Abstract
Bacterial tyrosyl-tRNA synthetases (TyrRS) possess a flexibly linked C-terminal domain of ∼80 residues, which has hitherto been disordered in crystal structures of the enzyme. We have determined the structure of Thermus thermophilus TyrRS at 2.0 Å reso lution in a crystal form in which the C-terminal domain is ordered, and confirm that the fold is similar to part of the C-terminal domain of ribosomal protein S4. We have also determined the structure at 2.9 Å resolution of the complex of T.thermophilus TyrRS with cognate tRNAtyr(GΨA). In this structure, the C-terminal domain binds between the characteristic long variable arm of the tRNA and the anti-codon stem, thus recognizing the unique shape of the tRNA. The anticodon bases have a novel conformation with A-36 stacked on G-34, and both G-34 and Ψ-35 are base-specifically recognized. The tRNA binds across the two subunits of the dimeric enzyme and, remarkably, the mode of recognition of the class I TyrRS for its cognate tRNA resembles that of a class II synthetase in being from the major groove side of the acceptor stem.
Keywords: class I aminoacyl-tRNA synthetase/ribosomal protein S4/tRNA recognition/tyrosyl-tRNA synthetase/X-ray crystallography
Introduction
Tyrosyl-tRNA synthetase (TyrRS) is a class I aminoacyl-tRNA synthetase, but is unusual in that it is a functional dimer, a feature only shared with tryptophanyl-tRNA synthetase amongst class I synthetases (Cusack, 1995). It was the first synthetase to have its crystal structure solved (Bhat et al., 1982), including its substrate complexes with tyrosine and tyrosyl-adenylate (Brick and Blow, 1987; Brick et al., 1989), and both the mechanism of tyrosyl-adenylate formation (reviewed in Fersht, 1987; First, 1997) and its interaction with cognate tRNAtyr (reviewed in Bedouelle, 1990; Bedouelle et al., 1993) have been the subject of intense biochemical study. However, there remain a number of significant areas where structural data that correlate with biochemical observations are lacking. Here we report the first crystal structure of a bacterial tyrosyl-tRNA synthetase complexed with cognate tRNAtyr. This permits us to visualize the mode of interaction of this synthetase for its cognate tRNA, which in prokaryotes, but not archaea or eukaryotes, is of the class 2 type, i.e. with a long variable arm. The enzyme subunit comprises an N-terminal Rossmann-fold catalytic domain, characteristic of class I synthetases, followed by a central α-helical domain and, finally, a putative tRNA-binding C-terminal domain of ∼80 residues, which has been invisible until now due to disorder in all crystal structures of TyrRS (Brick et al., 1989; Qiu et al., 2001). The structure described here reveals the exact role in specific tRNA recognition of the flexibly linked C-terminal domain and also shows how unique tertiary interactions in the core of tRNAtyr lead to a different orientation of the long variable arm, permitting discrimination from other long variable arm (class 2) tRNAs such as tRNAser. The structure confirms the expected cross-subunit binding of the tRNA (Bedouelle, 1990). Furthermore, the mode of recognition of TyrRS for its cognate tRNA is similar to that of a class II synthetase rather than a canonical class I synthetase (Rould et al., 1989; Ruff et al., 1991), a significant evolutionary anomaly.
Results and discussion
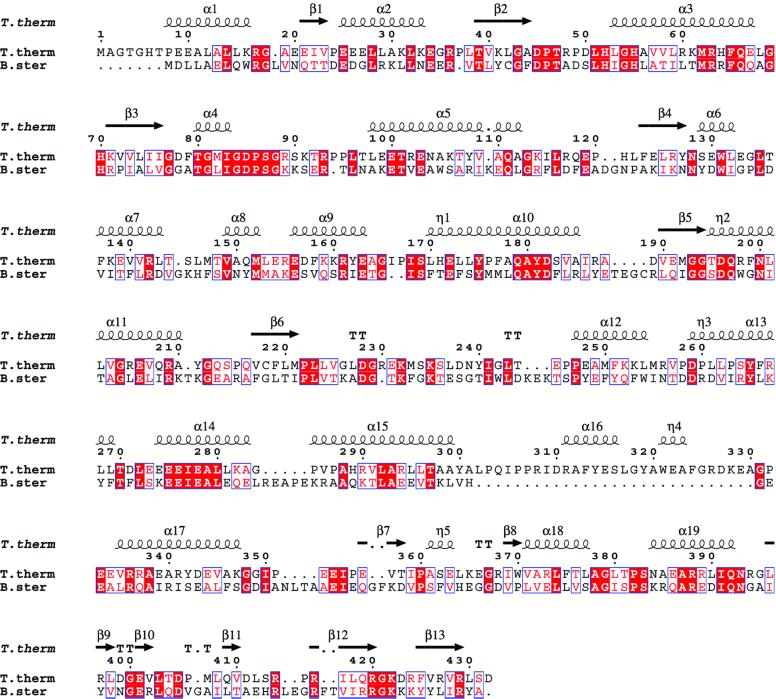
We have cloned and overexpressed TyrRS from the hyperthermophilic eubacterium Thermus thermophilus (TyrRSTT; strain HB27). It has a subunit of 432 residues and shares a branch in the phylogenetic tree of eubacterial TyrRS with the enzymes from Deinococcus radiodurans, Aquifex aeolicus, Haemophilus influenzae and Helico bacter pylori, distinct from the branch containing Bacillus stearothermophilus, Escherichia coli and eukaryotic mitochondrial TyrRS, for example (Wolf et al., 1999). Non-bacterial TyrRS, which recognize class I tRNAs without a long variable arm, are quite distinct and either lack (archaeal) or have an alternative (eukaryotic) C-terminal domain (Steer and Schimmel, 1999). This evolutionary divergence correlates with a systematically different first base pair of the cognate tRNAtyr acceptor stem, G1–C72 in prokaryotes and mitochondria, and C1–G72 in eukaryotes and archaea. This difference means that there is no cross-charging between eukaryote and prokaryote synthetases and tRNAtyrs, (e.g. Quinn et al., 1995). The sequence identity between TyrRSTT and B.stearothermophilus TyrRS (TyrRSBSt), whose atomic structure is known (Brick et al., 1989), is relatively low (28%; Figure 1) and the relative orientation of the domains is sufficiently different that the structure of TyrRSTT had to be solved de novo using the single isomorphous replacement with anomalous scattering (SIRAS) method (our unpublished results).
Fig. 1. Structure-based sequence alignment of tyrosyl-tRNA synthetase from T.thermophilus and B.stearothermophilus. The secondary structure of the TyrRSTT structure, calculated using DSSP (Kabsch and Sander, 1983), is superposed on top. Note the TyrRSTT-specific insertion between residues 302 and 330. The figure was prepared using ESPript (Gouet et al., 1999).
