Abstract
Enhancers can function over great distances and interact with almost any kind of promoter, but insulators or promoter competition generally limit their effect to a single gene. We provide in vivo evidence that retroelements may establish promoter competition with their neighboring genes and restrict the range of action of an enhancer. We report that the retroelement Idefix from Drosophila melanogaster inhibits white gene expression in testes by a promoter competition mechanism that does not occur in the eyes. The sequence specificity of the two TATA-less promoters of white and Idefix is a prime determinant in the competition that takes place in tissues where both are transcriptionally active. This study brings to light a novel mechanism whereby transcriptional interference by an active retrotransposon may perturb expression of neighboring genes. This capacity to interfere with the transcriptional regulation of their host, together with the facts that retroelements preferentially move within the germline and do not excise to replicate, suggest that these elements are cis-regulatory sequences able to imprint specific and heritable controls essential for eukaryotic gene regulation.
Keywords: Drosophila/Idefix/promoter competition/retrotransposons/transcriptional interference
Introduction
In eukaryotes, the tissue and spatial specificity of gene expression is regulated by interactions of proteins with short DNA sequences called enhancers. Enhancers exert long-distance effects independently of their position and orientation. Moreover, enhancers have been shown to be able to drive the transcription of a heterologous gene if located in its vicinity. This raises the question of how an enhancer specifically activates its target gene without affecting adjacent genes. Two main mechanisms have been described for regulating the interaction of a shared enhancer with a specific target promoter: insulator DNAs and promoter competition.
In the first mechanism, chromosomes are divided into distinct domains of gene action by insulator sequences: a given enhancer will then interact with a defined promoter only if both reside within the same domain. The elements scs and scs′ identified in the flanking regions of the Drosophila hsp70 locus were the first insulators to be identified (Udvardy et al., 1985; Kellum and Schedl, 1991, 1992). Additional insulator elements have since been identified, including the suppressor of Hairy wing insulator within the gypsy retroelement, the Fab-7 and Fab-8 insulators within the bithorax complex of Drosophila (Geyer and Corces, 1992; Hagstrom et al., 1996; Zhou et al., 1996, 1999; Barges et al., 2000) or, in vertebrates, the insulator present at the chicken β-globin locus (Chung et al., 1993, 1997). All the insulators described so far block gene expression only when located between a distal enhancer and a target promoter. Recently, a cis-regulatory element that permits an enhancer to bypass an insulator has been identified. This element, called promoter targeting sequence or PTS, allows distal enhancers to overcome the blocking activities of insulators (Zhou and Levine, 1999).
According to the promoter competition model, the inherent properties of promoters and enhancers allow only certain combinations to interact, while other combinations are inefficient. Consequently, a shared enhancer can activate multiple genes, but selects the promoter region of a single one. Activation of the preferred gene precludes expression of the neighboring genes. Promoter competition was first identified in the chicken globin gene cluster (Choi and Engel, 1988; Foley and Engel, 1992) and has since been implicated in the regulation of genes within the ANT-C in Drosophila (Ohtsuki et al., 1998) and in the regulation of mammalian Hox genes (Herault et al., 1997; Sharpe et al., 1998). Promoter competition is likely to be mediated by the promoter specificity. Three different core promoter elements located within a 50–60 bp sequence flanking the transcription start site have been identified: the TATA box, the initiator element (Inr) and the downstream promoter element (DPE) (Burke and Kadonaga, 1996). Promoters are subdivided into two classes: (i) TATA-less promoters, which contain conserved copies of Inr and/or DPE; and (ii) TATA promoters, which do not require Inr or DPE elements to initiate transcription (Burke and Kadonaga, 1996). It has been shown that some enhancers preferentially activate TATA promoters when given a choice between a linked TATA and a TATA-less promoter. In the ANT-C complex, the enhancer AE1 preferentially activates the TATA promoter of the fushi tarazu (ftz) gene over the TATA-less promoter of Sex combs reduced (Scr) (Ohtsuki et al., 1998). In contrast, some enhancers interact preferentially with DPE-containing promoters over promoters with a TATA box (Butler and Kadonaga, 2001). These findings suggest that proteins bound at the enhancer and promoter regulatory elements are probably implicated in the promoter competition mechanism.
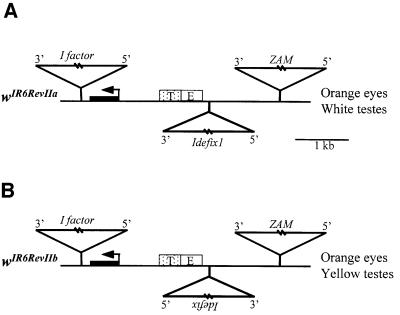
The arrival of additional enhancers and promoters due to recent insertions of transposable elements at a defined locus can affect the relationship established between the promoter of a nearby gene and its enhancers. Such alleles can thus be powerful tools to help understand the mechanisms by which enhancers activate promoters. We made use of recurrent insertions affecting the white locus of the Drosophila genome to examine the modifications of the enhancer–promoter interactions established after such insertions. Four alleles of the white gene from successive insertions of three transposable elements have already been described (Desset et al., 1999; Conte et al., 2000). The wIR6 allele is due to the insertion of an I factor within the first intron of the white gene. This insertion is responsible for an alteration of white gene splicing and thus a decrease in the wild-type transcripts of the gene, which leads to an orange instead of brick-red eye phenotype (Lajoinie et al., 1995). The wIR6RevI allele identified in a line referred to as RevI results from the insertion of the retroviral element ZAM 3 kb upstream from the wIR6 transcription start site (Leblanc et al., 1997). These flies display brick-red eyes. Spontaneous mutants with orange eyes among the red-eyed population were recovered in the RevI line and established as lines denoted RevII. These mutations arise from an insertion of the retroviral element, denoted Idefix, 1.7 kb upstream of the white gene promoter, leading to the wIR6RevII alleles (Figure 1A). Finally, an allele called wIR6RevIV, identified in a line denoted RevIV, derives from wIR6RevII through an additional insertion of Idefix at the 3′ end of the first Idefix and in the opposite orientation. Flies carrying this white allele display a full reversion of the wIR6RevII eye phenotype to brick-red (Desset et al., 1999).
