Abstract
Lariat formation has been studied intensively only with a few self-splicing group II introns, and little is known about how the numerous diverse introns in plant organelles are excised. Several of these introns have branch-points that are not a single bulge but are adjoined by A:A, A:C, A:G and G:G pairs. Using a highly sensitive in vivo approach, we demonstrate that all but one of the barley chloroplast introns splice via the common pathway that produces a branched product. RNA editing does not improve domain 5 and 6 structures of these introns. The conserved branch-point in tobacco rpl16 is chosen even if an adjacent unpaired adenosine is available, suggesting that spatial arrangements in domain 6 determine correct branch-point selection. Lariats were not detected for the chloroplast trnV intron, which lacks an unpaired adenosine in domain 6. Instead, this intron is released as linear molecules that undergo further polyadenylation. trnV, which is conserved throughout plant evolution, constitutes the first example of naturally occurring hydrolytic group II intron splicing in vivo.
Keywords: branch-point/intron circle/reverse transcription/RNA editing/trans-splicing
Introduction
Group II introns are found within mRNA, tRNA and rRNA genes of eukaryotic organelles and eubacteria (Michel and Ferat, 1995; Martínez-Abarca and Toro, 2000; Bonen and Vogel, 2001). Their removal from precursor transcripts (splicing) proceeds by two transesterification steps virtually identical to nuclear pre-mRNA splicing. The 2′ hydroxyl group of an intron-internal A residue attacks the 5′ splice site, followed by nucleophilic attack on the 3′ splice site by the 3′ OH of the upstream exon. The attacking adenosine (branch-point) is presented as a nucleotide bulged out of a short RNA helix, which is located –7 or –8 nucleotides from the 3′ splice site in intron domain 6 (D6) (Peebles et al., 1986; Schmelzer and Schweyen, 1986; van der Veen et al., 1986; Michel et al., 1989). The excised intron is a lariat with an internal 2′–5′ linkage and a short tail.
Group II intron splicing has been investigated mainly with self-splicing model introns that accumulate substantial amounts of lariat both in vivo and in vitro. Lariat formation, i.e. branch-point recognition, exhibits high fidelity in these introns, with cryptic branching being exceptional even if the wild-type branch-point is drastically altered (Schmelzer and Müller, 1987; Gaur et al., 1997; Chu et al., 1998; Podar et al., 1998). Deletions of single domains support the essential role of D6 in lariat formation (Jacquier and Jacquesson-Breuleux, 1991; Koch et al., 1992; Holländer and Kück, 1999). It is intriguing, however, that, except for the adenosine bulge, D6 is not highly conserved (Michel et al., 1989). Tertiary contacts otherwise important for intramolecular recognition of catalytic sites have not been mapped in the branch-point region nor does a simple model in which the bulged structure alone is sufficient to designate the branch-point seem to apply, since removal of the bulge by base pairing to G only modestly reduces lariat formation (Chu et al., 1998).
Mutational analyses employing mutant and modified nucleotides at the branch-site revealed that the chemical properties of adenosine contribute greatly to its recognition (Liu et al., 1997). Furthermore, the few self-splicing model introns share a tightly structured D6, in which the branch-point adenosine is adjoined by G:U base pairs. Studies with the yeast mitochondrial intron aI5γ indicated these wobble base pairs (Chu et al., 2001) as well as the linker length between D5 and D6 (Boulanger et al., 1996) to be further determinants for efficient branching.
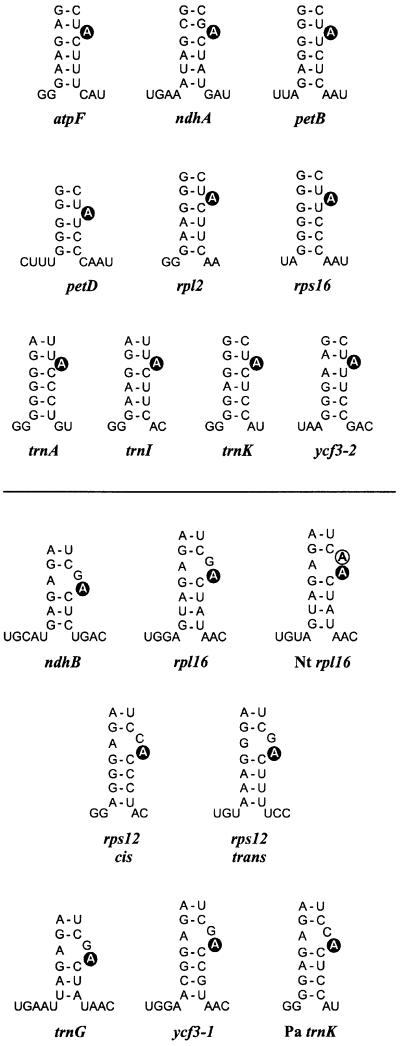
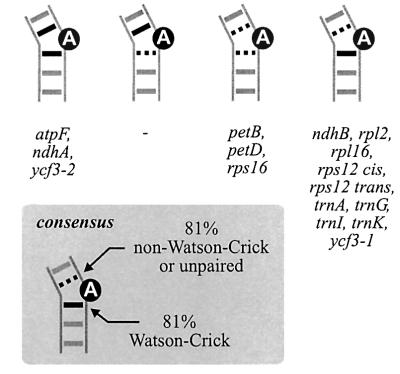
While this suggests an interplay of various factors in designating the branch-site in self-splicing introns, group II introns show wide variations in nature, and the generality of these observations remains unclear. In plant organelles, which provide the richest source of intron sequences to date, strong deviations of D6 structures are often seen (Michel et al., 1989; Carrillo et al., 2001). About one-third of the 17 group II introns found in barley chloroplasts differ from the group II consensus by having neither Watson–Crick nor G:U base pairs preceding the conserved branch-point (Figure 1, lower part). Moreover, the intron of trnV lacks a conserved bulged A (or other nucleotide) in all plant species examined so far (Learn et al., 1992), raising doubts about its capability of forming lariats. Mutations that block lariat formation in aI5γ were recently shown to activate an alternative splicing pathway via first-step hydrolysis, in which the intron is not released in lariat but in linear form (van der Veen et al., 1987; Jarrell et al., 1988; Podar et al., 1998). Kinetic analysis established that under permissive in vitro conditions, branching competes directly with hydrolysis even in wild-type allels of aI5γ (Daniels et al., 1996), suggesting that other group II introns might also compensate severe branch-point mutations by a switch to the intrinsic hydrolytic pathway.
Fig. 1. Proposed branch-point configurations of investigated barley and maize introns, and certain introns of other plants (Nt, tobacco; Pa, spruce) according to the group II consensus (Michel et al., 1989). The lower stem of D6, two or three positions upstream of the conserved bulged A (black circle), and the D5/D6 and D6/3′ss linker nucleotides are shown. Introns with mispairing upstream of the designated branch-point are given below the line. A second unpaired A is found in tobacco rpl16 (open circle).