We have also determined a series of structures of TyrRSTT complexed with various combinations of ATP and tyrosine, which address several questions relating to the mechanism of tyrosine activation. A full description of these results is beyond the scope of this paper; however, we can summarize the results in the following points. We observe that the TyrRSTT active site domain has a very plastic structure, which is remodelled, to a greater or lesser extent, at the level of side-chain orientations, backbone conformations and interdomain orientations, depending on which combination of substrates is bound (e.g. no substrates, tyrosine only, ATP only, ATP and the non-reactive analogue tyrosinol or tyrosyl-adenylate). The apo-enzyme has an open active site with a poorly ordered flexible loop (residues 80–100), and the catalytically important 51-HIGH and 233-KMSKS motifs are withdrawn from the active centre. In contrast, the ternary complex with ATP and the non-reactive tyrosine analogue tyrosinol is the most closed state, with all regions fully ordered and the KMSKS loop fully engaged with the ATP. Binding of tyrosine or tyrosinol alone only partially orders the active site (see below and Figure 2A). Binding of ATP or adenylate, in addition to ordering the KMSKS loop, also leads to a slight change in the relative orientation of the Rossmann-fold catalytic domain and the α-helical domain.
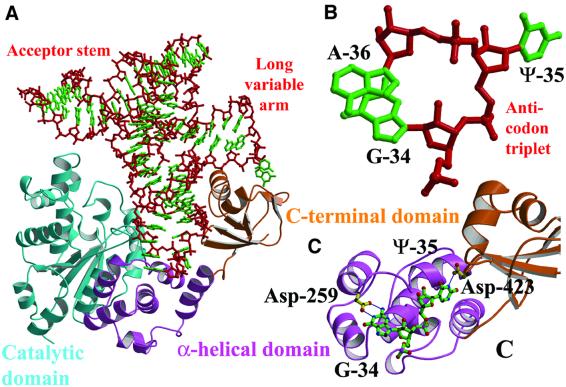
Fig. 2. Structure of T.thermophilus tyrosyl-tRNA synthetase and its tRNA complex. (A) Ribbon representation of the 2.0 Å resolution structure of T.thermophilus tyrosyl-tRNA synthetase complexed with tyrosinol. The catalytic domain is yellow (cyan), the α-helical domain red (pink) and the C-terminal domain green (orange), respectively, for each subunit in the dimer. The linkers between the α-helical and C-terminal domains, as well as some other loops involved in substrate binding, are disordered in this structure (see Table I). The small substrate tyrosinol is shown in solid-atom representation. (B) Same view, but shown as a stereoview, down the dimer 2-fold axis of the complex between T.thermophilus tyrosyl-tRNA synthetase, tRNAtyr(GΨA), tyrosinol and ATP. In this 2.9 Å resolution structure, all enzyme residues from 5–432 are visible, as are nucleotides 1–74 of the tRNA. Colours are as in (A), with the tRNA backbone in blue and the ATP and tyrosinol in solid-atom representation.
Crystal structure of T.thermophilus tyrosyl-tRNA synthetase including the C-terminal domain
One particular crystal form of TyrRSTT, co-crystallized with tyrosinol, and in which the dimer is the asymmetric unit (Table I), allows visualization for the first time of the complete enzyme including the C-terminal domain at 2.0 Å resolution. This domain (residues 352–432 in TyrRSTT) is connected to the preceding α-helical domain by a completely flexible peptide (residues 345–351), which is only partially visible in the structure. As a result, in the absence of bound tRNA, the two C-terminal domains of the dimer are observed in quite different orientations, as determined by crystal packing (Figure 2A). In a related crystal form, but obtained in the absence of any substrate, only one C-terminal domain is ordered (data not shown). In a third different crystal form, in which the monomer is the asymmetric unit, the C-terminal domain is invisible, as was the case for the TyrRS from B.stearothermophilus (Brick et al., 1989) and Staphylococcus aureus (Qiu et al., 2001). These observations are in agreement with conclusions from studies of the isolated C-terminal domain (Guez et al., 2000). Note that in the structure of TyrRSTT co-crystallized with tyrosinol, the presence of the tyrosinol substrate is not sufficient to fully order the active site. In particular, the loop 80–100 is poorly visible and other regions involved in ATP binding are not fully ordered (see above; Table I; Figure 2A).
Table I. Data collection and refinement statistics for TyrRSTT structures.
| Crystal | TyrRSTT + tyrosinol | TyrRSTT-tRNAtyr(GΨA) + tyrosinol + ATP |
|---|---|---|
| Beamline/detector | ID14-EH1, MarCCD | ID14-EH4, ADSC Quantum 4 |
| Wavelength | 0.934 Å | 0.939 Å |
| Exposure/image | 18 s/1° | 4 s/0.5° |
| Space group | P212121 | P3121 |
| Cell dimensions (Å) | a = 67.8, b = 111.1, c = 141.2 | a = 129.5, b = 129.5, c = 109.5, γ = 120 |
| Resolution | 25–2.0 Å | 23–2.9 Å |
| Total reflections | 464 280 | 76 961 |
| Unique reflections | 70 951 | 22 669 |
| Average redundancy (highest bin) | 6.5 (2.4) | 3.4 (1.8) |
| Completeness (%) (highest bin) | 97.6 (84.5) | 94.9 (63.8) |
|
R-merge (highest bin) |
0.089 (0.306) |
0.075 (0.509) |
| Refinement |
|
|
| Resolution (Å) | 20–2.0 | 20–2.9 |
| Solvent content (%) | 56 | 70 |
| Work reflections | 67 348 | 21 473 |
| Test reflections | 3557 (4.9%) | 1146 (4.8%) |
| R-free/R-work | 0.260/0.233 | 0.271/0.221 |
| Contents of asymmetric unit | TyrRSTT dimer (chains A/B), each monomer with bound tyrosinol | TyrRSTT monomer with one tRNA, ATP and tyrosinol |
| No. of protein atoms in model | 3231 (A) + 3256 (B) | 3403 |
| Poorly ordered regions | A: 1–4, 80–100, 348–351 | Nucleotides 74–76. Bases of . |
| B: 1–4, 84–96, 345–352 | nucleotides 16, 20, 474 | |
| No. of tRNA atoms | – | 1753 |
| No. of solvent molecules | 254 water, 5 sulfate | 13 water |
| No. of substrate atoms | 24 (2× tyrosinol) | 31 (ATP), 12 (tyrosinol) |
| <b> protein (Å2) | 31.3 | 65.3 |
| <b> tRNA (Å2) | - | 88.8 |
| <b> solvent (Å2) | 34.4 | 47.2 |
| <b> substrate (Å2) | 38.2 | 56.5 |
| Anisotropic B-factor correction (Å2) | B11 = 4.06, B22 = –8.77 | B11 = B22 = –10.0 |
| B33 = 4.72, B12 = 0.0 | B33 = 20.0, B12 = –24.5 | |
| B13 = B23 = 0.0 | B13 = B23 = 0.0 | |
| R.m.s.d. bonds (Å) | 0.006 | 0.008 |
| R.m.s.d. angles (°) | 1.19 | 1.33 |
| Ramachandran plot | ||
| Favourable | 94.7% | 88.5% |
| Additional | 5.3% | 11.0% |
| Generous | 0.0% | 0.3% |
| Disallowed | 0.0% | 0.3% (Asp-50) |
<b>, mean atomic B-factor.
