Abstract
Some protein-coding genes in metazoan genomes contain a minor class of introns that are excised by a distinct, low-abundance spliceosome. We have developed a quantitative RT–PCR assay that allows comparison of the relative rates of intron removal from the transcripts present in a pre-mRNA population. We show that the U12-type introns are more slowly spliced than the major-class (U2-type) introns from three endogenous pre-mRNAs in human tissue culture cells. In Drosophila melanogaster S2 cells, using minigene constructs designed to produce nearly identical mRNAs, we observe increased expression of fluorescent protein and mature mRNA upon mutation of a U12-type to a U2-type intron. These results provide evidence that the level of gene expression in vivo is lowered by the presence of a U12-type intron and implicate the U12-type spliceosome as a target in the post-transcriptional regulation of gene expression.
Keywords: pre-mRNA/regulation of gene expression/spliceosome/splicing rate/U12-type intron
Introduction
A rare, divergent class of introns was identified by Jackson (1991) and Hall and Padgett (1994); they noticed a few introns with unusual AU and AC dinucleotides at their 5′ and 3′ termini, differing from the nearly invariant GU and AG termini of canonical introns. These minor-class introns were further distinguishable from major-class introns based on highly conserved sequences at their 5′ splice site and branch site, as well as by the lack of a poly pyrimidine tract upstream of the 3′ splice site (Hall and Padgett, 1994). Sequence complementarity suggested that the 5′ splice sites and branch sites of these introns might interact with U11 and U12 snRNPs, respectively (Hall and Padgett, 1994). It was subsequently shown that the excision of minor-class introns is indeed mediated by a distinct low-abundance spliceosome. The U12-type spliceosome contains U11, U12, U4atac and U6atac snRNPs, which are the functional analogues of the major-class U1, U2, U4 and U6 snRNPs, while the U5 snRNP participates in the splicing of both U12-type and classical (U2-type) introns (Hall and Padgett, 1996; Tarn and Steitz, 1996a,b). Whereas AU–AC termini were initially considered to be a defining feature of minor-class introns, mutation to GU–AG termini does not interfere with splicing via the U12-dependent pathway (Dietrich et al., 1997). In fact, surveys of genomic databases have shown that the majority of naturally occurring U12-type introns possess GU–AG termini (Sharp and Burge, 1997; Burge et al., 1998).
Examples of U12-type introns are found in virtually all metazoan taxa, including vertebrates, insects, plants and cnidarians, but are absent from Caenorhabditis elegans, Saccharomyces cerevisiae, Schizosaccharomyces pombe and protists (Burge et al., 1998). A total of 60 non-redundant U12-type introns were identified in a search of all available genomic sequences in 1998 (Burge et al., 1998), and a more recent search of the human genome yielded 404 examples (Levine and Durbin, 2001). Thus, the frequency of occurrence of U12-type introns relative to U2-type introns is in the range of 0.15 to 0.34% in vertebrates, and lower in other taxa (Burge et al., 1998; Levine and Durbin, 2001). U12-type introns can be found at any position and almost always co-exist with U2-type introns within the same gene. In humans, the mean length of U12-type introns (3600 bp) is similar to that of U2-type introns (4130 bp) (Levine and Durbin, 2001), but does not yet include introns exceeding 20 kb.
By comparing introns at homologous positions (in terms of codon location and phase) in homologous genes from different species, Burge et al. (1998) concluded that U12-type introns occurred much more frequently in early evolutionary history and were either lost or converted to U2-type introns over time. Yet, Burge et al. (1998) identified several U12-type introns that are remarkably conserved, one example being the second intron of the sodium channel α subunit gene in humans and jellyfish, organisms that diverged at least 600–800 million years ago (Spafford et al., 1998). Perhaps even more surprising is the finding of U12-type introns at non-homologous positions in several paralogous genes (Burge et al., 1998).
These observations suggest that the few U12-type introns that have resisted conversion or loss may have been retained over evolutionary time because their presence is important to the genes that harbor them. Might they play an indispensable role in the regulation of gene expression? More specifically, if U12-type introns are spliced more slowly than neighboring U2-type introns in the same pre-mRNA, then their removal could represent the rate-determining step in the expression of that gene.
Here, we have directly tested the hypothesis that U12-type introns are excised more slowly than U2-type introns. We first developed a quantitative RT–PCR assay to assess relative splicing rates by measuring the levels of unspliced introns in the endogenous population of cellular pre-mRNAs. We examined the pre-mRNAs of three human genes, and observed that the U12-type intron was the most slowly spliced in each case. Secondly, we asked whether replacing the consensus sequences of a naturally occurring U12-type intron with those of a U2-type intron would affect the efficiency of gene expression. Using transiently transfected reporter constructs that monitor splicing by the production of different colored fluorescent proteins in Drosophila melanogaster tissue culture cells, we observed that mutational conversion of a U12-type intron to a U2-type intron does dramatically increase protein and mRNA levels.
Results
A quantitative RT–PCR method to assess relative rates of intron removal
We devised a method for evaluating the relative rates of intron removal from a particular transcript by determining the abundance of intron sequences within a steady-state population of partially processed cellular pre-mRNAs. If the excision of a U12-type intron occurs more slowly than that of major-class introns in the same pre-mRNA, then the amount of unspliced U12-type sequences should be higher than each of the other intron sequences. Indeed, it can be mathematically demonstrated that the amount of each unspliced intron within a steady-state pre-mRNA population is inversely proportional to the relative rate of removal of that intron (see Supplementary data available at The EMBO Journal Online). Because the cellular levels of unspliced pre-mRNAs were expected to be very low, the extreme sensitivity of quantitative RT–PCR was exploited to analyze total RNA isolated from growing human tissue culture cells (HeLa or SK Hep).
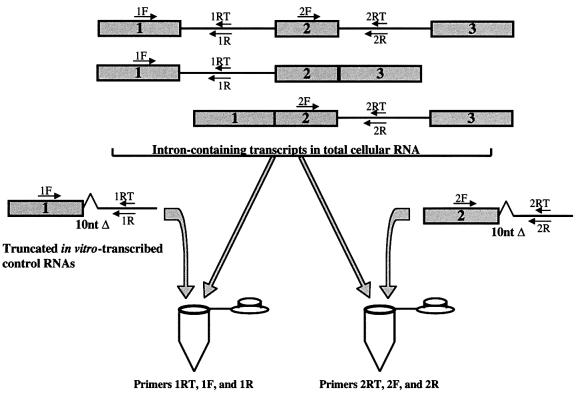
As schematized in Figure 1, reverse transcription was carried out using a sequence-specific primer for each intron. PCR was then performed using primer pairs that amplify only unspliced sequences: each forward primer was complementary to 5′ exonic sequences and each reverse primer was complementary to downstream intronic sequences. For each intron, a control RNA standard that possessed RT and PCR amplification efficiencies closely matched to those of the cellular RNA being analyzed was created. These control RNAs were identical to the target cellular sequences with the exception of an internal 10-nucleotide deletion. Because amplification variability can arise at the reverse transcription step due to RNA secondary structure impeding the polymerase (Freeman et al., 1999), we designed the RNA standards to mimic the local secondary structure of each target by including sequences from most of the upstream exon and >195 nt of the intron. Co-amplification of the cellular RNA population with a known quantity of each RNA standard therefore yielded amplicons that differed in size by 10 nt. By comparing their production after separation on a polyacrylamide gel, the amount of the cellular RNA sequence present was quantitated.