The set of plant chloroplast group II introns could help assess the general validity of the two-step transesterification pathway as well as of branching determinants dissected with model introns from yeast mitochondria. However, no chloroplast intron has been reported so far to self-splice in vitro, which is consistent with the fact that in vivo splicing in both algal and plant chloroplasts has been shown to be largely dependent on plastid- and nuclear-encoded factors (Choquet et al., 1988; Hess et al., 1994; Jenkins et al., 1997). Analysis of in vivo splicing intermediates, however, is challenging due to frequently low concentrations of released introns (Kim and Hollingsworth, 1993; Jenkins et al., 1997; Vogel et al., 1999). Indeed, branch-point mapping failed for two out of four spinach introns when a conventional method was employed, i.e. treatment with RNA debranching enzyme followed by primer extension (Kim and Hollingsworth, 1993). We recently developed a sensitive RT–PCR-based method that readily and specifically amplified lariat-derived cDNAs of the barley trnK intron (Vogel et al., 1997a). Using this approach, we report here on lariat formation of 16 plastid group II introns from barley as well as of selected examples from other plants. Since no lariats were detectable for trnV, we also investigated whether this and other introns were freed in linear form. Our results obtained with trnV provide the first evidence of a conserved hydrolytic splicing pathway in vivo and, furthermore, the first example of polyadenylation of an intron from any class.
Results
Strategy
Lariat formation was examined by a previously described method (Vogel et al., 1997a), which employed the ability of reverse transcriptases to read through 2′–5′ phosphodiesters (Lorsch et al., 1995). In several cases, chloroplast RNA was circularized with T4 RNA ligase prior to reverse transcription (Vogel and Hess, 2001) in order to amplify a possible linear form of the released intron. PCR bands of interest were excised from gels (approximately –10/+15 bp) and sequences were obtained from 6–18 independent Escherichia coli colonies after cloning.
Branch-points with adjacent Watson–Crick or G:U base pairs
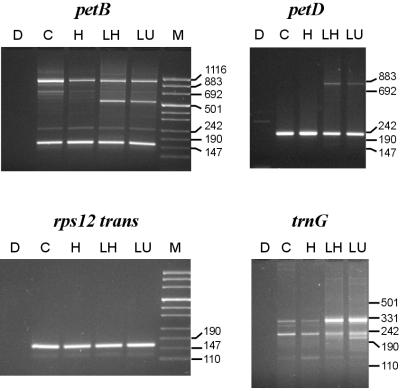
Ten out of 17 barley chloroplast introns (the trnL group I intron was omitted from this study) conform to the group II consensus in having the conserved bulged A adjoined by Watson–Crick and/or G:U wobble base pairs (Figure 1, upper part). PCR products of sizes expected for branching in D6 were readily obtained for all of these introns (shown for petB and petD in Figure 2, lanes C and H). Except for petB, no other major amplification products were detected. Sequencing of the additional petB band (∼1 kb) suggested that it arose by a template switch of reverse transcriptase within a 14 nucleotide region shared with the downstream-located petD gene (5′ss; data not shown).
Fig. 2. PCR products generated with lariat-specific primer sets using as template barley DNA (D; control) or cDNAs raised from barley RNA with various treatments [C, untreated RNA; prior incubation in ligase buffer without (LU) or with (H, LH) heat denaturation, and addition of RNA ligase (LH, LU)], and were separated on agarose gels along with marker DNA (M). Major bands from lanes C and H were of sizes estimated for lariat formation at the conserved adenosine in D6 (petB, 161 bp; petD, 231 bp; rps12 trans, 161 bp; trnG, 229 bp). Additional larger bands specific to lanes LH and LU proved successful RNA circularization (see text).
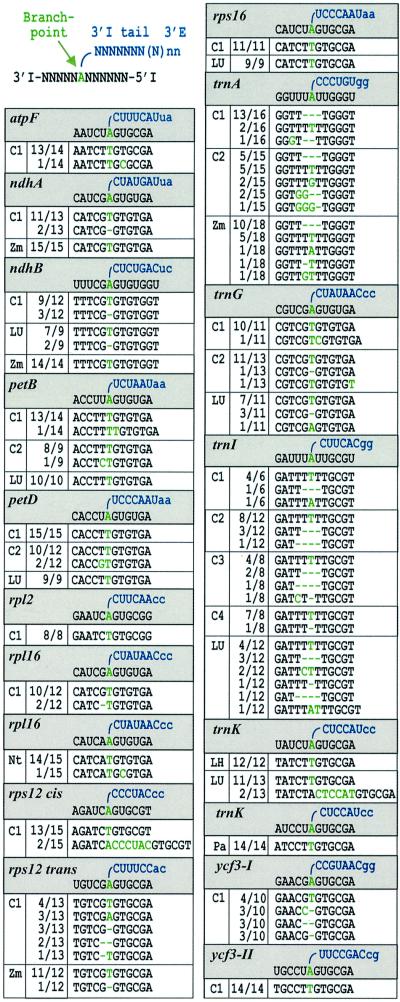
Analysis of cloned cDNAs demonstrated, for eight of these introns (for the remaining trnA and trnI, see below), that the lariat formed at the conserved bulged A. In the vast majority of clones, the 3′ intron sequence was at this position followed by the 5′ intron end (Table I). As observed earlier with trnK lariats (Vogel et al., 1997a), the 2′–5′-linked A itself is represented overwhelmingly as T in these cDNA sequences (sense strand), which implies misincorporation of adenosine as reverse transcriptase encounters the branched nucleotide. Nucleotide exchanges, deletions and insertions were found in a minor proportion of atpF, ndhA, petB and petD sequences, but their patterns may not be reproducible (compare petB clones from independently generated cDNAs C1 and C2). We also noted that the lack of the G preceding the branch-point A in two barley ndhA sequences was not repeated in ndhA clones from the closely related species Zea mays (maize; Table I). Moreover, recent experiments showed that reverse transcriptase incorporates G when passing a branched guanosine, resulting in C in the cDNA sense strand (T.Nyberg and P.S.Perlman, unpublished observation). Thus the observed deletion should not be considered as evidence that the upstream G serves as a minor branch-site in barley ndhA. While the frequency of the mutations shown in Table I did not significantly exceed that in the remaining part of covered intron sequences (see Materials and methods), a clustering close to the designated branch-point is obvious. We speculate that this relates to re-initiation of paused or terminated cDNA synthesis at the unusual 2′–5′ phosphodiester bond (Lorsch et al., 1995).
Table I. Analysis of lariat-specific cDNAs after cloning.
Branch-points are presented in grey boxes as shown to the upper left, and sequences obtained from cDNA clones (sense strand) are listed in white boxes below. The 5′ intron end (5′ I) is linked to the branch-point A in the intron 3′ part (3′ I). Nucleotides downstream of bulge A (3′ I tail) including the first two of the 3′ exon (3′ E; lower case) are in blue. The branch-point as well as positions that deviate from the expected lariat cDNA sequence are in green. Several independent cDNA pools raised from untreated (C1–C4) or ligase-treated barley RNA (LH and LU; as in Figure 2) were analysed for a number of introns including trnK which previously was shown to form lariats (Vogel etal., 1997a). Lariat cDNAs from other plants (Nt, tobacco; Pa, spruce; Zm, maize) were raised from untreated RNA. The number of inserts with a specific cDNA type is given out of the total analysed for each cloned PCR band (e.g. 13/14). The branch-point A was found to be represented overwhelmingly as T in the cDNA sequences.