As predicted from sequence alignments (Markus et al., 1998; Aravind and Koonin, 1999), the eubacterial C-terminal domain of TyrRS has a core fold with a similar topology to the C-terminal domain of ribosomal protein S4, with a short helical hairpin packed on a mixed β-sheet (see Figure 1 for secondary structure assignment). This structural motif has also been found in E.coli Hsp15, which binds the ribosome, and is denoted the αL motif (Staker et al., 2000). Using DALI (Holm and Sander, 1993), Hsp15 superposes with a Z-score of 6.7 (61 residues matched, 13 identities, r.m.s.d. on Cα positions of 1.74 Å) and S4 with a Z-score of 5.9 (57 residues matched, 10 identities, r.m.s.d. 1.47 Å). S4, however, differs in having a long insertion of 27 residues between the last two β-strands (Davies et al., 1998; Markus et al., 1998). The structure of the C-terminal domain of B.stearothermo philus TyrRS has recently been determined using NMR (Guijarro et al., 2002), and was found to have a very similar structure to that described here.
Crystal structure of the TyrRSTT–tRNAtyr complex
Overall structure. Five different crystal forms of the complex between TyrRSTT and native or transcript tRNAtyr have been analysed, three in which the monomer is in the asymmetric unit and two with the dimer in the asymmetric unit. In all crystal forms, there are two tRNAtyrs bound to each dimer in an essentially symmetrical fashion (Figure 2B), although previous experiments have suggested that TyrRS can only bind one tRNA per dimer (Dessen et al., 1982; Bedouelle, 1990). Importantly, co-crystals have only been obtained in the presence of small substrates either with ATP and tyrosinol, or the sulfamoyl analogue of tyrosyl-adenylate or pyrophosphate and tyrosyl-adenylate. The presence of these small substrates leads to a fully ordered, closed active site conformation of the enzyme in which the KMSKS loop interacts with the adenosine base (and triphosphate of ATP if present), as observed in other class I systems in the presence of ATP or adenylate such as GlnRS (Perona et al., 1993), TrpRS (Doublie et al., 1995) and LeuRS (Cusack et al., 2000). This contrasts with the previous reported structures of TyrRSBst in which the KMSKS loop is withdrawn some distance from the active site, in a presumably non-functional conformation, even in the presence of tyrosyl-adenylate (Brick et al., 1989). However, in none of our TyrRS–tRNAtyr complex structures is the tRNA 3′-end, beyond the phosphate of C-74, ordered and entering the active site. Nevertheless, the orientation and mode of binding of the tRNA to TyrRSTT, with the tRNA straddling both subunits, are remarkably similar, down to the level of some individual interactions, to the earlier model of the TyrRS–tRNAtyr complex proposed by Labouze and Bedouelle (1989) on the basis of extensive mutational, kinetic and biochemical studies. This correspondence strongly suggests that we have crystallized a functionally important state of the complex that also occurs in solution, if not a state active for aminoacylation. The apparent discrepancy between our symmetrical complexes with two tRNAs bound and the previously reported half-of-the-sites activity of TyrRS both for tyrosine activation (Fersht, 1987) and tRNA binding (Bedouelle, 1990) will be discussed in the conclusion.
The structure we describe at 2.9 Å resolution is that of the complex of TyrRSTT with native tRNAtyr(GΨA) (Egorova et al., 1998), in which ATP and tyrosinol are bound in the active site. Crystallographic data from this crystal form, in which the molecular dimer axis coincides with a crystallographic 2-fold axis (and hence the TyrRS dimer is perfectly symmetrical), are given in Table I.
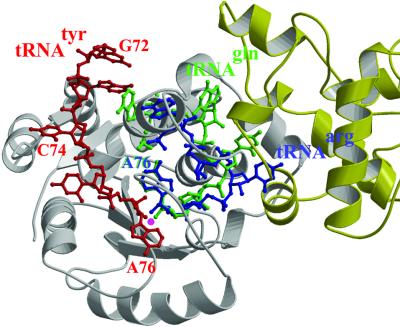
Role of the C-terminal domain in tRNA recognition. The C-terminal domain of TyrRSTT has a crucial role in the recognition of tRNAtyr, first by recognizing the tRNA’s unique shape, and secondly by participating in specific interactions with one of the anticodon bases. In the complex with cognate tRNA, the domain is stabilized in a fixed orientation by binding in the elbow between the long variable arm and the anti-codon stem, and the linker peptide connecting it to the last helix of the α-helical domain becomes ordered (Figure 3A). Other than these conformational changes, which are crucial for tRNA binding and recognition, there are no major changes in the backbone conformation of any of the domains of the enzyme upon tRNA binding, although some tRNA interacting side chains (e.g. Lys-251, Arg-255 and Arg-388) are re-orientated. Superposing 73 Cα positions (residues 358–430) from the bound and unbound C-terminal domains from the two structures reported here gives an r.m.s.d. of 0.53 or 0.62 Å (for the A and B chains of the uncomplexed structure, respectively). Similarly, superposing 340 Cα positions (residues 5–344) from the catalytic and α-helical domains of the ternary complex with ATP and tyrosinol (our unpublished data) onto the quaternary complex with tRNA gives an r.m.s.d. of 0.52 Å.
Fig. 3. Interactions between tyrosyl-tRNA synthetase and tRNAtyr. (A) The C-terminal domain (orange) binds in the elbow between the long variable arm and the anti-codon stem of the tRNA (red backbone, green bases). The anti-codon stem loop interacts with both the C-terminal domain and the α-helical domain (pink). The tRNA makes no contact with the catalytic domain of the same subunit (cyan). (B) The unusual conformation of the anti-codon triplet in which Ade-36 is stacked on Gua-34, while Psu-35 bulges out. (C) Base-specific interactions of Asp-259 from the α-helical domain with Gua-34 and Asp-423 from the C-terminal domain with Psu-35.
The elements of the C-terminal domain that contact the tRNA are limited largely to the helical hairpin (371–393), which contacts the long variable arm (nucleotides 46–472), and the β hairpin (420–423, notably Arg-420), which contacts the anti-codon stem (nucleotides 28–30) and anticodon (see below). The regions of the tRNA contacted agree very well with protection studies on the T.thermophilus system using ethyl-nitrosourea (Egorova et al., 1998) and correspond with the two C-terminal clusters of conserved basic residues identified as being in contact with the bound tRNA in the B.stearothermophilus system (Bedouelle, 1990; Nair et al., 1997). There are hydrogen bond interactions between C-terminal domain residues Trp-370, Arg-373 and Gln-409 and the bases of Gua-471 and Uri-472 in the long variable arm loop, but these are likely to be characteristic of the T.thermophilus system, as these residues and bases are not, in general, conserved. We note that the flexible connection of the tRNA-binding, C-terminal domain of bacterial type TyrRS may have been important in permitting the adaptation of certain mitochondrial TyrRSs to become cofactors in group I intron splicing (Myers et al., 2002).