Fig. 1. Assessing in vivo splicing rates by quantitative RT–PCR. Quantitation of unspliced introns from total cellular RNA containing pre-mRNAs of a hypothetical two intron-containing gene is schematized. In vitro-transcribed control RNA standards, truncated relative to the cellular RNA sequence by 10 nt, are added in equal amounts to separate reaction tubes containing a fixed amount of total cellular RNA. Reverse transcription is performed using short primers complementary to intronic sequences (1RT or 2RT). cDNAs are then PCR amplified using primer pairs that only amplify unspliced sequences (1F/1R or 2F/2R). The cellular and control RNA amplicons are then electrophoretically separated, and the quantity of each unspliced intron is inferred from the ratio of the amplicons produced.
For each gene studied, the concentration of unspliced U12-type intron within a fixed amount of total cellular RNA was determined by titration with varying amounts of RNA standard (data not shown). Then, that amount of RNA standard for each of the gene’s introns was added to separate tubes containing identical amounts of total RNA (Figure 1). If the U2-type introns are spliced more rapidly than the U12-type intron, then those unspliced sequences will be less abundant than the sequences of the corresponding RNA standards, as reflected in the ratio of the co-amplified RT–PCR products generated in each tube.
The most slowly spliced introns from three human pre-mRNAs in vivo are U12-type
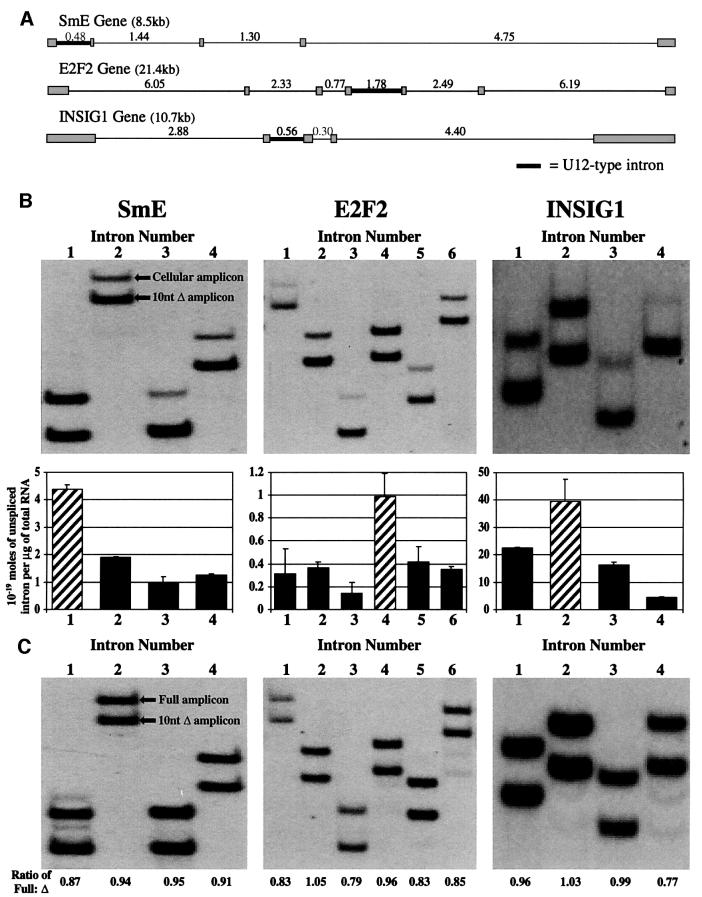
The three human U12-type intron-containing genes chosen for study had a relatively small number of total introns, making the analysis less cumbersome. These genes were: (i) SmE, a member of the core complex of proteins in spliceosomal snRNPs (Stanford et al., 1988; DDBJ/EMBL/GenBank accession No. AL356980); (ii) E2F2, a transcription factor involved in cell cycle regulation (Ivey-Hoyle et al., 1993; accession No. AL021154); and (iii) INSIG1, a gene of unknown function that may be important in liver regeneration (Peng et al., 1997; accession No. U96876). These genes contain four, six and four total introns, with the U12-type intron as the first, fourth and second intron, respectively (Figure 2A).
Fig. 2. U12-type introns are excised most slowly from three endogenous human pre-mRNAs. (A) The intron–exon structure is depicted for the three human genes, SmE, E2F2 and INSIG1. Shaded boxes represent exons, thin lines represent U2-type introns and thick lines represent U12-type introns, with intron lengths (in kb) indicated above. (B) Above are shown the gels from which the ratios of cellular to control amplicons (105–164 nt long with controls being 10 nt shorter) were measured. The amounts of each unspliced intron within the pre-mRNA population from growing HeLa cells (for SmE and E2F2) or SK Hep cells (for INSIG1) are shown graphically below. Solid bars represent U2-type introns and hatched bars represent U12-type introns, with error bars indicating the standard deviation of two experiments. 4.56 × 10–19 moles, 1.01 × 10–19 moles and 4.47 × 10–18 moles of control RNA standards were added per microgram of total cellular RNA for the analysis of introns from SmE, E2F2 and INSIG1, respectively. (C) Control co-amplification of equal amounts of in vitro-transcribed full-length and 10-nt truncated RNAs shows approximately equal production of amplicons (ratio of 0.91 ± 0.08). 10–19 moles of each RNA species were RT–PCR amplified.
RT–PCR quantitation of the unspliced introns from these three genes in the cellular RNA population is presented in Figure 2B. The upper band in each lane is the amplicon from the cellular RNA, while the lower band is that produced from the added RNA standard. Although the absolute intensities of the bands can vary from lane to lane depending on the amplification efficiencies of specific primer pairs, the ratio of intensities within any given lane provides an accurate measure of initial RNA quantities. Controls without reverse transcriptase added (data not shown) established that no undigested genomic DNA contributed to the amplicon intensity. Since equal amounts of the RNA standards are used in each reaction for a given gene, comparing the ratios across lanes provides a direct comparison of the relative amounts of each unspliced intron.
Graphical presentation of these intensity ratios (Figure 2B) shows that the unspliced U12-type intron sequences are two- to nine-fold more abundant than the neighboring U2-type intron sequences for all three pre-mRNAs analyzed. To confirm that the amplification efficiencies for each intron and its corresponding 10-nt deleted RNA standard were equivalent, equal amounts of in vitro transcribed full-length and truncated RNAs corresponding to each intron were co-amplified. The appearance of pairs of amplicons of approximately equal intensity in each lane (Figure 2C) allows us to conclude that the U12-type introns within the in vivo transcripts of three human genes are indeed spliced more slowly than their U2-type neighbors.
