Abstract
The developmental plasticity of transplanted adult stem cells challenges the notion that tissue-restricted stem cells have stringently limited lineage potential and prompts a re-evaluation of the stability of lineage commitment. Transformed cell systems are inappropriate for such studies, since transformation potentially dysregulates the processes governing lineage commitment. We have therefore assessed the stability of normal lineage commitment in primary adult haematopoietic cells. For these studies we have used prospectively isolated primary bipotent progenitors, which normally display only neutrophil and monocyte differentiation in vitro. In response to ectopic transcription factor expression, these neutrophil/monocyte progenitors were reprogrammed to take on erythroid, eosinophil and basophil-like cell fates, with the resultant colonies resembling the mixed lineage colonies normally generated by multipotential progenitors. Clone-marking and daughter cell experiments identified lineage switching rather than differential cell selection as the mechanism of altered lineage output. These results demonstrate that the cell type-specific programming of apparently committed primary progenitors is not irrevocably fixed, but may be radically re-specified in response to a single transcriptional regulator.
Keywords: haematopoiesis/lineage commitment/plasticity/stem cells/transcription factor GATA-1
Introduction
Lineage specification is a fundamental developmental process in multicellular organisms, reiterated throughout adulthood in stem cell-dependent tissues (Weissman et al., 2001). Stem cells can be most simply characterized as cells that can regenerate adult tissue by virtue of their capacity to both self-renew and to differentiate into more mature, lineage-restricted cells. Evidence has recently emerged that adult stem cells may be considerably more plastic in their developmental potential than previously thought, challenging the notion that tissue-specific stem cells have stringently limited lineage potential (reviewed in Weissman et al., 2001). Thus, bone marrow-derived cells were found to be able to differentiate into muscle, and cells that were thought to be restricted to neural development could differentiate into haematopoietic cells. Subsequent studies have reported interconversion of bone marrow to brain or liver, bone marrow stroma to brain, brain to heart, and more. Most strikingly, multi-organ, multi-lineage engraftment by a single bone marrow-derived stem cell has been described (Krause et al., 2001). The nature of stem cell plasticity in these experimental circumstances is unclear (see Vogel, 2002 and references therein) and has been challenged, raising interesting issues in terms of the mechanisms involved.
While the majority of studies in the plasticity area have focused on stem cells, some studies have particularly examined the stability or irreversibility of commitment decisions in progenitor cell populations. Glial progenitors from the rat optic nerve normally have the ability to produce only oligodendrocytes and some astrocytes. However, under appropriate in vitro culture conditions, these progenitors can give rise to neurospheres capable of generating neurons, astrocytes and oligodendrocytes (Kondo and Raff, 2000). In the haematopoietic system, committed T helper type 1 (Th1) cells can be induced to express Th2-specific cytokines by ectopic expression of GATA-3 (Lee et al., 2000). Common lymphoid progenitors normally produce only B and T lymphocytes, natural killer cells and antigen-presenting dendritic cells. However, when transduced with the GM-CSF receptor, these cells can generate macrophages and granulocytes in response to GM-CSF. Interestingly, culture in the presence of the lymphoid cytokine IL-7 can, after several days, render the cells resistant to this GM-CSF-induced transdifferentiation (Kondo et al., 2000). An interpretation of these data is that the genome is, for a limited period of time, sufficiently plastic in committed progenitors to be reprogrammed by external signalling cues. Plasticity of the lymphoid genome is also revealed by the alternative lineage potentials displayed by B-lymphoid lineage cells that are null for the lymphoid commitment factor Pax-5 (Nutt et al., 1999; Rolink et al., 1999).
Plasticity of the genome in committed progenitors has also been argued for on the basis of enforced expression experiments conducted in transformed haematopoietic cells and cell lines (Borzillo et al., 1990; Kulessa et al., 1995). Elegant as these studies are, transformation, by definition, alters the self-renewal versus differentiation potential of cells and thus undermines the use of transformed cell systems for the study of normal self-renewal and commitment processes. Thus, in these experiments, it remains unclear to what extent genome plasticity is a consequence of transformation, as opposed to an inherent property of the normal counterparts of the transformed cell in question. By the same token, the significance of the mixed lineage potentials of leukaemic cells has also been contentious (reviewed in Greaves et al., 1986). It is therefore essential to use primary progenitor cells, which are not transformed, in order to test the plasticity of normal haematopoietic cells. However, the use of primary progenitor cell populations poses a number of problems, not least in terms of distinguishing bona fide lineage switching from selective cell survival and/or expansion.
In this study we have examined the plasticity of primary myeloid progenitors, which normally produce only neutrophil or monocyte progeny in vitro (Williams et al., 1987). We show that, in response to ectopic transcription factor expression, these GM-committed cells may be reprogrammed to take on erythroid, eosinophil and basophil-like cell fates. Critically, the use of an inducible expression system has allowed us to conduct kinetic studies, which, together with direct clonal analyses, establish true lineage switching as opposed to differential cell survival or selection as the mechanism underlying the altered cell output.
Our results suggest that the committed state of lineage-restricted myeloid progenitors is not irrevocably fixed, but can be reprogrammed in response to a single transcriptional regulator. These data, taken together with those of Weissman and colleagues (Kondo et al., 2000), who showed that ectopic receptor-mediated signalling at the cell surface can initiate a programme of altered lineage output from lymphoid committed progenitors, argue for substantial lineage flexibility throughout the haematopoietic system, and indicate that aspects of stem cell plasticity persist or remain retrievable in more lineage-restricted progenitors.
Results
GATA-1-dependent lineage reprogramming of GM-CFC
Primary granulocyte–monocyte colony-forming cells (GM-CFC) were isolated from mouse bone marrow according to the scheme presented in Figure 1A. These cells were transduced with either a control retrovirus or a retrovirus containing a ligand-inducible form of the transcription factor GATA-1 (Figure 1B). GATA-1 was originally described as an erythroid lineage-affiliated transcription factor (Evans and Felsenfeld, 1989; Tsai et al., 1989) and has served as a paradigm for studies of tissue-specific transcription and lineage commitment (Orkin, 2000).
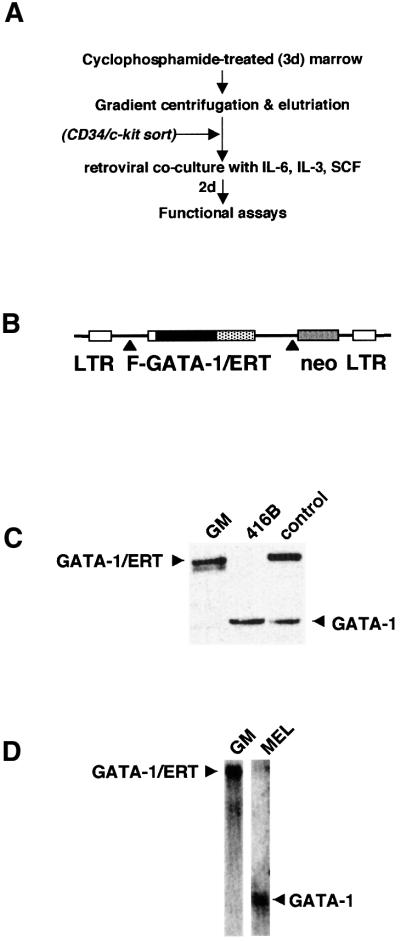
Fig. 1. Enforced expression of GATA-1/ERT in primary GM/CFC. (A) Purification and infection of GM-CFC from murine bone marrow. For some experiments, purified GM-CFC were further enriched on the basis of CD34+/c-kit+ antibody staining. (B) Retroviral construct. FLAG-tagged murine GATA-1 was fused in-frame to an ER ligand-binding domain mutagenized such that it is responsive to 4OHT rather than oestrogen (Littlewood et al., 1995; Heyworth et al., 1999) and inserted into the p-50-M-X-neo retroviral vector. Expression of both GATA-1/ERT and neo is under the control of the viral LTR and achieved through alternative splicing (▴). (C) Western blot analysis of the level of GATA-1/ERT and endogenous murine GATA-1 in retrovirally transduced GM-CFC (GM), compared with a myeloid progenitor cell line (416B) and a previously derived clone of FDCPmix cells stably infected with the same GATA-1/ERT retrovirus (control). (D) Northern blot analysis of GATA-1/ERT-transduced GM-CFC and a murine erythroleukaemia cell line (MEL) probed with a murine GATA-1 probe that recognizes both the retroviral GATA-1/ERT and endogenous GATA-1 mRNAs.