Conformation and recognition of the tRNAtyr anticodon. In the complex, the anticodon triplet of tRNAtyr(GΨA) takes up a novel conformation, in which Gua-34 and Ade-36 are stacked on top of each other and in which Psu-35 bulges out in the opposite direction (Figure 3B). This conformation is stabilized by the maintenance of the usual hydrogen bond between N3 of Uri-33 and the phosphate of Ade-36, and consecutive base stacking of purines 34, 36–38 (see Figure 7). There is base-specific recognition of Psu-35 by Asp-423 (on the last β-hairpin of the C-terminal domain), which hydrogen bonds to the N3 position, and also a hydrogen bond between the main-chain carbonyl of Tyr-342 and the N1 of the base, an interaction specific for pseudouridine (Figure 3C). Although the interactions made by Asp-423 would seem to be important, it is not a phylogenetically conserved residue. Tyr-342, on the last helix of the α-helical domain preceding the linker peptide, partially stacks with Psu-35 (Figure 7). An aromatic residue is generally conserved at this position in the TyrRS in members of the same phylogenetic branch as T.thermophilus (e.g. D.radiodurans, A.aeolicus, H.influenzae and H.pylori). Interestingly, it has recently been shown biochemically that in the B.stearothermo philus TyrRS system, another residue, Phe-323, also in the last helix of the α-helical domain and conserved in most TyrRSs, provides an essential aromatic function for tRNAtyr recognition (Gaillard and Bedouelle, 2001). However, Phe-323 in TyrRSBSt structurally aligns with Ala-346 in TyrRSTT (Figure 1), suggesting that the exact roles of aromatic residues at these distinct positions are different. In most TyrRSs, in members of the same phylogenetic branch as T.thermophilus (but not TyrRSTT itself), there is a conserved aromatic residue at both positions (see alignment in figure 2 of Gaillard and Bedouelle, 2001). Base-specific interactions also occur between the N1 and N2 positions of Gua-34 and the carboxyl group of Asp-259. This residue is conserved as a glutamate or aspartate in all known bacterial and mitochondrial TyrRSs, although it is not conserved in archaeal and eukaryotic sequences, which have a weaker interaction with the anti-codon bases (Fechter et al., 2001). In the same loop between two helices in the α-helical domain, Arg-256 closely contacts the backbone of Psu-35. The base Ade-36 makes no specific interactions, but makes van der Waal’s contact with the protein in the region of Pro-285 on its face opposite to that stacking with Gua-34.
Fig. 7. Simulated omit map showing electron density for the tRNAtyr anticodon loop. Stereoview showing electron density for the anticodon loop bases 34–38 and residues Tyr-342 and Asp-423, which hydrogen bond (dashed lines) to base Ψ-35. Note that purines Ade-34, Gua-36, Ade-37 and Ade-38 are stacked on top of each other. The map is a simulated annealing difference map, calculated using a standard CNS protocol and contoured at 2σ (Brünger et al., 1998).
Relatively few systematic experiments have been carried out on the identity elements of bacterial tRNAtyr, but the above observations on the T.thermophilus system are consistent with available results in other systems. The comparative study of phosphate protection upon tRNA binding to the synthetase in the T.thermophilus and E.coli systems shows that the mode of tRNA binding is very similar (Egorova et al., 1998). In the E.coli system, the change Gua-34→Cyt causes a 24-fold decrease in the catalytic efficiency of aminoacylation (Hou and Schimmel, 1989), and changing Uri-35→Gua causes a 200-fold decrease (Himeno et al., 1990). Thus, the anticodon bases 34–35 are shown both biochemically and structurally to be important recognition elements by the synthetase.
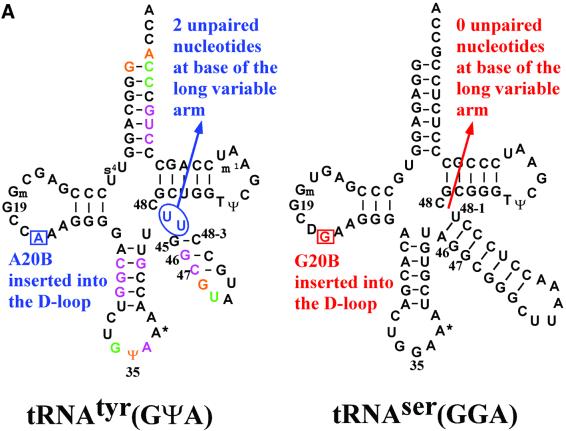
Conformation of the long variable arm of tRNAtyr compared with that of tRNAser. It has been proposed that a key determinant in the orientation of the long variable arm of class 2 tRNAs is the number of unpaired nucleotides at the 3′-end of the long variable arm. In the case of tRNAtyr, this is of critical importance as a positive identity element for recognition by TyrRS and as a negative identity element preventing mischarging of tRNAtyr by LeuRS and SerRS, which also have long variable arm cognate tRNAs (Himeno et al., 1990; Asahara et al., 1993). The two key differences in the secondary structures of tRNAtyr and tRNAser are: (i) that they have, respectively, a phylogenetically conserved adenosine or guanosine at position 20B in the D-loop; and (ii) that tRNAtyr has two unpaired uridines (U48-1 and U48-2) at the base of the long variable arm preceding nucleotide 48, whereas tRNAser has no unpaired bases at this position (Figure 4A). Elimination of the two unpaired uracils at the 3′ base of the variable arm in E.coli tRNAtyr leads to a 150-fold decrease in tyrosylation efficiency (Himeno et al., 1990).
Fig. 4. Structure of tRNAtyr compared with that of tRNAser. (A) Comparison of the secondary structures of T.thermophilus tRNAtyr(GΨA) (left) and tRNAtyr(GGA) (right), highlighting differences, conserved in other prokaryotic organisms, that determine the orientation of the long variable arm. tRNAtyr nucleotides with only backbone contacts to TyrRSTT are shown in purple, those with only base contacts are shown in green and those with backbone and base contacts are shown in orange. (B) Comparison of the 3D structures of the base of the long variable arm in T.thermophilus tRNAtyr and T.thermophilus tRNAser (Biou et al., 1994), based on the structural alignment in (C). In tRNAser, Gua-20B is unpaired and stacks against the first base pair of the long variable arm, which comprises A45:U48-1 (top). In tRNAtyr, U48-1 is unpaired and stacks against the first base pair of the long variable arm, which comprises A20B:U48–2 (bottom). (C) View looking down the anticodon stem-loop of the structural alignment of tRNAtyr (blue) and tRNAser (red) based on superposition of 46 phosphates from the acceptor stem, D- and T-loops (r.m.s.d. = 1.16 Å). The tRNA cores have a very similar structure, but the variable arms project at an angle differing by ∼50°.
