readily adapts itself by means of small changes in torsion angles to optimize protein–ssDNA contacts. Overall, the conformation of the bound ssDNA and the intermolecular interactions observed in the hnRNP K KH3 and FBP KH3 complexes are more similar to each other than to the FBP KH4 complex. This is due to the fact that the sequences recognized by hnRNP K KH3 and FBP KH3 are composed entirely of pyrimidines, whereas that bound by FBP KH4 is characterized by a purine (A) at position 2 of the core recognition sequence which has to be accommodated in the relatively narrow binding groove.
Discrimination of nucleotides 1 and 2 of the core recognition sequence
A key aspect of sequence discrimination and binding register involves the first two bases of the ssDNA core recognition site and the manner in which they interact with the residues of the first helix: specifically T5 and C6 for hnRNP K KH3, T5 and T6 for FBP KH3, and T2 and A3 for FBP KH4 (Figures 3B and 4). Particularly critical is the nature of the residue at position 25 (Ala, Val and Thr in hnRNP K KH3, FBP KH3 and FBP KH4, respectively), which appears to play an important role in the choice of the second base.
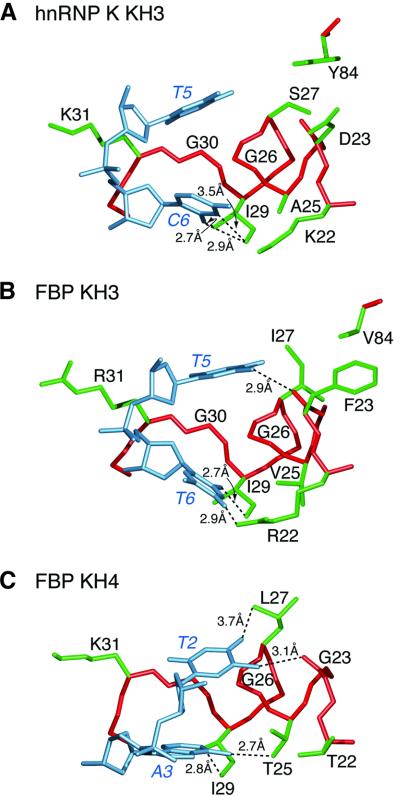
Fig. 4. Discrimination of the first two first bases of the ssDNA recognition site by KH domains of hnRNP K and FBP. (A) hnRNP K KH3 recognizes TC, (B) FBP KH3 recognizes TT and (C) FBP KH4 recognizes TA. The protein backbone and side chains are shown in red and green, respectively, and the DNA in light blue. The numbering scheme employed is that of the hnRNP K KH3–ssDNA complex. Dashed lines indicate intermolecular hydrogen bonds. The residue at position 25 plays a key role in selection of the first two bases of the site. The coordinates of the FBP KH3/KH4 complex (accession code 1J4W) are taken from Braddock et al. (2002).
In all three complexes, the first two bases grip helix 1, with Gly26 partially intercalated between them, but their exact positions differ subtly depending upon the nature of the interacting protein residues and the identity of the bases (Figures 3B and 4). In the view shown in Figure 4, the two bases shift progressively towards the right of the binding pocket as one proceeds from hnRNP K KH3 to FBP KH3 and finally to FBP KH4.
The first nucleotide is always a T with a 2′-endo sugar pucker and an anti-glycosidic bond torsion angle (χ) in the –105° to –110° range. In the case of hnRNP K KH3, T5 participates exclusively in hydrophobic interactions with the base packed against the α-methylene groups of Gly26 and Gly30, the β-methylene group of Ser27, and the aromatic ring of Tyr84, and the deoxyribose sugar bolstered by the aliphatic portion of the side chain of Lys31 (Figure 4A). For FBP KH3, the interactions are similar but not identical as a consequence of amino acid changes at positions 23 (Phe for Asp), 27 (Ile for Ser) and 84 (Val for Tyr). Thus, hydrophobic contacts with the base of T5 involve the δ-methyl group of Ile27, the methylene group of Phe23 and the methyl groups of Val84, as well as the α-methylenes of Gly26 and Gly30, and the deoxyribose sugar is still bolstered by the aliphatic portion of the side chain of Arg31 (Figure 4B). However, the methyl group of T5 donates a hydrogen bond to the backbone carbonyl of Phe23: the distance between donor and acceptor heavy atoms is 2.9 Å, compared with 5.4 Å for the equivalent atoms in the hnRNP K KH3 complex, as a consequence of a small right shift of the T bases in the KH domain binding groove (Figure 4A and B). In the FBP KH4 complex, the residue at position 84 is absent (since the FBP KH4 domain ends at position 81), and the residues at positions 23 and 27 are Gly and Leu, respectively. The methyl group of T2 donates a hydrogen bond to the backbone carbonyl of Gly23 and is also in van der Waals contact with the α-methylene group of Gly23; and the O4 atom of T2 accepts a hydrogen bond from the methyl group of Leu27 (Figure 4C). The further rightward shift of the T2 base in the binding site relative to the corresponding nucleotide in the FBP KH3 complex is such that the aliphatic portion of the side chain of Lys31 no longer contacts the deoxyribose sugar of T2 (Figure 4C).