Replacement of natural U12-type intron sequences with U2-type consensus sequences increases protein expression in vivo
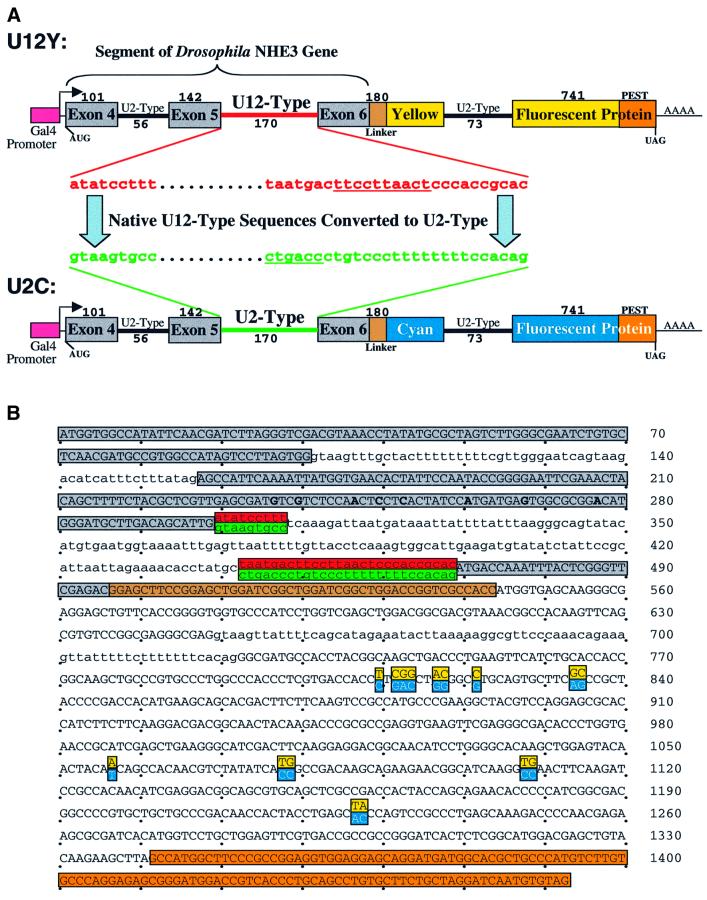
We next asked whether a U12- versus a U2-type intron in the same genetic context would lead to differential expression of the encoded protein. We designed plasmid constructs that would permit unambiguous correlation of reporter protein production with splicing efficiency. The constructs (Figure 3A) express either cyan (C) or yellow (Y) fluorescent protein (FP) only when completely spliced, dependent on removal of a U12-type versus a U2-type intron. Since the spliced mRNAs are nearly identical in sequence (Figure 3B), the relative intensity of the two fluorescent signals provides a readout of the relative efficiencies of the two splicing pathways within transfected tissue culture cells.
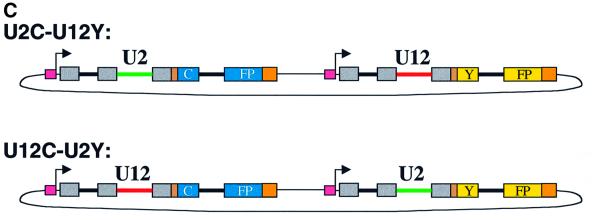
Fig. 3. Plasmids that report the efficiency of U12-type versus U2-type splicing. (A) A segment of the D.melanogaster NHE3 gene including introns 4 (U2-type) and 5 (U12-type) and their flanking exons was fused, via a linker, to a yellow (Y) or cyan (C) fluorescent protein coding sequence containing destabilizing ‘PEST’ sequences to form U12Y or U12C. Intron 6 (U2-type) of NHE3 was inserted into the CFP or YFP coding sequences. A Gal4-responsive promoter and a Kozak consensus translation start site were directly upstream of the fusion protein. Exon and intron lengths are indicated above and below (not drawn to scale). To form U2Y (or U2C), consensus splicing sequences of the U12-type intron were mutated to U2-type sequences derived from an adenovirus intron, keeping the intron length constant. (B) The sequences of the protein-coding regions of the fusion constructs are shown, with exon sequences in uppercase and intron sequences in lowercase and alternative sequences indicated. Red, U12-type consensus sequences; green, converted U2-type sequences; gray, NHE3 exon sequences; brown, linker sequences; orange, PEST sequences. The mutations converting cyan to yellow fluorescence are highlighted in cyan and yellow, respectively. The mutations introduced to change hydrophobic amino acids in the predicted transmembrane domain of exon 5 are in bold. (C) A plasmid containing the U2C and U12Y constructs in tandem (named U2C–U12Y) was created to ensure equal transfection of both constructs. The reciprocal plasmid (U12C–U2Y) with a reversed color scheme controlled for inherent differences in CFP and YFP fluorescence yield.
The fifth intron of the Drosophila sodium–hydrogen exchange channel gene, NHE3, was chosen for analysis because its adjacent exons and introns are small (see Figure 3A), facilitating the creation of small fusion constructs encoding fluorescent reporter proteins. Its sequences perfectly match the optimal U12-type consensus (C.Burge, personal communication). Since cooperativity between adjacent introns has been shown to influence the rate and fidelity of splicing (Robberson et al., 1990; Wu and Krainer, 1996), 496 nt of the NHE3 gene, spanning the 3′ portion of the fourth exon to the 5′ portion of the sixth exon, were included in the construct. A start codon with Kozak consensus (Kozak, 1987) was added to the 5′ end of the segment, and the segment was fused, via a short linker, upstream of either YFP or CFP cDNA (Miller et al., 1999). The fluorescent protein coding sequences were interrupted by insertion of the sixth intron from the NHE3 gene. Therefore, the U12-type intron in the construct was flanked by native exons and U2-type introns on both sides.
The consensus sequences within the U12-type intron were then mutated to U2-type consensus sequences derived from the first intron of the adenovirus major late transcription unit, an intron whose splicing has been well characterized (Solnick, 1985). Replacement with these particular sequences is reasonable since the adenovirus U2-type splice site sequences closely match the Drosophila U2-type consensus sequences (Mount et al., 1992). Changes were limited to mutations at the 5′ splice site, branch site, 3′ splice site and the insertion of a 3′ polypyrimidine tract (Figure 3A). The overall length of the intron was preserved, as were the internal, non-consensus sequences. Consequently, the mRNAs produced by splicing of the converted U2-type intron and the original construct containing the U12-type intron were identical, except for the nine codons that differ between YFP and CFP (Figure 3B).
Synthesis of pre-mRNA was driven by a Gal4-responsive promoter, so that expression could be induced by producing Gal4 protein from either a copper-responsive metallothionein or a heat shock promoter on a separate, co-transfected plasmid. Versions of YFP and CFP containing protein destabilizing ‘PEST’ sequences, which reduced the half-life of the fluorescent proteins from ∼24 to 2 h (Clontech product literature), were used so that the fluorescence signal would reflect differences in the rate of splicing rather than in protein accumulation. Also, eight hydrophobic amino acids within the predicted transmembrane region (exon 5) of NHE3 were mutated (shown in bold in Figure 3B) to avoid possible membrane-targeting of the fusion protein. Finally, to guarantee that each cell was transfected with equal amounts of each fluorescent protein gene, the two constructs were cloned in tandem into a single plasmid (Figure 3C). To normalize for inherent differences in the fluorescence intensities of YFP and CFP, a reciprocal construct was made with the reversed color scheme (Figure 3C).