The current structure and its comparison with the corresponding T.thermophilus seryl-tRNA synthetase– tRNAser complex (Biou et al., 1994), the only two crystal structures of long variable arm tRNAs, now permits us to examine this hypothesis further. In both tRNAs, the D-loop has the same number of nucleotides and a similar conformation to nucleotide 20A, forming a planar base-triple with the Levitt pair (G15–C48) and with the base 20B inserted into the tRNA core. This makes the backbone conformation of the two tRNAs, apart from the variable loop, rather similar (46 phosphates can be superimposed with an r.m.s.d. of 1.16 Å). However, the details of the core packing are significantly different (Figure 4B), resulting in an ∼50° change in orientation of the long variable arm helix (Figure 4C), which clearly permits shape discrimination between these two class 2 tRNAs by their respective synthetases. In tRNAser, the significant tilt of bases Ade-21 and Gua-9 (which stack in a parallel manner) allows deep penetration of Gua-20B into the core to stack against the first base pair of the long variable arm (A45–U48-1). In contrast, in tRNAtyr, the first base pair of the long variable is formed by a reverse Hoogsteen base pair between Ade-20B and Uri-48-2, against which the unpaired Uri-48-1 stacks. Ade-20B penetrates less far into the tRNA core, perhaps excluded by Ade-21, which forms a much more planar base-triple with Uri-8 and Ade-14 than in tRNAser. The N6 of Ade-21 also forms a single hydrogen bond to the O2 of Uri-48-1, helping to maintain this planarity and to position the latter. In tRNAtyr, the orientation of base Ade-21 is no longer strongly coupled to that of nucleotide 9, in contrast to the situation in tRNAser, where the strong stacking of Gua-9 on Ade-14 ensures that both bases remain markedly tilted (see Figure 4B). The lack of stacking, in turn, may be due to the presence of the pyrimidine Uri-9 in tRNAtyr, which has been noted by Nissan et al. (1999) and Nissan and Perona (2000) to be rather unusual in class 2 tRNAs and correlates with the occurrence of 3′ unpaired nucleotides in the variable stem, as in this case. In tRNAtyr, the base of Uri-9 makes a weak triple (one hydrogen bond) with Gua-13 and Ade-22, whereas in tRNAser, the base of Gua-9 makes two hydrogen bonds with Gua-13 and Ade-22, although in both cases the base of nucleotide 9 is tilted. A further correlation noted by Nissan and Perona (2000) is that tRNAtyr, with a pyrimidine at position 9, also conserves a C at position 20A. The base of 20A forms a triple with the reverse Watson–Crick G15–C48 (Levitt) base pair in both tRNAser and tRNAtyr, with the necessary slight shift in position to maintain two hydrogen bonds with G15 of either dihydrouridine-20A (tRNAser) or Cyt-20A (tRNAtyr). Further mutational studies, along the lines of the careful work of Nissan et al. (1999) and Nissan and Perona (2000), would be required to assess the relative importance of all these factors for determining the tRNA stability and variable arm orientation, although it is likely that the presence of Ade-20B and two 3′-unpaired uracils in the variable arm are the key features.
Acceptor stem recognition. The acceptor stem of the tRNA binds across the dimer interface onto the catalytic domain of the opposing subunit (Figure 2B). Two regions of this domain make important contacts: (i) residues 148–154, in particular Thr-148, Gln-151 and Glu-154, which partially overlap with the dimer interface; and (ii) the helix α11 formed by residues 198–211, in particular Arg-198 (Trp-196 in TyrRSBst; Figure 1), Leu-202, Arg-205, Glu-206 and Arg-209 (Arg-207 in TyrRSBst). These regions correspond to clusters 1 and 2, which were previously identified as contacting the tRNA (Bedouelle, 1990; Nair et al., 1997). Residues 21–27 also play an important secondary role in positioning side chains of directly interacting residues, notably Arg-198 and Arg-205, and in addition Val-23 (Thr-17 in TyrRSBst) packs against the ribose of Gua-1. Direct interactions are made to the tRNA backbone of nucleotides 67–69 on the 3′-strand and the 5′-phosphate of Gua-1 (Arg-205, Arg-209) as well as base-specific interactions in the major groove with Cyt-71 (Glu-154), Cyt-72 (Glu-154) and Ade-73 (Arg-198, Glu-154). Leu-202 packs on the base of Gua-1. Specific recognition of the discriminator base Ade-73 is made through a hydrogen bond between the N6 position and the main-chain carbonyl oxygen of Glu-154, an inter action that was predicted in the model for the B.stearothermophilus complex where Ala-150 is the equivalent residue (Labouze and Bedouelle, 1989). In addition, Arg-198 hydrogen bonds to the N3 position of Ade-73. These interactions give a rationale for the strong preference for adenosine as the discriminator base, with any other nucleotide reducing the catalytic efficiency by factors from 38 (A→G) to 8 (A→C) (Himeno et al., 1990) and causing loss of in vivo tyrosine specificity (Sherman et al., 1992). The interactions of Glu-154 with the N4 positions of Cyt-71 and Cyt-72, and Arg-198 with the N3 of Gua-1, also rationalize the phylogenetic preference of G–C as the first base pair in bacterial tRNAtyr, although it is not a strong identity element. Finally, the structure supports the important, but purely negative, discriminatory role proposed for Glu-152 in TyrRSBst, which helps reject non-cognate tRNAs by an electrostatic mechanism (Bedouelle and Nageotte, 1995); the equivalent residue in TyrRSTT is Glu-156, which is very close but does not touch the tRNA acceptor stem directly.
Conclusion
Extensive functional studies on wild-type and mutant TyrRSBSt by Ferhst and co-workers have characterized tyrosyl-tRNA synthetase as an intrinsically asymmetric homodimer with half of the site’s enzymatic activity (Fersht, 1987; Ward and Fersht, 1988a,b). In addition, various studies have shown that the TyrRSBSt or TyrRSEC dimer can only bind one tRNA in solution (Dessen et al., 1982; Bedouelle, 1990). In view of these results, it would seem contradictory to observe, as presented here, a perfectly symmetrical TyrRSTT– tRNAtyr–ATP–tyrosinol complex, and it raises questions about the functional significance of the structure. How ever, this is not a new problem, as many crystal structures of TyrRS, whether native or with small substrates, are either nearly (if the dimer is in the asymmetric unit) or perfectly symmetrical (Brick and Blow, 1987; Brick et al., 1989; our unpublished results) and the discrepancy has been discussed previously in the literature (e.g. Ward and Fersht, 1988a). Our understanding of this situation is the following. Careful reading of the literature of Fersht’s group shows that whereas the TyrRS enzyme exhibits strong negative co-operativity such that on short time scales tyrosyl-adenylate is only formed in one active site, on a longer time scale, tyrosyl-adenylate is formed in the second active site, converting the enzyme again to a symmetrical form. The half-life of formation of the first tyrosyl-adenylate is 39 ms (k = 17.8/s) and for the second it is 3.8 min (k = 0.003/s) for the case of TyrRSBSt (Mulvey and Fersht, 1977). Clearly, in a crystallization experiment, there is ample time for substrate binding to both active sites and there is quite likely preferential crystallization of a symmetrical form, as is frequently observed. In the case of tRNA binding, we believe the same arguments hold. In all our crystal forms of the complex, there are small substrates (ATP + tyrosinol or tyrosyl-adenylate) bound in both active sites, making a symmetrical dimer to which there is clearly no steric hinderance for binding of two tRNA molecules. However, it may require an asymmetric form of the dimer with one bound tRNA, a form we have not yet managed to crystallize, to observe the 3′ end of the tRNA functionally bound in the active site. We note that the small angle neutron scattering experiments of Dessen et al. (1982), which showed only one tRNA binding the dimer, were performed with no small substrates present. It would clearly be interesting to repeat these experiments in the presence of small substrates, corresponding to the crystallization conditions of the complex.