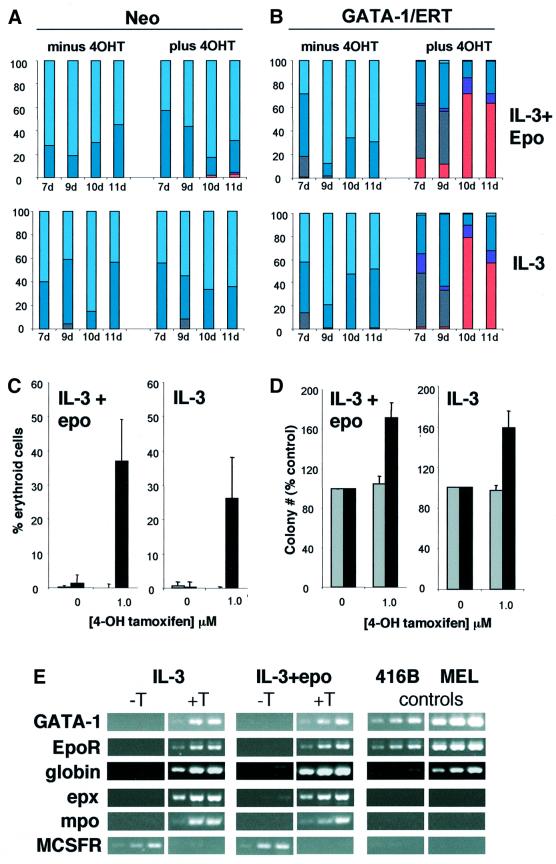
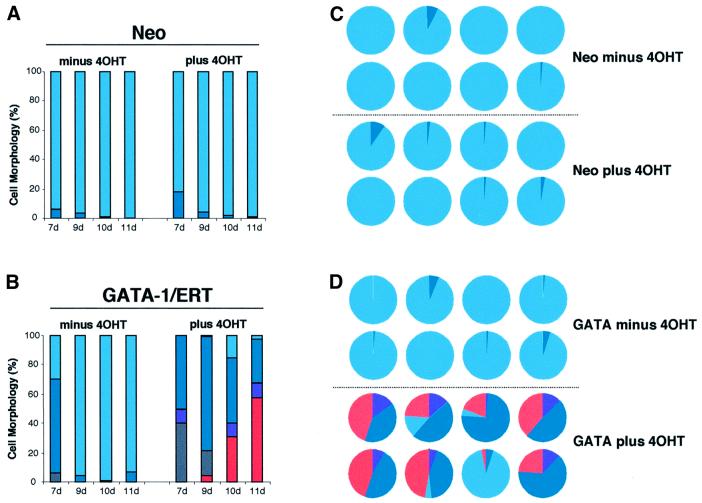
Western blot analysis (Figure 1C) shows that endogenous GATA-1 protein is not detectable in GM-CFC, and exogenous GATA-1/ERT levels in GATA-1/ERT-transduced GM-CFC fall within a physiological range, in that they roughly equate to the levels seen in two myeloid progenitor cell lines 416B and FDCPmix-A4. Northern blot analysis (Figure 1D) is consistent with this, but low levels of endogenous GATA-1 mRNA could be detected in GM-CFC populations by RT–PCR analysis (data not shown). Virally transduced cells were plated in soft agar in the presence of IL-3 or IL-3 + erythropoietin (epo) to promote erythroid lineage development. Colonies were sampled at 7, 9, 10 and 11 days of clonal culture, pooled and morphologically assessed. As expected, vector-only-transduced GM-CFC (Neo; Figure 2A) gave rise almost exclusively to GM colonies at all time points of the culture, and in the absence or presence of tamoxifen. GATA-1/ERT-transduced GM-CFC (Figure 2B) also gave rise to predominantly GM colonies in the absence of tamoxifen. In contrast, activation of exogenous GATA-1 activity resulted in the dramatic appearance of erythroblasts and mature erythroid cells from as early as day 7 of clonal culture (Figure 2B); the appearance of non-neutrophilic granulocytes of the eosinophil series and of basophil-like cells was also noted and is consistent with the normal expression of GATA-1 in these two lineages. However, megakaryocytes were rarely observed, possibly reflecting either the omission of thrombopoietin from the cultures or the level of GATA-1 expression achieved. The development of erythroid cells and/or non-neutrophilic granulocytes appeared to occur primarily at the expense of monocytes, with neutrophils largely unaffected. These results were confirmed in multiple subsequent day 8–9 clonal cultures (Figure 2C), which further revealed that induction of GATA-1/ERT activity resulted in an ∼50% increase in clonogenic efficiency (Figure 2D), perhaps as a consequence of the anti-apoptotic activity of GATA-1 (Weiss and Orkin, 1995; Gregory et al., 1999; Tanaka et al., 2000). Gene expression analysis of pooled colonies was also consistent with the development of erythroid cells and non-neutrophilic granulocytes upon activation of GATA-1/ERT (Figure 2E), with increases in the levels of mRNA for endogenous GATA-1 as well as EpoR and globin (erythroid), eosinophil peroxidase (epx; eosinophils and basophils) and myeloperoxidase (mpo; eosinophils). (The failure to detect MPO mRNA in control colonies presumably reflects the maturity of the cells, since MPO is optimally expressed at the promyelocyte stage.) In addition, the level of M-CSF receptor mRNA was seen to decrease, consistent with a reduction in the output of monocytes within the colonies.
Fig. 2. Colony formation by retrovirally transduced GM-CFC. GM-CFC transduced with neo-only (A) or GATA-1/ERT (B) retroviruses were plated in soft agar with G418 ± 1 µM 4OHT, in IL-3 + epo or IL-3 alone. The results show the average distribution of each cell type per colony (%). Grey, blasts; dark blue, neutrophils; light blue, monocytes; purple, immature granulocytes of the basophil and eosinophil lineages; pink, erythroid. Basophil-like cells were identified on the basis of morphology and dark blue staining of the granules by Toluidine Blue. (C) Virally transduced CD34+/c-kit+ GM-CFC were plated in IL-3 + epo or IL-3 alone and grown for 8–9 days. The number of erythroid cells generated is shown, expressed as a percentage of the total cell number on a per colony basis (average of 4–8 experiments ± SEM). Grey bars, neo-transduced cultures; black bars, GATA-1/ERT-transduced cultures. (D) Average ± SEM of total colony number expressed as a percentage of the number of colonies formed in 0 µM 4OHT (control) in the experiments described in Figure 2C. Grey bars, neo-transduced cultures; black bars, GATA-1/ERT-transduced cultures. (E) Semi-quantitative RT–PCR analysis of RNA from pooled colonies formed by GATA-1/ERT-transduced CD34+/c-kit+ GM-CFC cultured in IL-3 or IL-3 + epo, without and with 4OHT (T). Controls: 416B and MEL cell lines. Oligo-dT-primed reverse transcription was performed using 20, 60 and 180 ng of input RNA (left to right) for each sample, followed by 25 cycles of gene-specific PCR.