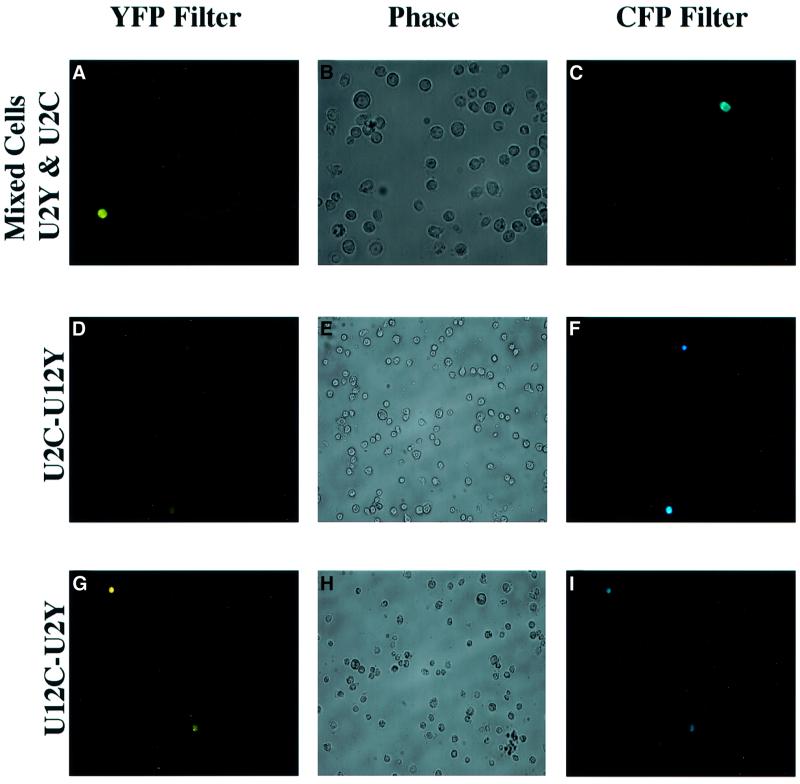
To confirm that the fluorescent signals from YFP and CFP were completely distinguishable (Miller et al., 1999), U2Y and U2C constructs (named on the basis of the middle intron and the fluorescent color) were separately transfected into Drosophila S2 cells with a metallothionein-Gal4 plasmid. Expression was induced 24 h later by adding cupric sulfate, and cells were viewed after 12 h. As expected, fluorescence from any single cell was detected with either the cyan or the yellow filter set, but never with both (Figure 4A–C). Fluorescence from cells transfected with either the U2Y or U2C construct was, on average, quite robust, whereas fluorescence from cells transfected with the U12Y or U12C construct appeared much weaker (data not shown).
Fig. 4. U2-dependent splicing produces brighter fluorescence than U12-dependent splicing in Drosophila S2 cells. (A–C) CFP and YFP fusion proteins are distinguishable. Drosophila S2 cells were separately co-transfected with either U2C or U2Y and metallothionein-Gal4, mixed and viewed 12 h after cupric sulfate induction using a YFP filter (A), phase contrast (B) or a CFP filter (C). (D–I) S2 cells co-transfected with U2C–U12Y (panels D–F) or U12C–U2Y (panels G–I) and metallothionein-Gal4 were similarly induced and viewed using a YFP filter (D and G), phase contrast (E and H) or a CFP filter (F and I). Similar results were obtained with constructs lacking ‘PEST’ sequences (data not shown).
Next, the tandem U2C–U12Y plasmid (Figure 3C) was co-transfected with a metallothionein-Gal4 plasmid, and gene expression induced. While different cells within the transfected population exhibited different fluorescence intensities, any particular cell consistently showed at least a 5-fold higher fluorescent signal with the cyan filter than with the yellow filter (based on comparison of integrated pixel intensities from CCD-captured digital images) (Figure 4D–F). Transfection of the reciprocal tandem U12C–U2Y plasmid (Figure 3C) yielded the opposite pattern: each transfected cell exhibited at least 5-fold more yellow fluorescence than cyan fluorescence (Figure 4G–I). Because all other differences between the constructs are controlled by the use of reciprocal plasmids, the altered expression levels can be ascribed to the identity of the middle intron.
To confirm that the fifth intron of the NHE3 gene is in fact spliced by the U12-type splicing machinery, excision of this intron was examined in endogenous pre-mRNA prepared from Drosophila larvae that were either homo- or heterozygous for a P-element disruption of U6atac (characterized by Otake et al., 2002). The homozygous U6atac-disrupted larvae die at the third instar because of a deficiency in U12-type splicing. Analysis of total RNA by RT–PCR revealed that the U6atac-deficient larvae (at ∼60 h post egg-laying) accumulated substantially higher amounts of unspliced intron 5 as compared with the control heterozygous larvae (data not shown), demonstrating excision of this intron by the U12-type spliceosome.
The 6- to 8-fold difference in fluorescent protein expression is not induced by saturation of the U12-type splicing machinery
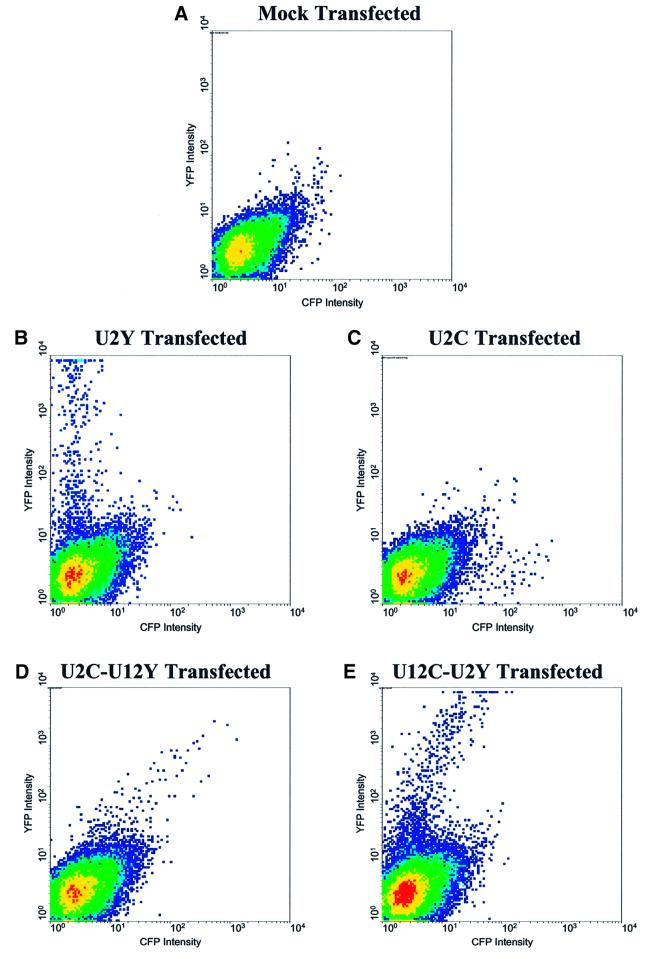
The results from fluorescence microscopy, showing a consistent and significant difference in the levels of YFP and CFP from the reciprocal constructs containing U12-type and U2-type introns (Figure 4D–I), were extended by fluorescence-activated cell sorting (FACS). We first verified that cyan and yellow fluorescence signals can be detected independently by FACS. Compared with mock-transfected cells, which exhibit minimal intensity on each channel (Figure 5A), S2 cells transfected with U2C or U2Y (and metallothionein-Gal4, induced as above) showed significant fluorescence on the cyan or yellow channel, respectively, and negligible fluorescence on the other channel (Figure 5B and C). The large range of fluorescence intensities seen (note the logarithmically scaled axes) probably resulted from cells being transfected with different numbers of plasmids or in different physiological states.