Fig. 1. Molecular structures of the wIR6RevII alleles and associated eye and testis phenotypes. The structure of the white alleles is represented as follows: the white gene transcription unit is indicated by a line, its first exon by a black box, and its transcription start site by an arrow. DNA upstream of the gene is shown as a line, and the white enhancers (E for eyes and T for testes) as boxes. The insertion sites of the I factor, ZAM and Idefix are indicated by triangles. The names of the white alleles are shown on the left, and the eye and testes colors of the flies bearing these alleles are indicated on the right.
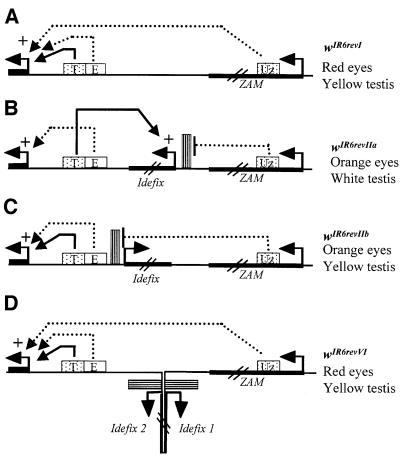
In a previous paper, we examined the molecular mechanisms by which ZAM and Idefix modulate the expression of the white gene in the eyes of the RevI, RevII and RevIV lines. We reported that ZAM contains an enhancer in its 5′-untranslated region (UTR) capable of activating downstream white gene expression in the eyes, while Idefix carries an insulator able to block the activation by the ZAM enhancer in this tissue (Conte et al., 2002; Figure 5A and B).
Fig. 5. Models of interactions between ZAM, Idefix and white in the different white alleles. (A), (B), (C) and (D) correspond to the wIR6RevI, wIR6RevIIa, wIR6RevIIb and wIR6RevIV alleles, respectively. The white gene is indicated by its promoter, and the beginning of its first exon is shown by a black box. DNA upstream of the gene is shown as a line. The regulatory sequences present in the different white alleles studied are represented as follows: enhancers of eyes (E) and testes (T) by boxes; Idefix insulator by a hatched rectangle; promoters by arrows; and the 5′-UTR of ZAM bearing an eye enhancer (Uz) by a dotted box. Pointed arrows indicate the activating effects of the eye enhancers. Plain lines indicate the activating effect of the testes enhancer. Names of the alleles and the eyes and testes colors of their corresponding lines are indicated on the right.
Since the white gene is controlled by two enhancers, one driving its expression in the eyes and the other in the testes, lending them a yellow color, we focused our attention on the testes color of these different lines. Here, we report that all the lines wIR6, RevI and RevIV display yellow testes, while the RevII lines, whose structure is shown in Figure 1A, display white testes. By transgenic experiments, we show that the upstream transcriptionally active Idefix is able to repress the downstream white gene, a phenomenon known as ‘transcriptional interference’. Our data indicate that in RevII, the promoter of Idefix is able to compete with the white gene promoter for the testes enhancer. This competition is orientation dependent, since an isolated novel RevII line with an Idefix insertion in the opposite orientation displays yellow testes. These results bring to light a novel mechanism by which transposable elements can interfere with the transcriptional regulation of their host, i.e. promoter competition. We report that this competition occurs between two TATA-less promoters that possess distinct properties enabling them to be selected or not by cis-regulatory sequences and establish promoter competition.
Results
Idefix interferes with white gene expression in testes in an orientation-dependent manner
The wild-type expression of white is driven by two enhancers located close together at 1.5 kb upstream of the white gene promoter (Figure 1). In a wild-type context, the testes enhancer leads to a yellow phenotype of the testes. When this enhancer is not functional, no coloration is observed and the testes are white. Focusing our attention on the testes color of the RevI, RevII and RevIV lines, we found that, as for wIR6, RevI and RevIV displayed a yellow testes phenotype, compared with a white one in RevII.
The yellow pigmentation of the RevI line potentially could be due to the activity of both the endogenous testes enhancer of white and an enhancer provided by the insertion of ZAM. The ZAM sequence is composed of two long terminal repeats (LTRs) flanking a long 5′-UTR and three open reading frames (ORFs) (Leblanc et al., 1997). Since the 5′-UTR and LTRs of diverse retroviral sequences have been found to be responsible for the deregulation of genes located close to their insertion, we tested the potential involvement of the 5′-UTR and the LTR of ZAM in their effects on the white gene. We analyzed the testes color of initially established transgenic lines to clarify the influence of ZAM on the eye color of the RevI line (Conte et al., 2002). Constructs called pUzW or pLzW, in which the 5′-UTR or the LTR of ZAM have been inserted upstream of the mini-white gene, respectively, were injected in the w1118 line. The latter displays a mutation of white leading to white eyes and testes. Among five independent transgenic lines analyzed for each of the transgenes, none displayed yellow testes (data not shown). This result indicates that ZAM does not act as an enhancer of the white gene that is able to direct its expression in the testes. The yellow phenotype of the RevI testes must therefore derive solely from activation of the white gene by its own testes enhancer.