Analysis of individual colonies
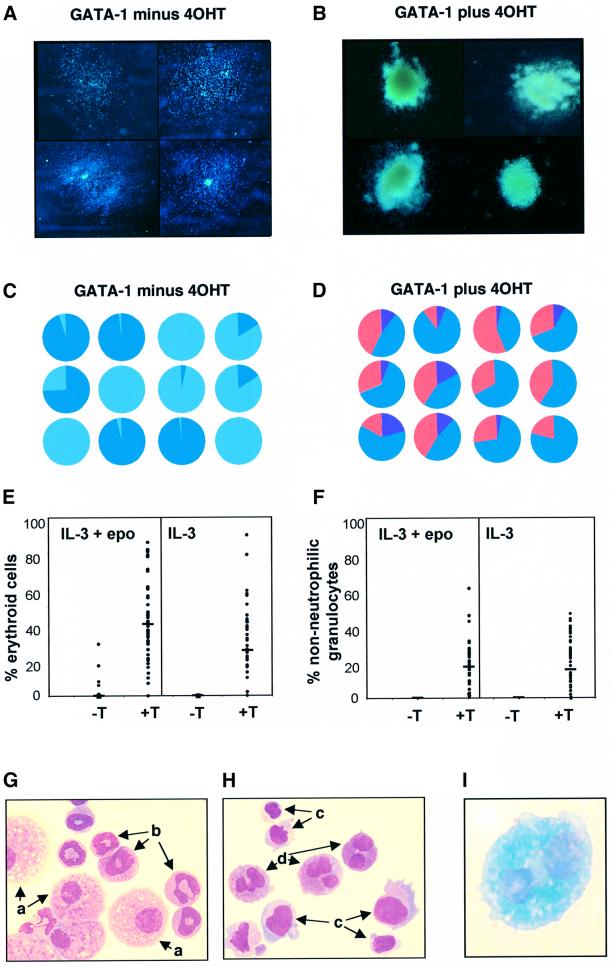
Analysis of individual colonies provided additional insight into the cellular origins of reprogramming of lineage output from GM-committed cells. GATA-1/ERT colonies cultured in the absence of 4-OH tamoxifen (4OHT) are typically small, mainly disperse in nature, and predominantly comprise monocytes and neutrophilic granulo cytes (Figure 3A). Ligand-dependent activation results in a dramatic alteration of gross colony morphology (Figure 3B), which now resemble mixed lineage colonies, being large, multicentric and pinkish in hue. The mixed lineage composition of individual colonies was confirmed by morphological scoring of cellular cytospins. In total >200 individual colonies were assessed. The relative cell compositions of 24 randomly selected individual GATA-1/ERT colonies are presented in Figure 3C and D, and the percentages of erythroid and non-neutrophilic-granulocytes recorded in each of the 200 colonies analysed are plotted in Figure 3E and F, respectively. Typical morphologies of cells from GATA-1/ERT colonies formed in the absence (Figure 3G) or presence (Figure 3H and I) of 4OHT are shown. These results indicate that: (i) the vast majority of colonies generated by GATA-1/ERT-transduced GM-CFC cultured in the presence of tamoxifen have the gross morphological appearance of mix-CFC-derived colonies, and (ii) >95% of these colonies contain both a significant erythroid and non-neutrophilic granulocyte component. Thus transcription factor activation endows neutrophil/monocyte progenitors with erythroid and non-neutrophilic granulocyte differentiation capacity in vitro. The development of the new lineage potentials appears to occur predominantly at the expense of monocytes rather than neutrophils. The spectrum of lineage outputs obtained is broadly consistent with the expression domain of GATA-1 within haematopoiesis (Zon et al., 1993), and with those seen when GATA-1 is expressed in transformed cell lines (Visvader et al., 1992; Kulessa et al., 1995; Seshasayee et al., 1998; Yamaguchi et al., 1998).
Fig. 3. Analysis of individual colonies. (A and B) Gross morphology of colonies at day 9 from GATA-1/ERT-transduced CD34+/c-kit+ GM-CFC plated in IL-3 in the absence (A) or presence (B) of 4OHT. (C and D) Cellular composition of 12 randomly selected GATA-1/ERT colonies plated in the absence or presence of 4OHT (as indicated). Each pie chart represents the distribution of different cell types within an individual colony. Colours as in Figure 2. (E and F) The percentage of erythroid cells (E) and non-neutrophilic granulocytes (F) within 50 colonies formed by GATA-1/ERT-transduced GM-CFC, with the means indicated by horizontal bars. Colonies were grown in IL-3 or IL-3 + epo, and in the absence (–T) or presence (+T) of 1 µM 4OHT. The minor erythroid component occasionally seen on plating in IL-3 + epo – T is presumably due to some leakiness of the GATA-1/ERT activity. Typical fields from cellular cytospins of individual GATA-1/ERT colonies cultured in the absence (G) or presence (H and I) of 4OHT. (G and H) Staining with O-dianisidine (haemoglobin) and counterstaining with May–Grunwald–Giemsa and (I) staining with Luxol Blue (eosinophilic granules). Cell types are indicated as follows: a, monocytes; b, neutrophils; c, erythroblasts; d, non-neutrophilic granulocytes.
Importantly, we confirmed that changes in lineage output could also be brought about by forced expression of a wild-type murine GATA-1 molecule, and thus were not dependent upon the chimeric nature of the GATA-1/ERT protein. Utilizing the same cell purification and retroviral infection strategy, forced expression of GATA-1 in GM progenitors resulted in the production of mixed lineage colonies, albeit at slightly lower frequency than previously observed (data not shown). Of 20 colonies cultured in IL-3 + epo and scored for morphology, 13 displayed altered lineage output with, on average, one-quarter of the cells within these colonies being of the eosinophilic or erythroid lineages. Basophil-like cells were rarely observed in these experiments. Similar frequencies of novel cell lineages were obtained when GATA-1-infected GM-CFC were cultured in IL-3 alone.
Effects of different cytokines on lineage output
Although the combination of cyclophosphamide treatment and elutriation provides GM lineage-restricted progenitors at high purity (Williams et al., 1987), we do observe an occasional colony containing a small number of cells of other lineages. Our elutriated cells are typically further enriched for colony-forming activity by isolating the CD34+/ckit+ fraction, and it has been previously shown that the CD34+/ckit+ fraction from untreated mouse bone marrow contains both common myeloid progenitors (CMP) and granulocyte/macrophage progenitors (Akashi et al., 2000). We therefore further characterized the developmental potential of our purified GM-CFC population by determining their lineage output on plating in a complex cocktail of cytokines that should support the development of all myelo-erythroid cell types (Akashi et al., 2000). The result of a representative experiment is shown in Figure 4. Under these conditions, neo-infected GM progenitors continued to produce, almost exclusively, colonies containing only granulocytes and macrophages. The single colony exhibiting a minor erythroid component (only 3% of cells) presumably reflects occasional inclusion in the purified population of a slightly less mature progenitor with a more multipotential phenotype. When GATA-1 was activated, every colony again developed a CFU-mix phenotype containing at least four cell types, with at least half of the cells within the colony exhibiting a non-GM morphology.
Fig. 4. Cellular composition of colonies generated by neo- or GATA-1/ERT-transduced CD34+/c-kit+ GM-CFC plated in the absence (–) or presence (+) of 40HT. Colonies were grown in stem cell factor (SCF), IL-3, IL-11, GM-CSF, epo and thrombopoietin (TPO). The mean average colony composition for each condition is shown, essentially as described by Akashi et al. (2000).
The GATA-1/ERT-dependent erythroid potential of GM-CFC could also be realized in the absence of added epo and in the presence of GM-CSF (Figure 5). Vector-only-transduced GM-CFC gave rise exclusively to GM-type colonies composed predominantly of monocytes at all time points of the culture (Figure 5A and C), whereas activation of exogenous GATA-1/ERT activity by 4OHT resulted in the appearance of erythroblasts from as early as day 9 of clonal culture, with mature erythroid forms appearing at day 10 and day 11 (Figure 5B and D). The emergence of non-neutrophilic granulocytes was also apparent, and the development of both these and erythroid cells again appeared to occur predominantly at the expense of monocytes rather than neutrophils. The apparent reduction in monocytic output as a consequence of GATA-1/ERT activation was not simply a function of altered cell ratios within the enlarged CFC-mix-like colonies but represented a real decrease of ∼10-fold in the average number of monocytes per colony (data not shown). Similar results to these were obtained when the CD34+/c-kit+ GM-CFC fraction was transduced with GATA-1/ERT and cultured in the presence of GM-CSF with or without 4OHT (data not shown). In this case, basophil-like cells and eosinophils, as opposed to erythroid cells, predominated in the colonies obtained after induction of GATA-1 expression, possibly reflecting the level of GATA-1 expression obtained in this particular experiment.