The chloroplast trnA and trnI introns provide a rare deviation from the group II consensus in that the highly conserved first nucleotide is not G but U (Michel et al., 1989). Here, clones from several independently generated cDNAs exhibited complex sequence patterns (Table I). In trnA pools, we found the expected configuration of branch-point A being changed to T (underlined), resulting in 5′-GGTTTTTTGGGT-3′ being limited to <25% of the sequences. In trnI sequences obtained from four different cDNA pools, the corresponding 5′-GATTTTTTGCGT-3′ configuration amounted to about two-thirds of the total number. Deletions of up to four nucleotides were seen in the vicinity of bulge A, as were up to three non-encoded nucleotides. It should be emphasized, however, that all these mutations were restricted to the TTTTTT region shared by the ‘correct’ lariat cDNAs of trnA and trnI. Our interpretation that they arose in a random fashion, possibly during re-initiation of cDNA synthesis at the branched nucleotide, was supported by the analysis of maize trnA, which has an identical configuration in the branch-point region (Michel et al., 1989). While the two major cDNAs matched in both species, the remaining three maize cDNAs were not found in barley (Table I).
Non-conventional base pairs upstream of the branch-point
About one-third of the barley introns have the conserved A neighboured by an adjacent A:C, A:G or G:G pair (Figure 1, lower part). Nonetheless, major PCR products were of sizes predicted for correct lariat formation (shown for rps12 trans and trnG in Figure 2). A predominant choice of the conserved adenosine residue was obvious from an A→T transversion in the major cDNA sequence of each intron (Table I). In addition, rps12 cis yielded two full-circle clones. Another cDNA sequence was frequent in introns with a G upstream of the designated branch-point, showing an exact deletion of that residue. We therefore tested lariat cDNAs of two maize introns with almost identical D6 structures. The 1 bp deletion was also found in maize rps12 trans, even though the overall cDNA pattern differed notably between barley and maize, but was not seen in maize ndhB cDNA sequences (as opposed to an ∼25% proportion in two independent barley cDNAs). As discussed earlier for ndhA, we do not consider this as evidence that the upstream guanosine serves as an alternative, minor branch-point since it should be represented as C in the cDNA sequences.
An unpaired G might not outcompete the conserved A residue as branch-point because of differences in chemistry (Gaur et al., 1997; Liu et al., 1997). A second unpaired A as provided by tobacco rpl16 (Figure 1), however, should outcompete it unless the exact position of a residue within D6 is a major determinant of branching. Analysis of 15 tobacco rpl16 clones (Table I) did not provide any evidence for branching at the upstream A, while the use of the conserved branch-point seemed even more fixed than in barley where the upstream nucleotide is an unpaired G (as in most other higher plants).
Linear intron RNA
We tested seven introns (ndhB, petB, petD, rps16, trnG, trnI and trnK) for products of an alternative hydrolytic pathway, i.e. linear intron molecules. RNA ligation prior to reverse transcription should lead to additional amplification of circularized precursor transcripts and full-length or degraded linear intron RNA. Additional more slowly migrating bands that could account for circularized precursors were observed with all seven introns (shown for petB, petD and trnG in Figure 2), regardless of whether the RNA was heat denatured prior to ligation (lanes LH and LU, respectively). Efficient ligation was obvious from trnG PCRs, as indicated by a strong band of ∼330 bp known to be the 5′ and 3′ matured tRNAGly(UCC) precursor (Vogel and Hess, 2001). Another band that migrated ahead of the lariat product resulted from ligation of the 5′ intron end to internally cleaved D6 (data not shown). In contrast, no linear intron PCR bands 6 or 7 bp larger than their respective lariat products were detected (even if run on gels with higher resolution; data not shown). However, when trnK bands were analysed after cloning, two sequences from LU RNA resembled a full-circle intron (Table I). As no such sequences were found in the corresponding LH product, only LU lariat products were cloned for the remaining six introns. None of them contained a circularized intron in a pool of 9–12 sequences determined in each case (Table I).
trnV lacking a bulged adenosine
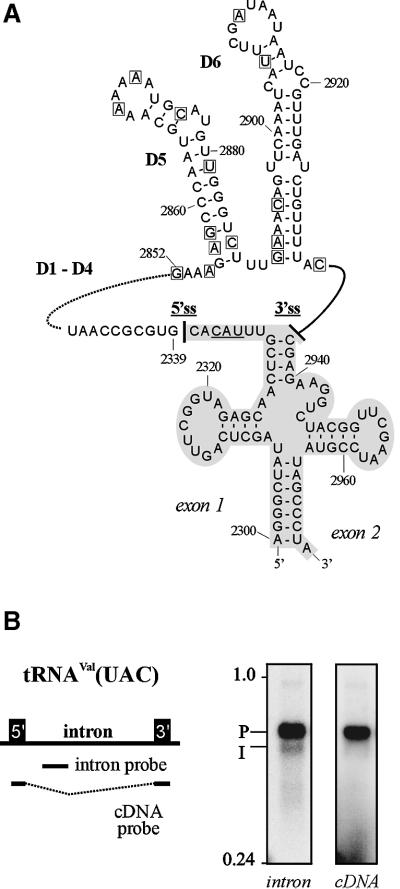
The trnV intron in tRNAVal(UAC) is strongly conserved in all plants for which a complete chloroplast genome sequence is available, and so is the lack of a bulged nucleotide in its D6 (see Figure 3A for barley; updated sequence alignment upon request). Barley trnV amplification products of sizes indicative of lariat formation anywhere in D6 were not seen when first visualized on ethidium bromide-stained agarose gels. In order to clarify whether this was due to an extremely low abundance of free intron molecules, northern hybridizations using either an exon- or an intron-specific probe were performed (Figure 3B). Signals for intron-containing precursor molecules (P) with a calculated length of ∼670 nucleotides for 5′ and 3′ matured exons (Vogel et al., 1999; Vogel and Hess, 2001) were obtained with both probes, whereas a set of smaller bands (I) was detected exclusively by the intron probe. Despite their smeary appearance, the migration of these intron-specific bands was in good agreement with a size of ∼600 nucleotides calculated for linear full-length intron molecules.
Fig. 3. Analysis of the barley trnV intron lacking a bulged A in D6. (A) Sequence of mature exons and intron domains 5 and 6. Base numbering follows DDBJ/EMBL/GenBank database entry X00408. The tRNAVal(UAC) anticodon is underlined. The 5′ss and 3′ss are marked by solid lines. A broken line indicates intron domains 1–4. Intron nucleotides that showed linkage to the 5′ intron end in attempts to amplify lariat-specific cDNAs are boxed. (B) Detection of trnV splice precursor and products on northern blots hybridized with riboprobes specific for spliced tRNAVal(UAC) (cDNA probe) or intron sequences (intron probe). Precursor signals (P) of ∼670 nucleotides for 5′ and 3′ matured tRNA exons (Vogel et al., 1999) were obtained with both probes, whereas a set of smaller bands (I) was detected with the intron probe alone. Despite a smeary appearance, these intron-specific bands migrated at ∼600 nucleotides as calculated for released trnV intron.