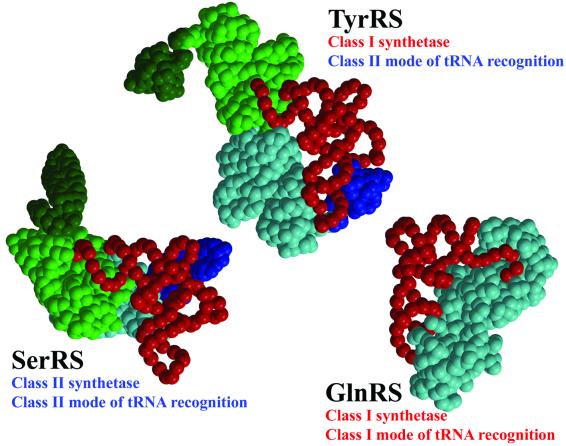
Despite having an unambiguous class I catalytic domain, tyrosyl-tRNA synthetase is in two important ways a non-canonical class I synthetase, as deduced previously from biochemical studies (Bedouelle et al., 1993) and visualized explicitly in the current structure. First, it is a functional dimer with cross-subunit tRNA binding (Figure 2B), a feature shared in class I synthetases only with its subclass Ic partner, tryptophanyl-tRNA synthetase. Secondly, it has a class II mode of tRNA recognition (Figure 5). This means that it interacts with tRNAtyr from the variable loop and acceptor stem major groove side as, for instance, in the case of class II AspRS (Ruff et al., 1991) and SerRS (Cusack et al., 1996), without requiring distortion of the 3′ end of the tRNA from its helical path to enter the active site. This is in strong contrast to canonical class I systems such as those of subclass Ib GlnRS (Rould et al., 1989; Rath et al., 1998) and subclass Ia ArgRS (Delagoutte et al., 2000), which approach cognate tRNA from the acceptor stem minor groove side and require a hairpin distortion of the tRNA 3′-end so that they can enter the active site. Despite this class II mode of tRNA recognition, TyrRS preferentially aminoacylates the 2′ OH of Ade-76 in accordance with other class I systems, although it can also apparently aminoacylate the 3′ OH (Cramer et al., 1975). The evolutionary scenario that led to these non-canonical features of TyrRS is not obvious, although the pairing of TyrRS and the class II synthetase PheRS (which anomalously aminoacylates at the 2′OH position) in a new hypothesis about the origin of the two synthetase classes is intriguing in this respect (Ribas de Pouplana and Schimmel, 2001). However, superposition of the GlnRS– tRNAgln or ArgRS–tRNAarg complex on TyrRS by means of the conserved Rossmann fold shows that the normal hairpin mode of class I tRNA binding is incompatible with dimerization of TyrRSTT (Figure 6). On the other hand, the observed class II mode of recognition of TyrRS for tRNAtyr, modelled to extend into the active site according to the model of Labouze and Bedouelle (1989), is clearly compatible with dimerization, at the same time as allowing the 2′ OH of the terminal ribose to be correctly positioned for aminoacylation (Figure 6). This suggests that dimerization and the class II mode of tRNA recognition in TyrRS may be evolutionarily linked, in that one may have necessitated the other. A crystal structure of the TyrRS– tRNAtyr complex, active for aminoacylation and perhaps necessarily asymmetric, is clearly required to give further insight into this still puzzling enigma.
Fig. 5. Schematic diagrams showing different modes of tRNA recognition by TyrRS, SerRS and GlnRS. TyrRS (class I) and SerRS (class 2) both have a class II mode of tRNA recognition approaching the tRNA acceptor stem from the major groove side. GlnRS has a canonical class I mode of tRNA recognition approaching the tRNA acceptor stem from the minor groove side. Notably, both SerRS and TyrRS have long variable arm (class 2) tRNAs, are dimeric and have cross-subunit tRNA binding.
Fig. 6. Model of the 3′ CCA end of tRNAtyr in the active site of TyrRSTT. The C74-C75-A76 3′ extremity of tRNAtyr (red) is modelled in the active site of TyrRSTT by matching the predicted model of Labouze and Bedouelle (1989) to the experimental structure (this work) of tRNAtyr, which extends only to A73. Due to the close agreement of the position of the tRNAtyr in the experimental and predicted models, this is possible with only minor adjustments to avoid steric clashes with enzyme side-chains. The backbone conformation of the CCA and the positions of the bases are by no means necessarily correct. The aim is to show that the observed class II mode of tRNA binding to TyrRSTT is compatible with unhindered entry of the 3′ end into the active site without major conformational changes (although it cannot be excluded that these occur), allowing the positioning of the 2′ OH of the terminal ribose (marked with a purple spot) adjacent to the carboxyl group of the substrate amino acid (not shown for clarity, since it is underneath in this view). This is clearly not the case for a class I mode of entry into the active site as exemplified by tRNAarg (blue) or tRNAgln (green). These tRNA 3′ ends were drawn after superposition of the Rossmann fold of TyrRSTT with that of yeast ArgRS (Delagoutte et al., 2000) or E.coli GlnRS (Rath et al., 1998) in the context of their respective tRNA complexes. Note that the bases of C75 in tRNAarg, and G73 and C75 in tRNAgln have been omitted for clarity. Although the 3′ hairpin conformations of tRNAarg and tRNAgln, from subclasses Ia and Ib, respectively, are somewhat different, the base of A76 and the position of the 2′ OH (purple spot) superpose very closely. It is clear that the CCA ends of these two class I mode tRNAs would both clash irremediably with the dimer interface of TyrRS (one subunit is grey, the other yellow), and A76 and C75 coincide with the position of the important active site loop residues 83–86 in TyrRS.