Fig. 5. Erythroid development in the absence of added erythropoietin. Time course of colony morphologies generated by GM-CFC infected with vector-only (A) or GATA-1/ERT (B) retroviruses. Cells were plated in GM-CSF and G418 in the absence or presence of 4OHT, and sampled at days 7, 9, 10 and 11 for morphological analysis. Relative cellular compositions are shown of 16 randomly selected individual colonies generated by (C) vector-only- and (D) GATA-1/ERT-infected GM-CFC plated with and without 4OHT. In this experiment, GM-CFC were not further purified by CD34+/c-kit+ sorting. Colours for cell morphology as in Figure 2.
Colony frequencies obtained in all experiments performed under the range of cytokine conditions tested are summarized in Table I. Taken together, our results indicate that the exogenous GATA-1-derived signal fundamentally alters the lineage output of the vast majority of individual GM-committed progenitor cells.
Table I. Summary of colony morphologies.
| Cells | Cytokines | % colony morphology |
|
|---|---|---|---|
| GM | Mixed lineage | ||
| Neo | IL-3 | 99 | 1 |
| IL-3 + epo | 98 | 2 | |
| GM-CSF | 100 | 0 | |
| Cocktail | 97 | 3 | |
| GATA-1 | IL-3 | 0 | 100 |
| IL-3 + epo | 2 | 98 | |
| GM-CSF | 2 | 98 | |
| Cocktail | 7 | 93 | |
The morphologies of all individual colonies assessed after culture in 4OHT are summarized. GM colonies contained monocytes and/or neutrophils only, whereas mixed lineage colonies contained these lineages as well as a combination of erythroid, eosinophilic and basophil-like cells. The following lists the number of independent experiments performed under each cytokine condition using GATA-1 cells, with the number of experiments in which neo controls were also performed given in parentheses afterwards: IL-3, 14 (9); IL-3 + epo, 10 (6); GM-CSF, 6 (4); cocktail, 2 (2). The initial population of CD34+/ c-kit+ GM-CFC was enriched ∼166-fold in GM colony-forming activity compared with normal bone marrow and the plating efficiency varied between 15 and 40%. Since each experiment represents an individual transfection, it is difficult to directly compare plating efficiencies in this context. This caveat aside, there was no obvious change in plating efficiency or colony size correlating with the cytokine used. The number of cells per colony was assessed for IL-3 and IL-3 + epo; GATA-1 colonies were generally 5- to 10-fold larger in the presence of 4OHT than in its absence.
Altering the timing and duration of GATA-1 activity
Since the activity of the exogenous GATA-1/ERT molecule is regulated by the addition of tamoxifen we could examine: (i) the signal duration required to promote alternative lineage output from GM-CFC; and (ii) the time scale in culture over which cells remain competent to respond. Importantly, these experiments also allowed us to address whether the changes in lineage output observed could have their origins in GATA-1-mediated effects on cell selection at the level of clonogenic cells.
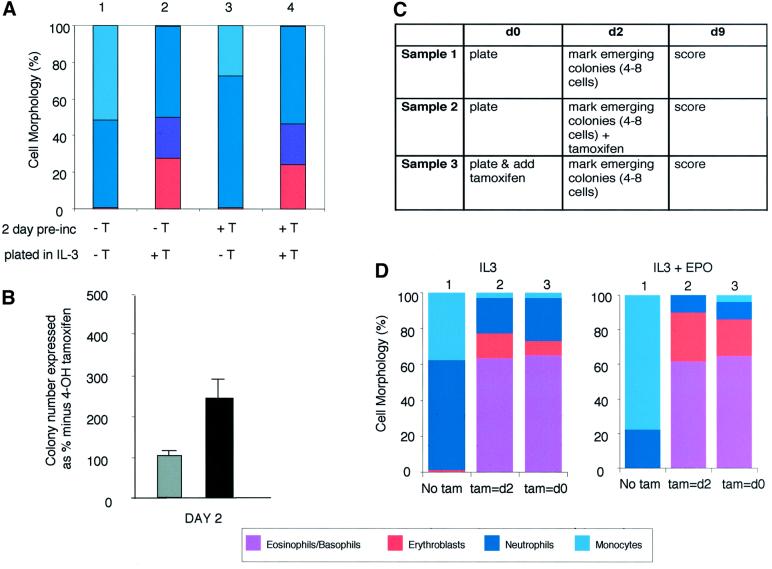
Vector-only and GATA-1/ERT-transduced GM-CFC were placed in suspension culture in IL-3 for 1, 2 or 3 days either in the presence or absence of 4OHT. Cells from each culture were subsequently plated in soft agar, again either with or without 4OHT, and in the presence of IL-3. The resultant colonies were pooled and scored for different cell lineage contributions. Cells cultured in the absence of 4OHT for 2 days retained the capacity to form mixed lineage colonies when GATA-1 was subsequently activated by addition of 4OHT to the plating mixture (Figure 6A, bar 2). In contrast, cells pre-exposed to 4OHT, and therefore exogenous GATA-1 activity, for 2 days in liquid culture did not go on to form mixed lineage colonies when plated in soft agar in the absence of 4OHT (Figure 6A, bar 3). Ideally we would have liked to have continued these kinetic experiments over longer time periods in culture, but were restricted by the fact that GM-CFC lose colony-forming potential after 3 days in liquid culture (data not shown). These results indicate: (i) that GM-CFC remain competent to respond to a subsequent GATA-1-derived erythrogenic signal for at least 2 days in culture; and (ii) that exposure to an exogenous GATA-1 signal for up to 2 days is insufficient to stably endow GM-CFC with erythroid differentiation potential. It is, however, sufficient to affect the behaviour of GM-CFC at the colony-forming level. GATA-1/ERT-transduced GM-CFC pre-incubated in tamoxifen for 2 days produce 2.5-fold more colonies than cells pre-cultured in the absence of tamoxifen when both are subsequently plated in soft agar lacking tamoxifen (Figure 6B), indicating that GATA-1 can act to increase, or at least maintain, colony-forming activity over the 2 day pre-culture period.