Analysis of the RevII lines revealed that the relationship established between the promoter of white and its testes enhancer was modified upon the insertion of Idefix 1.7 kb upstream of the white transcription start site. Testes are white in these lines instead of yellow. Two major observations prompted us to search for the molecular mechanisms responsible for this variation of white gene expression in RevII testes. First, the ability of Idefix to act as an insulator, described by Conte et al. (2002), could not account for the white phenotype of these testes, since Idefix is not positioned between the testes enhancer and the white promoter, but upstream of both (see the structure of the wIR6RevII allele, Figure 1A). Secondly, we recently established two new RevII lines from the RevI stock. The structure of their wIR6RevII allele is similar to that described in Desset et al. (1999) and in Figure 1A, but with one main difference: the Idefix insertion is in the opposite orientation. This novel allele is schematized in Figure 1B. Hence, two categories of RevII lines exist that differ in their Idefix orientation. Although both display an orange eye phenotype readily explained by the presence of the insulator able to counteract the enhancer effect of ZAM, they display, surprisingly, different testes colors. As reported above, the first RevII lines identified displayed white testes, while the last ones displayed yellow testes. These will be identified subsequently as wIR6RevIIa alleles in RevIIa lines when Idefix is inserted in the 5′–3′ orientation with regard to the white gene transcription start site or as wIR6RevIIb alleles in RevIIb lines when Idefix is inserted in the 3′–5′ orientation (Figure 1A and B, respectively). Thirdly, when two Idefix are present upstream of white and in the opposite orientation, as in RevIV, the testes are then yellow. These findings suggest that the molecular mechanism responsible for the mutagenic effect of the element present in RevIIa can be suppressed by an additional element in the other orientation.
From these observations, we conclude that the enhancer–promoter dialog established between the white gene and its testes enhancers may be modified upon arrival of Idefix. This modification is independent of the insulator function of Idefix. It depends on the orientation of Idefix and can be suppressed by an additional Idefix if inserted in the opposite orientation.
The LTR of Idefix is able to break the dialog between the testis enhancer and the white gene promoter
To elucidate the molecular mechanisms responsible for this variation in the testes color between ReIIa and b lines, we constructed transgenes designed to mimic the structure of these alleles and analyzed their expression in transgenic flies.
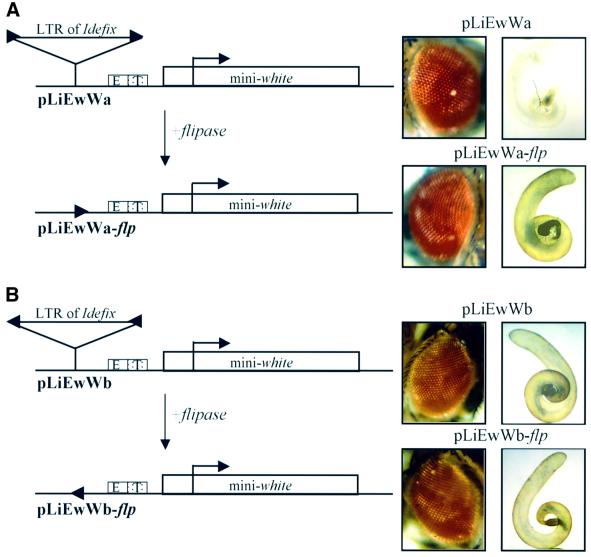
Idefix is composed of two LTRs flanking a 5′-UTR and three ORFs (Desset et al., 1999). Plasmids called pLiEwW were constructed to test the impact of the LTR of Idefix on white gene expression. They display the LTR (Li) placed upstream of the cis-regulatory sequences of white (Ew) and the mini-white gene (W). Additionally, the LTR is flanked by FRT elements, which are targets for the flp recombinase action (Figure 2A and B). We reasoned that if the LTR of Idefix is responsible for the phenotype of the RevII testes, then it should also interact in the dialog between the white enhancers (Ew) and the downstream mini-white gene (W). These artificial constructs injected into flies carrying the w1118 null mutation should lead to transgenic flies with white or yellow testes depending on Idefix orientation in the transgenes. Transgenic flies bearing pLiEwW constructs were identified by their eye color due to the expression of the mini-white gene in this tissue and established as lines denoted pLiEwW from the name of the injected plasmid. These lines were then subjected to the flp recombinase action. To this end, they were crossed to flies expressing the flp recombinase under the control of a heat shock promoter. Expression of flp in the descendants results in recombination between the two FRTs and deletion of the intervening LTR of Idefix. This yielded flies denoted pLiEwW-flp. This strategy permits an assessment of the white gene activity in the presence (pLiEwW flies) and absence (pLiEwW-flp flies) of Idefix LTR to avoid any complications in interpretation due to position effects. In addition, PCR experiments were performed on DNA extracts from heat-shocked and non-heat-shocked flies to check the excision of the FRT-flanked fragment in all the subsequent experiments (data not shown).
Fig. 2. The LTR of Idefix counteracts the activation effect of the downstream white testes enhancer. (A) The LTR of Idefix is inserted upstream of the white enhancers in the 5′–3′ orientation and (B) in the 3′–5′ orientation with regard to mini-white gene transcription. The structures of the P transformation vectors carried by the transgenic flies assayed for Idefix LTR inhibition effect are shown. The mini-white gene is represented by a white rectangle, its transcription start site by an arrowhead, and its eye and testes enhancers by boxes (E and T). The insertion site of the Idefix LTR upstream of the white enhancers is indicated by a triangle. The FRT sites, which flank the Idefix LTR, are shown by arrowheads. The names of the constructs are indicated below the figure, and the testes and eye colors of the flies bearing these constructs on the right. The second construct is the same as the first except that the LTR of Idefix has been removed via flp-mediated recombination.
Two series of pLiEwW transgenic flies were tested: (i) pLiEwWa flies bearing the LTR of Idefix in the 5′–3′ orientation according to the mini-white gene expression, as in RevIIa (Figure 2A); and (ii) pLiEwWb transgenic flies bearing the LTR of Idefix in the opposite orientation, as in RevIIb (Figure 2B). At least five independent transgenic lines were tested for each series of experiments.
Transgenic lines with the pLiEwWa construct displayed white testes (Figure 2A). When the LTR was excised after the flp recombinase action, the pLiEwWa-flp flies displayed yellow testes (Figure 2A). Thus, in this configuration, which mimics the wIR6RevIIa structure, Idefix is indeed able to block the activation of white gene expression in testes, and this effect is due to the presence of its LTR. In contrast, the pLiEwWb lines, in which Idefix LTR is inserted upstream of white and its enhancers, but in the opposite orientation, as in the wIR6RevIIb allele, displayed yellow testes (Figure 2B). When the LTR was excised after the flp recombinase action, the pLiEwWb-flp flies displayed yellow testes (Figure 2B).