The second nucleotide of the core recognition sequence is C for hnRNP K KH3 (3′-endo sugar pucker and χ approximately –150°), T for FBP KH3 (3′-endo sugar pucker and χ approximately –150°) and A for FBP KH4 (2′-endo sugar pucker and χ approximately –110°). The principal residues of the protein contacting the second nucleotide are located at positions 22, 25 and 29. As discussed above, the base of C6 in the hnRNP K KH3 complex accepts three hydrogen bonds from the methyl groups of Ile29 (δ-methyl to the N3 and O2 atoms, and the γ-methyl to the O2 atom). In the FBP KH3 and KH4 complexes, only the δ-methyl group of Ile29 donates a hydrogen bond, namely to the O2 atom of T6 and the N1 atom of A3, respectively. The guanidino group of Arg22 of FBP KH3 donates a hydrogen bond to the O4 atom of T6 (Figure 4B), but the shorter Lys and Thr side chains at this position in hnRNP K KH3 and FBP KH4 preclude any interaction with the second base (Figure 4A and C). In the hnRNP K KH3 complex, the methyl group of Ala25 packs against the N3 atom and the N4 amino group of C6 (Figure 4A); in the FBP KH3 complex, Ala is replaced by the larger Val side chain at position 25, which packs against the O2 atom and the N3 imino proton of T6 (Figure 4B); and in the FBP KH4 complex, the hydroxyl Oγ-atom of Thr25 of FBP KH4 accepts a hydrogen bond from the N6 amino group of A3 (Figure 4C).
This suggests that the presence of the small Ala side chain at position 25 of hnRNP K KH3 (which is conserved in all three KH domains of hnRNP K; Figure 3A) favors a C base over a T at position 2 of the core recognition sequence, presumably to permit optimal interactions with the larger N4 amino group of C versus the smaller O4 atom of T. The larger Val side chain at position 25 and the long Arg side chain at position 22 in FBP KH3 favor a T as a consequence of optimal hydrophobic and hydrogen- bonding interactions, respectively, which would not be available for a C base. Finally, a Thr at position 25 favors A as a consequence of optimal hydrogen-bonding interactions.
Recognition of nucleotides at the 3′-end of the core recognition sequence
The path of the DNA followed by nucleotides 3 and 4 (and in the case of FBP KH4, nucleotide 5 as well) of the core recognition sequence are probably driven to a large extent by the interactions set up by the first two bases. For all three complexes, there are extensive interactions with the bases of nucleotides 3 and 4, including numerous hydrogen bonds (Figure 3B). In addition, the sugar puckers and glycosidic bond torsion angles for these two nucleotides are 3′-endo and low anti (χ approximately –150°), respectively, in all three complexes.
Some important discriminatory interactions include optimization of CH---O hydrogen bonds involving Ile36 and Ile49 and the base of C7 at nucleotide 3 in the hnRNP K KH3 complex, together with a hydrogen bond between the backbone amide group of Ile49 and the O2 atom of C8 at nucleotide 4 (Figure 3B, left). In this regard, it is interesting to note that the amide proton of Ile49 resonates at 10.09 p.p.m., which is 1.34 p.p.m. downfield of its position in the free protein, indicative of a hydrogen-bonding interaction; by way of contrast, the amide protons of Phe49 and Leu49 in FBP KH3 and KH4 resonate at 9.2 and 8.6 p.p.m. and are only minimally shifted (0.09 p.p.m. downfield and 0.05 p.p.m. upfield, respectively) relative to their respective unbound states (D.T.Braddock and G.M.Clore, unpublished data). Thus, the path followed by the 3′-end of the ssDNA recognition sequence in the three complexes is clearly not identical.