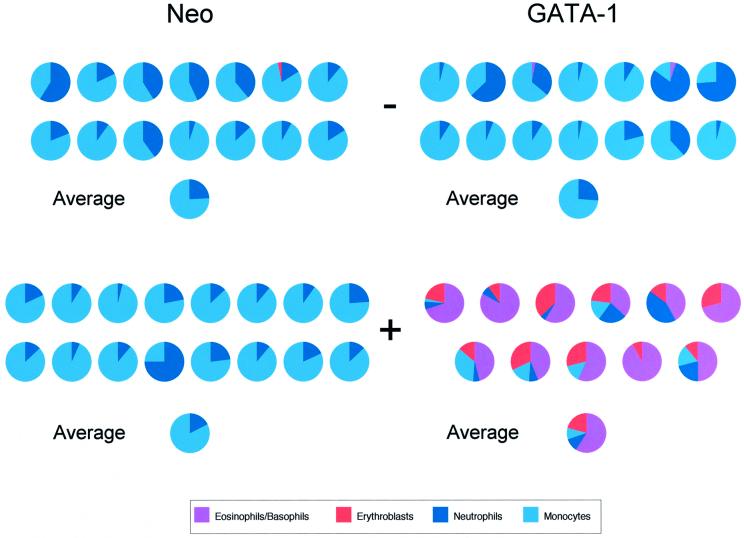
Fig. 6. Kinetic studies of transcription factor-mediated lineage reprogramming. (A) GATA-1/ERT-transduced GM-CFC were pre-incubated in liquid culture in IL-3 in the absence (–T) or presence (+T) of 4OHT (1 µM) for 2 days. The cells were then washed and counted before plating in soft agar in G418 selection and IL-3, again in the absence (–T) or presence (+T) of 4OHT (1 µM). Individual colonies were picked and their morphology assessed at day 8. The cellular composition of individual colonies was expressed as a percentage of the total number of cells in the colony. Approximately 20–24 colonies were assessed from each of three independent experiments; the results were averaged and plotted as bar charts. (B) The number of colonies formed by cells pre-incubated in 4OHT for 2 days and then plated in the absence of 4OHT [(A), condition 3], plotted as a percentage of the number of colonies formed by cells pre-incubated and plated in the absence of 4OHT [(A), condition 1]. The results are the average of three experiments ± SEM. Grey bars, neo-transduced cells; black bars, GATA-1/ERT-transduced cells. It should be noted that in the absence of GATA-1/ERT activation there is a gradual decrease in colony formation as the pre-incubation period is increased. One day of pre-incubation in 4OHT results in an actual increase in the absolute colony numbers (∼2-fold), whilst after 2 days there appears to be a maintenance of colony number compared with the initial input cells. (C) Experimental scheme for colony marking experiments. GATA-1/ERT-transduced cells were split into three samples and plated in methylcellulose in the presence of G418 and various growth factors (day 0). After 2 days, emerging colonies were marked, 4OHT was added to sample 2 and the culture was continued until day 9. Note that sample 3 is essentially a control where 4OHT is present throughout the plating, as in previous experiments. (D) Colony morphologies analysed at day 9 for samples 1, 2 and 3 [as described in (C) and indicated above the chart] cultured in IL-3 (left panel) or IL-3 + epo (right panel). The time of addition of 4OHT is indicated beneath the chart.
These data demonstrate that the exogenous GATA-1/ERT-derived signal has at least two distinct and separable effects on GM-CFC, at the levels of lineage output and colony formation. The results presented in Figures 6A and B further indicate that the exogenous GATA-1 signal does not effect alternative lineage output through simply allowing the survival and/or proliferation of cells within the purified GM-CFC population that (i) happen to retain intrinsic erythroid potential and (ii) normally fail to survive and form colonies. If it did, these CFC maintained during exposure to the GATA-1 signal in suspension culture should go on to produce erythroid lineage cells upon plating in the absence of 4OHT and in the presence of erythroid-conducive cytokine conditions such as IL-3 or epo and IL-3 (data not shown). Furthermore, to achieve the observed output of essentially all colonies having a ‘mixed lineage’ phenotype, GATA-1 activation would have to simultaneously compromise the survival or proliferation of the bipotent CFC which normally give rise to pure GM colonies. This is not the case, since exposure of the GM-CFC population to exogenous GATA-1 activity for 2 days does not diminish their GM-colony-forming potential or their ability to produce neutrophil or monocyte progeny when they are subsequently plated in the absence of the exogenous GATA-1/ERT signal.
Colony tracking and daughter cell experiments
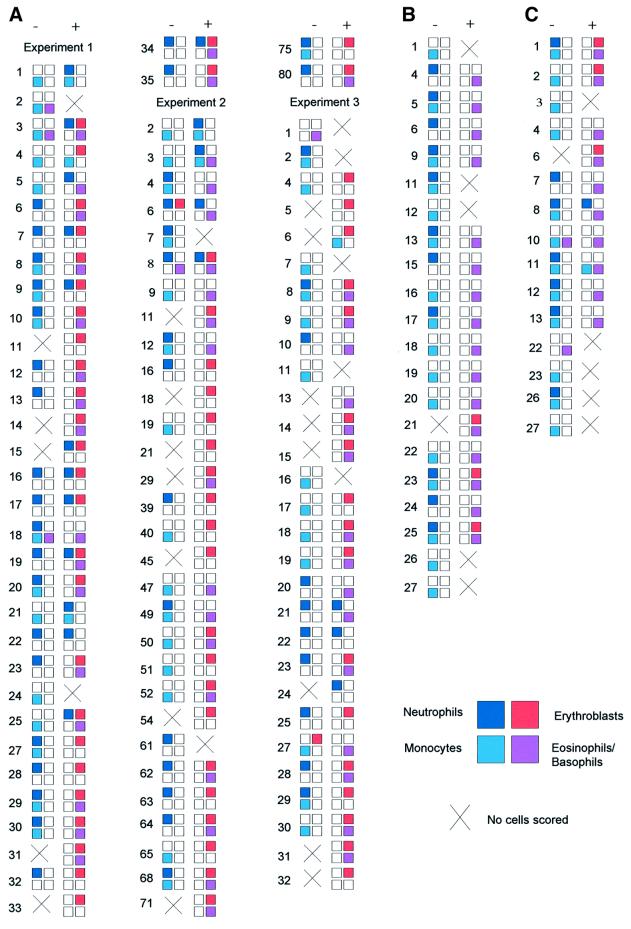
Rigorous demonstration that these changes in lineage output are mediated at the level of cell commitment, and not through GATA-1-mediated effects on cell selection at the level of clonogenic cells, requires direct clonal analysis. We have exploited the capacity of clonogenic assays to provide lineage analysis of individual cells in both colony marking or tracking experiments and in daughter cell assays. In colony marking or tracking experiments (Figure 6C and D), GATA-1/ERT-transduced cells were plated in the absence of tamoxifen and presence of IL-3 or IL-3 + epo and cultured for 2 days, at which point the location of the emerging colonies was marked. The clonal culture was continued in either the absence (sample 1) or presence (sample 2) of 4OHT and the colony morphologies analysed at day 9. In the continued absence of 4OHT, the colonies comprised only monocytes and neutrophils, as expected (Figure 6D, sample 1). However, the addition of 4OHT to the clonal culture at day 2 resulted in the equivalent colonies undergoing lineage switching with the de novo production of erythroid, basophilic and eosinophilic forms (Figure 6D, sample 2). Finally, we performed direct clone-splitting or daughter cell assays. Single cells were expanded in micro wells for 2 days and the resulting mini-clones (≥4 cells) split into parallel cultures with and without tamoxifen. These cultures were continued either in suspension cultures containing IL-3 (Figure 7A) or in methylcellulose containing the cytokine cocktail described previously (Figure 7B). We again observed that in the absence of tamoxifen we obtained almost exclusively granulocyte and macrophage output, but that exposure to tamoxifen in the latter stages, and thus to activated GATA-1/ERT, resulted in colonies with a multicentric morphology and a significant eosinophil, basophil and erythroid component. In this experiment we also confirmed that exposure to 4OHT for 2 days prior to plating in the absence of 4OHT is not usually sufficient to promote alternative lineage outputs (Figure 7C), as previously demonstrated by the experiment shown in Figure 6A. These daughter cell experiments conclusively demonstrate that the changes in lineage output observed do not result from GATA-1-mediated effects on cell selection at the level of clonogenic cells, but rather that sustained expression of GATA-1/ERT endows GM-committed clonogenic progenitors with alternative lineage differentiation capacity in vitro.