Fig. 5. FACS analysis of transfected S2 cells. Density plots of yellow versus cyan fluorescence intensity for S2 cells co-transfected with metallothionein-Gal4 and various plasmids, obtained 12 h after cupric sulfate induction. The flow cytometer detection optics are inherently less sensitive for CFP than for YFP. 50 000 cells were observed on each plot. (A) Mock-transfected S2 cells show minimal cyan or yellow fluorescence. (B) U2Y-transfected cells show only yellow fluorescence. (C) U2C-transfected cells show only cyan fluorescence. (D) U2C–U12Y-transfected cells show greater cyan than yellow fluorescence (correcting for low CFP detection sensitivity). (E) U12C–U2Y-transfected cells show greater yellow than cyan fluorescence.
Cells transfected with the tandem U2C–U12Y or U12C–U2Y plasmid displayed the same patterns of fluorescence as previously observed by microscopy: the intensity of the protein expressed from the U2-type intron containing gene was always several-fold greater than from that containing the U12-type intron (Figure 5D and E). This pattern manifests itself on the FACS plot as a roughly linear set of points through which a best-fit line can be drawn. As expected, the slope of the line for the U12C–U2Y transfected cells is greater than that for the U2C–U12Y transfected cells, because of increased relative expression of the protein from the mRNA generated by U2-dependent splicing only. The relative difference in expression can be calculated from the slopes of the two lines, taking into account the decreased CFP detection sensitivity and assuming that the nine amino acids that change the color of the fluorescent protein do not affect protein expression or stability. We conclude that the expression of fluorescent protein increases 6- to 8-fold when the U12-type intron is converted to a U2-type intron.
The linearity of the data points in Figure 5D and E also excludes the possibility that the observed differences result artificially from saturation of the U12-type splicing machinery by high levels of pre-mRNA expressed from the transfected constructs. Because the scale of the axes are logarithmic, the fluorescence emitted by individual cells in the population differed by several orders of magnitude. Over this entire range, the proportion of yellow to cyan fluorescence was roughly constant. For example, a strongly fluorescent cell making 50-fold more U2-type reporter protein also made ∼50-fold more U12-type reporter protein compared with a weakly fluorescent cell. This would not be the case if the capacity of the U12-type splicing machinery were saturated by high expression of the fluorescent protein pre-mRNA. Likewise, when different amounts of U2C–U12Y or U12C–U2Y were deliberately transfected into S2 cells (over a 20-fold range), the ratio of cyan to yellow fluorescence in each cell remained approximately constant (within detection limits) despite great differences in the absolute fluorescence intensity (data not shown).
Presence of a U12-type intron hinders production of mature mRNA
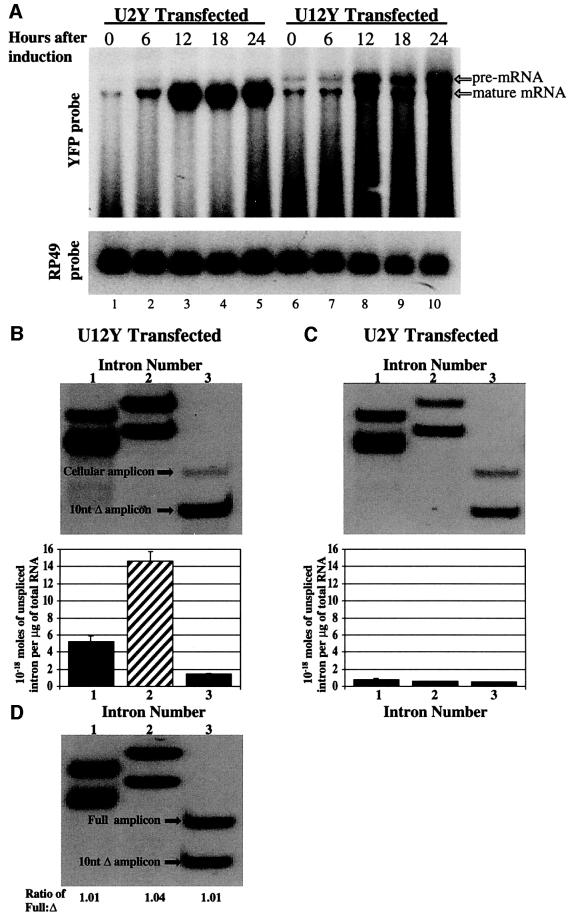
To confirm that the observed differences in fluorescent protein expression were caused by differences in the efficiency of U2-type versus U12-type splicing, the effects of intron conversion were examined directly at the RNA level. S2 cells were co-transfected with metallothionein-Gal4 and either U2Y or U12Y, expression was induced after 24 h, and total RNA at various times after induction was analyzed by northern blot. Cells transfected with the U2Y construct showed rapid accumulation of mature mRNA, whereas cells transfected with the U12Y construct accumulated less mature mRNA (Figure 6A). Also, a more slowly migrating band was observed in the U12Y-transfected cells. Hybridization of this band with a probe specific for the middle intron (data not shown) and further analysis by RT–PCR (see below) indicate that it consists of partially unspliced species predominantly containing the U12-type intron. Lanes corresponding to the U12Y-transfected cells also reproducibly showed a smear below the full-length bands (Figure 6A), suggesting that the pre-mRNA is prone to degradation if not efficiently spliced (Bousquet-Antonelli et al., 2000).
Fig. 6. mRNA production is slowed by inefficient splicing of a U12-type intron. (A) Northern blot of total RNA isolated from S2 cells co-transfected with U2Y (lanes 1–5) or U12Y (lanes 6–10) and metallothionein-Gal4 at the indicated times after induction of expression. The blot was successively hybridized with random-primed DNA probes complementary to the YFP coding sequence (top panel) and to a loading control, ribosomal protein 49 (bottom panel). The identity of the slowly migrating band as partially spliced pre-mRNA, was confirmed by hybridization to a probe specific for the middle intron (data not shown). (B) Quantitative RT–PCR analysis of unspliced introns (similar to Figure 2B) from total RNA prepared 12 h after induction of U12Y-transfected cells and amplified in the presence of a single in vitro transcribed RNA standard containing three separate 10-nt deletions, allowing quantitation of all three introns. 1.33 × 10–17 moles of control RNA standard was added per µg of total cellular RNA. Hatched bar, U12-type intron; solid bars, U2 type introns; error bars, standard deviation of two experiments. (C) Similar analysis of U2Y-transfected cells. Note that 10-fold less control RNA standard (1.33 × 10–18 moles per µg of total RNA) was used. (D) Confirmation of equal amplification efficiency of in vitro transcribed full-length and 10-nt truncated RNAs (amplicon ratio of 1.02 ± 0.02) (as in Figure 2C). 10–19 moles of each RNA species were RT–PCR amplified.