Very recently, we have determined a crystal structure at 1.95 Å resolution of a complex of TyrRSTT with a tRNAtyr transcript in the absence of small substrates. This new high resolution structure confirms the conclusions described here and shows more clearly the entry of the 3′ end of the tRNA into the active site, which is in an open conformation. The complex is crystallographically symmetrical with two tRNAs per tyrosyl-tRNA synthetase dimer.
Materials and methods
The gene for T.thermophilus (strain HB27) TyrRS was cloned and sequenced, and found to code for a protein subunit of 432 residues (A.Yaremchuk, S.Cusack and M.Tukalo, unpublished results). Over production and purification of TyrRSTT were carried out in a similar way to that of the T.thermophilus LeuRS (Yaremchuk et al., 2000). Thermus thermophilus tRNAtyr(GΨA) was purified from strain HB-27 as described previously (Egorova et al., 1998). Crystals of TyrRS alone and in complex with tyrosinol were obtained by the hanging drop vapour diffusion technique using equal volumes of the protein (12 mg/ml) and a reservoir solution that contained 1.2 M ammonium sulfate, 10 mM MgCl2, 0.5 mM dithiothreitol (DTT) and 50 mM MES pH 5.8. Crystals of TyrRS in complex with tyrosinol, ATP and tRNAtyr were grown at 293 K by the equilibration of 4 µl of protein/tRNA solution (4–5 mg/ml of TyrRSTT, molar ratio of protein:tRNA of 1:1 or 1:2.2, 5 mM tyrosinol, 10 mM MgCl2, 10 mM ATP, 50 mM HEPES pH 7.0 and 0.8 M ammonium sulfate) against 1 ml of reservoir solution containing 1.5– 1.6 M ammonium sulfate and 100 mM HEPES pH 7.0. Hexagonal bipyramid crystals grew to final dimensions of up to 500 µm within 2–4 weeks, depending on the protein/tRNA molar ratio.
Crystallography
Data collection statistics are given in Table I. Data were integrated with MOSFLM (Leslie, 1999) and subsequent processing was carried out using the CCP4 package (CCP4, 1994). Both structures were solved by molecular replacement using MOLREP. The search model employed was that of the catalytic and α-helical domain of TyrRSTT determined previously de novo at 2.1 Å resolution in a different crystal form (A.Yaremchuk, M.Tukalo and S.Cusack, unpublished results). For the 2.0 Å resolution structure of the complete enzyme, the ARP/wARP program (Perrakis et al., 1999) was used to automatically build the additional C-terminal domains and to add water molecules. Both structures were refined using CNS (Brünger et al., 1998). Due to the moderate resolution and high B-factors of the tRNA, base planarity, base pair and ribose pucker restraints were applied. Ribose puckers of nucleotides 7, 9, 18–20, 202 (20B), 34–35, 48, 58 and 60 were restrained to C2′ endo, and all others to C3′ endo. The refinement statistics and quality of structures are given in Table I. A simulated omit map of a portion of the tRNA electron density in the region of the anti-codon loop is shown in Figure 7.
Figures 2–5 were prepared with BOBSCRIPT (Esnouf, 1999) and rendered with RASTER3D (Merritt and Bacon, 1997).
Acknowledgments
Acknowledgements
The authors thank members of the ESRF-EMBL Joint Structural Biology Group for access to ESRF beamlines. This work was supported in part by an International Research Scholar’s award from the Howard Hughes Medical Institute (to M.T.), and in part by Human Frontiers Science Programme Research grant RGP0190/2001-M.
References
- Aravind L. and Koonin,E.V. (1999) Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol., 48, 291–302. [DOI] [PubMed] [Google Scholar]
- Asahara H., Himeno,H., Tamura,K., Hasegawa,T., Watanabe,K. and Shimizu,M. (1993) Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J. Mol. Biol., 231, 219–229. [DOI] [PubMed] [Google Scholar]
- Bedouelle H. (1990) Recognition of tRNAtyr by tyrosyl-tRNA synthetase. Biochimie, 72, 589–598. [DOI] [PubMed] [Google Scholar]
- Bedouelle H. and Nageotte,R. (1995) Macromolecular recognition through electrostatic repulsion. EMBO J., 14, 2945–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedouelle H., Guez-Ivanier,V. and Nageotte,R. (1993) Discrimination between transfer-RNAs by tyrosyl-tRNA synthetase. Biochimie, 75, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Bhat T.N., Blow,D.M., Brick,P. and Nyborg,J. (1982) Tyrosyl-tRNA synthetase forms a mononucleotide-binding fold. J. Mol. Biol., 158, 699–709. [DOI] [PubMed] [Google Scholar]
- Biou V., Yaremchuk,A., Tukalo,M.A. and Cusack,S. (1994) The 2.9Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNAser. Science, 263, 1404–1410. [DOI] [PubMed] [Google Scholar]
- Brick P. and Blow,D.M. (1987) Crystal structure of a deletion mutant of a tyrosyl-tRNA synthetase complexed with tyrosine. J. Mol. Biol., 194, 287–294. [DOI] [PubMed] [Google Scholar]
- Brick P., Bhat,T.N. and Blow,D.M. (1989) Structure of tyrosyl-tRNA synthetase refined at 2.3Å resolution. Interaction of the enzyme with the tyrosyl-adeylate intermediate. J. Mol. Biol., 208, 83–98. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallographic and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cramer F., Faulhammer,H., von der Haar,F., Sprinzl,M. and Sternbach,H. (1975) Aminoacyl-tRNA synthetases from baker’s yeast: reacting site of aminoacylation is not uniform for all tRNAs. FEBS Lett., 56, 212–214. [DOI] [PubMed] [Google Scholar]
- Cusack S. (1995) Eleven down and nine to go. Nat. Struct. Biol., 2, 824–831. [DOI] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (1996) The crystal structure of the ternary complex of T. thermophilus seryl-tRNA synthetase with tRNAser and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J., 15, 2834–2842. [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (2000) The 2Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J., 19, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C., Gerstner,R.B., Draper,D.E., Ramakrishnan,V. and White,S.W. (1998) The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif. EMBO J., 17, 4545–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delagoutte B., Moras,D. and Cavarelli,J. (2000) tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J., 19, 5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessen P., Zaccai,G. and Blanquet,S. (1982) Neutron scattering studies of Escherichia coli tyrosyl-tRNA synthetase and of its interaction with tRNAtyr. J. Mol. Biol., 159, 651–664. [DOI] [PubMed] [Google Scholar]
- Doublie S., Bricogne,G., Gilmore,C. and Carter,C.W.,Jr (1995) Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure, 3, 17–31. [DOI] [PubMed] [Google Scholar]
- Egorova S.P., Iaremchuk,A.D., Kriklivyi,I.A. and Tukalo,M.A. (1998) Comparative analysis of interaction sites of Thermus thermophilus and Escherichia coli tRNA(Tyr) with homologous aminoacyl-tRNA synthetases by means of chemical modification and nuclease hydrolysis. Russ. J. Bioorg. Chem., 24, 524–530. [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to Molscript version 1.4, including reading and contouring of electron density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Fechter P., Rudinger-Thirion,J., Tukalo,M. and Giege,R. (2001) Major tyrosine identity determinants in Methanococcus jannaschii and Saccharomyces cerevisiae tRNAtyr are conserved but expressed differently. Eur. J. Biochem., 268, 761–767. [DOI] [PubMed] [Google Scholar]
- Fersht A.F. (1987) Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry, 26, 8031–8037. [DOI] [PubMed] [Google Scholar]
- First E.A. (1997) Catalysis of tRNA aminoacylation by class I and II aminoacyl-tRNA synthetases. In Sinnott,M. (ed.), Comprehensive Biological Catalysis. Vol. 1. Academic Press, London, UK, pp. 573–607.