Fig. 7. Daughter cell assays. (A) Single GATA-1/ERT-transduced CD34+/c-kit+ GM-CFC were expanded for 2 days in IL-3 and the resulting mini-clones split into parallel suspension cultures in IL-3 without (–) and with (+) 4OHT. After a further 7 days, the cultures were cytospun and assessed for cell morphology. The compositions of all clones obtained in three independent experiments are shown. The presence of the major cell types is indicated by a filled box of the appropriate colour (see key). An open box indicates that cell type was not detected, although it should be noted that these cytospins often had relatively few cells. Failure to score cells (×) usually resulted from loss of cells at cytospin, although in some cases it was due to extensive cell death. The occasional culture displaying eosinophil, basophil or erythroid output in the absence of 4OHT may be due to leakiness of the GI/ERT activity, or to the presence of less mature cells at low frequency. (B) Mini-clones were generated as above, split and plated in methylcellulose containing SCF, IL-3, IL-11, GM-CSF, epo and TPO, in the absence (–) or presence (+) of 4OHT. After 10 days in clonal culture, individual colonies were picked, cytospun and assessed for cell morphology. The results are depicted as in (A). Clones generated in the presence of 4OHT displayed a multicentric morphology, consistent with a mixed lineage rather than GM composition; the failure to consistently detect neutrophils on cytospin analysis of the colonies reflects their initial low frequency and relative fragility in vitro. (C) The experiment described in (B) was repeated, but the initial mini-clones were generated in IL-3 + 4OHT. 4OHT was removed by washing the cells prior to plating in methylcellulose. Cell death accounted for the failure of scoring in clones 3, 22, 23, 26 and 27.
Discussion
In this study, we have examined the stability of lineage commitment in primary haematopoietic progenitor cells that are committed to the neutrophil/monocyte pathway. Ectopic expression of the erythroid affiliated transcription factor GATA-1 within these cells led to development, de novo, of erythroid as well as basophilic and eosinophilic cells in both suspension (data not shown) and clonal culture. The clonal assays also revealed that the experimental induction of lineage switching is a high frequency event as the vast majority of colonies (>95%) developed these alternative lineage components.
The kinetic, clone marking and daughter cell experiments in which exogenous GATA-1 activity is temporally controlled argue for a change in lineage programming of GM-CFC rather than an alteration in lineage output through changes in cell selection at the level of clonogenic progenitors. Ectopic transcription factor expression may alter differentiation potential by revealing, or retrieving, quiescent or residual lineage-affiliated programmes resident within bipotent neutrophil/monocyte progenitors. The detection using RT–PCR of low-level endogenous GATA-1 within cultured GM-CFC populations (data not shown) may reflect this potential and is consistent with a degree of remnant multilineage priming (Hu et al., 1997) within these cells. Similarly, the detection of eosinophil peroxidase expression in GATA-1/ERT-transduced GM-CFC prior to treatment with 4OHT may reflect ‘priming’ or alternatively result from a leakiness of GATA-1/ERT activity.
The fact that multiple, morphologically distinct cell types are elicited in these experiments also argues for a profound reprogramming of lineage choice. The larger size and mixed lineage composition of the colonies could most straightforwardly be explained by dedifferentiation of the GM-committed cell to a multipotential progenitor. However, our data do not categorically exclude a series of independent transdifferentiations of GM-CFC to erythroid, eosinophil and basophil progenitors. The spectrum of lineage outputs obtained in our experiments is broadly consistent with the expression domain of GATA-1 within haematopoiesis (Martin et al., 1990; Romeo et al., 1990; Zon et al., 1993) and those seen when GATA-1 is forcibly expressed in transformed cell lines (Visvader et al., 1992; Kulessa et al., 1995; Seshasayee et al., 1998; Yamaguchi et al., 1998). However, megakaryocytes were rarely observed, possibly reflecting sub-optimal culture conditions for megakaryocyte development or the level of GATA-1 expression achieved.
A striking feature of these GM-CFC experiments is the extent to which new lineage development predominantly occurs at the expense of monocytes as opposed to neutrophils. Intriguingly, a PU.1/ER chimera preferentially induces monocyte rather than neutrophil differentiation of PU.1-null myeloid progenitors at high tamoxifen concentrations (DeKoter and Singh, 2000). PU.1 activity has also been reported to be antagonized by GATA-1 (Rekhtman et al., 1999; Zhang et al., 1999; Nerlov et al., 2000), with the PU.1-induced formation of immature eosinophils by myb-ets-transformed multipotent progenitors (MEPs) involving a down-regulation of GATA-1 (Nerlov and Graf, 1998). While a real understanding of the differential effects on monocytes versus neutrophils must await a complete knowledge of their transcrip tional circuitry, it is interesting to speculate that the susceptibility of monocytes may reflect the hierarchical or evolutionary relationships of the various blood lineages. Dominant-default relationships have been postulated for B lymphocytes and monocytes (Rolink et al., 1999 and references therein), and while the relationship of monocytes to basophils and eosinophils is unclear (Akashi et al., 2000), it is perhaps interesting to note that the monocyte and eythroid lineages may share a common bipotent progenitor during embryonic mouse haematopoiesis (Rich, 1992).
The general issue of lineage flexibility within the haematopoietic system has previously been particularly emphasized in the context of transformed cells, with components of these ideas captured in the model of lineage promiscuity in leukaemic cells (Greaves et al., 1986). The ability of ectopically expressed transcription factors to modulate the cell fate of transformed haematopoietic cells also underscores their plasticity (Orkin, 2000), but it has remained unclear to what extent this flexibility is a consequence of transformation, and thus these data have been contentious. Our data showing plasticity in freshly isolated primary myeloid progenitors, taken together with those of Weissman and colleagues (Kondo et al., 2000), who showed that ectopic receptor-mediated signalling at the cell surface can initiate a programme of altered lineage output from lymphoid committed progenitors, argue for substantial lineage flexibility throughout the haematopoietic system and indicate that aspects of stem cell plasticity persist or remain retrievable in more lineage-restricted progenitors. These data provide strong support for the plasticity of the normal differentiated state and are topical in the context of recent observations demonstrating the hitherto unexpected plasticity of normal stem cell compartments (Anderson et al., 2001; Weissman et al., 2001). Key to the understanding of these apparent transformations in stem cell identity is an understanding of the global transcriptional profiles of specific stem cell compartments (Phillips et al., 2000) and different cell lineages as well as the molecular mechanisms by which cells change lineage programmes. Our observation that GATA-1 can re-specify the fate of committed myeloid progenitors invites comparison with MyoD, which has been shown to act as a master regulator, dictating lineage reprogramming to muscle of diverse phenotypes including primary fibroblasts, chondroblasts, smooth muscle and retinal pigmented epithelial cells (Choi et al., 1990; Lattanzi et al., 1998). Our results provide a mechanism for reprogramming of committed haematopoietic progenitors and an experimental entry point for directly identifying the key loci involved. Since manipulation of GATA-1 activity can also alter cell fate in multipotential progenitors that express endogenous GATA-1 and retain both erythroid and GM potential (our unpublished observations), the same mechanisms and loci are likely to also be directly involved in normal uni-lineage differentiation from multipotent progenitor cells.
Materials and methods
Western blotting
GM-CFC (2 × 106) were harvested, washed in PBS, resuspended in protein sample buffer (10% v/v glycerol, 3% w/v SDS, 5% v/v 2ME, 62.5 mM Tris–HC1 pH 6.8, 0.001% w/v Bromophenol Blue) and boiled for 5 min. Protein extracts were resolved by SDS–PAGE and transferred onto PVDF membrane. The membrane was probed with an anti-GATA-1 monoclonal antibody (N6; Santa Cruz Biotechnology) and a horse radish peroxidase-conjugated donkey anti-rabbit polyclonal antibody (Amersham), and visualized by chemiluminescence (ECL; Amersham).
Northern blotting
Total RNA was prepared from 105–106 GM-CFC using Trizol (Gibco-BRL) and quantitated by spectrophotometry. Four micrograms of each RNA were electrophoresed under standard denaturing conditions before transfer in 20× SSC to HybondN+ membrane. A murine GATA-1 cDNA was radiolabelled by random oligonucleotide priming, heat denatured and hybridized with the membrane at 65°C overnight. The membrane was washed with 0.5× SSC, 0.5% SDS at 65°C and visualized by autoradiography.