In the case of the FBP KH3 complex, discriminatory interactions that favor Ts at nucleotides 3 and 4 include hydrogen bonds from the guanidino group of Arg59 and the carboxyamide of Gln48 to the O4 atom of T7 and the O2 atom of T8, respectively (Figure 3B, right).
The length of the bound ssDNA in the FBP KH4 complex is more extensive than in the other two complexes, and discriminatory contacts involve hydrogen bonds from the methyl group of Leu49 and the guanidino group of Arg 51 to the O4 atom of T4 at nucleotide 3, a hydrogen bond from the γ-methyl of Ile36 to the O2 atom of T5 at nucleotide 4, and outside the core recognition sequence, hydrogen bonds from the carboxylate of Glu48 to the N4 amino group of C7 at nucleotide 6 and from the guanidino group of Arg46 to the O2 atom of C8 at nucleotide seven (Figure 3B, center). These more extensive interactions outside the core recognition sequence are probably a consequence of the relative locations of the FBP KH4 and KH3 binding sites: the former is 5′ of the latter and the two sites are separated by five nucleotides (Braddock et al., 2002). Thus the path followed by the 3′-end of the ssDNA in the binding groove of FBP KH4 may in part be dictated by topological and stereochemical considerations that ensure that the first two bases of the KH3 core recognition sequence are correctly positioned to optimally interact with the KH3 domain.
Concluding remarks
The structure of the hnRNP K KH3–ssDNA complex, together with the previously determined structures of the FBP KH3 and KH4 complexes (Braddock et al., 2002), provide a framework for understanding the molecular basis of ssDNA sequence-specific recognition and illustrate the ability of ssDNA to structurally conform to subtle amino acid changes in the binding sites of KH proteins, differences that confer on the KH domains their unique base specificity. The structures also highlight the role and versatility of weak methyl CH---O hydrogen bonds in mediating specific interactions between KH domains and ssDNA.
hnRNP K has been implicated in the regulation of transcription as an activator and as a repressor, as a translational repressor, and as a participant in a variety of signaling systems (Bomsztyk et al., 1997; Ostareck-Lederer et al., 1998). hnRNP K receives signals from different protein kinases that modify both its nuclear– cytoplasmic distribution and its activity in transcription or translation (Schullery et al., 1999; Ostrowski et al., 2000, 2001; Habelhah et al., 2001a,b). hnRNP K may also serve as a platform to dock otherwise separated molecules (Bomsztyk et al., 1997; Ostareck-Lederer et al., 1998). What insight does the present structure provide to understand the role of hnRNP K in the cell? The stereotypical folding of the polypeptide backbone of KH domains creates an elongated groove on the surface of the protein for single-stranded nucleic acids, DNA or RNA. Upon binding ssDNA, only relatively minor adjustments in amino acid side chains are required for base recognition. The dense network of interactions between the floor and ridges of the groove that recognize specific sequences is relatively indifferent to the features distinguishing RNA and ssDNA. Base recognition is sufficiently robust so that SELEX of random RNA (Thisted et al., 2001) delivers the same sequence of bases that occurs naturally in the DNA (i.e. the CT element) upstream of the c-myc promoter (Michelotti et al., 1996a). Once aligned via hydrogen bonds between the bases and the side chains, van der Waals contacts may impart a slight preference for DNA consistent with biochemical observations (Tomonaga and Levens, 1995). hnRNP K is sufficiently abundant and binds target nucleic acids with sufficiently high affinity to ensure interaction with most available targets. In the case of DNA, the array of candidate target sequences is stringently filtered by the requirement that these sequences be melted. Therefore, DNA binding by hnRNP K must be obligatorily coupled with some nuclear process stably or dynamically stressing B-DNA. In the case of RNA, secondary structure may also provide an impediment for the recognition of some candidate targets. The physiological regulation layered upon hnRNP K by protein phosphorylation and dephosphorylation most likely modifies hnRNP K’s ability to engage protein partners, and to partition between the nucleus and cytoplasm (Schullery et al., 1999; Ostrowski et al., 2000, 2001; Habelhah et al., 2001a,b). None of the known sites of hnRNP K phosphorylation occurs within KH domains and so phosphorylation is unlikely to directly augment or interfere with sequence recognition. The role of hnRNP K is therefore likely to be dictated by parameters governing its partners and location. As such, hnRNP K has features particularly well suited to coordinating and integrating steps in gene expression.