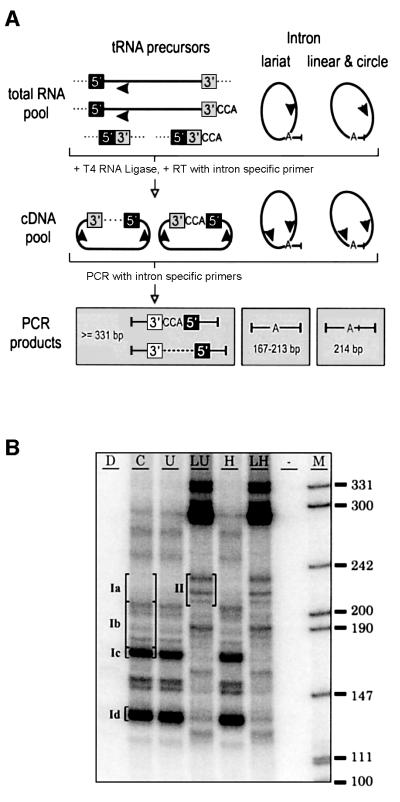
To increase the sensitivity of our assay, PCR products were radiolabelled and separated on polyacrylamide gels (Figure 4B). According to the primer design, PCR products of lariats formed between the 5′ intron end and any residue in D6 down to the 3′ss were expected in a 167–213 bp range. The two major bands from untreated RNA (cDNA reaction C; bands Ic and Id), however, migrated close to or below this size range and revealed predominant products of 169 and 132 bp, respectively, upon cloning. In the former, the mature 5′ end of tRNAVal(UAC) (A2300, Figure 3A) was linked to G2852 located between D4 and D5. In the latter, C2344 of D1 was followed by C2859 of D5. Notably, C2344 is the first nucleotide of a 5′-CGUAACC-3′ stretch that also precedes C2859. Since the 132 bp band was also seen with in vitro transcripts of trnV, which did not self-splice during cDNA synthesis (data not shown), it may have originated from either RNA recombination or a template switch by reverse transcriptase.
Fig. 4. Detection of linear trnV intron molecules. (A) Protocol of tRNAVal(UAC) precursor and intron circularization followed by RT–PCR. The sizes of expected PCR products are given in bp. (B) Radiolabelled amplification products of trnV separated on a non- denaturing polyacrylamide gel. PCRs used templates as outlined in Figure 1, and included an additional cDNA raised from RNA mock- treated in ligase buffer without prior heat treatment (U). Bands from five different gel regions (Ia–d, untreated RNA; II, ligase-treated RNA) were cut and cloned after elution. Region II almost exclusively contained circularization products of linear, mostly polyadenylated trnV intron RNAs (≥214 bp).
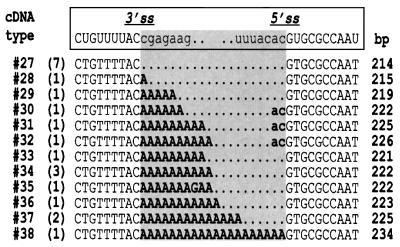
Most of the 52 clones from gel regions Ia and Ib (untreated RNA) contained inserts of smaller sizes than estimated from band Ic and co-migrating size markers. These included variations of the band Id product as well as cDNA sequences in which the first intron nucleotide (G2339) was not linked to a residue in the 3′ part of the intron (for a full listing of trnV cDNA sequences see Supplementary data available at The EMBO Journal Online). Twelve clones showed linkage of G2339 to individual residues scattered across D5 and D6 (Figure 3A). None of the U and A residues among them indicated an A→T transversion at the cDNA level. Thus, taking our other data into account, it can be excluded that any of these cDNAs arose from lariat RNAs. Surprisingly, three additional clones represented full-circle intron molecules, i.e. linkage of C2936 to G2339.
The absence of a major lariat-derived cDNA suggested release of the trnV intron in linear form. To test this hypothesis, chloroplast RNA was treated with T4 RNA ligase prior to reverse transcription. Reactions that started with RNA mock-treated in T4 RNA ligase buffer (U and H) resulted in congruent band patterns as compared with reactions from untreated RNA (C). T4 RNA ligase treatment (lanes LU and LH) yielded strong bands >300 bp that resulted from circularization of trnV precursor transcripts (Vogel and Hess, 2001), and three bands in the 200–240 bp region that were absent without prior ligation. As opposed to those from regions Ia and Ib, 21 of 22 clones from region II contained sequences of full-circle intron (Table II). Seven clones showed precise ligation of 5′ and 3′ intron termini (cDNA type 27). In the remaining 14 clones, the intron termini were found unexpectedly to be separated by non-encoded A residues, which most probably resulted from 3′ polyadenylation of intron molecules. The dinucleotide AC found upstream of the 5′ intron end in cDNA types 30, 31 and 32 was assigned tentatively to the 5′ exon, thus implying miscleavage at the 5′ss, while the possibility remains that intron polyadenyl ation was terminated by incorporation of cytidine.
Table II. Sequence analysis of cloned trnV PCR products with complete intron termini from gel region II (Figure 4B).
The nucleotides surrounding the 3′ss and 5′ss are shown (exons in lower case). The respective cDNA type number according to a listing of all cDNA types obtained in trnV experiments (see Supplementary data available at The EMBO Journal Online) is given to the left. Numbers in parentheses indicate the total clones obtained for each cDNA type. Insert sizes (in bp) are given in the right column. Nucleotides that separate the 3′ and 5′ termini of circularized intron and most probably stem from 3′ polyadenylation are in bold.
Lack of RNA editing in intron core structures
The group II intron domain 5 (essential for catalysis) and the lower stem of D6 exhibit a large degree of structural conservation (Michel et al., 1989; Costa et al., 1998). In plant mitochondria, some A:C mismatches in these domains are restored to A:U base pairs by an RNA editing activity, thereby improving overall secondary structure (Wissinger et al., 1991; Carrillo and Bonen, 1997; Farré and Araya, 1999). Despite the presence of a C→U editing activity in plant chloroplasts, no such editing of A:C mismatches in D5/D6 helices was seen in several intron-containing cDNAs from maize and tobacco plastids (Maier et al., 1996; Hirose et al., 1999). However, corrections of A:C mismatches could be rate limiting for splicing, rendering them hard to detect in cDNA populations loaded with splice precursor. In contrast to the aforementioned studies, our cDNA pools were raised from intron molecules that had already undergone at least the first step of splicing.
The sole intronic editing site reported in plant chloroplasts so far is located in the matK maturase reading frame of the trnK intron (Vogel et al., 1997b). In several control experiments, a C→T exchange at this position was limited to ∼75% of the cDNA clones from total RNA that contained both trnK-matK splice precursor and lariat, whereas 100% editing was seen in cDNAs raised from trnK lariats alone (data not shown). These experiments proved that our lariat-specific cDNA pools were not generated from a fraction of chloroplast RNAs that was excluded from RNA editing.
We then inspected all other cDNA sequences with regard to putative C/U editing sites in D5 and D6 regions (D5, barley petB, rpl2, rpl16, trnV, ycf3-i1 and tobacco rpl16; D6, barley atpF, ndhB, petD, rps12 cis, rps16, maize ndhB and spruce trnK). No substitutions of C by T at the designated candidate positions were found. In particular, in barley rps12 cis and spuce trnK, where editing could repair the A:C mismatch preceding bulge A to a Watson–Crick pair, lariats strictly formed at the conserved A residue without correction of the upstream C to U (Figure 1; Table I). Infrequent C→U changes seemed to be random and did not exceed mutation rates of other nucleotides. Likewise, no other consistent nucleotide exchanges were observed, which for example could have resolved an A:A pair in the upper helix of trnK (conserved throughout higher plants but paired in liverwort and pine). Taken together, our data reflect well the overall difference in editing frequencies in plant organellar exon sequences, with an observed ∼30 sites in ∼150 chloroplast genes (Maier et al., 1996; Tsudzuki et al., 2001) as opposed to >441 sites within and outside 56 mitochondrial genes of Arabidopsis thaliana (Giegé and Brennicke, 1999).