RT–PCR analysis
Total RNA was prepared from GM-CFC and various haemopoietic cell lines as for northern blotting, quantitated by spectrophotometry, and checked for equal concentration and integrity between samples by gel electrophoresis. Fifty, 150 and 600 ng of each RNA sample were used in 10 µl reverse transcription reactions with oligo-dT priming and M-MLV RT enzyme (Gibco-BRL). Gene-specific PCR was then performed on an aliquot of each RT reaction (between 0.25 and 1.0 µl, depending on the expected abundance of the transcript) using primers that spanned an intron. In analysing expression of GATA-1 itself, primers were designed from the 3′ untranslated portion of the mRNA which is not present in the GATA-1 retroviral construct. The PCR programme was 94°C for 5 min followed by 25 cycles of 94°C for 20 s, 55°C for 15 s, 72°C for 30 s, and then 72°C for 7 min, although for actin the number of cycles had to be reduced to 22 to remain within the linear range. PCR products were analysed by agarose gel electrophoresis.
Constructs and retrovirus producer cell lines
A murine GATA-1 cDNA (kind gift of Stuart Orkin, Harvard Medical School, Boston, MA) was tagged at the N-terminus with the FLAG epitope. PCR was used to replace the stop codon with a novel valine and introduce a BglII site. This was ligated in-frame to an ER ligand-binding domain that had been mutagenized such that it was reponsive to tamoxifen rather than oestrogen (G525; kind gift of Dr C.Marshall, ICR, London, UK). The entire FLAG-GATA-1/ERT fusion was transferred into the MESV-based retroviral vector p50-M-X-neo and used to generate retrovirus producer cell lines. The wild-type murine GATA-1 cDNA was expressed from the same retroviral vector. Stable retrovirus producer cell lines were made by transfection of GP + E-86 cells (Markowitz et al., 1988) using lipofection (Gibco) as described previously (Fairbairn and Spooncer, 1993). The packaging cell lines were fed daily by removing the medium [DMEM supplemented with 10% v/v newborn calf serum (NBCS)] and replacing it with fresh medium for the 3 days prior to use.
Purification of GM-CFC from bone marrow
GM-CFC were obtained from normal murine bone marrow as described previously (Cook et al., 1989) based on the method described by Williams et al. (1987). Briefly, femoral bone marrow cells were harvested from 6- to 8-week-old B6D2F1 mice 3 days after administration of a single dose of cyclophosphamide (cyclo; 200 mg/kg i.p.; supplied by ASTA Medica AG, Halle-Kunsebeck, Germany). The cell suspension was layered onto metrizaminde gradients (density 1.079), which were then spun at 800 g for 15 min at 4°C. The cells at the interface were harvested, washed and resuspended in 3 ml of Fischer’s medium. These cells were then loaded into the elutriation centrifuge via the loading chamber. Elutriation medium (Fischer’s medium supplemented with 5% v/v horse serum) was passed through the elutriator and the flow rate increased sequentially to 14, 16.5 and 19.5 ml/min, collecting 100 ml for each fraction; these fractions could be collected together and discarded. As the flow rate was again increased to 22, 25.5, 29 and 32 ml/min, 100 ml fractions were collected and pooled. These represented the GM-CFC-rich fraction. Once pelleted, the cells were resuspended in PBS supplemented with 1% heat-inactivated fetal calf serum (HI-FCS), counted and stained for CD34 and c-kit as follows. A few cells were spun in an Eppendorf centrifuge, resuspended in PBS/HI-FCS, and the appropriate FITC- and PE-labelled isotype control antibodies added at a concentration of 1 µg/106 cells. The remaining cells were spun in an Eppendorf centrifuge, resuspended in PBS/HI-FCS and FITC-conjugated CD34 along with PE-conjugated c-kit antibodies added at a concentration of 1 µg/106 cells. The cells were labelled for 30 min at 4°C. Unbound antibody was washed off using PBS/HI-FCS. The cells were then resuspended in PBS/HI-FCS and FACS sorted, collecting all CD34+/c-kit+ cells into DMEM supplemented with 10% NBCS.
Retroviral infection and culture of GM-CFC
GM-CFC were co-cultured with GP + E-86 packaging cell lines, which had been previously irradiated (3000 rad), for 48 h. Loosely adherent and non-adherent cells were harvested and placed either in suspension or colony culture. For suspension culture, an aliquot of cells was seeded at 1 × 105 cells/ml in Iscove’s medium supplemented with 20% v/v FCS, 0.1% w/v bovine serum albumin, 1.4 mg/ml G418 (Sauvageau et al., 1995), various growth factor cocktails as indicated [10 ng/ml recombinant murine interleukin-3, 2 U/ml recombinant human epo (Boehringer Mannheim), 250 U/ml recombinant murine GM-CSF (a kind gift from Biogen, Switzerland)] and various doses of 4OHT (kind gift from Dr C.Marshall, ICR, UK). These cultures were incubated for the time indicated at 37°C in 5% O2/5% CO2 prior to harvesting. The cells were counted, cytospun and stained.
For colony assays, cells were directly plated into soft agar at 3000 cells/ml as described previously for 5-fluorouracil-treated bone marrow (Heyworth and Spooncer, 1993). These plating assays contained the same medium as the suspension cultures, but agar was also added. These assays were incubated at 37°C for 10 days normally in 5% O2/5% CO2, and the colonies scored as described previously (Heyworth and Spooncer, 1993). The transduction efficiency was determined by comparison with the number of colonies obtained on plating in the absence of G418; an efficiency of 30–40% was normally achieved. Individual colonies were picked from the plates, cytospun, stained with O-dianisidine, counterstained with May–Grunwald–Giemsa and assessed for cellular morphology as described previously (Heyworth et al., 1995). Luxol Fast Blue staining was performed as described previously (Johnson and Metcalf, 1980). In the colony marking experiments, the developing colonies were marked at day 2 as described previously (Heyworth et al., 1990) and the plates were overlaid with 100 µl of the appropriate concentration of 4OHT.
Acknowledgments
Acknowledgements
We are indebted to Mel Greaves and Mike Dexter for helpful discussions and reading the manuscript, to Karin Gale and Dorothy Gagen for technical assistance, and to Janine Harris for secretarial assistance. We are also grateful to Mike Hughes and Jeff Barry for help with FACS sorting, and to Steve Bagley for help with microscopy. This work was supported by a Specialist Leukaemia Research Fund Programme Award and Cancer Research UK.
References
- Akashi K., Traver,D., Miyamoto,T. and Weissman,I.L. (2000) A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature, 404, 193–197. [DOI] [PubMed] [Google Scholar]
- Anderson D.J., Gage,F.H. and Weissman,I.L. (2001) Can stem cells cross lineage boundaries? Nat. Med., 7, 393–395. [DOI] [PubMed] [Google Scholar]
- Borzillo G.V., Ashmun,R.A. and Sherr,C.J. (1990) Macrophage lineage switching of murine early pre-B lymphoid cells expressing transduced fms genes. Mol. Cell. Biol., 10, 2703–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Costa,M.L., Mermelstein,C.S., Chagas,C., Holtzer,S. and Holtzer,H. (1990) MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc. Natl Acad. Sci. USA, 87, 7988–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook N., Dexter,T.M., Lord,B.I., Cragoe,E.J.,Jr and Whetton,A.D. (1989) Identification of a common signal associated with cellular proliferation stimulated by four haemopoietic growth factors in a highly enriched population of granulocyte/macrophage colony-forming cells. EMBO J., 8, 2967–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKoter R.P. and Singh,H. (2000) Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science, 288, 1439–1441. [DOI] [PubMed] [Google Scholar]
- Evans T. and Felsenfeld,G. (1989) The erythroid-specific transcription factor Eryf1: a new finger protein. Cell, 58, 877–885. [DOI] [PubMed] [Google Scholar]
- Fairbairn L.J. and Spooncer,E. (1993) Retroviral gene transfer into haemopoietic cells. In Testa,N. and Molineux,G. (eds), Haemopoiesis: A Practical Approach. IRL Press, Oxford, UK, pp. 175–188.