- Gaillard C. and Bedouelle,H. (2001) An essential residue in the flexible peptide linking the two idiosyncratic domains of bacterial tyrosyl-tRNA synthetases. Biochemistry, 40, 7192–7199. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Guez V., Nair,S., Chaffotte,A. and Bedouelle,H. (2000) The anticodon-binding domain of tyrosyl-tRNA synthetase: state of folding and origin of the crystallographic disorder. Biochemistry, 39, 1739–1747. [DOI] [PubMed] [Google Scholar]
- Guijarro J.I., Pintar,A., Prochnicka-Chalufour,A., Guez,V., Gilquin,B., Bedouelle,H. and Delepierre,M. (2002) Structure and dynamics of the anticodon-arm binding domain of Bacillus stearothermophilus tyrosyl-tRNA synthetase. Structure, 10, 311–317. [DOI] [PubMed] [Google Scholar]
- Himeno H., Hasegawa,T., Ueda,T., Watanabe,K. and Shimizu,M. (1990) Conversion of aminoacylation specificity from tRNATyr to tRNASerin vitro. Nucleic Acids Res., 18, 6815–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Hou Y.M. and Schimmel,P. (1989) Modeling with in vitro kinetic parameters for the elaboration of transfer RNA identity in vivo. Biochemistry, 28, 4942–4947. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Labouze E. and Bedouelle,H. (1989) Structural and kinetic bases for the recognition of tRNATyr by tyrosyl-tRNA synthetase. J. Mol. Biol., 205, 729–735. [DOI] [PubMed] [Google Scholar]
- Leslie A.G. (1999) Integration of macromolecular diffraction data. Acta Crystallogr. D, 55, 1696–1702. [DOI] [PubMed] [Google Scholar]
- Markus M.A., Gerstner,R.B., Draper,D.E. and Torchia,D.A. (1998) The solution structure of ribosomal protein S4 Δ41 reveals two sub-domains and a positively charged surface that may interact with RNA. EMBO J., 17, 4559–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt M.A. and Bacon,D.J. (1997) Raster 3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Mulvey R.S. and Fersht,A.R. (1977) Ligand binding stoichiometries, subunit structure and slow transitions in aminoacyl-tRNA synthetases. Biochemistry, 16, 4005–4013. [DOI] [PubMed] [Google Scholar]
- Myers C.A., Kuhla,B., Cusack,S. and Lambowitz,A.M. (2002) tRNA-like recognition of group I introns by a tyrosyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 99, 2630–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S., Ribas de Pouplana,L., Houman,F., Avruch,A., Shen,X. and Schimmel,P. (1997) Species-specific tRNA recognition in relation to tRNA synthetase contact residues. J. Mol. Biol., 269, 1–9. [DOI] [PubMed] [Google Scholar]
- Nissan T.A. and Perona,J.J. (2000) Alternative designs for construction of the class II transfer RNA tertiary core. RNA, 6, 1585–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T.A., Oliphant,B. and Perona,J.J. (1999) An engineered class I transfer RNA with a class II tertiary fold. RNA, 5, 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona J.J., Rould,M.A. and Steitz,T.A. (1993) Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry, 32, 8758–8771. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Qiu X. et al. (2001) Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors. Protein Sci., 10, 2008–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn C.L., Tao,N. and Schimmel,P. (1995) Species-specific microhelix aminoacylation by a eukaryotic pathogen tRNA synthetase dependent on a single base-pair. Biochemistry, 34, 12489–12495. [DOI] [PubMed] [Google Scholar]
- Rath V.L., Silvian,L.F., Beijer,B., Sproat,B.S. and Steitz,T.A. (1998) How glutaminyl-tRNA synthetase selects glutamin. Structure, 6, 439–449. [DOI] [PubMed] [Google Scholar]
- Ribas de Pouplana L. and Schimmel,P. (2001) Two classes of tRNA synthetases suggested by sterically compatible dockings on tRNA acceptor stem. Cell, 104, 191–193. [DOI] [PubMed] [Google Scholar]
- Rould M.A., Perona,J.J., Söll,D. and Steitz,T.A. (1989) Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8Å resolution. Science, 246, 1135–1142. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy,S., Boeglin,M., Poterszman,A., Mitschler,A., Podjarny,A., Rees,B., Thierry,J.C. and Moras,D. (1991) Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science, 252, 1682–1689. [DOI] [PubMed] [Google Scholar]
- Sherman J.M., Rogers,K., Rogers,M.J. and Soll,D. (1992) Synthetase competition and tRNA context determine the in vivo identify of tRNA discriminator mutants. J. Mol. Biol., 228, 1055–1062. [DOI] [PubMed] [Google Scholar]
- Staker B.L., Korber,P., Bardwell,J.C. and Saper,M.A. (2000) Structure of Hsp15 reveals a novel RNA-binding motif. EMBO J., 19, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer B.A. and Schimmel,P. (1999) Major anticodon-binding region missing from an archaebacterial tRNA synthetase. J. Biol. Chem., 274, 35601–35606. [DOI] [PubMed] [Google Scholar]
- Ward W.H. and Fersht,A.R. (1988a) Asymmetry of tyrosyl-tRNA synthetase in solution. Biochemistry, 27, 1041–1049. [DOI] [PubMed] [Google Scholar]
- Ward W.H. and Fersht,A.R. (1988b) Tyrosyl-tRNA synthetase acts as an asymmetric dimer in charging tRNA. A rationale for half-of-the-sites activity. Biochemistry, 27, 5525–5530. [DOI] [PubMed] [Google Scholar]
- Wolf Y.I., Aravind,L., Grishin,N.V. and Koonin,E.V. (1999) Evolution of aminoacyl-tRNA synthetases-analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res., 9, 689–710. [PubMed] [Google Scholar]
- Yaremchuk A., Cusack,S., Gudzera,O., Grøtli,M. and Tukalo,M. (2000) Crystallization and preliminary crystallographic analysis of Thermus thermophilus leucyl-tRNA synthetase and its complexes with leucine and a non-hydrolysable leucine-adenylate analogue. Acta Crystallogr. D, 56, 667–669. [DOI] [PubMed] [Google Scholar]