Discussion
Lariat formation is well established as a key feature of group II intron splicing (Peebles et al., 1986; Schmelzer and Schweyen, 1986; van der Veen et al., 1986). The formation of a branched RNA intermediate at an intron-internal bulged adenosine is also an evolutionary link with group III and nuclear pre-mRNA introns (Sharp, 1985; Copertino et al., 1994). This intermediate was demonstrated previously for several group II introns in vitro and/or in vivo by biochemical and electron microscopy analysis and, furthermore, is known to act as a key player in group II intron mobility (Zimmerly et al., 1995; Belfort et al., 2001). These data, in combination with the vast majority of known intron sequences that contain a bulged A in D6, have led to the notion of lariat formation being the common splicing pathway. Such a general validity was challenged, however, when two other mechanisms of intron release—either as linear molecules (van der Veen et al., 1987; Jarrell et al., 1988; Daniels et al., 1996; Podar et al., 1998) or as circles (Murray et al., 2001)—were shown to operate in yeast mitochondrial introns that normally form lariats. Besides, a bacterial intron with a bulged A was reported recently to splice solely by hydrolysis (when tested in vitro; Granlund et al., 2001). Furthermore, work on self-splicing introns dissected several determinants of the branching reaction (Schmelzer and Müller, 1987; van der Veen et al., 1987; Chu et al., 1998, 2001; Podar et al., 1998), e.g. G:U base pairs adjacent to the bulged A, mutations of which in aI5γ gradually activate the alternative hydrolysis pathway. It is therefore possible that many plant group II introns with atypical D6 structures (Michel et al., 1989; Carrillo et al., 2001) do not rely on the lariat pathway but have evolved splicing via different routes.
The chloroplast intron trnV splices via first-step hydrolysis
The chloroplast trnV intron was proposed to splice by first-step hydrolysis when such a pathway was shown to be viable in yeast mitochondria (Podar et al., 1998). In contrast to all other introns investigated here, trnV failed to yield cDNAs indicative of lariats. Linear RNAs liberated by hydrolytic splicing should possess 5′ monophosphates and 3′ hydroxyl groups (Michel and Ferat, 1995), which serve as substrates for T4 RNA ligase (Walker et al., 1975). Since ligase treatment led to a dramatic increase of circularized trnV molecules as compared with mock reactions, we argue that these circular RNAs were generated by joining of free termini of linear molecules and not by an intramolecular rearrangement of possible trnV lariats. Unexpectedly, non-encoded adenosine residues were observed in a large fraction of the intron-derived cDNAs. Polyadenylation at the 3′ end of mRNAs occurs in chloroplasts and has been reported to stimulate RNA degradation by endo- and exonucleases (Hayes et al., 1999). It could here explain the relatively low abundance of released trnV introns, as it should target linear trnV introns for rapid degradation.
As the available plant chloroplast sequences implicate the conservation of a hydrolytic pathway for trnV throughout plastid evolution (Learn et al., 1992), it raises the intriguing question of how this intron evolved initially. Group II intron mobility is initiated through cleavage of RNA or DNA targets by a reverse splicing reaction (Zimmerly et al., 1995; Eskes et al., 1997; Cousineau et al., 1998; Dickson et al., 2001) that lariat but not linear intron RNAs are able to perform (Michel and Ferat, 1995). At first glance, it seems likely that trnV underwent a severe branch-point mutation resulting in a block of the lariat pathway only after introduction in an ancestral plant chloroplast genome, or was transmitted through a lariat-independent mobility pathway. However, a minor proportion of trnV cDNAs from unligated RNA resembled full-length intron. Unless these cDNAs stem from infrequent excision of trnV as circles (Murray et al., 2001), they could reflect reverse splicing into tRNAVal(UAC) precursor transcripts, resulting in a transient intron RNA duplicate in vivo. Such an assumed ability of linear intron RNAs to reverse-splice, even if at low level, could have sufficed transposition of the trnV intron in the same fashion as established for group II introns that form lariats.
Mass production of lariat RNAs in plant chloroplasts
We present here the first survey of lariat formation of a complete and diverse intron set from an organellar genome in vivo. Lariats were detected for all but one of the barley chloroplast introns, and for several of their homologues from other plants. The higher plant barley has retained almost all known plant plastid introns, and two of the three introns missing (in clpP and rpoC1 genes of other species) do not possess D6 structures more unusual than those of the introns investigated here (Michel et al., 1989). We therefore conclude that the lariat pathway has been maintained throughout the course of plant evolution despite the loss of autocatalytic activity and the frequent lack of features previously shown to be important for efficient branching in self-splicing introns.
Plant chloroplasts are peculiar in having a third of their introns in tRNAs, which are here shown to splice in a group II intron manner. This mirrors the origin of the chloroplast trnL intron in a self-splicing group I intron (Xu et al., 1990) and proves the independence of plastid tRNA splicing from the plant nuclear tRNA endonuclease/ligase activities (Stange and Beier, 1987). Moreover, the strong transcription of the plastid host tRNA genes implies a mass production of branched RNAs, which nonetheless appear to be degraded quickly. So far, only the nuclear-encoded RNA debranching enzyme (DBR) has been reported to degrade lariats (Ooi et al., 2001), while homologous genes have not been found in eukaryotic organelles. Furthermore, mitochondrial intron metabolism appeared to be unaffected by disruption of the single nuclear DBR locus in yeast (Podar et al., 1998). Since A.thaliana (Initiative, 2000) also encodes only a single nuclear DBR, the intriguing question of how organellar RNA lariats are degraded remains to be resolved.
Mechanism of branch-point recognition
On the theme of branch-point choice, we observed nearly exclusively selection of the adenosine predicted by sequence analysis (Michel et al., 1989), even for those introns that lack proper base pairing upstream of the assigned branch site (ndhB, rpl16, rps12 cis, rps12 trans, trnG, spruce trnK, ycf3-1; Figure 1). These data corroborate earlier results obtained with aI5γ that implicated the chemical properties of the adenine base as an important factor to designate the site of branching (Liu et al., 1997).
Base pairs surrounding the branch-point. We expected the plastid C/U RNA editing activity (Maier et al., 1996) to restore A:C mismatches adjacent to the branch-point to Watson–Crick pairing (Figure 1). In contrast to plant mitochondria, where RNA editing sometimes improves secondary structures of D5 and D6 helices (Wissinger et al., 1991; Carrillo and Bonen, 1997; Farré and Araya, 1999), such corrections were not observed at the rps12 cis or at the spruce trnK branch-point, nor at any other position in the lariat cDNAs sequences. Hence, while base pairing downstream of the branch-point seems to be fixed (Figure 1; Michel et al., 1989; Chu et al., 2001), upstream Watson–Crick or G:U base pairs can be substituted by A:A, A:C, A:G and G:G pairs.
Relaxed base pairing in the close vicinity of the branch-point previously was implicated as a structural determinant that governs branch-site selection in aI5γ. When the two neighbouring G:U wobble pairs were mutated to G:C, branching efficiency dropped dramatically (Chu et al., 1998; Podar et al., 1998). More recent experiments with an elaborate set of D6 mutants of this intron indicated that the sensitivity of these two wobble pairs towards mutations rests in the upstream G:U pair (Chu et al., 2001).