From these findings, we conclude that the LTR of Idefix is able to block the dialog established between the testis enhancer and its white target promoter when inserted upstream. In addition, this interference with the transcriptional regulation of white is orientation dependent and only occurs when the LTR is in the same orientation as white gene transcription.
Both cis-regulatory sequences of white driving its expression in testes and in the eyes are present in our constructs. Surprisingly, no pLiEwW lines tested displayed any modification of the eye color on removal of the Idefix LTR (Figure 2A and B). This result therefore indicates that the enhancer-blocking activity of the LTR of Idefix on downstream enhancers may be specific to some enhancers and ineffective on others.
The enhancer-blocking activity of the Idefix LTR is dependent on its transcription
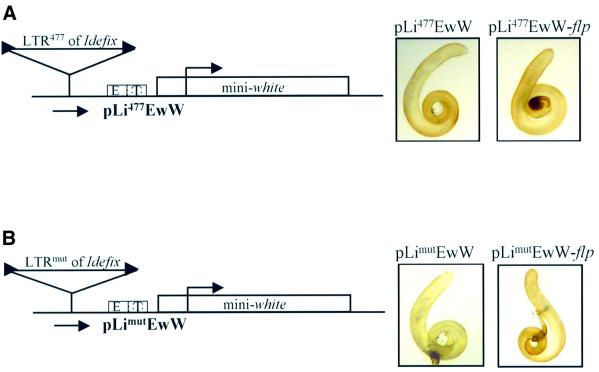
Since the promoter of Idefix is contained within its LTR, we wondered whether the phenotypes observed in the transgenic lines were related or not to the transcriptional activity of Idefix within the transgenes. Previous experiments with a mutated LTR (Li477) displaying a point mutation in the Inr sequence of the Idefix promoter had shown that this LTR is unable to initiate transcription when fused to the lacZ reporter gene (see Materials and methods and Conte et al., 2002). A series of experiments additional to those reported above was performed with the wild-type LTR of the constructs replaced by the mutated LTR. Li477 was inserted in the 5′–3′ orientation upstream of the enhancers of white and the mini-white gene in constructs denoted pLi477EwW (Figure 4A). No modification of the eye color was observed before or after the removal of this LTR477 within the transgenic lines (data not shown). Additionally, the presence of this mutated LTR inserted upstream of white and its enhancers did not prevent the testis pigmentation, i.e. testes were yellow (Figure 4A). As expected, on removal of the Idefix sequence, pLi477EwW-flp lines also displayed yellow testes due to the activation of white gene expression by its testes enhancer.
Fig. 4. Promoter competition between white and Idefix depends on Idefix promoter sequence. (A) The mutated LTR477 of Idefix (Li477) does not block the dialog between the white gene promoter and its testes enhancer when inserted upstream. Li477 is inserted in the 5′–3′ orientation with regard to the mini-white gene orientation. This orientation and the corresponding name of the P transformation vector are indicated below the constructs. The testes color of the flies bearing the P transformation vectors is shown on the right before and after the flp recombinase action. (B) The mutated LTRmut of Idefix (Limut) does not inhibit white gene expression driven by its testes enhancer, although it initiates transcription. The P transformation vector assayed for the effect of Limut on white gene expression is the same as in (A) except that Li477 of Idefix has been replaced by Limut. The orientation of Limut is indicated by an arrowhead below the construct. The testes color of the flies bearing the P transformation vectors is shown on the right before and after the flp recombinase action.
These results indicate that the Idefix LTR may exert an enhancer-blocking activity on specific downstream enhancers, but this interference with nearby genes depends on both its orientation at the locus and its transcriptional activity.
According to these results, the testis phenotype of the RevIIa flies (wIR6RevIIa allele) is readily explained. In a 5′–3′ orientation (according to the downstream white gene transcription), the Idefix promoter blocks interactions between the testes enhancer and the white gene promoter, resulting in a white testis phenotype. In contrast, Idefix inserted in the opposite orientation is not able to compete with the white gene promoter for its enhancer, and testes are yellow as observed in the RevIIb flies (wIR6RevIIb allele).
Competition between Idefix and white promoters depends on their sequence
Two main mechanisms potentially can explain the transcriptional interference observed between Idefix and white in RevII lines: the readthrough transcription model or the promoter competition model.
In the readthrough transcription model, interference with white expression would result from readthrough transcription from the upstream Idefix promoter. In three series of RT–PCR experiments performed on the pLiEwWa lines, we searched for a chimeric transcript that would result from such a trancriptional interference and thus contain Idefix sequences plus white sequences. No chimeric transcript was ever detected in these lines.
In the promoter competition model, the inherent properties of the promoters and enhancers allow only certain combinations to interact, other combinations remaining inefficient. For example, it was shown that TATA promoters are activated preferentially over TATA-less promoters by some enhancers (Ohtsuki et al., 1998). In our system, the promoters of white and Idefix are both TATA-less promoters. Consensus sequences have been reported for Inr and DPE sequences that function co-operatively to direct accurate and efficient initiation of transcription in TATA-less promoters. The Inr and DPE motif of white and Idefix do not conform completely to the described consensus. The transcription start site of white conforms to the consensus of the Inr element (TCAGTT), which is sufficient to direct initiation in the absence of a TATA element. However, its DPE motif (GAAG) does not conform completely to the consensus G-T/A-C-G (Kutach and Kadonaga, 2000). For the Idefix transcription sites, its Inr (TCAGAG) at nucleotide 472 according to the Idefix sequence does not conform to the Inr consensus, while its DPE (GTCG) at nucleotide 502 does (Tcheressiz et al., 2002).