- Greaves M.F., Chan,L.C., Furley,A.J., Watt,S.M. and Molgaard,H.V. (1986) Lineage promiscuity in hemopoietic differentiation and leukemia. Blood, 67, 1–11. [PubMed] [Google Scholar]
- Gregory T., Yu,C., Ma,A., Orkin,S.H., Blobel,G.A. and Weiss,M.J. (1999) GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood, 94, 87–96. [PubMed] [Google Scholar]
- Heyworth C.M. and Spooncer,E. (1993) In vitro clonal assays for murine multipotential and lineage restricted myeloid progenitor cells. In Testa,N.G. and Molineux,G. (eds), Haemopoiesis: A Practical Approach. IRL Press, Oxford, UK, pp. 37–53.
- Heyworth C.M., Dexter,T.M., Kan,O. and Whetton,A.D. (1990) The role of hemopoietic growth factors in self-renewal and differentiation of IL-3-dependent multipotential stem cells. Growth Factors, 2, 197–211. [DOI] [PubMed] [Google Scholar]
- Heyworth C.M., Alauldin,M., Cross,M.A., Fairbairn,L.J., Dexter,T.M. and Whetton,A.D. (1995) Erythroid development of the FDCP-Mix A4 multipotent cell line is governed by the relative concentrations of erythropoietin and interleukin 3. Br. J. Haematol., 91, 15–22. [DOI] [PubMed] [Google Scholar]
- Heyworth C., Gale,K., Dexter,M., May,G. and Enver,T. (1999) A GATA-2/estrogen receptor chimera functions as a ligand-dependent negative regulator of self-renewal. Genes Dev., 13, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Krause,D., Greaves,M., Sharkis,S., Dexter,M., Heyworth,C. and Enver,T. (1997) Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev., 11, 774–785. [DOI] [PubMed] [Google Scholar]
- Johnson G.R. and Metcalf,D. (1980) Detection of a new type of mouse eosinophil colony by Luxol-fast-blue staining. Exp. Hematol., 8, 549–561. [PubMed] [Google Scholar]
- Kondo M., Scherer,D.C., Miyamoto,T., King,A.G., Akashi,K., Sugamura,K. and Weissman,I.L. (2000) Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature, 407, 383–386. [DOI] [PubMed] [Google Scholar]
- Kondo T. and Raff,M. (2000) Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science, 289, 1754–1757. [DOI] [PubMed] [Google Scholar]
- Krause D.S., Theise,N.D., Collector,M.I., Henegariu,O., Hwang,S., Gardner,R., Neutzel,S. and Sharkis,S.J. (2001) Multi-organ, multi-lineage engraftment by a single bone marrow-derived stem cell. Cell, 105, 369–377. [DOI] [PubMed] [Google Scholar]
- Kulessa H., Frampton,J. and Graf,T. (1995) GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev., 9, 1250–1262. [DOI] [PubMed] [Google Scholar]
- Lattanzi L. et al. (1998) High efficiency myogenic conversion of human fibroblasts by adenoviral vector-mediated MyoD gene transfer. An alternative strategy for ex vivo gene therapy of primary myopathies. J. Clin. Invest., 101, 2119–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Takemoto,N., Kurata,H., Kamogawa,Y., Miyatake,S., O’Garra,A. and Arai,N. (2000) GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J. Exp. Med., 192, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood T.D., Hancock,D.C., Danielian,P.S., Parker,M.G. and Evan,G.I. (1995) A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res., 23, 1686–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz D., Goff,S. and Bank,A. (1988) A safe packaging line for gene transfer: separating viral genes on two different plasmids. J. Virol., 62, 1120–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.I., Zon,L.I., Mutter,G. and Orkin,S.H. (1990) Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature, 344, 444–447. [DOI] [PubMed] [Google Scholar]
- Nerlov C. and Graf,T. (1998) PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev., 12, 2403–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C., Querfurth,E., Kulessa,H. and Graf,T. (2000) GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood, 95, 2543–2551. [PubMed] [Google Scholar]
- Nutt S.L., Heavey,B., Rolink,A.G. and Busslinger,M. (1999) Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature, 401, 556–562. [DOI] [PubMed] [Google Scholar]
- Orkin S.H. (2000) Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet., 1, 57–64. [DOI] [PubMed] [Google Scholar]
- Phillips R.L., Ernst,R.E., Brunk,B., Ivanova,N., Mahan,M.A., Deanehan, J.K., Moore,K.A., Overton,G.C. and Lemischka,I.R. (2000) The genetic program of hematopoietic stem cells. Science, 288, 1635–1640. [DOI] [PubMed] [Google Scholar]
- Rekhtman N., Radparvar,F., Evans,T. and Skoultchi,A.I. (1999) Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev., 13, 1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich I.N. (1992) The developmental biology of hemopoiesis: effect of growth factors on the colony formation by embryonic cells. Exp. Hematol., 20, 368–370. [PubMed] [Google Scholar]
- Rolink A.G., Nutt,S.L., Melchers,F. and Busslinger,M. (1999) Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature, 401, 603–606. [DOI] [PubMed] [Google Scholar]
- Romeo P.H., Prandini,M.H., Joulin,V., Mignotte,V., Prenant,M., Vainchenker,W., Marguerie,G. and Uzan,G. (1990) Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature, 344, 447–449. [DOI] [PubMed] [Google Scholar]
- Sauvageau G., Thorsteinsdottir,U., Eaves,C.J., Lawrence,H.J., Largman,C., Lansdorp,P.M. and Humphries,R.K. (1995) Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev., 9, 1753–1765. [DOI] [PubMed] [Google Scholar]
- Seshasayee D., Gaines,P. and Wojchowski,D.M. (1998) GATA-1 dominantly activates a program of erythroid gene expression in factor-dependent myeloid FDCW2 cells. Mol. Cell. Biol., 18, 3278–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H. et al. (2000) GATA-1 blocks IL-6-induced macrophage differentiation and apoptosis through the sustained expression of cyclin D1 and bcl-2 in a murine myeloid cell line M1. Blood, 95, 1264–1273. [PubMed] [Google Scholar]
- Tsai S.F., Martin,D.I., Zon,L.I., D’Andrea,A.D., Wong,G.G. and Orkin,S.H. (1989) Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature, 339, 446–451. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Elefanty,A.G., Strasser,A. and Adams,J.M. (1992) GATA-1 but not SCL induces megakaryocytic differentiation in an early myeloid line. EMBO J., 11, 4557–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G. (2002) Stem cell research. Studies cast doubt on plasticity of adult cells. Science, 295, 1989–1991. [DOI] [PubMed] [Google Scholar]
- Weiss M.J. and Orkin,S.H. (1995) Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc. Natl Acad. Sci. USA, 92, 9623–9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman I.L., Anderson,D.J. and Gage,F. (2001) Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu. Rev. Cell Dev. Biol., 17, 387–403. [DOI] [PubMed] [Google Scholar]
- Williams D.E., Straneva,J.E., Shen,R.N. and Broxmeyer,H.E. (1987) Purification of murine bone-marrow-derived granulocyte–macrophage colony-forming cells. Exp. Hematol., 15, 243–250. [PubMed] [Google Scholar]
- Yamaguchi Y., Zon,L.I., Ackerman,S.J., Yamamoto,M. and Suda,T. (1998) Forced GATA-1 expression in the murine myeloid cell line M1: induction of c-Mpl expression and megakaryocytic/erythroid differentiation. Blood, 91, 450–457. [PubMed] [Google Scholar]
- Zhang P., Behre,G., Pan,J., Iwama,A., Wara-Aswapati,N., Radomska,H.S., Auron,P.E., Tenen,D.G. and Sun,Z. (1999) Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl Acad. Sci. USA, 96, 8705–8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon L.I., Yamaguchi,Y., Yee,K., Albee,E.A., Kimura,A., Bennett,J.C., Orkin,S.H. and Ackerman,S.J. (1993) Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: potential role in gene transcription. Blood, 81, 3234–3241. [PubMed] [Google Scholar]