Our notion that relaxed or non-pairing upstream with concomitant tight pairing downstream could be an important feature of local structure surrounding the branch-site of group II introns gains support from a branch-point consensus structure drawn from all lariat-forming introns of barley (Figure 5). Thirteen out of 16 introns lack Watson–Crick pairing upstream of the confirmed branching adenosine. In the same number of introns, either G:C or A:U pairs are located at the downstream position, with G:C being the downstream pair wherever there is an upstream A:C, A:G or G:G configuration. Notably, there is no intron in which upstream Watson–Crick pairing coincides with downstream G:U or other weak pairs. The consensus based on the barley introns is fully consistent with their homologous introns from eight sequenced plant chloroplast genomes (alignment available upon request) and finds further support in previous alignments of D6 structures from a broad range of group II introns (Michel et al., 1989; Chu et al., 2001). Thus, the phylogeny data imply that in a number of introns the branch-point adenosine is not a single bulge but is part of a two-nucleotide bulge that includes the nucleotide preceding it.
Fig. 5. Classification of all 16 lariat-forming group II introns of barley chloroplasts according to base pairs neighbouring the determined branch-site adenosine (black circle; cf. Figure 1). Watson–Crick pairs are represented by black thick lines. Dashed black lines represent wobble or non-pairing. A consensus structure drawn from this intron set suggests weak or non-pairing upstream, with concomitant tight Watson–Crick pairing downstream of the branch-site (lower left).
While this manuscript was in preparation, a two-nucleotide bulge around the branch-site was reported for aI5γ, which, however, includes the downstream nucleotide (Zhang and Doudna, 2002). Since the latter proposal is derived from structural mapping in vitro, most importantly in an isolated D5/D6 context with an incomplete D6, and seems to be in stark contrast to the aforementioned phylogeny data, its generality remains unclear. A two-nucleotide bulge with an unpaired downstream nucleotide could be limited to certain subgroup IIB introns with extended guanosine stretches in D6 that could adopt alternative structures. More detailed in vivo and in vitro studies of appropiate combinatorial D6 mutants of self-splicing model introns similar to the set of aI5γ mutants investigated by Chu et al. (2001) will be required to establish the substructure of the branch-point.
Spatial positioning of the branch-point in D6. In tobacco rpl16, lariats form solely at the evolutionarily conserved adenosine even though another unpaired A is available. This intron shows considerable variation of D6 structure among different plant species, but mutations do not greatly affect the length of the lower D6 helix and the linkers with D5 and the 3′ss. Thus, we conclude that in rpl16 branch-point recognition, the bulged A structure plays a minor role, while the major determinant is found in the spatial arrangement of the adenosine to become the branch-site. Such a mechanism that measures distances within the lower D6 region was deduced recently from derivates of aI5γ in which the branch-point had been moved systematically (Chu et al., 2001). It could operate, furthermore, in other introns with relaxed D6 structures, e.g. those in Figure 1, lower part, and several plant mitochondrial introns (Carrillo et al., 2001).
Faithful detection of lariats and branched RNAs in vivo
While lariat-specific cDNAs were the major fraction obtained in our experiments, minor cDNA fractions of trnV and rps12 cis with joint 3′ and 5′ intron termini could account for yet another pathway, i.e. excision as circles (Murray et al., 2001). Alternatively, these cDNAs might have arisen by jumping of reverse transcriptase on the intron template (Tuschl et al., 1998) or through reverse splicing into precursor RNA in vivo, as proposed for yeast group II introns (Mueller et al., 1993). Nonetheless, the lariat amplification method generally proves of high sensitivity and fidelity. Mapping of the petB branch-point previously failed in spinach, while petD proved successful in the same study (Kim and Hollingsworth, 1993). Since PCR signals of similar intensity were observed here for barley petB and petD, lariat splicing may occur even if branch-sites cannot be detected in conventional approaches. The method also allows simple detection of branched RNA species generated by split group II introns. Intron 1 of rps12 represents the sole example of trans-splicing in plant chloroplasts. While previous studies demonstrated exon ligation, the nature of the intron splicing intermediate remained unclear (Koller et al., 1987; Hildebrand et al., 1988; Kohchi et al., 1988). The approach used here unequivocally identified a branched (most probably Y-shaped) RNA intermediate composed of intron parts from different loci, suggesting that rps12 trans-splicing is truly mediated by the split group II intron.
The minute amounts of RNA required allow lariat detection and branch-point mapping even if starting material is extremely limited. Moreover, branching at adenosine is marked by a specific A→T exchange during cDNA synthesis, which recently has been confirmed with plant mitochondrial group II introns (Carrillo et al., 2001), nuclear pre-mRNA introns (e.g. Hirose and Steitz, 2001) and trypanosomal trans-splicing introns (A.Bindereif, personal communication). The method is thus open to a broad application, e.g. evaluation of alternative branch-points, characterization of intron intermediates in trans-splicing, and when branch-point mapping by primer extension is hindered by exons that are short or quickly degraded.
Mixed pathways in group II intron splicing
In summary, two splicing pathways are shown to operate in higher plant chloroplasts. The vast majority of the introns follows the typical lariat pathway, while hydrolytic splicing is demonstrated for trnV and appears to co-exist with branching in trnK. The lack of a branch-point adenosine in all trnV sequences examined (Learn et al., 1992), including that of liverwort as the earliest known land plant (Qiu et al., 1998), suggests that a hydrolytic splicing pathway has been maintained for trnV throughout the entire course of plant evolution.
Evidence in favour of hydrolytic group II intron splicing first came from in vitro experiments with mutants that were altered at the branch-site (van der Veen et al., 1987) and the observation of a linear intron–3′ exon intermediate under certain salt conditions (Jarrell et al., 1988). It was demonstrated later for aI5γ that under permissive in vitro conditions, branching and hydrolysis obey classical models of parallel kinetics (Daniels et al., 1996), and that mutants with severely reduced lariat formation are viable in yeast mitochondria (Podar et al., 1998). These data along with the example of naturally occurring hydrolytic trnV splicing here presented lead us to believe that first-step hydrolysis is more than a mere side reaction and could be a pathway of equal choice for certain group II introns.
Materials and methods
Nucleic acid preparation and hybridizations
Total RNA from barley (Hordeum vulgare L., cultivar ‘Haisa’), maize (Zea mays) and tobacco (Nicotiana tobacco) leaves was prepared with TRIzol reagent (Gibco), as described by the manufacturer, and freed of DNA with double DNase I (Boehringer Mannheim) treatment. Total plant DNA was extracted according to Rogers and Bendich (1985). Norway spruce (Picea abies) RNA and DNA were a gift from Mathieu Ingouff (SLU, Uppsala, Sweden). For detection of trnV, 20 µg of total barley RNA were run on a 1.5% agarose–formaldehyde gel, blotted and hybridized with riboprobes specific for spliced tRNAVal(UAC) (probe trnV cDNA as described in Vogel et al., 1999) or intron sequences [a 128 bp fragment spanning the 2554–2682 region according to DDBJ/EMBL/GenBank accession No. X00408 was cloned in pGEM-T (Promega) and transcribed from the plasmid’s T7 promoter after linearization]. Hybridization signals were visualized on a Bio-Rad phosphoimager.