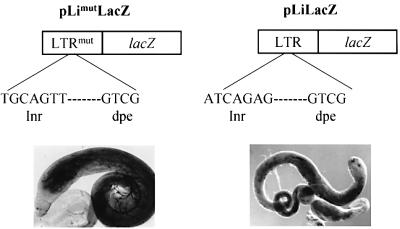
Thus we wondered whether the specificity of the Idefix promoter sequence was important for competition with the white gene promoter. To test this, transgenic experiments were conducted with a novel mutated LTR, called Limut. In this Limut, the Inr sequence of Idefix has been changed in such a way that a novel Inr sequence conforms to the consensus GCAGTT (Figure 3; Kutach and Kadonaga, 2000). We verified that this mutated LTR fused to a lacZ reporter gene is able to initiate Idefix transcription in testes like the wild-type LTR (see Figure 3, LimutLacZ compared with LiLacZ; Tcheressiz et al., 2002). We then tested the ability of this LTR to compete with the white gene promoter. For this purpose, Limut was inserted in the 5′–3′ orientation upstream of the endogenous enhancers of white and the mini-white gene in a construct called pLimutEwW (Figure 4B). The five pLimutEwW lines tested displayed yellow testes, indicating that Limut does not inhibit interactions between the testes enhancer and the white gene. Thus this mutated promoter does not compete with the white gene promoter for the shared testes enhancer, although it is transcriptionally active.
Fig. 3. The mutated LTR of Idefix, Limut, is able to initiate transcription. An Limut construct was subjected to in vivo transcription analysis by transgenic experiments. The name of the construct containing a Limut–lacZ fusion gene and its structure are indicated at the top. The lacZ reporter gene expression is detected in testes of flies bearing the Limut–lacZ construct. LacZ staining observed in a similar experiment with the wild-type LTR of Idefix (LiLacZ) is shown on the right (Tcheressiz et al., 2002).
These findings show that transcription of competing promoters is necessary but not sufficient for promoter competition. It is the sequence specificity of the core promoters and their inherent properties that form the central machinery mediating competition.
Discussion
The present work describes a new mechanism by which transposable elements can interfere with the host genome in D.melanogaster. We have reported elsewhere that the insertions of two retrotransposons, ZAM and Idefix, upstream of the transcription start site of white bring novel regulatory sequences such as an enhancer and an insulator that modify the proper regulation of white in the eyes (Conte et al., 2002; Figure 5). This work reports another type of regulation, namely promoter competition as a mechanism of transcriptional interference, potentially afforded by such mobile elements. It places transposable elements as regulatory elements able to impose functional restraints on nearby genes.
Idefix promoter competes with the white gene promoter for the same enhancer
The present study was initiated after the recovery of novel mutations affecting the eye color of the RevI stock of flies. RevI flies display brick-red eyes due to the presence of a ZAM element 3 kb upstream of wIR6 (Leblanc et al., 1999). These new and independent mutations were found to be due to the insertion of an Idefix element, 1.7 kb upstream of wIR6 between the transcription start site of white and the insertion of ZAM. Such insertions had already been reported as responsible to wIR6RevII alleles; however, in these new mutational events, Idefix was inserted in the opposite orientation, i.e. 3′–5′ with regard to the white gene transcription instead of 5′–3′ in the 12 lines already reported in Desset et al. (1999). We denoted the alleles wIR6RevIIa when Idefix is inserted 5′–3′, and wIR6RevIIb when it is in the opposite orientation. Lines established were called RevIIa and RevIIb, respectively. Interestingly, all these Idefix insertions are responsible for a similar modification of the eye phenotype whatever their orientation, going from brick-red to orange; however, they have a different mutagenic impact on the expression of the downstream white gene in testes. Flies bearing wIR6RevIIa display white testes while flies bearing the wIR6RevIIb allele display yellow testes (Figure 1A and B).
Transgenic assays enabled us to characterize the molecular mechanisms responsible for the mutagenic impact of Idefix in testes. We have tested the influence of different fragments of Idefix potentially involved in regulation of the wIR6RevII alleles and found that when its LTR is present in transgenes designed as follows: ‘5′LTR3′-enhancers-5′mini-white3′’, the Idefix LTR prevents the mini-white promoter from being activated by its testes enhancer. Transgenic lines bearing this construct that mimics the wIR6RevIIa allele display white testes. However, the LTR in this orientation does not block the white gene being activated by its eye enhancer also present in the constructs. In the presence or absence of the LTR, no modification of the eye color was observed in any of the transgenic lines obtained.
In RevIIa, as in the transgenic lines, the enhancer-blocking activity of white observed in testes cannot be explained by the presence of an insulator sequence within the LTR since insulators block gene expression only when located between a distal enhancer and a target gene. In RevIIa and the tested constructs, the Idefix LTR is located upstream of both the enhancer and the target gene (see Figures 2A and 5B). Since the promoter of Idefix is present within its LTR, promoter competition established in this tissue offers a better explanation for such a transcriptional interference (as defined by Villemure et al., 2001) that leads to the repression of the downstream white gene by the upstream Idefix unit. An alternative explanation could be that the LTR possesses a silencer element downstream of the insulator and is responsible for the repressive effect. However, this explanation is very unlikely because the observed transcriptional interference can be prevented by two point mutations, Limut and Li477, one of them overlapping with the initiator element (see below).
Transgenic experiments performed with the mutated LTR of Idefix (LTR477), which suffered a single nucleotide alteration at the promoter start site of Idefix, revealed that this inactive promoter is no longer able to interfere in the regulation of the downstream mini-white gene in testes. This result shows that the promoter of Idefix is engaged directly in the regulation of the wIR6RevIIa allele by competing with the promoter of white. Since both white and Idefix initiate their transcription from a TATA-less promoter, these data indicate that the promoter of Idefix displays specific properties allowing its interaction with the testes enhancer and precluding expression of the white promoter in this tissue. This specificity can be lost through a single mutation affecting its core promoter such as the LTR477 mutation that is sufficient to render the interaction inefficient. Additionally, the specific properties of this promoter do not allow the competition to occur in all the tissues where white is expressed since it does not take place in the eyes. The reason for this is still unknown; however, it is interesting to note that the promoter of Idefix appears to be unable to establish the competition when it is inactive. This observation derives from two main results. First, the promoter present in LTR477 is unable to direct Idefix transcription and it is then not involved in the competition. Secondly, experiments described above indicate that the challenge between the two promoters only occurs in testes where Idefix is transcriptionally active and not in the eyes where Idefix is inactive (Tcheressiz et al., 2002; Figure 2A).