Finally, to examine differences in the splicing rate of the middle intron relative to its neighboring introns for both the U2Y and U12Y constructs, we carried out quantitative RT–PCR experiments, as described for the endogenous human pre-mRNAs (Figure 1). S2 cellular RNA prepared 12 h after induction of expression was RT–PCR amplified in the presence of a single in vitro transcribed RNA standard containing three separate 10-nt deletions, allowing quantitation of all three introns. The levels of unspliced introns from the U12Y construct indicate that the U12-type middle intron was more abundant than its U2-type neighbors, confirming that it is spliced more slowly (Figure 6B). Conversion of the consensus sequences to U2-type dramatically increased the splicing of that intron as evidenced by the lower relative abundance of its unspliced form (Figure 6C). Interestingly, conversion to U2-type sequences also appeared to increase the splicing rate of the two adjacent introns, suggesting cooperativity in the excision of these introns. As before, to verify that the amplification efficiencies of the full-length and truncated RNAs were equal, equimolar amounts of full-length and truncated in vitro generated transcripts were co-amplified and observed to produce amplicons of equal intensity (Figure 6D).
Discussion
Our data support the hypothesis that U12-type introns are excised from pre-mRNA transcripts more slowly than canonical U2-type introns. We demonstrate that this is the case for the transcripts of three human genes. Additionally, mutation of native U12-type to U2-type intron consensus sequences within a D.melanogaster minigene results in significantly increased levels of mRNA and protein expression. Together, these observations suggest that persistence of U12-type introns over evolutionary time could result from their importance as post-transcriptional regulators of the genes that harbor them.
Although we analyzed endogenous splicing rates for the transcripts of only three human genes, we argue that our results are generalizable. These genes were chosen because they had a small number of total introns, whereas the average number of introns in U12-type intron containing genes is ∼15 (Burge et al., 1998). However, approximately one-third of these genes have <10 introns, and there is no obvious reason why U12-type introns would be selectively spliced more slowly from genes with only a few introns. Extrapolation to other U12-type intron containing genes is warranted if coincidental observation of the same pattern within the studied sample is highly improbable. The likelihood of finding by random chance that the U12-type intron is spliced most slowly from genes containing 4, 4 and 6 total introns is 1/4, 1/4 and 1/6, respectively. However, the probability of independently finding this pattern for all three genes is the product of the individual probabilities, 1/96. Therefore, the consistency we observe is unlikely to occur randomly and is thus probably representative of other genes containing U12-type introns. It is even conceivable that if the introns we analyzed had been those that are extremely highly conserved through evolution, more dramatic differences in excision rates might have been observed.
Indeed, another U12-type intron in the ribosomal protein L1a gene of Xenopus laevis was initially characterized as the ‘regulated third intron’ (Bozzoni et al., 1984; Caffarelli et al., 1987, 1992; Pierandrei-Amaldi et al., 1988; Fragapane et al., 1992) because of its low efficiency of splicing. Not only did partially spliced pre-mRNAs containing this intron (and to a lesser extent, the second intron) accumulate and produce little extra protein upon microinjection of cloned RPL1a genomic sequences into Xenopus oocytes, but endogenous partially spliced RPL1a pre-mRNAs were detected at low levels in uninjected oocytes (Bozzoni et al., 1984). Using our sensitive RT–PCR assay, we have confirmed that unspliced U12-type intron sequences are indeed present at higher levels than those of the other eight RPL1a U2-type introns in total RNA prepared from oocytes (data not shown). However, interpretation of these experiments is complicated by (i) the presence of snoRNAs, which can be removed via a pathway that competes with splicing (Cafarelli et al., 1994), within several RPL1a introns (including the U12-type intron), and (ii) potential amplification of transcript sequences from both gene copies in the pseudo-tetraploid genome of X.laevis. Thus, human genes were chosen for subsequent analyses.
For the three human genes analyzed, there is no obvious correlation of intron position or length with rate of excision (Figure 2). While the U12-type introns are, on average, shorter than the U2-type introns, in two out of three genes they are not the shortest intron. Also, we did not observe slower splicing rates for the longer U2-type introns within these genes, consistent with previous findings (Gardner et al., 1988; Gudas et al., 1990; Kessler et al., 1993; Wetterberg et al., 1996).
Our D.melanogaster splicing constructs produced mRNAs differing by only nine codons for CFP versus YFP. It was possible that these changes affect the efficiency of either splicing, polyadenylation, export or translation. Therefore, the reciprocal constructs with reversed color schemes were important to control for differences not only in fluorescence yield, but also expression. Their use confirmed that the differences in fluorescent protein expression are attributable to the changed intron type.
Why should U12-type introns be excised more slowly than U2-type introns? In vitro, the splicing of U12-type introns has consistently been reported to be extremely slow, even for several different splicing substrates (Tarn and Steitz, 1996b; Wu and Krainer, 1996). The ∼100-fold lower abundance of the minor-class snRNPs relative to the major-class snRNPs in mammalian cell nuclei and nuclear extracts (Montzka and Steitz, 1988; Tarn and Steitz, 1996a) may well reduce the rate of assembly of the U12-dependent spliceosome. However, since splicing components are thought to be assembled co-transcriptionally in vivo (Misteli and Spector, 1999; reviewed in Hirose and Manley, 2000), the effects of snRNP concentrations on in vivo splicing rates are not easily predictable. Alternatively, it is possible that slower catalysis of one or both splicing reactions by the spliceosome underlies the lower rate of excision of U12-type introns. It will be interesting to explore whether the low abundance of minor-class splicing components might cause RNA polymerase II to stall at U12-type introns.
The existence of genes whose expression might be regulated by the presence of inefficiently spliced introns hints at a commonality among these genes. Burge et al. (1998) note that a large proportion of U12-type intron-containing genes can be categorized as ‘information processing’, including genes involved in DNA replication/repair, transcription, RNA processing and translation; only a few perform operational functions such as energy metabolism or biosynthesis. Could U12-type introns in these genes be acting as post-transcriptional bottlenecks, preventing overexpression that might be harmful to the organism? Alternatively, since expression of such genes is upregulated in proliferating compared with quiescent cells, the activity of the U12-type spliceosome may increase during proliferation. It is also possible that U12-type splicing activity is modulated in a tissue- or a developmental stage-specific manner. This could occur through up- (or down-) regulation of any snRNA or protein component specific to the U12-type spliceosome. The fluorescent protein-coding constructs described here provide a means of investigating these questions in the developmentally well-studied organism, D.melanogaster.