Lariat RT–PCR and sequencing
Lariat cDNAs were generated and amplified as outlined in the original description of the method (Vogel et al., 1997a), using Superscript II (Gibco) and a mixture of oligonucleotide primers (see Supplementary data available at The EMBO Journal Online) specific for the 5′ part of each intron for cDNA synthesis. T4 RNA ligase treatment of chloroplast RNA was carried out as described in Vogel and Hess (2001). For heat denaturation, RNA samples were boiled in water for 5 min, followed by quick-chill on ice.
PCRs employed Hotstar DNA polymerase (Qiagen) along with an intron-specific primer pair (facing outwards in the 5′ and 3′ part of the intron, respectively; see Supplementary data). PCRs were separated on 3% Nusieve agarose gels, and bands of interest were excised. Cloning was performed in pGEM-T vector (Promega). Inserts of E.coli transformants grown without isopropyl-β-d-thiogalactopyranoside (IPTG) induction were obtained by colony-PCR and sequenced on an ABI DNA sequencer. Inspection of sequences in regions outside those shown in Tables I and II, and excluding those covered by the amplification primers, revealed overall mutation rates (deletions, insertions and nucleotide exchanges) of 0.1–0.5%. All of these mutations appeared to be random.
For trnV, PCRs contained 5 µl of [α-32P]dCTP (50 µl total volume) and were separated on 5% 1× TBE polyacrylamide gels. Following autoradiography, DNA was eluted from excised sections by overnight shaking in 1× TBE, and ethanol precipitated. Qiagen products were used to purify gel fragments and PCR products.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to L.Bonen, W.R.Hess, D.Hughes, A.M.Pyle and E.G.H.Wagner for helpful comments on the manuscript, and to M.Ingouff for providing material. This work was supported by the Fond der Chemischen Industrie, Frankfurt, and a grant from the Deutsche Forschungsgemeinschaft, Bonn, Germany. J.V. acknowledges the support of an EMBO long-term fellowship.
References
- Belfort M., Derbyshire,V., Parker,M.M., Cousineau,B. and Lambowitz,A.M. (2001) Mobile introns: pathways and proteins. In Craig,N.L., Craigie,M., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. ASM Press, pp. 761–783.
- Bonen L. and Vogel,J. (2001) The ins and outs of group II introns. Trends Genet., 17, 322–331. [DOI] [PubMed] [Google Scholar]
- Boulanger S.C., Faix,P.H., Yang,H., Zhuo,J., Franzen,J.S., Peebles,C.L. and Perlman,P.S. (1996) Length changes in the joining segment between domains 5 and 6 of a group II intron inhibit self-splicing and alter 3′ splice site selection. Mol. Cell. Biol., 16, 5896–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo C. and Bonen,L. (1997) RNA editing status of nad7 intron domains in wheat mitochondria. Nucleic Acids Res., 25, 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo C., Chapdelaine,Y. and Bonen,L. (2001) Variation in sequence and RNA editing within core domains of mitochondrial group II introns among plants. Mol. Gen. Genet., 264, 595–603. [DOI] [PubMed] [Google Scholar]
- Choquet Y., Goldschmidt-Clermont,M., Girard-Bascou,J., Kuck,U., Bennoun,P. and Rochaix,J.D. (1988) Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C.reinhardtii chloroplast. Cell, 52, 903–913. [DOI] [PubMed] [Google Scholar]
- Chu V.T., Liu,Q., Podar,M., Perlman,P.S. and Pyle,A.M. (1998) More than one way to splice an RNA: branching without a bulge and splicing without branching in group II introns. RNA, 4, 1186–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V.T., Adamidi,C., Liu,Q., Perlman,P.S. and Pyle,A.M. (2001) Control of branch-site choice by a group II intron. EMBO J., 20, 6866–6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copertino D.W., Hall,E.T., Van Hook,F.W., Jenkins,K.P. and Hallick,R.B. (1994) A group III twintron encoding a maturase-like gene excises through lariat intermediates. Nucleic Acids Res., 22, 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Christian,E.L. and Michel,F. (1998) Differential chemical probing of a group II self-splicing intron identifies bases involved in tertiary interactions and supports an alternative secondary structure model of domain V. RNA, 4, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousineau B. et al. (1998) Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell, 94, 451–462. [DOI] [PubMed] [Google Scholar]
- Daniels D.L., Michels,W.J.,Jr and Pyle,A.M. (1996) Two competing pathways for self-splicing by group II introns: a quantitative analysis of in vitro reaction rates and products. J. Mol. Biol., 256, 31–49. [DOI] [PubMed] [Google Scholar]
- Dickson L., Huang,H.R., Liu,L., Matsuura,M., Lambowitz,A.M. and Perlman,P.S. (2001) Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites. Proc. Natl Acad. Sci. USA, 98, 13207–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskes R., Yang,J., Lambowitz,A.M. and Perlman,P.S. (1997) Mobility of yeast mitochondrial group II introns: engineering a new site specificity and retrohoming via full reverse splicing. Cell, 88, 865–874. [DOI] [PubMed] [Google Scholar]
- Farré J.C. and Araya,A. (1999) The mat-r open reading frame is transcribed from a non-canonical promoter and contains an internal promoter to co-transcribe exons nad1e and nad5III in wheat mitochondria. Plant Mol. Biol., 40, 959–967. [DOI] [PubMed] [Google Scholar]
- Gaur R.K., McLaughlin,L.W. and Green,M.R. (1997) Functional group substitutions of the branchpoint adenosine in a nuclear pre-mRNA and a group II intron. RNA, 3, 861–869. [PMC free article] [PubMed] [Google Scholar]
- Giegé P. and Brennicke,A. (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA, 96, 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granlund M., Michel,F. and Norgren,M. (2001) Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J. Bacteriol., 183, 2560–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes R., Kudla,J. and Gruissem,W. (1999) Degrading chloroplast mRNA: the role of polyadenylation. Trends Biochem. Sci., 24, 199–202. [DOI] [PubMed] [Google Scholar]
- Hess W.R., Hoch,B., Zeltz,P., Hübschmann,T., Kössel,H. and Börner,T. (1994) Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell, 6, 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand M., Hallick,R.B., Passavant,C.W. and Bourque,D.P. (1988) Trans-splicing in chloroplasts: the rps 12 loci of Nicotiana tabacum. Proc. Natl Acad. Sci. USA, 85, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T. and Steitz,J.A. (2001) Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 98, 12914–12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Kusumegi,T., Tsudzuki,T. and Sugiura,M. (1999) RNA editing sites in tobacco chloroplast transcripts: editing as a possible regulator of chloroplast RNA polymerase activity. Mol. Gen. Genet., 262, 462–467. [DOI] [PubMed] [Google Scholar]
- Holländer V. and Kück,U. (1999) Group II intron splicing in chloroplasts: identification of mutations determining intron stability and fate of exon RNA. Nucleic Acids Res., 27, 2345–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Initiative T.A.G. (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Jacquier A. and Jacquesson-Breuleux,N. (1991) Splice site selection and role of the lariat in a group II intron. J. Mol. Biol., 219, 415–428. [DOI] [PubMed] [Google Scholar]