Materials and methods
Sample preparation
hnRNP K KH3 comprising the last 85 C-terminal residues of hnRNP K (residues 379–463, which correspond to residues 5–89 in the current numbering scheme) together with an additional four residues at the N-terminus were cloned, expressed and purified by affinity chromatography using standard procedures as described previously (Baber et al., 2000). The 10mer ssDNA was purchased from Midlands Certified Reagent Company (Midland, TX) and purified by anion-exchange chromatography. Samples for NMR contained ∼1:1.2 complexes of protein (15N, or 15N/13C) to ssDNA in 20 mM sodium phosphate, 20 µM EDTA and 0.02% NaN3 pH 6.8.
NMR spectroscopy
All NMR experiments were carried out at 35°C on Bruker 600, 750 and 800 MHz spectrometers. 1H, 15N and 13C backbone and side-chain resonances of the protein were assigned by 3D double and triple resonance NMR experiments, and 1H resonances for the ssDNA by 2D 12C-filtered experiments (Clore and Gronenborn, 1998a). In addition, 31P resonances of the ssDNA were assigned from a 2D 1H-31P single quantum coherence correlation spectrum in which the 31P resonance of residue (i) is correlated to H3′(i – 1) via a 3J coupling and to the H4′ via a 4J coupling (Gorenstein, 1995). The observation of strong 31P(i)–H4′(i) correlations and the absence of 31P(i)–H5′/H5”(i) correlations indicates that the β- and γ-sugar–phosphate torsion angles are in the t and g+ conformations, respectively; and the very narrow dispersion of the 31P resonances (between 4 and 4.5 p.p.m.) indicates that the overall conformation of the sugar–phosphate backbone is characteristic of conventional right-handed DNA (Gorenstein, 1995). Interproton distance restraints (classified into ranges; Clore and Gronenborn, 1998a) within the protein were derived from 3D and 4D 15N- and 13C-separated NOE experiments; within the DNA from 2D 12C-filtered NOE and ROE experiments; and between the protein and DNA from 3D 13C-separated/12C-filtered and 15N-separated/12C-filtered NOE experiments (Clore and Gronenborn, 1998a). Backbone φ,ψ torsion angle restraints were derived from backbone chemical shifts using the program TALOS (Cornilescu et al., 1999). Broad torsion angle restraints for the DNA sugar–phosphate backbone were derived as described previously (Murphy et al., 2001): α = –70 ± 50°, β = 180 ± 50°, γ = 60 ± 35°, ε = 180 ± 50° and ζ = –85 ± 50°. Restraints for the δ (C5′–C4′–C3′–O3) torsion angle, which is related to the sugar pucker, were deduced from 2D 12C-filtered ROE, short (20 ms) mixing time 12C-filtered NOE, and out (H1′ and H3′) and back (H2′/H2”) 12C-filtered COSY spectra (Murphy et al., 2001). Heteronuclear 3J couplings were measured by quantitative J-correlation spectroscopy (Bax et al., 1994). Dipolar couplings were measured in dilute liquid-crystalline medium of 18 mg/ml phage pf1 (Clore et al., 1998b; Hansen et al., 2000) and 5% C12E5 polyethylene glycol/hexanol mixture (with a molar ratio of surfactant to alcohol of 0.96) (Rückert and Otting, 2000).
Structure calculations
Structures were calculated using well-established procedures (Clore and Gronenborn, 1998b) from the experimental NMR restraints by simulated annealing in torsion angle space (Schwieters and Clore, 2001a) using the program XPLOR-NIH (Clore et al., 2001). The non-bonded contacts in the target function are represented by a quartic van der Waals repulsion term supplemented by a torsion angle database potential of mean force (Clore and Kuszewski, 2002). Structure figures were generated with the programs VMD-XPLOR (Schwieters and Clore, 2001b) and GRASP (Nicholls et al., 1991).
The coordinates have been deposited in the Protein Data Bank (accession code 1J5K).

Acknowledgments
Acknowledgements
This work was supported in part by the AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G.M.C.)