Materials and methods
Preparation of RNA standards
Segments of genomic sequences from human E2F2, SmE and INSIG1 were PCR-amplified from genomic clones RPCI1-15005 (Sanger Centre, Hinxton, Cambs, UK), RPCI11-397P13 (Sanger Centre), and HP-5 (Peng et al., 1997), respectively. The segments (between 301 and 853 nt) included most of the upstream exon and >195 nt of intronic sequence surrounding each 5′ splice site. To create an RNA standard for the Drosophila constructs, a single 872-nt segment including all three introns was PCR-amplified from the U12Y plasmid (construction of U12Y described below). XhoI and NotI restriction sites, introduced by the PCR primers, were used to insert the amplified segments into pBluescript SK+ vectors (Stratagene) downstream of a T7 promoter. Ten-nucleotide deletions were introduced by either PCR mutagenesis or using the QuickChange Mutagenesis system (Stratagene). Oligonucleotides for PCR cloning and mutagenesis are listed in the Supplementary data. RNA standards were prepared by in vitro transcription from the T7 promoter essentially as described by Tarn and Steitz (1996b), internally trace-labelling with [α-32P]UTP. Gel-purified transcripts were quantified by liquid scintillation counting and stored at –70°C in aqueous solution containing 20 µg/ml yeast total carrier RNA.
Quantitative RT–PCR analysis
RNA for analysis of E2F2 and SmE was prepared, using Trizol (Gibco-BRL) according to the manufacturer’s directions, from log-phase HeLa cells grown in suspension in RPMI (Gibco-BRL) containing 10% fetal bovine serum (FBS; Gibco-BRL). RNA for analysis of INSIG1 was similarly prepared from ∼70% confluent SK Hep cells grown in 35 mm dishes in Dulbecco’s modified Eagle’s medium (Gibco-BRL) with 10% FBS. RNA for analysis of U12Y and U2Y was isolated from transiently transfected Drosophila S2 cells 12 h after induction of expression as described below. Contaminating genomic DNA was removed by treating with RQ1 DNase (Promega) in 5 mM MgCl2 and 50 mM Tris pH 8.0 at 37°C for 1 h, followed by heat inactivation at 65°C for 10 min. Reverse transcription was then performed using Thermoscript RT (Invitrogen) according to the manufacturer’s directions, with 0.5 µM gene-specific primers (see Supplementary data), 7.5 ng/µl (for SmE, E2F2 and INSIG1) or 0.188 ng/µl (for U12Y and U2Y) total cellular RNA, and various concentrations (between 3.36 × 10–20 and 7.60 × 10–22 moles/µl) of in vitro transcribed RNA standards. RT reactions were incubated at 37°C for 15 min, then 55°C for 30 min, then 65°C for 30 min and finally 85°C for 10 min. Escherichia coli RNase H (0.1 U/µl; Invitrogen) were added and the samples incubated at 37°C for 30 min. PCR was then performed using Platinum Taq polymerase (Invitrogen) in the buffer supplied with 1.5 mM MgCl2, 0.4 mM dNTPs, 250 nM unlabelled primers (see Supplementary data) doped with 5′-radiolabelled forward primer (to label only one strand of amplicon), and 10% (by volume) of reverse-transcribed cDNA. Between 24 and 28 cycles of PCR were performed with 2 min extension times and annealing temperatures of 62.5, 60, 61.5 and 61°C for the E2F2, SmE, INSIG1 and U12Y/U2Y samples, respectively (based on lowest predicted Tm). Amplicons were separated on a 10% polyacrylamide sequencing gel, and quantitated using a Molecular Dynamics PhosphorImager.
Construction of Drosophila splicing reporter plasmids
Reporter plasmids contained a Drosophila NHE3 gene segment fused to CFP or YFP coding sequences (destabilized with ‘PEST’ sequences) (Clontech) with modifications as schematized in Figure 3 (sequences available upon request). The NHE3 segment spanning exon 4 through exon 6 was PCR-amplified from genomic clone DS07134 (Berkeley Drosophila Genome Database) with primers that introduced an upstream translation start site, a downstream linker sequence and mutations to remove hydrophobic amino acids from the predicted transmembrane domain (see Supplementary data). Alternative primers were used to mutate U12-type consensus sequences to U2-type sequences. PCR products were inserted between the BglII and AgeI sites of pd2-ECFP-N1 and pd2-EYFP-N1 plasmids (Clontech). Synthetic DNA including the entire NHE3 intron 6 sequence (see Supplementary data) was inserted into CFP and YFP coding sequences via a BcgI site. A Gal4-responsive UAS promoter was excised from pUAST (Brand and Perrimon, 1993) using BamHI and BglII sites, and was inserted within a BamHI site upstream of the NHE3 gene segment. Finally, tandem constructs were created by inserting sequences from one plasmid, cut with XhoI and NgoMIV, into the XhoI and XmaI sites of the second plasmid.
Transfection of Drosophila S2 cells
S2 cells were grown at 25°C in Shields and Sang M3 medium (Sigma) containing 12.5% FBS (Gibco-BRL) in 35 mm dishes and transiently transfected using 25 µl of Lipofectin reagent (Gibco-BRL) per dish, according to the manufacturer’s directions. For most experiments, 0.5 µg of reporter plasmids were co-transfected with 1.5 µg of either pmt-Gal4 (metallothionein expression, gift of S.Artavanis-Tsakonas) or heat-shock-Gal4 plasmid (gift of T.Xu). Experiments to investigate splicing saturation used between 1 µg and 0.05 µg reporter plasmid, supplementing with unrelated plasmid to keep the total transfected DNA constant. Expression was induced 24 h after transfection either by adding 0.7 mM cupric sulfate (Sigma) or by heat shock at 37°C for 30 min.
Fluorescence microscopy
Cells were visualized 12–24 h after induction using a Zeiss Axiophot II fluorescence microscope with CFP (Ex: D436/20; Em: D480/40; BS: 455dclp) and YFP (Ex: HQ500/20; Em: HQ535/30; BS: Q515lp) filter sets (Chroma). Images were captured using a Quantix CCD camera (Photometrics) and analyzed with IP Lab software (Scanalytics).
FACS analysis
Cells were analyzed 12–24 h after induction with a FACS Vantage dual laser flow cytometer (Becton-Dickinson). CFP was excited at 457 nm with a Spectra-Physics 2025 argon laser and the fluorescence emission was collected through a 480/30 nm band pass filter. YFP was excited at 514 nm using a Coherent Innova 70 argon–krypton laser and the fluorescence collected through a 550/30 nm band pass filter. Fifty thousand cells were observed for each sample. Statistical analysis was performed using WINMIDI software (Joseph Trotter, Scripps Clinic, CA).
Northern blot analysis
Growth medium was removed from S2 cells at various times after induction, and cells were disrupted in 1 ml of Trizol (Gibco-BRL) for RNA isolation according to the manufacturer’s instructions. Total RNA (∼10 µg) was run per lane on a formaldehyde–agarose gel for northern blotting and was probed with internally 32P-labelled DNA probes covering either the entire open reading frame of YFP, or ribosomal protein 49 (gift of L.Cooley), or the non-consensus internal sequences (nucleotides 319–436, Figure 3B) of NHE3 intron 5.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Chris Burge for communicating unpublished findings which identified the Drosophila NHE3 gene for incorporation into our reporter plasmids. We are also grateful to Rocco Carbone for assistance with the FACS experiments, and to Kazio Tycowski, Michael Koelle and Carl Hashimoto for critical reading of the manuscript. This work was supported by grant GM 26154 from the National Institutes of Health to J.A.S., who is an investigator at the Howard Hughes Medical Institute.