- Jarrell K.A., Peebles,C.L., Dietrich,R.C., Romiti,S.L. and Perlman,P.S. (1988) Group II intron self-splicing. Alternative reaction conditions yield novel products. J. Biol. Chem., 263, 3432–3439. [PubMed] [Google Scholar]
- Jenkins B.D., Kulhanek,D.J. and Barkan,A. (1997) Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell, 9, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.K. and Hollingsworth,M.J. (1993) Splicing of group II introns in spinach chloroplasts (in vivo): analysis of lariat formation. Curr. Genet., 23, 175–180. [DOI] [PubMed] [Google Scholar]
- Koch J.L., Boulanger,S.C., Dib-Hajj,S.D., Hebbar,S.K. and Perlman,P.S. (1992) Group II introns deleted for multiple substructures retain self-splicing activity. Mol. Cell. Biol., 12, 1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohchi T., Umesono,K., Ogura,Y., Komine,Y., Nakahigashi,K., Komano,T., Yamada,Y., Ozeki,H. and Ohyama,K. (1988) A nicked group II intron and trans-splicing in liverwort, Marchantia polymorpha, chloroplasts. Nucleic Acids Res., 16, 10025–10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller B., Fromm,H., Galun,E. and Edelman,M. (1987) Evidence for in vivo trans splicing of pre-mRNAs in tobacco chloroplasts. Cell, 48, 111–119. [DOI] [PubMed] [Google Scholar]
- Learn G.H. Jr, Shore,J.S., Furnier,G.R., Zurawski,G. and Clegg,M.T. (1992) Constraints on the evolution of plastid introns: the group II intron in the gene encoding tRNA-Val(UAC). Mol. Biol. Evol., 9, 856–871. [DOI] [PubMed] [Google Scholar]
- Liu Q., Green,J.B., Khodadadi,A., Haeberli,P., Beigelman,L. and Pyle,A.M. (1997) Branch-site selection in a group II intron mediated by active recognition of the adenine amino group and steric exclusion of non-adenine functionalities. J. Mol. Biol., 267, 163–171. [DOI] [PubMed] [Google Scholar]
- Lorsch J.R., Bartel,D.P. and Szostak,J.W. (1995) Reverse transcriptase reads through a 2′–5′ linkage and a 2′-thiophosphate in a template. Nucleic Acids Res., 23, 2811–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R.M., Zeltz,P., Kossel,H., Bonnard,G., Gualberto,J.M. and Grienenberger,J.M. (1996) RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol., 32, 343–365. [DOI] [PubMed] [Google Scholar]
- Martínez-Abarca F. and Toro,N. (2000) Group II introns in the bacterial world. Mol. Microbiol., 38, 917–926. [DOI] [PubMed] [Google Scholar]
- Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono,K. and Ozeki,H. (1989) Comparative and functional anatomy of group II catalytic introns—a review. Gene, 82, 5–30. [DOI] [PubMed] [Google Scholar]
- Mueller M.W., Allmaier,M., Eskes,R. and Schweyen,R.J. (1993) Transposition of group II intron aI1 in yeast and invasion of mitochondrial genes at new locations. Nature, 366, 174–176. [DOI] [PubMed] [Google Scholar]
- Murray H.L., Mikheeva,S., Coljee,V.W., Turczyk,B.M., Donahue,W.F., Bar-Shalom,A. and Jarrell,K.A. (2001) Excision of group II introns as circles. Mol. Cell, 8, 201–211. [DOI] [PubMed] [Google Scholar]
- Ooi S.L., Dann,C.,III, Nam,K., Leahy,D.J., Damha,M.J. and Boeke,J.D. (2001) RNA lariat debranching enzyme. Methods Enzymol., 342, 233–248. [DOI] [PubMed] [Google Scholar]
- Peebles C.L., Perlman,P.S., Mecklenburg,K.L., Petrillo,M.L., Tabor, J.H., Jarrell,K.A. and Cheng,H.L. (1986) A self-splicing RNA excises an intron lariat. Cell, 44, 213–223. [DOI] [PubMed] [Google Scholar]
- Podar M., Chu,V.T., Pyle,A.M. and Perlman,P.S. (1998) Group II intron splicing in vivo by first-step hydrolysis. Nature, 391, 915–918. [DOI] [PubMed] [Google Scholar]
- Qiu Y.L., Cho,Y., Cox,J.C. and Palmer,J.D. (1998) The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature, 394, 671–674. [DOI] [PubMed] [Google Scholar]
- Rogers S. and Bendich,A. (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol., 5, 69–76. [DOI] [PubMed] [Google Scholar]
- Schmelzer C. and Müller,M.W. (1987) Self-splicing of group II introns in vitro: lariat formation and 3′ splice site selection in mutant RNAs. Cell, 51, 753–762. [DOI] [PubMed] [Google Scholar]
- Schmelzer C. and Schweyen,R.J. (1986) Self-splicing of group II introns in vitro: mapping of the branch point and mutational inhibition of lariat formation. Cell, 46, 557–565. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (1985) On the origin of RNA splicing and introns. Cell, 42, 397–400. [DOI] [PubMed] [Google Scholar]
- Stange N. and Beier,H. (1987) A cell-free plant extract for accurate pre-tRNA processing, splicing and modification. EMBO J., 6, 2811–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsudzuki T., Wakasugi,T. and Sugiura,M. (2001) Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol., 53, 327–332. [DOI] [PubMed] [Google Scholar]
- Tuschl T., Sharp,P.A. and Bartel,D.P. (1998) Selection in vitro of novel ribozymes from a partially randomized U2 and U6 snRNA library. EMBO J., 17, 2637–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen R., Arnberg,A.C., van der Horst,G., Bonen,L., Tabak,H.F. and Grivell,L.A. (1986) Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell, 44, 225–234. [DOI] [PubMed] [Google Scholar]
- van der Veen R., Kwakman,J.H. and Grivell,L.A. (1987) Mutations at the lariat acceptor site allow self-splicing of a group II intron without lariat formation. EMBO J., 6, 3827–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. and Hess,W.R. (2001) Complete 5′ and 3′ end maturation of group II intron-containing tRNA precursors. RNA, 7, 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Hess,W.R. and Börner,T. (1997a) Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res., 25, 2030–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Hübschmann,T., Börner,T. and Hess,W.R. (1997b) Splicing and intron-internal RNA editing of trnK-matK transcripts in barley plastids: support for MatK as an essential splice factor. J. Mol. Biol., 270, 179–187. [DOI] [PubMed] [Google Scholar]
- Vogel J., Börner,T. and Hess,W.R. (1999) Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res., 27, 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G.C., Uhlenbeck,O.C., Bedows,E. and Gumport,R.I. (1975) T4-induced RNA ligase joins single-stranded oligoribonucleotides. Proc. Natl Acad. Sci. USA, 72, 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissinger B., Schuster,W. and Brennicke,A. (1991) Trans splicing in Oenothera mitochondria: nad1 mRNAs are edited in exon and trans-splicing group II intron sequences. Cell, 65, 473–482. [DOI] [PubMed] [Google Scholar]
- Xu M.Q., Kathe,S.D., Goodrich-Blair,H., Nierzwicki-Bauer,S.A. and Shub,D.A. (1990) Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science, 250, 1566–1570. [DOI] [PubMed] [Google Scholar]
- Zhang L. and Doudna,J.A. (2002) Structural insights into group II intron catalysis and branch-site selection. Science, 295, 2084–2088. [DOI] [PubMed] [Google Scholar]
- Zimmerly S., Guo,H., Eskes,R., Yang,J., Perlman,P.S. and Lambowitz,A.M. (1995) A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell, 83, 529–538. [DOI] [PubMed] [Google Scholar]