It must be noted that a mechanism other than promoter competition potentially can explain our experiments: interference of white expression resulting from readthrough transcription from the upstream Idefix promoter. Readthrough transcription may occlude transcription factor-binding sites in a downstream promoter. This mechansim has been suggested previously to explain transcriptional interference at other loci (Proudfoot, 1986; Greger et al., 1998, 2000). Activation of Idefix transcription by testis enhancer could therefore result in a reduction of white expression in the testes, as is observed. According to a series of data, this mechanism appears unlikely to explain the white color of the RevIIa testes. First, in constructs bearing Limut, the LTR is able to initiate transcription and thus a potential readthrough of the downstream mini-white. Nevertheless, testes of these transgenic lines are yellow as if the mini-white gene expression was not occluded by the upstream Limut transcription. Secondly, in a series of three independent experiments, we searched to detect a potential chimeric transcript initiated in the Idefix LTR and ending in the mini-white gene. No such transcript was ever detected. Thirdly, in RevIIa, which displays white testes, a full-length Idefix is inserted upstream of white. All signals necessary to start transcription in the 5′ LTR and then to stop it within the 3′ LTR are present in Idefix. Thus, in the wIR6RevIIa alleles, Idefix transcription does not pass through the downstream white promoter. Nevertheless, testes are white.
All these data stress the fact that promoter competition is the molecular mechanism responsible for wIR6RevII phenotypes in testes.
The specificity of the TATA-less core promoter of Idefix is a prime determinant in the competition
As reported above, competition occurs in tissues where both promoters are transcriptionally active; however, we have found that this necessary condition is not sufficient to establish the competition. Transgenic experiments were performed with constructs that display a modification of its promoter so that the sequence was changed but its activity remained. Two main motifs characterized TATA-less promoters: an Inr sequence and a downstream DPE sequence (Kutach and Kadonaga, 2000). They function co-operatively for the binding of TFIID. Both Idefix and white contain these motifs but they do not conform to the optimal consensus sequence recently reported by Kutach and Kadonaga (2000). Testing an alteration of the Idefix TATA-less promoter (Limut) to create a synthetic promoter conforming to the consensus sequence of Inr core promoters indicated that although Limut is still active and able to drive Idefix expression in testes, it is unable to compete with the white gene for its testes enhancer. It is thus clear that the signals necessary for active transcription of a TATA-less promoter such as that created in Limut are not sufficient to establish competition. The wild-type promoter of Idefix appears to be optimal for selective interactions with the white enhancer of testes, while a mutated promoter, although able to initiate transcription in testes, is not. The sequence of the active promoter by itself is an additional essential parameter. These findings strengthen the fact that different TATA-less core promoters exist and possess distinct properties that enable them to be activated preferentially by cis-regulatory sequences. In that context, it will be interesting in the future to test the strength of promoters and enhancers involved in this transcriptional interference. We suggest that sequences of the Inr and DPE motifs act in concert with factors somehow to restrict the action of an enhancer. The binding of specific proteins to these motifs may be a necessary step in defining the competition.
Competition depends on the orientation of the promoters
The testes phenotypes compared between RevIIa and RevIIb lines reveal that the competition depends on the orientation of the Idefix promoter. When the Idefix LTR was placed upstream of the white enhancers and the mini-white gene but in the orientation ‘3′LTR5′-enhancers-5′mini-white3′’, it was no longer involved in the competition and thus testes were yellow. In this configuration, the insulator sequence identified in Idefix may explain the orien tation dependence since it has been identified in the U3 part of the LTR and is thus upstream of the initiation start site of transcription (Conte et al., 2002). In these constructs that mimic wIR6RevIIb, the insulator is interposed between the testes enhancer and the promoter of Idefix. Therefore, it is able to prevent the enhancer from activating the Idefix promoter, as illustrated in Figure 5C. The orientation of promoters involved in competition is certainly an essential determinant for the competition to occur. Several studies have reported that many promoters possess an intrinsic enhancer-blocking activity essential for their proper regulation (O’Donnell et al., 1994; Arkhipova, 1995; Ohtsuki and Levine, 1998). Such an insulator activity coupled with promoters might be a level of control of promoter competition within the genome. Insulators have been described to protect gene expression from chromosomal position effect by restricting the domain of action of enhancers. These data show that insulators potentially are also required to restrict promoter competition. Recently, Cai et al. (2001) have shown that the suHw-mediated blockage of the AE1 enhancer from a downstream promoter depends on the ability of the promoter to compete for AE1. Promoters that are highly competitive for the enhancer are blocked less effectively. In our experiments, the Idefix promoter is competitive enough to turn away the testes enhancer from its proper white gene. Nevertheless, when interposed, the insulator of Idefix is effective to restore the activation of white by its enhancer. These results suggest that the ability of a promoter to compete for an enhancer may not be the only parameter keeping the insulator activity through promoter competition.
Two adjacent identical core promoters can block promoter competition
Conditions exist whereby such competition can be suppressed. For example, the presence of two Idefix inserted close together and in the opposite orientation, as in the RevIV line, disrupts this promoter competition and leads to flies with yellow testes. RevIV derives from a RevIIa line with the addition of a novel Idefix close to the first one but in the opposite orientation (Desset et al., 1999). Two main mechanisms can be proposed to explain this suppressive effect. First, the insulator brought by the second Idefix potentially can interfere with the competition since its LTR is positionned in the 3′–5′ orientation, as in the wIR6RevIIb alleles; its insulator sequence present between the first Idefix and the testes enhancer is able to block the action of the enhancer on the Idefix promoter. Secondly, it has been shown recently that two adjacent insulators are ineffective in blocking enhancers from a downstream promoter, and we have already reported that this must be why RevIV displays brick-red eyes (Conte et al., 2002). A model for this suppression of insulation is that such insulators may interact with each other through protein complexes bound to them, forming chromatin loop domains that annul their insulator properties (Cai and Shen, 2001; Muravyova et al., 2001). It is tempting to think that such structures might also trap adjacent promoters, preventing them from competing with nearby genes for an enhancer. Further insights into the molecular mechanisms underlying this competition and its suppression by two Idefix are necessary to discriminate between these two models. It will be particularly interesting to elucidate whether two Idefix insertions (or, in a more general way, two identical sequences) present at two different genomic sites can either allow or restrict transcription of neighboring genes depending on their respective position toward the regulatory sequences of a transcriptional unit.