References
- Bousquet-Antonelli C., Presutti,C. and Tollervey,D. (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell, 102, 765–775. [DOI] [PubMed] [Google Scholar]
- Bozzoni I., Fragapane,P., Annesi,F., Pierandrei-Amaldi,P., Amaldi,F. and Beccari,E. (1984) Expression of two Xenopus laevis ribosomal protein genes in injected frog oocytes. A specific splicing block interferes with the L1 RNA maturation. J. Mol. Biol., 180, 987–1005. [DOI] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Padgett,R.A. and Sharp,P.A. (1998) Evolutionary fates and origins of U12-type introns. Mol. Cell, 2, 773–785. [DOI] [PubMed] [Google Scholar]
- Caffarelli E., Fragapane,P., Gehring,C. and Bozzoni,I. (1987) The accumulation of mature RNA for the Xenopus laevis ribosomal protein L1 is controlled at the level of splicing and turnover of the precursor RNA. EMBO J., 6, 3493–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli E., Fragapane,P. and Bozzoni,I. (1992) Inefficient in vitro splicing of the regulatory intron of the L1 ribosomal protein gene of X.laevis depends on suboptimal splice site sequences. Biochem. Biophys. Res. Commun., 183, 680–687. [DOI] [PubMed] [Google Scholar]
- Caffarelli E., Arese,M., Santoro,B., Fragapane,P. and Bozzoni,I. (1994) In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol. Cell. Biol., 14, 2966–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich R.C., Incorvaia,R. and Padgett,R.A. (1997) Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell, 1, 151–160. [DOI] [PubMed] [Google Scholar]
- Fragapane P., Caffarelli,E., Lener,M., Prislei,S., Santoro,B. and Bozzoni,I. (1992) Identification of the sequences responsible for the splicing phenotype of the regulatory intron of the L1 ribosomal protein gene of Xenopus laevis. Mol. Cell. Biol., 12, 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman W.M., Walker,S.J. and Vrana,K.E. (1999) Quantitative RT–PCR: pitfalls and potential. Biotechniques, 26, 112–122. [DOI] [PubMed] [Google Scholar]
- Gardner D.G., Cathala,G., Lan,N.Y., David-Inouye,Y. and Baxter,J.D. (1988) Processing of the primary transcript for the rat growth hormone gene in vivo. DNA, 7, 537–544. [DOI] [PubMed] [Google Scholar]
- Gudas J.M., Knight,G.B. and Pardee,A.B. (1990) Ordered splicing of thymidine kinase pre-mRNA during the S phase of the cell cycle. Mol. Cell. Biol., 10, 5591–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S.L. and Padgett,R.A. (1994) Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol., 239, 357–365. [DOI] [PubMed] [Google Scholar]
- Hall S.L. and Padgett,R.A. (1996) Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science, 271, 1716–1718. [DOI] [PubMed] [Google Scholar]
- Hirose Y. and Manley,J.L. (2000) RNA polymerase II and the integration of nuclear events. Genes Dev., 14, 1415–1429. [PubMed] [Google Scholar]
- Ivey-Hoyle M., Conroy,R., Huber,H.E., Goodhart,P.J., Oliff,A. and Heimbrook,D.C. (1993) Cloning and characterization of E2F-2, a novel protein with the biochemical properties of transcription factor E2F. Mol. Cell. Biol., 13, 7802–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson I.J. (1991) A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res., 19, 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O., Jiang,Y. and Chasin,L.A. (1993) Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol. Cell. Biol., 13, 6211–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1987) At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol., 196, 947–950. [DOI] [PubMed] [Google Scholar]
- Levine A. and Durbin,R. (2001) A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res., 29, 4006–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.M. 3rd, Desai,N.S., Hardin,D.C., Piston,D.W., Patterson,G.H., Fleenor,J., Xu,S. and Fire,A. (1999) Two-color GFP expression system for C.elegans. Biotechniques, 26, 914–918. [DOI] [PubMed] [Google Scholar]
- Misteli T. and Spector,D.L. (1999) RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell, 3, 697–705. [DOI] [PubMed] [Google Scholar]
- Montzka K.A. and Steitz,J.A. (1988) Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl Acad. Sci. USA, 85, 8885–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S.M., Burks,C., Hertz,G., Stormo,G.D., White,O. and Fields,C. (1992) Splicing signals in Drosophila: intron size, information content and consensus sequences. Nucleic Acids Res., 20, 4255–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otake L.R., Scamborova,P., Hashimoto,C. and Steitz,J.A. (2002) The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol. Cell, 9, 439–446. [DOI] [PubMed] [Google Scholar]
- Peng Y., Schwarz,E.J., Lazar,M.A., Genin,A., Spinner,N.B. and Taub,R. (1997) Cloning, human chromosomal assignment and adipose and hepatic expression of the CL-6/INSIG1 gene. Genomics, 43, 278–284. [DOI] [PubMed] [Google Scholar]
- Pierandrei-Amaldi P., Bozzoni,I. and Cardinali,B. (1988) Expression of the gene for ribosomal protein L1 in Xenopus embryos: alteration of gene dosage by microinjection. Genes Dev., 2, 23–31. [DOI] [PubMed] [Google Scholar]
- Robberson B.L., Cote,G.J. and Berget,S.M. (1990) Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol., 10, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.A. and Burge,C.B. (1997) Classification of introns: U2-type or U12-type. Cell, 91, 875–879. [DOI] [PubMed] [Google Scholar]
- Solnick D. (1985) Trans splicing of mRNA precursors. Cell, 42, 157–164. [DOI] [PubMed] [Google Scholar]
- Spafford J.D., Spencer,A.N. and Gallin,W.J. (1998) A putative voltage-gated sodium channel α subunit (PpSCN1) from the hydrozoan jellyfish, Polyorchis penicillatus: structural comparisons and evolutionary considerations. Biochem. Biophys. Res. Commun., 244, 772–780. [DOI] [PubMed] [Google Scholar]
- Stanford D.R., Perry,C.A., Holicky,E.L., Rohleder,A.M. and Wieben,E.D. (1988) The small nuclear ribonucleoprotein E protein gene contains four introns and has upstream similarities to genes for ribosomal proteins. J. Biol. Chem., 263, 17772–17779. [PubMed] [Google Scholar]
- Tarn W.Y. and Steitz,J.A. (1996a) Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT–AC introns. Science, 273, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Tarn W.Y. and Steitz,J.A. (1996b) A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT–AC) intron in vitro. Cell, 84, 801–811. [DOI] [PubMed] [Google Scholar]
- Wetterberg I., Bauren,G. and Wieslander,L. (1996) The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA, 2, 641–651. [PMC free article] [PubMed] [Google Scholar]
- Wu Q. and Krainer,A.R. (1996) U1-mediated exon definition interactions between AT–AC and GT–AG introns. Science, 274, 1005–1008. [DOI] [PubMed] [Google Scholar]