References
- Baber J.L., Libutti,D., Levens,D. and Tjandra,N. (1999) High-precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein J, a c-myc transcription factor. J. Mol. Biol., 289, 949–962. [DOI] [PubMed] [Google Scholar]
- Baber J.L., Levens,D., Libutti,D. and Tjandra,N. (2000) Chemical shift mapped DNA-binding sites and the 15N relaxation analysis of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K. Biochemistry, 39, 6022–6032. [DOI] [PubMed] [Google Scholar]
- Bax A., Vuister,G.W., Grzesiek,S., Delaglio,F., Wang,A.C., Tschudin,R. and Zhu,G. (1994) Measurement of homo- and heteronuclear J-couplings from quantitative J correlation spectroscopy. Methods Enzymol., 239, 79–106. [DOI] [PubMed] [Google Scholar]
- Bomsztyk K., van Seuningen,I., Suzuki,H., Denisenko,O. and Ostrowski,J. (1997) Diverse molecular interactions of the hnRNP K protein. FEBS Lett., 403, 113–115. [DOI] [PubMed] [Google Scholar]
- Braddock D.T., Louis,J.M., Baber,J.L., Levens,D. and Clore,G.M. (2002) Structure and dynamics of KH domains from FBP bound to ss DNA. Nature, 415, 1051–1056. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Garrett,D.S. (1999) R-factor, free R and complete cross-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc., 121, 9008–9012. [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1998a) Determining structures of larger proteins and protein complexes by NMR. Trends Biotechnol., 16, 22–34. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1998b) New methods of structure refinement for macromolecular structure determination by NMR. Proc. Natl Acad. Sci. USA, 95, 5891–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G.M. and Kuszewski,J. (2002) χ1 rotamer populations and angles of mobile surface side chains are accurately predicted by a torsion angle database potential of mean force. J. Am. Chem. Soc., 124, 2866–2867. [DOI] [PubMed] [Google Scholar]
- Clore G.M., Gronenborn,A.M. and Bax,A. (1998a) A robust method for determining the magnitude of the fully asymmetric alignment tensor of oriented macromolecules in the absence of structural information. J. Magn. Reson., 131, 159–162. [DOI] [PubMed] [Google Scholar]
- Clore G.M., Straich,M.R. and Gronenborn,A.M. (1998b) Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J. Am. Chem. Soc., 120, 10571–10572. [Google Scholar]
- Clore G.M., Kuszewski,J., Schwieters,C.D. and Tjandra,N. (2001) XPLOR-NIH version 1.1.2. http://nmr.cit.nih.gov/xplor-nih
- Cornilescu G., Delaglio,F. and Bax,A. (1999) Protein backbone angle restraints from searching a database of protein chemical shift and sequence homology. J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- Derewenda Z.S., Lee,L. and Derewenda,U. (1995) The occurence of CH---O hydrogen bonds in proteins. J. Mol. Biol., 252, 248–262. [DOI] [PubMed] [Google Scholar]
- Duncan R., Bazar,L., Michelotti,G.A, Tomonaga,T., Krutzsh,H., Avigan,M. and Levens,D. (1994) A sequence-specific, single-stranded binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev., 8, 465–480. [DOI] [PubMed] [Google Scholar]
- Gorenstein D.G. (1995) Conformation and dynamics of DNA and protein–DNA complexes by 31P NMR. Chem. Rev., 94, 1315–1338. [Google Scholar]
- Habelhah H., Shah,K., Huang,L., Ostareck-Lederer,A., Burlingam,A.L., Shokat,K.M., Hentze,M.W. and Ronai,Z. (2001a) ERK phos phorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol., 3, 325–330. [DOI] [PubMed] [Google Scholar]
- Habelhah H., Shah,K., Huang,L., Burlingame,A.L., Shokat,K.M. and Ronai,Z. (2001b) Identification of new JNK substrate using ATP pocket mutant JNK and a corresponding ATP analogue. J. Biol. Chem., 276, 18090–18095. [DOI] [PubMed] [Google Scholar]
- Hansen M.R., Hanson,P. and Pardi,A. (2000) Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Methods Enzymol., 317, 220–240. [DOI] [PubMed] [Google Scholar]