Concluding remarks
This study brings to light a novel mechanism by which retroelements can interfere with and thus contribute to the regulation of their host. Insertion of mobile elements influences genomic structure and function through several mechanisms that have been widely depicted in the literature. In addition to disrupting exons, they can insert into non-coding regions and modify gene regulation by adding transcriptional enhancers or insulator sequences delimiting novel autonomous domains of transcription. Our data provide in vivo evidence that they can also establish promoter competition with genes located in the vicinity of their insertion and then disrupt endogenous enhancer–promoter communications.
Transcriptional interference involving insulator function and/or promoter competition imprinted by retroelements may disrupt enhancer–promoter communication of neighboring genes and thus contribute to their control in specific tissues and/or developmental stages. It must be stressed that such controls can be found in a line or subgroups of a population, when an insertion recently occurred at a defined locus. However, they can also be established in a whole species if the insertion occurred a long time ago in any ancestor. Identifying vestiges of transposable elements dispersed throughout the genome that have been highly degenerated through evolution but which retained their capacity to initiate transcription or their insulator function should reveal novel cis-regulatory elements contributing to eukaryotic gene regulation.
Materials and methods
Plasmid constructs
The regulatory sequences of white are located between –1084 and –1465 relative to the transcription start site of the gene (Qian et al., 1992). A 890 bp fragment containing these sequences was amplified by PCR using the primers W1 (ATGCGGATCCGAATTCACGCCTCAGTTCAAGTT AC) and W2 (GTACGGATCCGAATTCTACCATTTTCACGGACG AT). The EcoRI fragment was inserted in the single EcoRI site of pCaSpeR4 (Pirrotta, 1988) to give a P transformation vector called pEwR. The LTR of Idefix amplified by PCR using the primers I1: GTCGACGTGACATATCCATAAG (starting at nucleotide 1 according to the Idefix sequence) and I2: CTTCAGTTGATCAGTACCGTAC (starting at nucleotide 659) was cloned in pGEM-T (Promega). Two different PCR products were obtained, the wild-type LTR and the LTR477 (or Li477) containing a T→C mutation at nucleotide 477 of Idefix (Conte et al., 2002). Also, site-directed mutagenesis was performed as described by Jarrell et al. (1988) to introduce an Inr sequence that fits the consensus. The mutated LTR obtained, called Limut, was identical to the Idefix LTR except that its transcription start site ATCAGAG is replaced by TGCAGTT (Figure 3).
The NotI–BamHI fragments containing the LTR, the Li477 or the Limut were inserted between the FRT in the pKB345 plasmid. The KpnI FRT-flanked LTR, Li477 and Limut of Idefix fragments were inserted into the KpnI sites of pEwR to give, respectively, pLi477EwW (Figure 4A) and pLimutEwW (Figure 4B) transformation vectors.
The KpnI FRT-flanked Limut was inserted into the KpnI site of pW6AUGβgal (provided by A.Pélisson) to construct the pLimutLacZ (Figure 3). Staining of testes to measure lacZ expression was performed as follows: testes were dissected, fixed with glutaraldehyde and stained with X-gal according to the method of Glaser et al. (1986).
P transformation vector
P element transformation vectors containing the white reporter gene were introduced into the Drosophila germline by injecting w1118 embryos as described previously (Rubin and Spradling, 1982). At least five independent transformants were obtained and analyzed for each recombinant P element transformation vector.
Fly strains and heat-shock regimes
Fly stocks were maintained on cornmeal–glucose–yeast media at 20°C. The hsFLP flies (w1118 70FLP; cu kar2 Sb/TM6, Ubx es), kindly provided by Kent Golic, express the flp recombinase under the heat shock promoter. Virgin hsFLP females were crossed with transgenic males for 24 h on cornmeal–glucose–yeast media at 20°C. Heat shocks of embryos <24 h old were performed as described by Ahmad and Golic (1996). For each recombinant P element transformation vector injected, five independent transgenic lines were heat shocked to compare the eye color of heat-shocked and non-heat-shocked flies.
Acknowledgments
Acknowledgements
We thank K.Golic for providing hs-FLP strains, K.Bassler for pKB345 vector, B.Jourde, E.Goy and Françoise Pélissier for excellent technical assistance, and members of our laboratory for helpful suggestions and insights. We are especially grateful to an anonymous reviewer for valuable comments. This work was supported by grants from the INSERM (U384), CNRS (GDR 2157) and by a project grant from the ARC (Association pour la Recherche contre le Cancer) to C.V. C.C. received a grant from Ministère de l’Enseignement Supérieur et de la Recherche (MESR).