- He L., Liu,J., Collins,I., Sanford,S., O’Connel,B., Benham,C.J. and Levens,D. (2000) Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J., 19, 1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuszewski J., Gronenborn,A.M. and Clore,G.M. (1996) Improvements and extensions in the conformational database potential for the refinement of NMR and X-ray structures of proteins and nucleic acids. J. Magn. Reson., 125, 171–177. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lewis H.A., Musunuru,K., Jensen,K.B., Edo,C., Chen,H., Darnell,R.B. and Burley,S.K. (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell, 100, 323–332. [DOI] [PubMed] [Google Scholar]
- Liu J. et al. (2001) Defective interplay of activators and repressors with TFIIH in xeroderma pegmentosum. Cell, 104, 353–363. [DOI] [PubMed] [Google Scholar]
- Mandel-Gutfreund Y., Margalit,H., Jernigan,R.L. and Zhurkin,V.B. (1998) A role for CH---O interactions in protein–DNA recognition. J. Mol. Biol., 277, 1129–1140. [DOI] [PubMed] [Google Scholar]
- Matunis M.L., Michael,W.M. and Dreyfuss,G. (1992) Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol., 12, 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti E.F., Michelotti,G.A., Aronsohn,A.I. and Levens,D. (1996a) Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol., 16, 2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti G.A., Michelotti,E.F., Pullner,A., Duncan,R.C., Eick,D. and Levens,D. (1996b) Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol., 16, 2656–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti E.F., Sanford,S. and Levens,D. (1997) Marking of active genes on mitotic chromosomes. Nature, 388, 895–899. [DOI] [PubMed] [Google Scholar]
- Murphy E.C., Zhurkin,V.B., Louis,J.M., Cornilescu,G. and Clore,G.M. (2001) Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol., 312, 481–499. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights into interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Ostareck D.H., Ostareck-Lederer,A., Wilm,M., Thiele,B.J., Mann,M. and Hentze,M.W. (1997) mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell, 89, 597–606. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A., Pstaredk,D.H. and Hentze,M.W. (1998) Cyto plasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci., 23, 409–411. [DOI] [PubMed] [Google Scholar]
- Ostrowski J., Schullery,D.S., Denisenko,O.N., Higaki,Y., Watts,J., Aebersold,R., Stempka,L., Gschwendt,M. and Bomsztyk,K. (2000) Role of tyrosine phosphorylation in the regulation of the interaction of heterogeneous ribonucleoprotein protein K with its protein and RNA partners. J. Biol. Chem., 275, 3619–3628. [DOI] [PubMed] [Google Scholar]
- Ostrowski J., Kawata,Y., Schullery,D.S., Denisenko,O.N., Higaki,Y., Abrass,C.K. and Bomsztyk,K. (2001) Insulin alters heterogeneous nuclear ribonucleoprotein K protein binding to DNA and RNA. Proc. Natl Acad. Sci. USA, 98, 9044–9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückert M. and Otting,G. (2000) Alignment of biological macro molecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc., 122, 7793–7797. [Google Scholar]
- Schullery D.S., Ostrowski,J., Denisenko,O.N., Stempka,L., Shnyereva, M., Suzuki,H., Gswendt,M. and Bomsztyk,K. (1999) Regulated interaction of protein kinase Cδ with the heterogeneous nuclear ribonucleoprotein K protein. J. Biol. Chem., 274, 15101–15109. [DOI] [PubMed] [Google Scholar]
- Schwieters C.D. and Clore,G.M. (2001a) Internal coordinates for molecular dynamics and minimization in structure determination and refinement. J. Magn. Reson., 152, 288–302. [DOI] [PubMed] [Google Scholar]
- Schwieters C.D. and Clore,G.M. (2001b) The VMD-XPLOR visualization package for NMR structure refinement. J. Magn. Reson., 149, 239–244. [DOI] [PubMed] [Google Scholar]
- Thisted T., Lyakhov,D.L. and Liebhaber,S.A. (2001) Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and αCP-2KL, suggest distinct modes of RNA recognition. J. Biol. Chem., 276, 17474–17496. [DOI] [PubMed] [Google Scholar]
- Tomonaga T. and Levens,D. (1995) Keterogeneous nuclear ribo nucleoprotein K is a DNA-binding transactivator. J. Biol. Chem., 270, 4875–4881. [DOI] [PubMed] [Google Scholar]
- Tomonaga T. and Levens,D. (1996) Activating transcription from single-stranded DNA. Proc. Natl Acad. Sci. USA, 93, 5830–5835. [DOI] [PMC free article] [PubMed] [Google Scholar]