References
- Ahmad K. and Golic,K.G. (1996) Somatic reversion of chromosomal position effects in Drosophila melanogaster. Genetics, 144, 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova I.R. (1995) Promoter elements in Drosophila melanogaster revealed by sequence analysis. Genetics, 139, 1359–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges S., Mihaly,J., Galloni,M., Hagstrom,K., Muller,M., Shanower,G., Schedl,P., Gyurkovics,H. and Karch,F. (2000) The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development, 127, 779–790. [DOI] [PubMed] [Google Scholar]
- Burke T.W. and Kadonaga,J.T. (1996) Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev., 10, 711–724. [DOI] [PubMed] [Google Scholar]
- Butler J.E. and Kadonaga,J.T. (2001) Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev., 15, 2515–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H.N. and Shen,P. (2001) Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science, 291, 493–495. [DOI] [PubMed] [Google Scholar]
- Choi O.R. and Engel,J.D. (1988) Developmental regulation of β-globin gene switching. Cell, 55, 17–26. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Bell,A.C. and Felsenfeld,G. (1997) Characterization of the chicken β-globin insulator. Proc. Natl Acad. Sci. USA, 94, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte C., Calco,V., Desset,S., Leblanc,P., Dastugue,B. and Vaury,C. (2000) Impact of multiple insertions of two retroelements, ZAM and Idefix, at an euchromatic locus. Genetica, 109, 53–59. [DOI] [PubMed] [Google Scholar]
- Conte C., Dastugue,B. and Vaury,C. (2002) Coupling of enhancer and insulator properties identified in two retrotransposons modulate their mutagenic impact on nearby genes. Mol. Cell. Biol., 22, 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desset S., Conte,C., Dimitri,P., Calco,V., Dastugue,B. and Vaury,C. (1999) Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol. Biol. Evol., 16, 54–66. [DOI] [PubMed] [Google Scholar]
- Foley K.P. and Engel,J.D. (1992) Individual stage selector element mutations lead to reciprocal changes in β- vs. ε-globin gene transcription: genetic confirmation of promoter competition during globin gene switching. Genes Dev., 6, 730–744. [DOI] [PubMed] [Google Scholar]
- Geyer P.K. and Corces,V.G. (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev., 6, 1865–1873. [DOI] [PubMed] [Google Scholar]
- Glaser R.L., Wolfner,M.F. and Lis,J.T. (1986) Spatial and temporal pattern of hsp26 expression during normal development. EMBO J., 5, 747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger I.H., Demarchi,F., Giacca,M. and Proudfoot,N.J. (1998) Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res., 26, 1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger I.H., Aranda,A. and Proudfoot,N. (2000) Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 8415–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K., Muller,M. and Schedl,P. (1996) Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev., 10, 3202–3215. [DOI] [PubMed] [Google Scholar]
- Herault Y., Fraudeau,N., Zakany,J. and Duboule,D. (1997) Ulnaless (Ul), a regulatory mutation inducing both loss-of-function and gain-of-function of posterior Hoxd genes. Development, 124, 3493–3500. [DOI] [PubMed] [Google Scholar]
- Jarrell K.A., Dietrich,R.C. and Perlman,P.S. (1988) Group II intron domain 5 facilitates a trans-splicing reaction. Mol. Cell. Biol., 8, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R. and Schedl,P. (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell, 64, 941–950. [DOI] [PubMed] [Google Scholar]
- Kellum R. and Schedl,P. (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol., 12, 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach A.K. and Kadonaga,J.T. (2000) The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol., 20, 4754–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoinie O., Drake,M.E., Dastugue,B. and Vaury,C. (1995) Aberrant pre-mRNA maturation is caused by LINE insertions into introns of the white gene of Drosophila melanogaster. Nucleic Acids Res., 23, 4015–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc P., Desset,S., Dastugue,B. and Vaury,C. (1997) Invertebrate retroviruses: ZAM a new candidate in D.melanogaster. EMBO J., 16, 7521–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc P., Dastugue,B. and Vaury,C. (1999) The integration machinery of ZAM, a retroelement from Drosophila melanogaster, acts as a sequence-specific endonuclease. J. Virol., 73, 7061–7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E., Golovnin,A., Gracheva,E., Parshikov,A., Belenkaya,T., Pirrotta,V. and Georgiev,P. (2001) Loss of insulator activity by paired Su(Hw) chromatin insulators. Science, 291, 495–498. [DOI] [PubMed] [Google Scholar]
- O’Donnell K.H., Chen,C.T. and Wensink,P.C. (1994) Insulating DNA directs ubiquitous transcription of the Drosophila melanogaster α1-tubulin gene. Mol. Cell. Biol., 14, 6398–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S. and Levine,M. (1998) GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev., 12, 3325–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S., Levine,M. and Cai,H.N. (1998) Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev., 12, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. (1988) Vector for P-mediated transformation in Drosophila. Biotechnology, 10, 437–456. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J. (1986) Transcriptional interference and termination between duplicated α-globin gene constructs suggests a novel mechanism for gene regulation. Nature, 322, 562–565. [DOI] [PubMed] [Google Scholar]
- Qian S., Varjavand,B. and Pirrotta,V. (1992) Molecular analysis of the zeste–white interaction reveals a promoter-proximal element essential for distant enhancer–promoter communication. Genetics, 131, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Sharpe J., Nonchev,S., Gould,A., Whiting,J. and Krumlauf,R. (1998) Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J., 17, 1788–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcheressiz S., Calco,V., Arnaud,F., Arthaud,L., Dastugue,B. and Vaury,C. (2002) Expression of the Idefix retrotransposon in early follicle cells in the germarium of Drosophila melanogaster is determined by its LTR sequences and a specific genomic context. Mol. Genet. Genomics, 267, 133–141. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Maine,E. and Schedl,P. (1985) The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol., 185, 341–358. [DOI] [PubMed] [Google Scholar]
- Villemure J.F., Savard,N. and Belmaaza,A. (2001) Promoter suppression in cultured mammalian cells can be blocked by the chicken β-globin chromatin insulator 5′HS4 and matrix/scaffold attachment regions. J. Mol. Biol., 312, 963–974. [DOI] [PubMed] [Google Scholar]
- Zhou J. and Levine,M. (1999) A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell, 99, 567–575. [DOI] [PubMed] [Google Scholar]
- Zhou J., Barolo,S., Szymanski,P. and Levine,M. (1996) The Fab-7 element of the bithorax complex attenuates enhancer–promoter interactions in the Drosophila embryo. Genes Dev., 10, 3195–3201. [DOI] [PubMed] [Google Scholar]
- Zhou J., Ashe,H., Burks,C. and Levine,M. (1999) Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development, 126, 3057–3065. [DOI] [PubMed] [Google Scholar]